Abstract
To generate transgenic mice that express Cre-recombinase exclusively in the megakaryocytic lineage, we modified a mouse bacterial artificial chromosome (BAC) clone by homologous recombination and replaced the first exon of the platelet factor 4 (Pf4), also called CXCL4, with a codon-improved Cre cDNA. Several strains expressing the transgene were obtained and one strain, Q3, was studied in detail. Crossing Q3 mice with the ROSA26-lacZ reporter strain showed that Cre-recombinase activity was confined to megakaryocytes. These results were further verified by crossing the Q3 mice with a strain containing loxP-flanked integrin β1. Excision of this conditional allele in megakaryocytes was complete at the DNA level, and platelets were virtually devoid of the integrin β1 protein. The Pf4-Cre transgenic strain will be a valuable tool to study megakaryopoiesis, platelet formation, and platelet function.
Introduction
Deletion of ubiquitously expressed genes often results in severe phenotypes that interfere with the analysis of particular cell types or tissues.1 To overcome this limitation, conditional gene inactivation with the loxP-Cre system has been widely used. Several genes that are expressed in the megakaryocytic lineage have been described, including GpIIb (CD41), GpIIIa (CD61), and platelet factor 4 (Pf4).2,3 The promoters of these genes can be used to direct expression of transgenes to megakaryocytes and precursors.4-7 Classical transgenes with relatively short promoters are known to be prone to silencing and position effect variegation, resulting in mosaic expression, whereas copy number-dependent and position-independent transgene expression was reported when large genomic fragments that contain the promoter of choice were used.8-10 Here, we describe the generation of transgenic mice that exclusively express Cre-recombinase in megakaryocytes. To achieve specific Cre expression, we used a bacterial artificial chromosome (BAC) clone containing the entire mouse Pf4 promoter.
Materials and methods
Transgenic mice
The Pf4-Cre transgene construct was generated by homologous recombination in bacteria (for details, see legend to Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).11 BAC DNA from the final construct was purified using the Nucleobond AX 500 DNA purification kit (Macherey-Nagel, Düren, Germany). An approximately 100-kb (kilobase) NotI fragment harboring the Cre cDNA was separated from the vector by pulsed-field gel electrophoresis prior to microinjection into C57BL/6 oocytes. Genotyping was performed using a forward primer located in the Pf4-promotor (CCCATACAGCACACCTTTTG) and a reverse primer in the Cre-cDNA (TGCACAGTCAGCAGGTT) yielding a 450-bp polymerase chain reaction (PCR) product. ROSA26-lacZ mice were purchased from Jackson Laboratories (Bar Harbor, ME).12 Integrin β1 lox mice were received from Dr Ulrich Müller (Scripps Research Institute, La Jolla, CA).13
RNA
Total RNA was isolated with TriFast (PeqLab, Erlangen, Germany) and treated with Turbo DNAse (Ambion, Austin, TX). Reverse transcription was performed with Omniscript reverse transcriptase (Qiagen, Hilden, Germany). Real-time PCR was performed using SYBR Green PCR master mix on an ABI Prism 7000 (Applied Biosystems, Foster City, CA) with the primers TCCTGTACCTGCAAGCCAGA and GGTGCTGTTGGATGGTCTTCA (codon-improved Cre-recombinase) and ATCCGCAAGCCTGTGACTGT and TCGGGCCAGGGTGTTTTT (mouse Rpl19, for normalization).14 Relative expression values were calculated by the ΔΔCT method using one bone marrow sample as a calibrator that was set to the value of 1.15,16
Western blot
Cre protein was detected with a polyclonal rabbit anti-Cre antibody (Novagen, Madison, WI). Membranes were reprobed using the monoclonal anti–β-actin antibody AC-15 (Sigma-Aldrich, St Louis, MO).
Cells, tissues, and staining
Platelets were obtained from platelet-rich plasma by centrifugation at 1300g for 5 minutes. Purity was assessed on an Advia 120 hematology analyzer (Bayer, Leverkusen, Germany). Leukocytes were purified from buffy coat by red cell lysis. Femurs and tibiae were flushed with CATCH buffer to collect bone marrow cells.17 Megakaryocyte isolation is described in the legend to Figure S2. Purity of megakaryocytes was assessed by May-Grünwald-Giemsa staining and by acetylcholine esterase staining.18,19 Cryosections of organs were stained for β-galactosidase activity as described.20 Images of lacZ- or acetylcholine esterase-stained cells and tissue sections were taken using a Leica DM LS microscope (Leica, Wetzlar, Germany) equipped with an N PLAN 40×/0.65 numerical aperture objective and a mounted Leica DFC480 R2 digital camera. Images were acquired using Leica FireCam software version 1.5.
Flow cytometry
Platelets and megakaryocytes were stained with FITC anti–mouse CD41 antibody and biotin-conjugated hamster anti–rat CD29 (integrin β1) antibody (both Becton Dickinson, Franklin Lakes, NJ), followed by streptavidin conjugated to PE or Cy5 and analyzed on a fluorescence-activated cell sorter (FACSCalibur; Becton Dickinson). No fixatives were used. Megakaryocytes were incubated in hypotonic citrate buffer prior to FACS analysis.21
Results and discussion
A Cre-recombinase cDNA optimized for codon-usage in eukaryotic cells and containing a nuclear localization signal and a myc tag was inserted by homologous recombination in bacteria into an approximately 100-kb mouse BAC clone containing the Pf4 gene (Figure 1A; for details, see Figure S1).11,22 Hereby, the first exon of Pf4 was replaced by the Cre cDNA. The BAC clone also contained 4 other genes, CXCL5, CXCL7, CXCL15, and Gm1960 (Document S1). Pronuclear injections yielded 6 transgenic founders in the C57BL/6 inbred background. Five of the 6 transgenic strains expressed the transgene, but only 2 strains showed a restricted expression pattern (Table S1). One of these strains, Q3, with a single copy transgene was selected for detailed analysis.
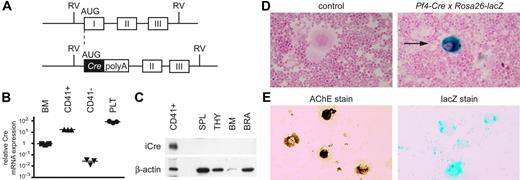
Selective expression of Cre in megakaryocytes and platelets in Pf4-Cre transgenic mic. (A) Pf4-Cre transgene construct. A codon-improved cDNA for Cre-recombinase was inserted by homologous recombination into bacterial artificial chromosome DNA containing the mouse Pf4 gene. Numbered boxes indicate Pf4 exons; polyA, bovine growth hormone polyadenylation signal; RV, EcoRV restriction sites. Note that exon 1 of Pf4 is deleted in the transgene construct. (B) Quantification of Cre mRNA expression by real-time PCR in Pf4-Cre mice. RNA from unfractionated bone marrow (BM), bone marrow cells separated into CD41+ or CD41− fractions, and platelets (PLT) were analyzed. Relative expression levels calculated by the ΔΔCT method are shown. One bone marrow sample was set to the value of 1. Horizontal lines mark the mean of the values from 3 mice. (C) Expression of the Cre protein in tissues from Pf4-Cre mice. Western blot probed with an anti-Cre antibody (top) and reprobed with an anti–β-actin antibody (bottom) is shown. SPL indicates, spleen; THY, thymus; BM, bone marrow; BRA, brain. (D) Detection of Cre-mediated recombination in Pf4-Cre mice using the ROSA26-lacZ reporter. Bone marrow cytospins from Pf4-Cre transgenic mice and controls are shown. Excision of a stop cassette flanked by loxP sites results in β-galactosidase activity that can be visualized by X-Gal staining (blue color). The samples were counterstained with nuclear fast red to visualize nuclei. Note that the blue staining is limited to cells with megakaryocyte morphology (arrows). (E) CD41+ magnetic cell sorting (MACS)–purified megakaryocytes from Pf4-Cre × ROSA26-lacZ mice were stained for acetylcholine esterase (left) or β-galactosidase activity (right).
Selective expression of Cre in megakaryocytes and platelets in Pf4-Cre transgenic mic. (A) Pf4-Cre transgene construct. A codon-improved cDNA for Cre-recombinase was inserted by homologous recombination into bacterial artificial chromosome DNA containing the mouse Pf4 gene. Numbered boxes indicate Pf4 exons; polyA, bovine growth hormone polyadenylation signal; RV, EcoRV restriction sites. Note that exon 1 of Pf4 is deleted in the transgene construct. (B) Quantification of Cre mRNA expression by real-time PCR in Pf4-Cre mice. RNA from unfractionated bone marrow (BM), bone marrow cells separated into CD41+ or CD41− fractions, and platelets (PLT) were analyzed. Relative expression levels calculated by the ΔΔCT method are shown. One bone marrow sample was set to the value of 1. Horizontal lines mark the mean of the values from 3 mice. (C) Expression of the Cre protein in tissues from Pf4-Cre mice. Western blot probed with an anti-Cre antibody (top) and reprobed with an anti–β-actin antibody (bottom) is shown. SPL indicates, spleen; THY, thymus; BM, bone marrow; BRA, brain. (D) Detection of Cre-mediated recombination in Pf4-Cre mice using the ROSA26-lacZ reporter. Bone marrow cytospins from Pf4-Cre transgenic mice and controls are shown. Excision of a stop cassette flanked by loxP sites results in β-galactosidase activity that can be visualized by X-Gal staining (blue color). The samples were counterstained with nuclear fast red to visualize nuclei. Note that the blue staining is limited to cells with megakaryocyte morphology (arrows). (E) CD41+ magnetic cell sorting (MACS)–purified megakaryocytes from Pf4-Cre × ROSA26-lacZ mice were stained for acetylcholine esterase (left) or β-galactosidase activity (right).
Cre mRNA expression by real-time PCR was detectable in bone marrow, but not in other organs (Figure 1B; data not shown). The highest expression was found in platelets and purified CD41+ bone marrow cells, whereas CD41− cells had levels indistinguishable from background (Figure 1B). The majority of the CD41+ cells consisted of large megakaryocytes, as determined by morphology of May-Grünwald-Giemsa–stained samples and by acetylcholine esterase staining (Figure S2). Expression of Cre protein by Western blot was strong in CD41+ cells but was below detection in bone marrow and other tissues (Figure 1C). To demonstrate that the Cre protein is functional and can mediate lineage-specific excision, we crossed the Q3 strain with ROSA26-lacZ reporter mice.12 In this strain, expression of β-galactosidase is activated on Cre-mediated deletion of premature polyadenylation signals. Bone marrow cryosections of Pf4-Cre × ROSA26-lacZ mice stained with X-Gal revealed that Cre activity was restricted to megakaryocytes in bone marrow (Figure 1D) and spleen (not shown). Even though large differences in lacZ staining intensities between individual megakaryocytes were noted, the majority of megakaryocytes displayed at least small amounts of blue staining. This finding was confirmed by LacZ staining of cytospin slides with more than a hundred purified megakaryocytes (Figure 1E). Cre activity was absent from all other inspected organs (data not shown). Thus, Cre expression and functional activity follows the lineage distribution expected for the PF4 promoter.
We used an integrin β1 lox allele as a reporter to further investigate the lineage specificity of Cre-mediated excision (Figure 2A).13 We obtained live offspring with the expected Mendelian ratios (data not shown), demonstrating that the embryonic lethal phenotype of the complete integrin β1 knockout is prevented by lineage-restricted expression of Cre. In addition, no alterations in blood counts were detected (Table S2), confirming previous data from an inducible Mx-Cre × integrin β1 lox knockout.23 This also indicates that the additional copy of the chemokine genes CXCL5, CXCL7, CXCL15 present in the BAC construct has no effect on the blood counts from the Q3 mice.
Efficiency and specificity of Cre-mediated excision in Pf4-Cre × integrin β1 lox/lox mice. (A) Diagram of integrin β1 floxed and excised alleles. Numbered black boxes represent coding exons, loxP sites are depicted as gray triangles. A thick line indicates the position of the Southern probe. (B) Southern blot analysis of genomic DNA from mouse tissues digested with PvuII and hybridized to the probe shown above. A composite picture of 2 blots is shown. Whole indicates unfractionated bone marrow cells; MEG, highly pure megakaryocytes obtained from bone marrow by FACS sort and subsequent TPO culture; CD41−, bone marrow depleted of CD41+ cells using antibodies and magnetic beads; LEU, peripheral blood leukocytes; LIV, liver; SPL, spleen; KID, kidney; INT, intestine; OVA, ovary; THY, thymus; HEA, heart; LUN, lung; BRA, brain. (C) Cell-surface expression of integrin β1 protein on platelets (top) or megakaryocytes (bottom) from mice with the indicated genotypes was assessed by flow cytometry. Megakaryocytes obtained from bone marrow cell cultures grown in the presence of recombinant TPO were used. Note that integrin β1 protein expression is virtually absent on platelets and markedly reduced on megakaryocytes from Pf4-Cre × integrin β1 lox/lox mice.
Efficiency and specificity of Cre-mediated excision in Pf4-Cre × integrin β1 lox/lox mice. (A) Diagram of integrin β1 floxed and excised alleles. Numbered black boxes represent coding exons, loxP sites are depicted as gray triangles. A thick line indicates the position of the Southern probe. (B) Southern blot analysis of genomic DNA from mouse tissues digested with PvuII and hybridized to the probe shown above. A composite picture of 2 blots is shown. Whole indicates unfractionated bone marrow cells; MEG, highly pure megakaryocytes obtained from bone marrow by FACS sort and subsequent TPO culture; CD41−, bone marrow depleted of CD41+ cells using antibodies and magnetic beads; LEU, peripheral blood leukocytes; LIV, liver; SPL, spleen; KID, kidney; INT, intestine; OVA, ovary; THY, thymus; HEA, heart; LUN, lung; BRA, brain. (C) Cell-surface expression of integrin β1 protein on platelets (top) or megakaryocytes (bottom) from mice with the indicated genotypes was assessed by flow cytometry. Megakaryocytes obtained from bone marrow cell cultures grown in the presence of recombinant TPO were used. Note that integrin β1 protein expression is virtually absent on platelets and markedly reduced on megakaryocytes from Pf4-Cre × integrin β1 lox/lox mice.
To detect excision of the loxP-flanked integrin β1 exon, we analyzed DNA from several tissues of Pf4-Cre × integrin β1 lox/lox mice by Southern blot (Figure 2B). A faint band of excised DNA was observed in unfractionated bone marrow but not in other mouse tissues (Figure 2B). This excised band was not detectable in bone marrow cells depleted of CD41 cells by magnetic beads (CD41−). To obtain highly purified megakaryocytes, we isolated CD41+/Gr1− bone marrow cells by FACS and cultured the cells for 2 additional days in medium containing thrombopoietin (TPO).24 This procedure yielded a pure population of large, acetylcholine esterase-positive cells (Figure S2), and in DNA from these cells we detected complete excision of integrin β1 (Figure 2B).
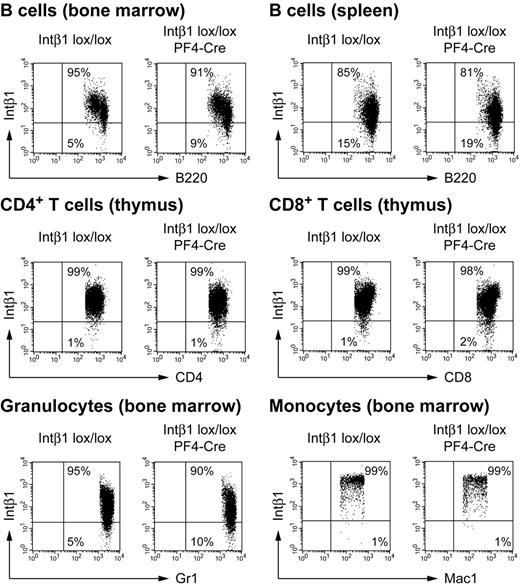
To determine the efficiency of Cre-mediated excision in megakaryocytes and platelets, we measured the loss of integrin β1 protein on the cell surface by flow cytometry (Figure 2C). Platelets from Pf4-Cre × integrin β1 lox/lox mice showed an almost complete disappearance of the integrin β1 protein, indicating that Pf4-Cre is highly active in terminally differentiated megakaryocytes. To obtain sufficient numbers of megakaryocytes for flow cytometry, we cultured bone marrow cells for 7 days in the presence of TPO. In cultured megakaryocytes from Pf4-Cre × integrin β1 lox/lox mice, integrin β1 protein was still detectable, although at clearly reduced levels (Figure 2C). This finding seems to be partially at odds with the complete excision of the floxed integrin β1 gene in Southern blot analysis, but this discrepancy could reflect a long half-life of the integrin β1 protein in megakaryocytes. Lineage specificity of Pf4-Cre transgene expression was further demonstrated by the lack of Cre-mediated excision in myeloid and lymphoid cells. We detected no or only minimal decrease in integrin β1 protein surface expression in B cells, T cells, granulocytes, and monocytes from Pf4-Cre × integrin β1 lox/lox mice (Figure 3), confirming the absence of excision in leukocytes assessed by Southern analysis (Figure 2B).
Three-color flow cytometry of different hematopoietic lineages from bone marrow, spleen, and thymus. After lysis of red blood cells, leukocytes were triple-stained with antibodies against the respective lineage markers (x-axis, PE) integrin β1 (y-axis, APC) and CD41 (not shown, FITC). CD41+ cells were excluded from the analysis by gating to minimize contamination by adhering platelets. A second gate was set to select cells positive for the respective lineage markers. The quadrants were set based on isotype controls. Analysis of peripheral blood leukocytes with a gate on CD41− cells led to similar results (not shown).
Three-color flow cytometry of different hematopoietic lineages from bone marrow, spleen, and thymus. After lysis of red blood cells, leukocytes were triple-stained with antibodies against the respective lineage markers (x-axis, PE) integrin β1 (y-axis, APC) and CD41 (not shown, FITC). CD41+ cells were excluded from the analysis by gating to minimize contamination by adhering platelets. A second gate was set to select cells positive for the respective lineage markers. The quadrants were set based on isotype controls. Analysis of peripheral blood leukocytes with a gate on CD41− cells led to similar results (not shown).
The BAC-derived Pf4-Cre transgene allowed efficient and lineage-restricted excision of a loxP target gene. We did not observe unforeseen effects of the transgene insertion or the presence of the additional copy of chemokine genes present in the BAC construct, but such effects may need to be controlled for, depending on the future specific applications. Given the lineage specificity and efficiency of excision, the Pf4-Cre mice promise to be a very useful tool to study megakaryopoiesis, platelet formation, and platelet function.
Authorship
Contribution: R.T. designed and performed the research, analyzed the data, and wrote the paper; T.S. designed and performed the research and analyzed data, H.H.-S. performed the research; and R.C.S. designed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
R.T. and T.S. contributed equally to this study.
Correspondence: Radek C. Skoda, Department of Research and Experimental Hematology, University Hospital Basel, Hebelstrasse 20, 4031 Basel, Switzerland; e-mail: radek.skoda@unibas.ch.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Dr Ulrich Müller (The Scripps Research Institute, La Jolla, CA) for the integrin β1 lox mice, Dr Günther Schütz and Dr Erich Greiner (German Cancer Research Center, Heidelberg, Germany) for the codon-improved Cre cDNA, and Verena Jäggin and Dr Christian Kalberer (Department of Research, University Hospital Basel, Switzerland) for help with flow cytometry.
This work was supported by the Swiss National Science Foundation (grant 310 000-108 006/1).


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal