Abstract
The chemokine stromal cell–derived factor-1 (SDF-1) is constitutively expressed by bone marrow stromal cells and plays key roles in hematopoiesis. Fibroblast growth factor 2 (FGF2), a member of the FGF family that plays important roles in developmental morphogenic processes, is abnormally elevated in the bone marrow from patients with clonal myeloid disorders and other disorders where normal hematopoiesis is impaired. Here, we report that FGF2 reduces SDF-1 secretion and protein content in bone marrow stromal cells. By inhibiting SDF-1 production, FGF2 compromises stromal cell support of hematopoietic progenitor cells. Reverse-transcriptase–polymerase chain reaction (RT-PCR) analysis revealed that bone marrow stromal cells express 5 FGF receptors (FGFRs) among the 7 known FGFR subtypes. Blocking experiments identified FGFR1 IIIc as the receptor mediating FGF2 inhibition of SDF-1 expression in bone marrow stromal cells. Analysis of the mechanisms underlying FGF2 inhibition of SDF-1 production in bone marrow stromal cells revealed that FGF2 reduces the SDF-1 mRNA content by posttranscriptionally accelerating SDF-1 mRNA decay. Thus, we identify FGF2 as an inhibitor of SDF-1 production in bone marrow stromal cells and a regulator of stromal cell supportive functions for hematopoietic progenitor cells.
Introduction
Stromal cell–derived factor-1 (SDF-1) is a highly conserved CXC chemokine (CXCL12) originally cloned from cDNA libraries constructed from mouse bone marrow,1 activated mouse embryo,2 and a mouse stromal cell line.3 SDF-1 mRNA is detected in many organs and tissues, and is especially abundant in the bone marrow, lymph nodes, spleen, lung, and liver.4,5 SDF-1 and its CXCR4 receptor serve as critical regulators of hematopoiesis during development and after birth.6-9 Stromal cells, which constitutively express SDF-1, are a principal source of the chemokine in the bone marrow.10-12 Hematopoietic progenitor cells and pre-B cells express CXCR4 and physically interact with SDF-1–positive stromal cells.12 This SDF-1/CXCR4 interaction serves as a retention signal for bone marrow cells to the bone marrow, preventing their release to the peripheral blood.7 CXCR4 or SDF-1 inactivation promotes the mobilization of hematopoietic progenitors to the peripheral blood.13-17 SDF-1 is a growth factor for pre-B cells6 and a survival factor for myeloid progenitor cells.10,18,19 Acting cooperatively with other growth factors, SDF-1 can promote the proliferation of CD34+ hematopoietic progenitor cells.18 Thus, it is predictable that regulation of SDF-1 expression in bone marrow stromal cells can play important roles in hematopoiesis. However, there are only a limited number of studies investigating the patterns of SDF-1 expression in bone marrow stromal cells,20 and little is currently known about transcriptional and posttranscriptional regulation of SDF-1 gene expression.21
FGF2 and other structurally related polypeptides are potent inducers of growth, survival, chemotaxis, and differentiation in a variety of cell types, and play key roles in morphogenesis, development, angiogenesis, bone formation, and wound healing.22-24 Members of the FGF superfamily exert their activities by binding to heparan sulfate proteoglycans and FGF receptors (FGFRs).25,26 The FGFR superfamily consists of 4 members, designated FGFR1, FGFR2, FGFR3, and FGFR4.26 Alternative splicing events in FGFR1, FGFR2, and FGFR3 increase the number of principal FGFRs to 7 (FGFR1-IIIb, FGFR1-IIIc, FGFR2-IIIb, FGFR2-IIIc, FGFR3-IIIb, FGFR3-IIIc, and FGFR4).26-33 Structurally, FGFRs consist of an extracellular region containing 3 immunoglobulin (Ig)–like domains (D1-D3), a single transmembrane helix, and a cytoplasmic domain with protein tyrosine kinase activity.34
FGF2 is present in bone marrow, but the cell types that produce FGF2 in bone marrow remain undefined. Bone marrow stromal cells,35 megakaryocytes, and platelets36 have been reported to contain FGF2. Several studies reported that FGF2 variously modulates hematopoiesis in vitro, and suggested that FGF2 may play a role in normal and pathological hematopoiesis.37 In long-term bone marrow cultures, FGF2 at low concentrations (0.2-2 ng/mL) increased the number of progenitor cells of myeloid lineage, but the mechanisms underlying this action are not clear.38 FGF2 stimulated megakaryocytopoiesis in various culture systems, acting indirectly through IL-6, IL-1, or perhaps IL-3.37,39-41 Genetic defects of FGF receptors have been linked to a set of diseases affecting the musculoskeletal system, and deregulated FGF2 has been linked to atherosclerosis.37 FGF2 is found at abnormally high concentrations in the bone marrow of patients with various clonal chronic myeloproliferative diseases, including myeloid metaplasia with myelofibrosis,42 which are often associated with reduced bone marrow hematopoiesis, myelofibrosis, release of immature cells to the peripheral blood, and the development of extramedullary hematopoiesis.43,44
Since reduced bone marrow hematopoiesis, premature release of immature hematopoietic progenitor cells from the bone marrow, and extramedullary hematopoiesis are potential consequences of long-term SDF-1 reduction in the bone marrow, we investigated the possibility that FGF2 might down-regulate SDF-1 production in bone marrow stromal cells. In this study, we identify FGF2 as an inhibitor of SDF1 expression in bone marrow stromal cells, and provide evidence that FGF2 can subvert the supportive properties of stromal cells for hematopoiesis.
Materials and methods
Cells and reagents
The mouse bone marrow–derived stromal cell lines MS-545 and S-1746 were gifts from Dr A. C. Berardi (Ospedale Bambin Gesu, Rome, Italy) and Dr K. Dorshkind (UCLA, Riverside, CA). Stromal cell lines were maintained in αMEM containing 10% FBS (Invitrogen, Carlsbad, CA). Primary bone marrow stromal cell cultures were established according to the method of Dexter et al47,48 with modifications. Briefly, bone marrow cells were harvested by flushing the femoral bone cavity of 6-week-old female C57BL/6 mice (National Cancer Institute, Frederick, MD) in αMEM containing 20% FBS. Cultures were fed weekly by removal of the supernatant medium containing suspension cells and replacement with fresh medium. When the adherent cells grew to confluency, they were trypsinized and subcultured. Human recombinant FGF2 was from Peprotech (Rocky Hill, NJ) and R&D Systems (Minneapolis, MN); human recombinant SDF-1α, FGFR/Fc chimeras (FGFR1 IIIb/Fc, FGFR1 IIIc/Fc, FGFR3 IIIb/Fc, and FGFR3 IIIc/Fc), anti–FGFR2 IIIc neutralizing antibody (rat antimouse mAb), FGF2, FGF4, FGF7, FGF9, FGF16, and FGF19 were from R&D Systems. AMD 3100 was from the NIH AIDS Research and Reference Reagent Program (Germantown, MD).
Reverse-transcriptase–polymerase chain reaction (RT-PCR) analysis
Total RNA was extracted using TRIzol Reagent (Molecular Research Center, Cincinnati, OH). Semiquantitative SDF-1 RT-PCR was carried out as described.11 Amounts of cDNA for each amplification reaction were based on the results of PCR for GAPDH showing equivalent amounts of product amplified from all samples. Primer sequences for detection of FGFR expression, predicted sizes of the amplified product, and positive controls for the expression of mRNA are shown in Table 1
Primers, PCR product sizes, and positive controls for FGFR PCR analysis
| Gene . | PCR size, bp . | Positive control . | Primer sequences . | |
|---|---|---|---|---|
| Forward . | Reverse . | |||
| FGFR1 IIIb | 320 | Brain | 5′-CTTGACGTCGTGGAACGATCT-3′ | 5′-CACGCAGACTGGTTAGCTTCAC-3′ |
| FGFR1 IIIc | 344 | Brain | 5′-CTTGACGTCGTGGAACGATCT-3′ | 5′-AGAACGGTCAACCATGCAGAG-3′ |
| FGFR2 IIIb | 216 | Lung | 5′-CCCATCCTCCAAGCTGGACTG-3′ | 5′-CAGAGCCAGCACTTCTGCATTG-3′ |
| FGFR2 IIIc | 330 | Lung | 5′-CCCATCCTCCAAGCTGGACTG-3′ | 5′-TCTCACAGGCGCTGGCAGAAC-3′ |
| FGFR3 IIIb | 336 | Lung | 5′-CAAGTTTGGCAGCATCCGGCAGAC-3′ | 5′-TCTCAGCCACGCCTATGAAATTGGTG-3′ |
| FGFR3 IIIc | 138 | Brain (E13) | 5′-GGCGCTAACACCACCGAC-3′ | 5′-TGGCAGCACCACCAGCCAC-3′ |
| FGFR4 | 243 | Liver | 5′-GTACCCTCGGACCGCGGCACATAC-3′ | 5′-GCCGAAGCTGCTGCCGTTGATG-3′ |
| GAPDH | 446 | NA | 5′-GCCACCCAGAAGACTGTGGATGGC-3′ | 5′-CATGTAGGCCATGAGGTCCACCAC-3′ |
| Gene . | PCR size, bp . | Positive control . | Primer sequences . | |
|---|---|---|---|---|
| Forward . | Reverse . | |||
| FGFR1 IIIb | 320 | Brain | 5′-CTTGACGTCGTGGAACGATCT-3′ | 5′-CACGCAGACTGGTTAGCTTCAC-3′ |
| FGFR1 IIIc | 344 | Brain | 5′-CTTGACGTCGTGGAACGATCT-3′ | 5′-AGAACGGTCAACCATGCAGAG-3′ |
| FGFR2 IIIb | 216 | Lung | 5′-CCCATCCTCCAAGCTGGACTG-3′ | 5′-CAGAGCCAGCACTTCTGCATTG-3′ |
| FGFR2 IIIc | 330 | Lung | 5′-CCCATCCTCCAAGCTGGACTG-3′ | 5′-TCTCACAGGCGCTGGCAGAAC-3′ |
| FGFR3 IIIb | 336 | Lung | 5′-CAAGTTTGGCAGCATCCGGCAGAC-3′ | 5′-TCTCAGCCACGCCTATGAAATTGGTG-3′ |
| FGFR3 IIIc | 138 | Brain (E13) | 5′-GGCGCTAACACCACCGAC-3′ | 5′-TGGCAGCACCACCAGCCAC-3′ |
| FGFR4 | 243 | Liver | 5′-GTACCCTCGGACCGCGGCACATAC-3′ | 5′-GCCGAAGCTGCTGCCGTTGATG-3′ |
| GAPDH | 446 | NA | 5′-GCCACCCAGAAGACTGTGGATGGC-3′ | 5′-CATGTAGGCCATGAGGTCCACCAC-3′ |
NA indicates not applicable.
SDF-1 enzyme-linked immunosorbent assay (ELISA) and Western blotting
The ELISA for detection of murine SDF-1 was performed as described.11 The lower detection limit for SDF-1 was 33 pg/mL. Western blotting for murine SDF-1 was performed as described,11 using anti–SDF-1 antibody (1 μg/mL; Peprotech) and an affinity-purified, peroxidase-linked, donkey anti–rabbit IgG antibody (Amersham Pharmacia Biotech, Piscataway, NJ).
In vitro stromal cell stimulation with FGFs
MS-5 cells and S-17 cells (80%-90% confluent) were incubated in culture medium (αMEM containing 10% FBS) alone, or with FGF2, FGF4, FGF7, FGF9, FGF16, or FGF19 (50 ng/mL) for the indicated time, and then cell-free culture supernatants were collected. Primary bone marrow stromal monolayers were established in 6-well plates (80%-90% confluent) and stimulated with or without FGF2 (50 ng/mL in αMEM containing 20% FBS) for the indicated times. SDF-1 content in the culture supernatant and/or in the cell lysates was measured by a specific ELISA and Western blotting, respectively.
In vitro cell proliferation studies
Cells were washed twice with PBS, suspended in culture medium (αMEM containing 10% FBS), plated (2000 cells/well in 0.2 mL culture medium) in triplicate with the addition of FGFs (50 ng/mL), FGFR/Fc chimeras (2000 ng/mL), or anti–FGFR2 IIIc antibody (2000 ng/mL) onto 96-well plates, and incubated for 64 hours. DNA synthesis was measured by 3 H thymidine deoxyribose uptake (0.5 μCi [0.0185 MBq]/well; New England Nuclear, Boston, MA) during the last 16 hours of culture as described previously.49 The results are expressed as mean cpm (SD)/culture.
In vitro inhibition assays using FGFR/Fc chimeras and anti–FGFR2 IIIc neutralizing antibody
FGFR Fc/chimeras (0-2000 ng/mL in αMEM containing 10% FBS) were preincubated with/without FGF2 (10 ng/mL) for 30 minutes, and then applied onto stromal layers (∼ 90% confluent). Anti–FGFR2 IIIc antibody (0-2000 ng/mL final concentration) was first applied onto stromal monolayers and incubated for 30 minutes, and then FGF2 (10 ng/mL final concentration) was added. A specific ELISA was used to measure SDF-1 in 72-hour culture supernatants. The percent SDF-1 secretion was calculated as follows: ([SDF-1 secretion in FGFR/Fc group]/[SDF-1 secretion in control group with no additive]) × 100 (%). All experiments were performed at least in triplicate and repeated 4 times.
Flow cytometry
Cells were washed twice in PBS, and stained with fluorescein isothiocyanate (FITC)–labeled mouse monoclonal antibodies to human CD33 and CD45, or with FITC-labeled isotype-matched antibodies (2 μg/mL at 4°C) for 45 minutes (all antibodies from Becton Dickinson, Franklin Lakes, NJ). Surface antigens were evaluated from 1 × 104 viable cells using a FACScalibur cytofluorometer (Becton Dickinson) and analyzed using CELLQuest software (Becton Dickinson). Background fluorescence was assessed through staining with isotype-matched antibodies.
Coculture of CD34+ peripheral blood progenitor cells with MS-5 and S-17 cells
Cocultures were carried out as described50 with modifications. MS-5 and S-17 stromal cell feeders (80%-90% confluent established on 0.5% gelatin precoated 12-well plates) were pretreated with FGF2 (50 ng/mL) or medium alone for 72 hours. Pretreated stromal cell monolayers were incubated with SDF-1α (2 hours, 500 ng/mL) or medium alone. G-CSF–mobilized CD34+ peripheral blood stem cells (PBSCs; 1.0 × 105 cells) were applied in 1 mL long-term culture medium (αMEM containing 12.5% horse serum, 12.5% FBS, 1 μM hydrocortisone, and 50 μM 2-ME) onto stromal cell monolayers, and the cocultures were incubated for 3 weeks in medium alone, AMD3100 (5 μg/mL), FGF2 (10 ng/mL), or FGF2 (10 ng/mL) plus SDF-1α (500 ng/mL) with replenishment of 0.5 mL culture medium alone or with the original additives twice/week. After 3-week incubation, nonadherent and loosely adherent cells were counted and analyzed by fluorescence-activated cell sorting (FACS); percent cell growth was calculated as follows: (CD45+ cell number with AMD3100, with FGF2, or with FGF2 plus SDF-1/CD45+ cell number with no additive) × 100 (%). To evaluate whether FGFR1 IIIc/Fc reconstitutes the supportive property of stromal cells for CD34+ progenitor cells in the presence of FGF2, MS-5 and S-17 stromal cells (80% confluent, established 0.5% gelatin precoated 24-well plate, 4 wells per group) were pretreated with medium alone, with FGFR1 IIIc/Fc (800 ng/mL), with FGF2 (10 ng/mL), with FGFR1 IIIc/Fc (800 ng/mL) plus FGF2 (10 ng/mL), or with FGFR1 IIIb/Fc (800 ng/mL) plus FGF2 (10 ng/mL) for 72 hours. The mixture of FGFR/Fc and FGF2 was preincubated (30 minutes) before addition to stromal layers. After 72-hour incubation, culture supernatants were removed, and PBSCs were applied onto pretreated stromal layers. PBSCs (6 × 104 cells /well) were suspended (360 μL) with long-term culture medium alone, with FGFR1 IIIc/Fc (800 ng/mL), with FGF2 (10 ng/mL), with FGFR1 IIIc/Fc (800 ng/mL) plus FGF2 (10 ng/mL), or with FGFR1 IIIb/Fc (800 ng/mL) plus FGF2 (10 ng/mL). Cocultures were replenished with 180 μL medium alone or containing the original additives twice/week. After 3-week coculture, nonadherent and loosely adherent cells were counted and analyzed by FACS. The percent growth of PBSCs was calculated as ([CD45+ cell number with FGFR1 IIIc, with FGF2, with FGF2 plus FGFR1 IIIc/Fc, or with FGF2 plus FGFR1 IIIc/Fc]/[cell number with no additive]) × 100 (%).
Isolation of SDF-1 upstream sequences and plasmid construction
An approximately 2.2-kb DNA fragment containing SDF-1 upstream sequences and the partial coding sequence (−2030 to +149 relative to the transcription start site) was amplified by PCR using primers SDF-1-I (5′-GTAGGAGGACCTACTGAACG-3′) and SDF-1-II (5′-TGAACTCACCGTCACTGATGC-3′). The primer design was based on the database of mouse genome sequence (UCSC Genome Bioinformatics, downloadable at http://genome.UCSC.edu/); genomic DNA extracted from MS-5 cells using QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA) was used as a template for PCR. The PCR-amplified product was subcloned into PCR2.1(+) cloning vector using TOPO TA Cloning Kits (Invitrogen, Carlsbad, CA) and sequenced at the NCI sequencing core facility NCI (Bethesda, MD) to verify sequence identity to the mouse genome database, except for a 2-T deletion in 35 Ts in a row (−1693 to −1659). The SDF-1 upstream sequence (−2030 to +41) was isolated by PCR using primers SDF-1-I and SDF-1-III (5′-CCTGTCGTTCTCCTGTGGCTC-3′). The PCR2.1 (+) vector containing the 2.2-kb DNA-amplified fragment was used as a template. The amplified 2071-bp PCR product was subcloned into PCR2.1 (+) cloning vector and designated as PCR2.1+2071 bp. PCR2.1+2071 bp, which possesses a unique Kpn1 and Xho1 site, was digested with Kpn1 and Xho1, and the fragment inserted into the promoterless reporter vector pGL3.0-Basic (Promega, Madison, WI) was designated pGL3.0+2071 bp.
Transient transfection and luciferase assay
MS-5 cells and S-17 cells (70%-80% confluent in 48-well plates) were transfected with pGL3.0-basic or pGL3.0+2071bp (250 ng/well) with 0.3 μL/well lipofectamine 2000 reagent (Invitrogen). phRL-SV40 (2.5 ng/well, Renilla luciferase reference control plasmid; Promega) was transfected as a control. All assays were carried out in at least 4 independent wells. After 24-hour incubation, pGL3.0+2071 and phRL-SV40–transfected cells were stimulated with/without FGF2 (50 ng/mL). Firefly luciferase activity and Renilla luciferase activity were measured using the Dual-Glo Luciferase assay system following the manufacturer's instructions (Promega). All experiments were performed at least in triplicate. Relative luciferase activity is defined as the ratio of firefly luciferase activity to Renilla luciferase activity.
PCR measurement of SDF-1 mRNA decay
To evaluate the stability of SDF-1 mRNA, S-17 cells (80%-90% confluent) were treated with/without 5 μg/mL α-amanitin (Sigma, St Louis, MO) for 8, 16, 24, 32, 40, or 48 hours at 37°C. mRNA was extracted and subjected to semiquantitative RT-PCR analysis, as described. Semiquantitative RT-PCR analysis for flt3-L gene51 was performed in parallel. The number of amplification cycles was determined experimentally for flt3-L primer pair to fit the linear part of the sigmoid curve, reflecting the relationship between the number of amplification cycles and the amount of PCR product. To evaluate the effect of FGF2 on SDF-1 mRNA decay, S-17 cells were pretreated with/without FGF2 (50 ng/mL) for 6 hours, and then α-amanitin (5 μg/mL) was added to cultures. FGF2-pretreated cells were also tested without α-amanitin addition. After incubation (1.5, 3, 4.5, 6, 7.5, 10, and 24 hours), SDF-1 mRNA was measured by semiquantitative PCR. Amounts of cDNA used for each amplification reaction were based on the results of PCR for GAPDH showing equivalent amounts of product amplified. GAPDH half-life has been reported to be considerably long.52,53 Relative SDF-1 mRNA level (SDF-1/GAPDH) was calculated by densitometric scanning using NIH image software (http://rsb.info.nih.gov/nih-image/). Percent relative SDF-1 expression is defined as percent of the initial (−6-hour time point) relative SDF-1 expression. Experiments were repeated 3 times.
Statistical analysis
Statistical significance of group differences was evaluated by the Student t test using Excel software (Microsoft, Redmond, WA).
Results
FGF2 down-regulates SDF-1 production in stromal cells and impairs stromal cell supportive properties
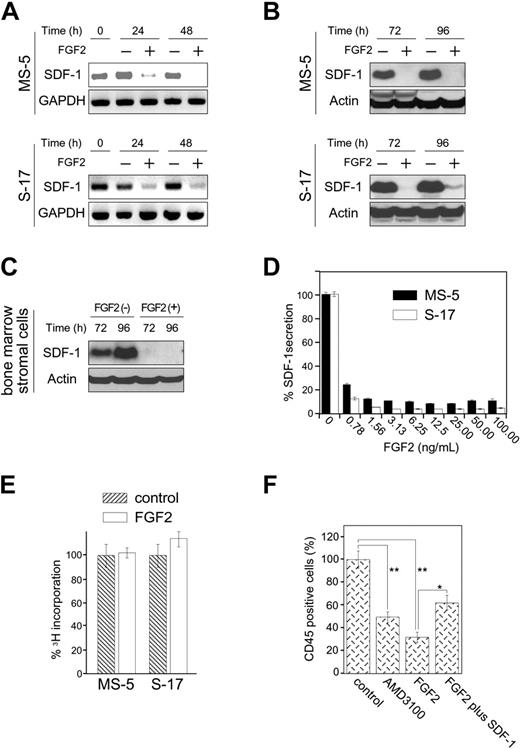
We tested whether FGF2 can modulate SDF1 expression in the bone marrow–derived stromal cell lines MS-5 and S-17 and primary bone marrow stromal cells. By semiquantitative RT-PCR, FGF2 (50 ng/mL) reduced SDF-1 mRNA levels in MS-5 and S-17 bone marrow stromal cells after 24 and 48 hours (Figure 1A). By immunoblotting, FGF2 decreased the SDF-1 content in MS-5 and S-17 cells after 72 and 96 hours (Figure 1B). Similarly, FGF2 decreased the SDF-1 content in primary, bone marrow–derived stromal cells (Figure 1C). A dose-response analysis using a specific ELISA showed that FGF2 concentrations equal to or higher than 2.5 ng/mL produced maximal inhibition of SDF-1 secretion by MS-5 and S-17 cells after 72 hours (Figure 1D). FGF2 (50 ng/mL) minimally altered the proliferation of MS-5 and S-17 cells after 64-hour incubation under culture conditions associated with reduced SDF-1 content and secretion (Figure 1E).
Effects of FGF2 on stromal cell production of SDF-1. (A) Semiconfluent cultures of MS-5 and S-17 cells were incubated in medium alone or with FGF2 (50 ng/mL). At the indicated time points, total RNA was extracted and tested by semiquantitative RT-PCR for SDF-1 and GAPDH mRNAs (representative experiment of 3). (B) Semiconfluent cultures of MS-5 and S-17 cells were incubated with or without FGF2 (50 ng/mL) for the indicated times. SDF-1 content was measured by Western blotting in cell lysates. After immunoblotting for SDF-1, the membranes were reprobed with antibodies to actin (representative experiment of 3 performed). (C) Semiconfluent cultures of primary bone marrow stromal cells were incubated with or without FGF2 (50 ng/mL) for the indicated times. SDF-1 content was measured by Western blotting in cell lysates. After immunoblotting for SDF-1, the membranes were reprobed with antibodies to actin (representative experiment of 3 performed). (D) MS-5 and S-17 cells were cultured with FGF2 at various concentrations (0-100 ng/mL) for 72 hours; levels of SDF-1 were measured in the culture supernatants. The results reflect the means (± SD) of triplicate determinations (representative experiment of 3 performed). (E) Effects of FGF2 (50 ng/mL) on MS-5 and S-17 cell proliferation. MS-5 cells and S-17 cells were detached, washed twice with PBS, suspended in culture medium (αMEM containing 10% FBS), and incubated (2000 cells/well in 0.2 mL culture medium; triplicate cultures in 96-well plates) with the addition of medium only or medium with FGF2 (50 ng/mL). After 64-hour incubation, DNA synthesis was measured by 3 H thymidine deoxyribose uptake. The results, expressed as mean (± SD) cpm/culture, from 3 independent experiments are shown. (F) Effects of exogenous SDF-1 on hematopoietic progenitor cell growth onto stromal cells. MS-5 feeders (12-well plate) were pretreated for 72 hours with or without FGF2 (50 ng/mL). After washing, FGF2-treated feeders were incubated with medium only or with recombinant human SDF-1 (500 ng/mL) for 2 hours. After removal of culture supernatants from all wells, human CD34+ peripheral blood progenitor cells (PBSCs, 1.0 × 105 cells/well, 3 wells/group) were added onto treated and untreated MS-5 cells. CD34+ cells were added in medium alone or medium with AMD3100 (5 μg/mL) onto untreated feeders; and CD34+ cells were added in medium with FGF2 (10 ng/mL) alone or FGF2 plus SDF-1 (500 ng/mL) onto FGF2-treated only and FGF2-treated and SDF-1–replenished feeders, respectively. Cocultures were incubated for 3 weeks with replenishment of culture medium alone (0.5 mL) or with the appropriate additives (10 ng/mL FGF2, 5 μg/mL AMD3100, 10 ng/mL FGF2 plus 500 ng/mL SDF-1) twice/wk. Nonadherent viable cells were counted and analyzed by FACS at the end of incubation. Control indicates untreated MS-5 cells plus untreated CD34+ cells; AMD3100, untreated MS-5 cells plus untreated CD34+ cells plus AMD3100; FGF2, MS-5 feeders treated with FGF2 plus untreated CD34+ cells; and FGF2 plus SDF-1, MS-5 feeders treated with FGF2 plus SDF-1–treated CD34+ cells. The percent growth of PBSCs was calculated as (CD45+ cell number with AMD3100, FGF2, or FGF2 plus SDF-1/CD45+ cell number with no additive) × 100 (%). The results reflect the means (± SD) of 3 independent experiments. The asterisk denotes statistical significance (**P < .01; *P < .05).
Effects of FGF2 on stromal cell production of SDF-1. (A) Semiconfluent cultures of MS-5 and S-17 cells were incubated in medium alone or with FGF2 (50 ng/mL). At the indicated time points, total RNA was extracted and tested by semiquantitative RT-PCR for SDF-1 and GAPDH mRNAs (representative experiment of 3). (B) Semiconfluent cultures of MS-5 and S-17 cells were incubated with or without FGF2 (50 ng/mL) for the indicated times. SDF-1 content was measured by Western blotting in cell lysates. After immunoblotting for SDF-1, the membranes were reprobed with antibodies to actin (representative experiment of 3 performed). (C) Semiconfluent cultures of primary bone marrow stromal cells were incubated with or without FGF2 (50 ng/mL) for the indicated times. SDF-1 content was measured by Western blotting in cell lysates. After immunoblotting for SDF-1, the membranes were reprobed with antibodies to actin (representative experiment of 3 performed). (D) MS-5 and S-17 cells were cultured with FGF2 at various concentrations (0-100 ng/mL) for 72 hours; levels of SDF-1 were measured in the culture supernatants. The results reflect the means (± SD) of triplicate determinations (representative experiment of 3 performed). (E) Effects of FGF2 (50 ng/mL) on MS-5 and S-17 cell proliferation. MS-5 cells and S-17 cells were detached, washed twice with PBS, suspended in culture medium (αMEM containing 10% FBS), and incubated (2000 cells/well in 0.2 mL culture medium; triplicate cultures in 96-well plates) with the addition of medium only or medium with FGF2 (50 ng/mL). After 64-hour incubation, DNA synthesis was measured by 3 H thymidine deoxyribose uptake. The results, expressed as mean (± SD) cpm/culture, from 3 independent experiments are shown. (F) Effects of exogenous SDF-1 on hematopoietic progenitor cell growth onto stromal cells. MS-5 feeders (12-well plate) were pretreated for 72 hours with or without FGF2 (50 ng/mL). After washing, FGF2-treated feeders were incubated with medium only or with recombinant human SDF-1 (500 ng/mL) for 2 hours. After removal of culture supernatants from all wells, human CD34+ peripheral blood progenitor cells (PBSCs, 1.0 × 105 cells/well, 3 wells/group) were added onto treated and untreated MS-5 cells. CD34+ cells were added in medium alone or medium with AMD3100 (5 μg/mL) onto untreated feeders; and CD34+ cells were added in medium with FGF2 (10 ng/mL) alone or FGF2 plus SDF-1 (500 ng/mL) onto FGF2-treated only and FGF2-treated and SDF-1–replenished feeders, respectively. Cocultures were incubated for 3 weeks with replenishment of culture medium alone (0.5 mL) or with the appropriate additives (10 ng/mL FGF2, 5 μg/mL AMD3100, 10 ng/mL FGF2 plus 500 ng/mL SDF-1) twice/wk. Nonadherent viable cells were counted and analyzed by FACS at the end of incubation. Control indicates untreated MS-5 cells plus untreated CD34+ cells; AMD3100, untreated MS-5 cells plus untreated CD34+ cells plus AMD3100; FGF2, MS-5 feeders treated with FGF2 plus untreated CD34+ cells; and FGF2 plus SDF-1, MS-5 feeders treated with FGF2 plus SDF-1–treated CD34+ cells. The percent growth of PBSCs was calculated as (CD45+ cell number with AMD3100, FGF2, or FGF2 plus SDF-1/CD45+ cell number with no additive) × 100 (%). The results reflect the means (± SD) of 3 independent experiments. The asterisk denotes statistical significance (**P < .01; *P < .05).
We tested whether, by reducing SDF-1 expression, FGF2 impairs stromal cell support of CD34+ cells. Using a previously described in vitro coculture system in which purified human CD34+ hematopoietic cells are incubated for 3 weeks onto murine stromal cell monolayers,50 we found that recovery of viable human CD45+ cells was significantly reduced if the cocultures were supplemented with FGF2 (50 ng/mL). Recovery of human CD45+ cells was also reduced by culture supplementation with AMD3100 (5 μg/mL), a specific inhibitor of the SDF-1 receptor CXCR4. Addition of SDF-1 to the FGF2-supplemented cocultures (by preincubation of the MS-5 stromal cells with SDF-1 and periodic addition of SDF-1) significantly improved the recovery of human CD45+ cells (Figure 1E). All CD45+ cells recovered from 3-week coculture were also CD33+. Similar results were derived from coculture of human CD34+ cells onto S-17 stromal cells (not shown). AMD3100 did not affect the proliferation or SDF-1 secretion of MS-5 and S-17 stromal cells (not shown). FGF2 did not reduce viability or CXCR4 expression in CD34+ cells (not shown). These results provide evidence that FGF2 compromises stromal cell support of hematopoietic progenitor cells by reducing SDF-1 expression.
Analysis of FGF receptor expression and function in stromal cells
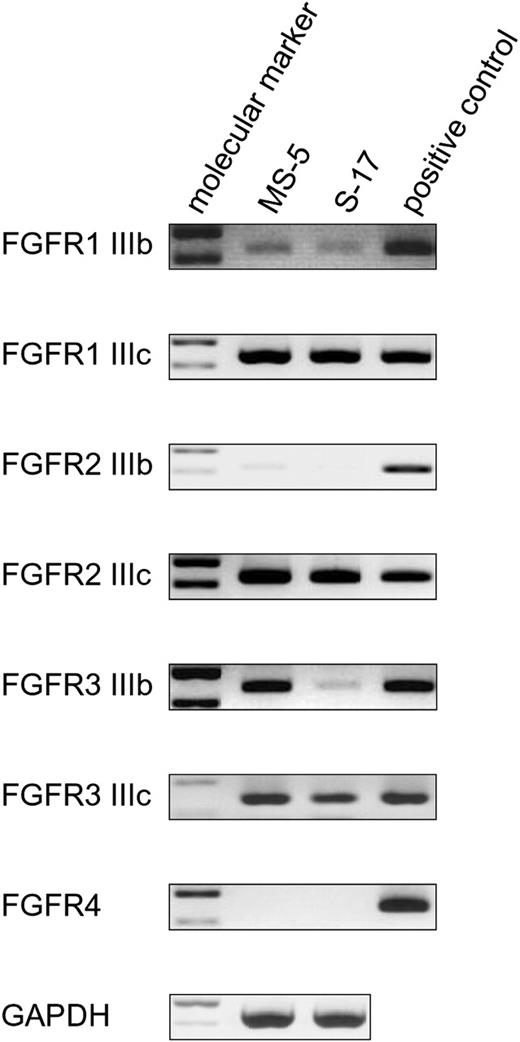
Since the FGFR expression profile in stromal cells is incompletely described, we tested MS-5 and S-17 cells for the presence of specific transcripts for each of the 7 receptors. Using specific primer pairs and appropriate control mRNAs from mouse tissues (Table 1), we detected similar expression of FGFR1 IIIb, FGFR1 IIIc, FGFR2 IIIc, and FGFR3 IIIc in MS-5 and S-17 cells (Figure 2). Compared with control mRNA from brain tissue, FGFR1 IIIb levels were somewhat lower in both MS-5 and S-17 cells. FGFR3 IIIb was expressed strongly in MS-5 cells but only weakly in S-17 cells, and FGFR2 IIIb and FGFR4 were minimally expressed in both cell lines (Figure 2B).
Expression of FGF receptors in stromal cells. Expression of FGFRs in MS-5 and S-17 cells was detected by RT-PCR. Total RNA extracted from MS-5 cells and S-17 cells was subjected to RT-PCR using specific primer pairs. RNA quality was evaluated in all samples by parallel RT-PCR for GAPDH. Absence of contaminating genomic DNA was ensured by RNA-PCR. PCR products were separated on a 2% agarose gel prestained with 1 μg/mL ethidium bromide and visualized under UV light. Representative results from at least 2 independent experiments are shown.
Expression of FGF receptors in stromal cells. Expression of FGFRs in MS-5 and S-17 cells was detected by RT-PCR. Total RNA extracted from MS-5 cells and S-17 cells was subjected to RT-PCR using specific primer pairs. RNA quality was evaluated in all samples by parallel RT-PCR for GAPDH. Absence of contaminating genomic DNA was ensured by RNA-PCR. PCR products were separated on a 2% agarose gel prestained with 1 μg/mL ethidium bromide and visualized under UV light. Representative results from at least 2 independent experiments are shown.
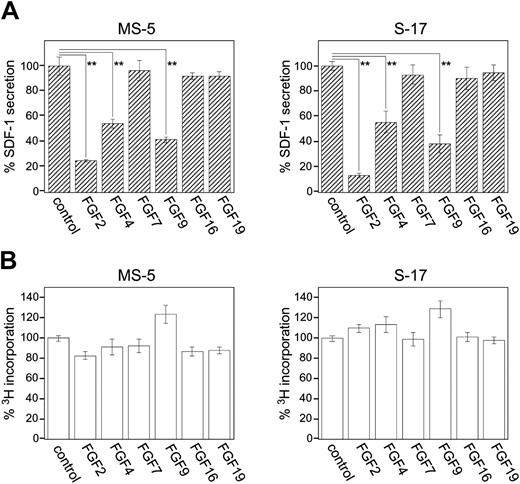
FGF2 has been reported to bind to FGFR1 IIIc and FGFR2 IIIc, but not to FGFR2 IIIb or FGFR3 IIIc.26 The binding of FGF2 to FGFR1 IIIb, FGFR3 IIIb, or FGFR4 has not been studied. The FGF superfamily consists of 22 members in humans and mice,54 each member possessing different binding patterns to FGFR subtypes.26 To identify the receptor that mediates FGF2 inhibition of SDF-1 expression in stromal cells, we examined the effects of other FGF family members (FGF4, FGF9, FGF16, and FGF19) selected on the basis of their reported26,32,33,55,56 preferential binding to individual FGFRs (Table 2)Using a specific ELISA, we measured SDF-1 secretion by MS-5 and S-17 cells cultured (72 hours) in the presence of 50 ng/mL FGF2, FGF4, FGF7, FGF9, FGF16, or FGF19. In addition to FGF2, FGF4 and FGF9 significantly decreased SDF-1 production by MS-5 and S-17 cells compared with control medium alone (Figure 3A). FGF7, which binds only to FGFR2 IIIB; FGF16, which binds only to FGFR2 IIIc; and FGF19, which binds only to FGFR4, minimally affected SDF-1 secretion by the MS-5 and S-17 stromal cells, suggesting that FGFR2 IIIb, FGFR2 IIIc, and FGFR4 are unlikely involved in FGF2-induced SDF-1 down-regulation. FGF9 binds weekly to FGFR2 IIIc and may not induce SDF-1 depletion through this receptor. None of the FGFs tested here significantly affected the proliferation of MS-5 and S-17 stromal cells under the conditions used to study SDF-1 modulation (Figure 3B). Thus, the patterns of FGF receptor subtype expression and the results of SDF-1 modulation by FGF family members restricted the candidate to FGFR1 IIIb, FGFR1 IIIc, and FGFR3 IIIc, and less likely to FGFR2 IIIc and FGFR3 IIIb.
Patterns of FGF family members binding to FGFR
| FGF . | FGFR1 IIIb . | FGFR1 IIIc . | FGFR2 IIIb . | FGFR2 IIIc . | FGFR3 IIIb . | FGFR3 IIIc . | FGFR4 . |
|---|---|---|---|---|---|---|---|
| FGF2 | ND | +++ | NB | +++ | M(−) | NB | ND |
| FGF4 | M(+) | ++ | ++ | +++ | M(−) | NB | M(+) |
| FGF7 | M(−) | NB | + | NB | M(−) | NB | M(−) |
| FGF9 | M(−) | + | NB | + | M(+) | NB | M(+) |
| FGF16 | ND | ND | ND | + | ND | ND | ND |
| FGF19 | ND | NB | ND | NB | ND | NB | ++ |
| FGF . | FGFR1 IIIb . | FGFR1 IIIc . | FGFR2 IIIb . | FGFR2 IIIc . | FGFR3 IIIb . | FGFR3 IIIc . | FGFR4 . |
|---|---|---|---|---|---|---|---|
| FGF2 | ND | +++ | NB | +++ | M(−) | NB | ND |
| FGF4 | M(+) | ++ | ++ | +++ | M(−) | NB | M(+) |
| FGF7 | M(−) | NB | + | NB | M(−) | NB | M(−) |
| FGF9 | M(−) | + | NB | + | M(+) | NB | M(+) |
| FGF16 | ND | ND | ND | + | ND | ND | ND |
| FGF19 | ND | NB | ND | NB | ND | NB | ++ |
NB indicates negligible binding; ND, not determined; M(+), mitogenic response in FGFR-overexpressing BaF3 cells; M(−), no mitogenic response of FGFR-overexpressing BaF3 cells; +++, strong binding; ++, intermediate binding; and +, weak binding.
Effects of FGF family members on stromal cell secretion of SDF-1. (A) MS-5 cells and S-17 cells (80%-90% confluent in 96-well plate) were incubated in culture medium (αMEM containing 10% FBS, 200 μL/well) alone, or with FGF2, FGF4, FGF7, FGF9, FGF16, or FGF19 (50 ng/mL) for 72 hours; cell-free culture supernatants were collected and SDF-1 content was measured by a specific ELISA. All experiments were performed in triplicate and means (± SD) were calculated. The percent SDF-1 secretion was calculated as follows: (SDF-1 secretion in FGF group/SDF-1 secretion in control group with no additive) × 100 (%). The results reflect the means (± SD) of 3 to 4 independent experiments. The asterisk denotes statistical significance (**P < .01). (B) MS-5 cells and S-17 cells were detached and washed twice with PBS. Cells were suspended in culture medium (αMEM containing 10% FBS), plated (2000 cells/well in 0.2 mL culture medium) in triplicate wells (96-well plates) with the addition of FGFs (50 ng/mL), and incubated for 64 hours. DNA synthesis was measured by 3 H thymidine deoxyribose uptake during the last 16 hours of culture. The percent 3 H incorporation was calculated as follows: (3 H incorporation in FGF group/3 H incorporation in control group with no additive) × 100 (%). The results reflect the means (± SD) of 3 to 4 independent experiments.
Effects of FGF family members on stromal cell secretion of SDF-1. (A) MS-5 cells and S-17 cells (80%-90% confluent in 96-well plate) were incubated in culture medium (αMEM containing 10% FBS, 200 μL/well) alone, or with FGF2, FGF4, FGF7, FGF9, FGF16, or FGF19 (50 ng/mL) for 72 hours; cell-free culture supernatants were collected and SDF-1 content was measured by a specific ELISA. All experiments were performed in triplicate and means (± SD) were calculated. The percent SDF-1 secretion was calculated as follows: (SDF-1 secretion in FGF group/SDF-1 secretion in control group with no additive) × 100 (%). The results reflect the means (± SD) of 3 to 4 independent experiments. The asterisk denotes statistical significance (**P < .01). (B) MS-5 cells and S-17 cells were detached and washed twice with PBS. Cells were suspended in culture medium (αMEM containing 10% FBS), plated (2000 cells/well in 0.2 mL culture medium) in triplicate wells (96-well plates) with the addition of FGFs (50 ng/mL), and incubated for 64 hours. DNA synthesis was measured by 3 H thymidine deoxyribose uptake during the last 16 hours of culture. The percent 3 H incorporation was calculated as follows: (3 H incorporation in FGF group/3 H incorporation in control group with no additive) × 100 (%). The results reflect the means (± SD) of 3 to 4 independent experiments.
FGF2 down-regulates SDF-1 production and impairs stromal cell supportive functions through FGFR1 IIIc
To identify which of these receptor subtypes is responsible for mediating FGF2 inhibition of SDF-1 expression in stromal cells, we used soluble, Fc-fusion chimeras of the extracellular domain for each of the 4 candidate receptors (FGFR1 IIIb/Fc, FGFR1 IIIc/Fc, FGFR3 IIIb/Fc, and FGFR3 IIIc/Fc; all from R&D Systems) and a specific neutralizing antibody for FGFR2 IIIc. Stromal cells were incubated in medium alone, with FGF2 (10 ng/mL), or FGF2 (10 ng/mL) plus each of the FGFR/Fc chimeras or the anti–FGF2 IIIc antibody (0-2000 ng/mL) for 72 hours; we measured the SDF-1 content in culture supernatants by a specific ELISA. As expected, FGF2 (10 ng/mL) significantly reduced levels of SDF-1 in the culture supernatants of MS-5 and S-17. Addition of FGFR1 IIIc/Fc chimera to FGF2-supplemented cultures dose-dependently restored SDF-1 levels in culture supernatants. At the highest FGFR1 IIIc/Fc chimera concentration used (2000 ng/mL), levels of SDF-1 were similar to those detected in supernatants of MS-5 and S-17 cells cultured in medium alone, without FGF2 (Figure 4A). By contrast, FGFR1 IIIb/Fc, FGFR3 IIIb/Fc, FGFR3 IIIc/Fc, or the anti–FGFR2 IIIc neutralizing antibody did not enhance SDF-1 levels in stromal cell cultures supplemented with FGF2 (Figure 4A). None of FGFR/Fc chimeras and the anti–FGFR2 IIIc antibody (2000 ng/mL) added alone to MS-5 and S-17 cell cultures (72-hour incubation) significantly affected SDF-1 secretion (Figure 4B) and proliferation (Figure 4C) in these cells. Of note, FGF4 and FGF9, which like FGF2 significantly reduce SDF-1 production in MS-5 and S-17 stromal cells (Figure 2B), were previously reported to bind FGFR1 IIIc (Table 2), providing additional evidence that FGFR1 IIIc mediates FGF-induced down-regulation of SDF-1 production in stromal cells.
Effects of FGFR/Fc chimeras on stromal cell secretion of SDF-1 and supportive function for hematopoietic progenitor cells. (A) FGFR/Fc chimeras (0-2000 ng/mL in αMEM containing 10% FBS) were incubated in medium only or with FGF2 (10 ng/mL) for 30 minutes, and then added to stromal cell (MS-5 and S-17) monolayers (approximately 90% confluent). A specific anti–FGFR2 IIIc neutralizing mAb (0-2000 ng/mL) was first applied onto stromal monolayers for 30 minutes, and then FGF2 (10 ng/mL) was added. After 72-hour incubation, culture supernatants were collected and used to measure SDF-1 content by a specific SDF-1 ELISA. The percent SDF-1 secretion was calculated as follows: ([SDF-1 secretion in FGFR/Fc or anti–FGFR2 IIIc mAb group]/[SDF-1 secretion in control group with no additive]) × 100 (%). All experiments were performed at least in triplicate. Representative results from 4 independent experiments are shown. (B) MS-5 cells and S-17 cells (90% confluent, in 200 μL, 96-well plates) were incubated in medium alone, with FGFR/Fc chimeras, or with anti–FGFR2 IIIc mAb (2000 ng/mL) for 72 hours. The SDF-1 content was determined by a specific ELISA, and the results were expressed as percent SDF-1 secretion. All experiments were performed at least in triplicate and repeated 4 times. The results reflect the means (± SD) of 4 independent experiments. (C) MS-5 cells and S-17 cells were detached and washed twice with PBS. Cells were suspended in culture medium (αMEM containing 10% FBS), plated in triplicate (2000 cells/well in 0.2 mL culture medium) with the addition of FGFR/chimeras (2000 ng/mL) or with anti–FGFR2 IIIc mAb (2000 ng/mL) onto 96-well plates, and incubated for 64 hours. DNA synthesis was measured by 3 H thymidine deoxyribose uptake during the last 16 hours of culture. The results are expressed as percent 3 H incorporation. All experiments were performed at least in triplicate and repeated 4 times. The results reflect the means (± SD) of 4 independent experiments. (D) MS-5 cells (80%-90% confluent on gelatin-coated 24-well plates) were incubated in medium alone, with FGFR1 IIIc/Fc chimera (800 ng/mL), with FGF2 (10 ng/mL), with FGFR1 IIIc/Fc chimera (800 ng/mL) plus FGF2 (10 ng/mL), or FGFR1 IIIb (800 ng/mL) plus FGF2 (10 ng/mL) for 72 hours, and then washed twice with PBS. Human CD34+ peripheral blood progenitor cells (PBSCs: 5 × 104 cells in 0.36 mL) in medium alone, with FGFR1 IIIc/Fc chimera (800 ng/mL), with FGF2 (10 ng/mL), with FGFR1 IIIc/Fc chimera (800 ng/mL) plus FGF2 (10 ng/mL), or FGFR1 IIIb/chimera (800 ng/mL) plus FGF2 (10 ng/mL) were applied onto pretreated stromal layers (4 wells/subgroup). Cocultures were incubated for 3 weeks with replenishment of culture medium alone (0.18 mL) or with the appropriate original additives (800 ng/mL FGFR1 IIIc, 10 ng/mL FGF2, 800 ng/mL FGFR1 IIIc plus 10 ng/mL FGF2, or 800 ng/mL FGFR1 IIIb plus 10 ng/mL FGF2) twice/wk. After 3 weeks, nonadherent viable cells were counted and analyzed by FACS. The percent growth of PBSCs was calculated as ([CD45+ cell number with FGFR1 IIIc/Fc, FGF2, FGF2 plus FGFR1 IIIc/Fc, or FGF2 plus FGFR1 IIIb/Fc]/[CD45+ cell number with no additive]) × 100 (%). The results reflect the means (± SD) of 3 independent experiments. The asterisk denotes statistical significance (**P < .01; *P < .05).
Effects of FGFR/Fc chimeras on stromal cell secretion of SDF-1 and supportive function for hematopoietic progenitor cells. (A) FGFR/Fc chimeras (0-2000 ng/mL in αMEM containing 10% FBS) were incubated in medium only or with FGF2 (10 ng/mL) for 30 minutes, and then added to stromal cell (MS-5 and S-17) monolayers (approximately 90% confluent). A specific anti–FGFR2 IIIc neutralizing mAb (0-2000 ng/mL) was first applied onto stromal monolayers for 30 minutes, and then FGF2 (10 ng/mL) was added. After 72-hour incubation, culture supernatants were collected and used to measure SDF-1 content by a specific SDF-1 ELISA. The percent SDF-1 secretion was calculated as follows: ([SDF-1 secretion in FGFR/Fc or anti–FGFR2 IIIc mAb group]/[SDF-1 secretion in control group with no additive]) × 100 (%). All experiments were performed at least in triplicate. Representative results from 4 independent experiments are shown. (B) MS-5 cells and S-17 cells (90% confluent, in 200 μL, 96-well plates) were incubated in medium alone, with FGFR/Fc chimeras, or with anti–FGFR2 IIIc mAb (2000 ng/mL) for 72 hours. The SDF-1 content was determined by a specific ELISA, and the results were expressed as percent SDF-1 secretion. All experiments were performed at least in triplicate and repeated 4 times. The results reflect the means (± SD) of 4 independent experiments. (C) MS-5 cells and S-17 cells were detached and washed twice with PBS. Cells were suspended in culture medium (αMEM containing 10% FBS), plated in triplicate (2000 cells/well in 0.2 mL culture medium) with the addition of FGFR/chimeras (2000 ng/mL) or with anti–FGFR2 IIIc mAb (2000 ng/mL) onto 96-well plates, and incubated for 64 hours. DNA synthesis was measured by 3 H thymidine deoxyribose uptake during the last 16 hours of culture. The results are expressed as percent 3 H incorporation. All experiments were performed at least in triplicate and repeated 4 times. The results reflect the means (± SD) of 4 independent experiments. (D) MS-5 cells (80%-90% confluent on gelatin-coated 24-well plates) were incubated in medium alone, with FGFR1 IIIc/Fc chimera (800 ng/mL), with FGF2 (10 ng/mL), with FGFR1 IIIc/Fc chimera (800 ng/mL) plus FGF2 (10 ng/mL), or FGFR1 IIIb (800 ng/mL) plus FGF2 (10 ng/mL) for 72 hours, and then washed twice with PBS. Human CD34+ peripheral blood progenitor cells (PBSCs: 5 × 104 cells in 0.36 mL) in medium alone, with FGFR1 IIIc/Fc chimera (800 ng/mL), with FGF2 (10 ng/mL), with FGFR1 IIIc/Fc chimera (800 ng/mL) plus FGF2 (10 ng/mL), or FGFR1 IIIb/chimera (800 ng/mL) plus FGF2 (10 ng/mL) were applied onto pretreated stromal layers (4 wells/subgroup). Cocultures were incubated for 3 weeks with replenishment of culture medium alone (0.18 mL) or with the appropriate original additives (800 ng/mL FGFR1 IIIc, 10 ng/mL FGF2, 800 ng/mL FGFR1 IIIc plus 10 ng/mL FGF2, or 800 ng/mL FGFR1 IIIb plus 10 ng/mL FGF2) twice/wk. After 3 weeks, nonadherent viable cells were counted and analyzed by FACS. The percent growth of PBSCs was calculated as ([CD45+ cell number with FGFR1 IIIc/Fc, FGF2, FGF2 plus FGFR1 IIIc/Fc, or FGF2 plus FGFR1 IIIb/Fc]/[CD45+ cell number with no additive]) × 100 (%). The results reflect the means (± SD) of 3 independent experiments. The asterisk denotes statistical significance (**P < .01; *P < .05).
The results shown in Figure 1D demonstrated that FGF2 impairs MS-5 stromal cell–supportive functions for human CD34+ progenitor cells, such that recovery of human cells is markedly reduced after 3-week coculture in the presence of FGF2 (10 ng/mL). We now tested whether this inhibitory function of FGF2 is mediated through FGFR1 IIIc. The addition of FGFR1 IIIc/Fc (800 ng/mL) added to cocultures of MS-5 stromal cells and human CD34+ cells fully reversed FGF2 (10 ng/mL) inhibition and permitted recovery of human cells in similar numbers as those recovered from cocultures without FGF2 (Figure 4D). FGFR1 IIIc/Fc alone, without FGF2, minimally affected CD45+ cell recovery from the cocultures, and the control FGFR1 IIIb/Fc (800 ng/mL) was ineffective at reversing FGF2 inhibition (Figure 4D). These results demonstrate that FGFR1 IIIc is the receptor that mediates FGF2 inhibition of SDF-1 production and of hematopoietic cell supportive function in stromal cells.
Analysis of the mechanisms underlying FGF2-induced down-regulation of SDF-1 expression in stromal cells
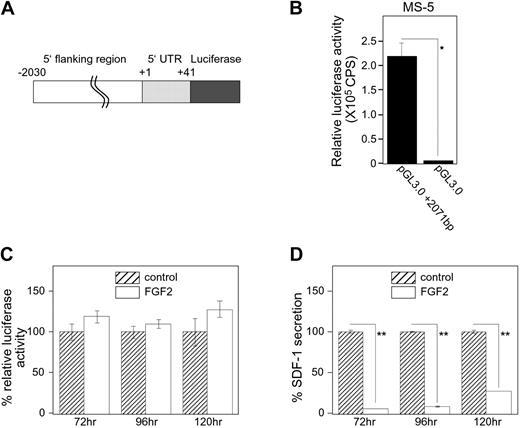
Since we established that FGF2 reduced SDF-1 mRNA levels (Figure 1A), we examined whether FGF2 regulates the SDF1 promoter activity. The SDF1 gene transcription regulatory region was recently mapped.21 We constructed a luciferase vector containing the SDF1 promoter region (−2030 base pairs) including the first 41 intronic base pairs (+41) of the SDF1 gene, and assessed FGF2 regulation of the promoter activity using luciferase assays in transiently transfected stromal cells. We designated this vector as pGL3.0+2071 bp (Figure 5A). As shown in Figure 5B, pGL3.0+2071 bp produced significant luciferase activity compared with the control (no promoter) luciferase vector (pGL3.0) 24 hours after transfection into MS-5 cells. A time-course experiment showed persistence of significant luciferase activity for 120 hours after transfection of the pGL3.0+2071bp vector into MS-5 cells (not shown). FGF2 (50 ng/mL) did not reduce luciferase activity induced by the pGL3.0+2071bp vector in transfected MS-5 cells after 72, 96, or 120 hours (Figure 5C), even though the SDF-1 content in the culture supernatant from the same FGF2-stimulated cultures was significantly decreased (Figure 5D). We also confirmed that FGF2 did not modulate the promoter activity at earlier time points (24 and 48 hours, not shown). Similar results were obtained when S-17 cells were transfected with pGL3+2071bp vector and subsequently treated with FGF2 (data not shown). These observations provide evidence that FGF2 does not regulate SDF-1 transcription in stromal cells.
Effects of FGF2 on SDF-1 promoter activity. (A) Schematic representation of the SDF-1 promoter-reporter construct pGL3.0+2071bp. An approximately 2.2-kb DNA fragment containing SDF-1 upstream sequences and the partial coding sequence (−2030 to +149 relative to the transcription start site) was amplified by PCR, and the fragment (−2030 to +41) was inserted into the reporter vector pGL3.0-Basic. (B) MS-5 cells were transfected with pGL3.0-Basic (250 ng/well) plus phRL-SV40 (Renilla luciferase reference control plasmid, 2.5 ng/well) or with pGL3.0+2071 bp (250 ng/well) plus phRL-SV40 (2.5 ng/well). All assays were carried out at least in triplicate sets and repeated twice. After 24-hour incubation, firefly luciferase activity was evaluated. Transfection efficiency was adjusted by Renilla luciferase activities (relative luciferase activity). CPS indicates counts per second. (C-D) pGL3.0+2071 bp-transfected MS-5 cells were stimulated with or without FGF2 (50 ng/mL) for a period of 72, 96, or 120 hours. Cell lysates and culture supernatants were obtained for luciferase assay and SDF-1 ELISA, respectively. All experiments were performed at least in triplicate and repeated 3 times. The percent relative luciferase activity was defined as follows: (relative luciferase activity in FGF2-stimulated group/relative luciferase activity in control group) × 100 (%). The percent SDF-1 secretion was calculated as follows: (SDF-1 secretion in FGF2-stimulated group/SDF-1 secretion in control group) × 100 (%). Error bars denote standard deviations of 3 experiments; asterisks denote statistical significance (*P < .05; **P < .001).
Effects of FGF2 on SDF-1 promoter activity. (A) Schematic representation of the SDF-1 promoter-reporter construct pGL3.0+2071bp. An approximately 2.2-kb DNA fragment containing SDF-1 upstream sequences and the partial coding sequence (−2030 to +149 relative to the transcription start site) was amplified by PCR, and the fragment (−2030 to +41) was inserted into the reporter vector pGL3.0-Basic. (B) MS-5 cells were transfected with pGL3.0-Basic (250 ng/well) plus phRL-SV40 (Renilla luciferase reference control plasmid, 2.5 ng/well) or with pGL3.0+2071 bp (250 ng/well) plus phRL-SV40 (2.5 ng/well). All assays were carried out at least in triplicate sets and repeated twice. After 24-hour incubation, firefly luciferase activity was evaluated. Transfection efficiency was adjusted by Renilla luciferase activities (relative luciferase activity). CPS indicates counts per second. (C-D) pGL3.0+2071 bp-transfected MS-5 cells were stimulated with or without FGF2 (50 ng/mL) for a period of 72, 96, or 120 hours. Cell lysates and culture supernatants were obtained for luciferase assay and SDF-1 ELISA, respectively. All experiments were performed at least in triplicate and repeated 3 times. The percent relative luciferase activity was defined as follows: (relative luciferase activity in FGF2-stimulated group/relative luciferase activity in control group) × 100 (%). The percent SDF-1 secretion was calculated as follows: (SDF-1 secretion in FGF2-stimulated group/SDF-1 secretion in control group) × 100 (%). Error bars denote standard deviations of 3 experiments; asterisks denote statistical significance (*P < .05; **P < .001).
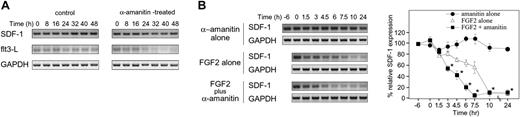
We evaluated whether FGF2 can alter SDF-1 mRNA stability. Since little is currently known about SDF-1 mRNA stability, we evaluated SDF-1 mRNA levels in S-17 cells treated with α-amanitin, a specific RNA polymerase II inhibitor. Semiquantitative RT-PCR analysis revealed that SDF-1 mRNA is detectable at similar levels at time 0, before α-amanitin treatment, and at several time points during 48-hour treatment with α-amanitin (5 μg/mL) (Figure 6A). Cell viability was 80% to 85% at the 48-hour time point after exposure to α-amanitin. In contrast to SDF-1, the mRNA for the flt3-L, a growth factor for hematopoietic progenitor cells,57 was markedly reduced in S-17 cells after 24-hour exposure to 5 μg/mL α-amanitin (Figure 6A). A similar difference between SDF-1 and flt3-L mRNA stability was observed when actinomycin-D (10 μg/mL) was used to block transcription (not shown).
Effects of FGF2 on SDF-1 mRNA stability. (A) Analysis of SDF-1, Flt3-L, and GAPDH mRNA stability. S-17 cells (80%-90% confluent) were cultured in medium alone or in medium supplemented with 5 μg/mL α-amanitin for 8, 16, 24, 32, 40, or 48 hours at 37°C. Levels of specific mRNAs were measured by semiquantitative RT-PCR. Amplified products were separated on agarose gels. (B) To evaluate the effect of FGF2 on SDF-1 mRNA decay, S-17 cells were preincubated in medium alone or with FGF2 (50 ng/mL) for 6 hours, and then α-amanitin (5 μg/mL) was added to cultures. FGF2-pretreated cells were also tested without α-amanitin addition. After incubation for 1.5, 3, 4.5, 6, 7.5, 10, and 24 hours, SDF-1 and GAPDH mRNAs were measured by semiquantitative PCR. The amount of cDNA used for each amplification reaction was based on the results of PCR for GAPDH showing equivalent amounts of product amplified from all samples. PCR products of SDF-1 and GAPDH were separated on a 3% and 2% agarose gel, respectively, and visualized under a UV light (left panel; representative experiment of 3). Relative SDF-1 mRNA level (SDF-1/GAPDH) was calculated by densitometric scanning using NIH image software. Percent relative SDF-1 expression is defined as percent of the initial ratio of relative SDF-1 expression (right panel). The results reflect the means (± SD) of 3 independent experiments. The asterisk denotes statistical significance (amanitin alone vs FGF2 plus amanitin, P < .05).
Effects of FGF2 on SDF-1 mRNA stability. (A) Analysis of SDF-1, Flt3-L, and GAPDH mRNA stability. S-17 cells (80%-90% confluent) were cultured in medium alone or in medium supplemented with 5 μg/mL α-amanitin for 8, 16, 24, 32, 40, or 48 hours at 37°C. Levels of specific mRNAs were measured by semiquantitative RT-PCR. Amplified products were separated on agarose gels. (B) To evaluate the effect of FGF2 on SDF-1 mRNA decay, S-17 cells were preincubated in medium alone or with FGF2 (50 ng/mL) for 6 hours, and then α-amanitin (5 μg/mL) was added to cultures. FGF2-pretreated cells were also tested without α-amanitin addition. After incubation for 1.5, 3, 4.5, 6, 7.5, 10, and 24 hours, SDF-1 and GAPDH mRNAs were measured by semiquantitative PCR. The amount of cDNA used for each amplification reaction was based on the results of PCR for GAPDH showing equivalent amounts of product amplified from all samples. PCR products of SDF-1 and GAPDH were separated on a 3% and 2% agarose gel, respectively, and visualized under a UV light (left panel; representative experiment of 3). Relative SDF-1 mRNA level (SDF-1/GAPDH) was calculated by densitometric scanning using NIH image software. Percent relative SDF-1 expression is defined as percent of the initial ratio of relative SDF-1 expression (right panel). The results reflect the means (± SD) of 3 independent experiments. The asterisk denotes statistical significance (amanitin alone vs FGF2 plus amanitin, P < .05).
The experiment shown in Figure 6A indicates that in the presence of transcriptional inhibition, SDF-1 mRNA levels in S-17 cells remain fairly stable over a 48-hour period; the experiment shown in Figure 1A indicates that in the presence of FGF2 (no transcription inhibitors) SDF-1 mRNA is significantly reduced after 24-hour exposure. We then tested the effects of FGF2 (50 ng/mL) on SDF-1 mRNA levels in the presence of the transcriptional inhibitor α-amanitin (5 μg/mL), and compared them with FGF2 alone (50 ng/mL) and α-amanitin (5 μg/mL) alone (Figure 6B). In S-17 cells, levels of SDF-1 mRNA were found to be significantly lower after the cells were incubated for 3 hours with FGF2 plus α-amanitin in comparison with time 0, whereas as expected from the experiment shown in Figure 6A, levels of SDF-1 mRNA were not decreased even after the cells were incubated for 24 hours with α-amanitin alone. FGF2, added alone without the transcriptional inhibitor, also reduced SDF-1 mRNA levels after 4.5-hour incubation, but the level of reduction was smaller than that observed with the α-amanitin inhibitor (Figure 6B), attributable to the absence of newly synthesized mRNA. These results provide evidence that FGF2 down-regulates SDF-1 expression by accelerating SDF-1 mRNA decay.
Discussion
We show that FGF2 inhibits SDF1 expression in bone marrow stromal cells acting through FGFR1 IIIc, and by this mechanism FGF2 reduces stromal cell support for CD34+ hematopoietic progenitor cells. Previous studies have described FGF2 as an inducer of growth, survival, chemotaxis, and differentiation in a variety of cell types, and a key regulator of prenatal development and angiogenesis.25,37 However, neither FGF2 nor activated FGFRs have previously been linked to negative regulation of SDF1 expression in bone marrow stromal cells or any other cell type. SDF-1 has emerged as a critical modulator of hematopoiesis that supports the growth and survival of hematopoietic/myeloid progenitor cells and pre-B cells, and serves to retain immature blood cells in the bone marrow.6,7,9,10,18,19 Stromal cells are a principal source of SDF-1 in the bone marrow.10,12 Our observation linking FGF2 to regulation of SDF-1 expression in bone marrow stromal cells raises the possibility that FGF2 can also play important roles in hematopoiesis.
Earlier studies in vitro have concluded that under defined experimental conditions FGF2 can promote myelopoiesis and megakaryocytopoiesis.37-41 FGF2 knock-out mice are viable and display only mild cardiovascular, skeletal, and neuronal defects, suggesting that FGF2 is not critical to normal development of the hematopoietic systems,58-60 but there is much redundancy among the 22 FGF species that may compensate for the absence of FGF2. When administered systemically to mice and rats, FGF2 promoted bone formation, but hematopoiesis was not investigated.61 Preclinical studies in cynomologous monkeys indicated that FGF2 consistently caused anemia attributable to decreased erythropoiesis.62 Of note, abnormally high levels of FGF2 were detected in the bone marrow of patients with certain clonal myeloid disorders that are characteristically associated with severe defects of normal hematopoiesis.42
FGFs exert their biological activities through the tyrosine kinase FGF receptors expressed on various cell types.63 An essential feature of FGFRs is the existence of 2 alternative exons, IIIb and IIIc, which encode a different C-terminal portion of domain 3. In the case of FGFR2, the choice between IIIb and IIIc exons is mutually exclusive so that the 2 receptor subtypes are not expressed together on the cell surface.28,64 In the mouse skin, for example, only IIIc transcripts are detected in the dermis, while IIIb transcripts are found mainly in the epidermis.30,65 Consistently, we found that MS-5 cells and S-17 cells express FGFR2 IIIc, but do not or minimally express FGFR2 IIIb. In the case of FGFR3, the choice between the exons IIIb and IIIc appears not to be strictly tissue specific.66 In the current experiments, we found that FGFR3 IIIb and FGFR3 IIIc are both expressed in stromal cells. Similarly, we found that stromal cells express both FGFR1 IIIb and IIIc. Importantly, mice with targeted disruption of the whole FGFR1 gene or the FGFR1 IIIc alone die perinatally,67,68 whereas mice with an inactive exon IIIb were viable and fertile, providing evidence that the IIIc isoform of FGFR1 is responsible for most of the biological functions of FGFR1, whereas the IIIb isoform plays a minor role.69 Consistently with FGFR1 IIIc playing a dominant role over FGFR1 IIIb, we found here that FGFR1 IIIc is exclusively responsible for mediating FGF2-induced SDF-1 depletion and reduced supportive function in stromal cells.
The expression of many chemokines is transcriptionally induced by exogenous signals but is otherwise low or absent. Instead, SDF1 expression is constitutive in many tissues and cell types.1,5 Degradation has been identified as a critical mechanism for regulation of SDF-1 function. CD26/dipeptidyl peptidase, neutrophil elastase, cathepsin G, and several metalloproteinases have been identified as proteolytic enzymes that functionally inactivate SDF-1.70-74 In the circulation, SDF-1 is present at nanogram per milliliter levels but is cleaved and functionally inactive.70 Nonetheless, VEGF-A, gamma irradiation, 5-fluouracyl, and IL-1β were reported to induce SDF1 expression in some cell types,10,11 and G-CSF treatment decreased SDF-1 mRNA and protein in bone marrow.75 Here, we report that FGF2 reduces SDF1 mRNA and protein in bone marrow stromal cells. Recent studies showed that VEGF-A, 5-Fu, and IL-1β did not induce SDF-1 promoter activity in lung fibroblast and astrocytoma cells, in spite of their ability to promote SDF-1 expression, and suggested a contribution by posttranscriptional regulatory mechanisms such as mRNA stability.21 Similarly, we show here that FGF2 reduces SDF-1 mRNA and protein levels without substantially regulating SDF1 promoter activity.
mRNA stability is a highly controlled process still incompletely characterized. Nevertheless, several sequence motifs frequently located in the 3′-UTR of the message have been identified that can regulate the rate of mRNA turnover. Among the most studied sequences are the adenine/uridine (AU)–rich elements (AREs) found in mRNAs encoding growth factors and cytokines, which confer instability to mRNA.76-78 A number of proteins can bind to AREs, including TTP, AUF1, and KSRP, and promote mRNA destabilization.79-82 Various signaling pathways, including the c-Jun amino-terminal kinase (JNK) signaling pathway and the p38 mitogen–activated protein-kinase (MAPK) pathway, have been linked to regulation of specific mRNA decay by modulation ARE-dependent mRNA stability.78 When activated by FGF and heparan sulfate proteoglycans, the FGFRs undergo phosphorylation and subsequent stimulation of the Ras/MAP kinase pathway that includes ERK1/2, p38, and JNK kinases; the P-I-3 kinase–AKT pathway; and the PLCγ pathway83,84 ; the activation of ERK1/2 and p38 in response to FGF has been observed in all cell types.63 SDF-1 mRNA possesses 12 AREs within its 3′-UTR.3 It is thus possible that such sequences may mediate FGF-induced SDF-1 mRNA decay in stromal cells. Consistently with our observation that FGF2 does not modulate SDF1 transcription, the SDF1 promoter that contains several putative transcription factor–binding sites21 does not contain binding sites for ERKs, p38, or JNKs.
Collectively, the current study shows that FGF2 acting through FGFR1 IIIc accelerates SDF-1 mRNA decay in bone marrow stromal cells, thereby decreasing SDF-1 expression in these cells and reducing supportive functions for hematopoietic progenitor cells.
Authorship
Contribution: T.N. and G.T. designed research; T.N. and N.M. performed research; T.N. and G.T. analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: G. Tosato, Basic Research Lab, Center for Cancer Research, National Cancer Institute, Bldg 10, Rm 12C207, 10 Center Dr, Bethesda, MD 20892; e-mail:tosatog@mail.nih.gov.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the Intramural Research Program of the NIH, NCI, Center for Cancer Research, and a grant from the Japan Society for the Promotion of Science.
The authors wish to thank Drs M. C. Berardi, K. Dorskind, P. Kincade, and M. Narazaki for their contributions to various aspects of this work, and the Clinical Center Blood Bank personnel for cell purification. We also express appreciation to Dr Z. Zheng for helpful suggestions.

![Figure 4. Effects of FGFR/Fc chimeras on stromal cell secretion of SDF-1 and supportive function for hematopoietic progenitor cells. (A) FGFR/Fc chimeras (0-2000 ng/mL in αMEM containing 10% FBS) were incubated in medium only or with FGF2 (10 ng/mL) for 30 minutes, and then added to stromal cell (MS-5 and S-17) monolayers (approximately 90% confluent). A specific anti–FGFR2 IIIc neutralizing mAb (0-2000 ng/mL) was first applied onto stromal monolayers for 30 minutes, and then FGF2 (10 ng/mL) was added. After 72-hour incubation, culture supernatants were collected and used to measure SDF-1 content by a specific SDF-1 ELISA. The percent SDF-1 secretion was calculated as follows: ([SDF-1 secretion in FGFR/Fc or anti–FGFR2 IIIc mAb group]/[SDF-1 secretion in control group with no additive]) × 100 (%). All experiments were performed at least in triplicate. Representative results from 4 independent experiments are shown. (B) MS-5 cells and S-17 cells (90% confluent, in 200 μL, 96-well plates) were incubated in medium alone, with FGFR/Fc chimeras, or with anti–FGFR2 IIIc mAb (2000 ng/mL) for 72 hours. The SDF-1 content was determined by a specific ELISA, and the results were expressed as percent SDF-1 secretion. All experiments were performed at least in triplicate and repeated 4 times. The results reflect the means (± SD) of 4 independent experiments. (C) MS-5 cells and S-17 cells were detached and washed twice with PBS. Cells were suspended in culture medium (αMEM containing 10% FBS), plated in triplicate (2000 cells/well in 0.2 mL culture medium) with the addition of FGFR/chimeras (2000 ng/mL) or with anti–FGFR2 IIIc mAb (2000 ng/mL) onto 96-well plates, and incubated for 64 hours. DNA synthesis was measured by 3H thymidine deoxyribose uptake during the last 16 hours of culture. The results are expressed as percent 3H incorporation. All experiments were performed at least in triplicate and repeated 4 times. The results reflect the means (± SD) of 4 independent experiments. (D) MS-5 cells (80%-90% confluent on gelatin-coated 24-well plates) were incubated in medium alone, with FGFR1 IIIc/Fc chimera (800 ng/mL), with FGF2 (10 ng/mL), with FGFR1 IIIc/Fc chimera (800 ng/mL) plus FGF2 (10 ng/mL), or FGFR1 IIIb (800 ng/mL) plus FGF2 (10 ng/mL) for 72 hours, and then washed twice with PBS. Human CD34+ peripheral blood progenitor cells (PBSCs: 5 × 104 cells in 0.36 mL) in medium alone, with FGFR1 IIIc/Fc chimera (800 ng/mL), with FGF2 (10 ng/mL), with FGFR1 IIIc/Fc chimera (800 ng/mL) plus FGF2 (10 ng/mL), or FGFR1 IIIb/chimera (800 ng/mL) plus FGF2 (10 ng/mL) were applied onto pretreated stromal layers (4 wells/subgroup). Cocultures were incubated for 3 weeks with replenishment of culture medium alone (0.18 mL) or with the appropriate original additives (800 ng/mL FGFR1 IIIc, 10 ng/mL FGF2, 800 ng/mL FGFR1 IIIc plus 10 ng/mL FGF2, or 800 ng/mL FGFR1 IIIb plus 10 ng/mL FGF2) twice/wk. After 3 weeks, nonadherent viable cells were counted and analyzed by FACS. The percent growth of PBSCs was calculated as ([CD45+ cell number with FGFR1 IIIc/Fc, FGF2, FGF2 plus FGFR1 IIIc/Fc, or FGF2 plus FGFR1 IIIb/Fc]/[CD45+ cell number with no additive]) × 100 (%). The results reflect the means (± SD) of 3 independent experiments. The asterisk denotes statistical significance (**P < .01; *P < .05).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/4/10.1182_blood-2006-06-028217/4/m_zh80040708410004.jpeg?Expires=1769363718&Signature=tXVPQxSAz~66ri~p31LsQBmqaIl5ScT3p4DCBVk1dX30~7Kld06upoXoFlUhdCX2ehxMInSrn8vNdb1qhXatKUN0f4KTTPo2UKRh16izKvzV6J2HSSSBl7dJDqU0~MZbtVMCaj2F0gMtwqvYVkt6ox0Oi9UMPiQOROtd48G8m-tX9yoxk9Z9uKs64O4SduQ0PJrZZks4MKh4s7SewKyMWtcOnQkRaKzNZqPrnP2Oo3b7VW4GM5bDpICEztxHY4CiaEb1TZiVuNzaqKRdjMaM66-51Ie2ns7bOIy7NXp0fyU30OiEl4aSu0YGINDwd4Mv9M~64qNW-ePASHwRyZghig__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal