Abstract
Promyelocytic leukemia nuclear bodies (PML NBs), the structural domains of the eukaryotic cell nucleus, play a role in cancer and apoptosis, and their involvement in antiviral mechanisms mediated by interferons (IFNs) is proposed. IFNs dramatically increase the transcription of the PML gene. In this study, we have shown that the response of 2 structural PML NB components, PML and Sp100, to interferon-α (IFNα) was suppressed in cells simultaneously treated with histone deacetylase (HDAC) inhibitors (trichostatin A, sodium butyrate, MS-275, SAHA, and valproic acid). Trichostatin A (TSA) blocked the increase of PML NB number and suppressed up-regulation of PML mRNA and protein levels in several human cell lines and in normal diploid skin fibroblasts. Moreover, IFNα induction of IRF-1 was also inhibited by TSA, although incompletely. Analysis of cellular fractions did not show any defects in cytoplasmic-nuclear transport of STAT2, a component of transcription factor ISGF3 responsible for IFNα/β-dependent gene transcription. Moreover, chromatin immunoprecipitation showed that after IFNα stimulation STAT2 binds to ISRE element of PML promoter even in the presence of TSA and thus excluded STAT2-dependent mechanism of TSA effect. These results indicate that the action of histone deacetylases is necessary for the full transcriptional activation of IFNα-stimulated genes.
Introduction
Acute promyelocytic leukemia (APL) is a subtype of acute myeloid leukemia (AML) that comprises about 10% of AMLs.1 The molecular basis of APL is characterized by a chromosomal translocation invariably involving retinoic acid receptor alpha (RARα) gene on chromosome 17. The translocation partner for RARα is in 98% of APL cases represented by promyelocytic leukemia protein (PML) gene located on chromosome 15. This t(15;17) translocation leads to the production of reciprocal fusion proteins PML-RARα and RARα-PML.2,3 Other APL-associated gene rearrangements were reported with promyelocytic leukemia zinc finger protein (PLZF4 ), nucleophosmin (NPM5 ), nuclear mitotic apparatus protein (NuMA6 ), and signal transducer and activator of transcription 5b (STAT5b7 ). The resulting fusion proteins are considered responsible for a differentiation block of myeloid line and for the subsequent accumulation of immature cells in bone marrow.
The molecular mechanism underlying APL was described previously.8-10 It was shown that RARα in normal cells binds to retinoic acid response elements (RAREs) located in the promoters of retinoic acid (RA) target genes and modulates transcription through an interaction with various specific cofactors. In the absence of ligand (RA), RARα associates with corepressor molecules such as N-CoR and SMRT, recruiting histone deacetylases to the target genes. Resulting histone deacetylation leads to chromatin reorganization and repression of transcription. On the other hand, in the presence of RA, corepressor complex dissociates, and a number of transcriptional coactivator proteins bind to RARα leading to the activation of RA-inducible gene transcription. In APL cells, the RARα portion of fusion proteins retains functional DNA- and RA-binding domains. Thus, fusion proteins can still bind to RARE in gene promoters and recruit a corepressor complex comprising histone deacetylases. In the case of PML-RARα, the repressor complex is aberrantly not released after the treatment with physiological levels of RA (10−9-10−8 M) due to the attached fusion partner that causes a stronger interaction of fusion protein with corepressor complex. Thus, higher pharmacological doses of RA (10−7-10−6 M) are required to induce dissociation of corepressor complex and to activate transcription.11 Based on this model, it has been suggested that in addition to RA, HDAC inhibitors could be used for APL treatment in order to eliminate the repressing effect of HDACs. The observation that trichostatin A (TSA), a specific inhibitor of HDAC, caused reactivation of RA-inducible genes in APL cells,9 strongly supports this hypothesis.
Several HDAC inhibitors have been shown to arrest cancer cell growth and/or induce apoptosis in doses that have little toxicity to normal cells. HDAC inhibitors have been tested as potent anticancer reagents in different types of solid tumor cell lines and in hematopoietic transformed cell lines. Several of them are in initial phases of clinical trials (for reviews, see Marks et al12 and Kelly et al13 ). HDAC inhibitors also have the ability to overcome a differentiation block in promyelocyte maturation—therefore, they are intensively tested for a potential treatment of APL.
In the APL studies, translocation partners of RARα have to be taken into account, since their functional impairment due to the fusion could also play an important role in cancer development. The most frequent translocation partner of RARα, tumor suppressor PML, is involved in several stress response pathways to nongenotoxic and genotoxic stresses such as DNA damage sensing and repair (for a review, see Dellaire and Bazett-Jones14 ), and cell cycle checkpoint control and its outcomes such as cellular senescence15 and apoptosis.16 All these processes are frequently accompanied by dynamic changes in PML gene expression and in the structure of PML NBs. The role of PML in viral infections is also documented.17 The implication of PML in antiviral defense was first suggested after the finding that interferons of both type I and type II induce PML expression and dramatically increase number and average size of PML NBs18,19 for whose formation PML protein is essential. IFNs activate PML gene transcription through interferon-responsive elements, interferon-stimulated response element (ISRE) and gamma-interferon–activated site (GAS), located in its promoter.20 In addition, the expression of other structural components of PML NBs, Sp10021 and ISG20,22 is also induced by IFN treatment, which contributes to the observed enlargement and multiplication of PML NBs. Ablation of PML gene in mice decreases resistance to specific infections and shortens the lifespan of animals.23
In this study, we describe for the first time the effect of HDAC inhibitors, including TSA, sodium butyrate, valproic acid, SAHA, and MS-275, on IFNα-induced expression of PML and 2 other IFN-stimulated genes (ISGs), Sp100 and interferon regulatory factor 1 (IRF-1). All these compounds suppressed the IFNα-mediated induction of PML in normal and transformed human cells at the transcriptional level. As can be implicated from the known functions of PML NBs in cellular stress and from the role of ISGs in antiviral mechanisms, this suppressive effect should be taken into account as a potential cause of therapeutic adverse effects of HDAC inhibitors currently tested or prepared in the future for clinical use.
Materials and methods
Antibodies
The following antibodies were used: mouse mAb 5E10 against PML (a gift from R. Van Driel24 ), mouse anti-PML, rabbit anti-PML, anti-Stat2, anti–IRF-1 (all from Santa Cruz Biotechnology, Santa Cruz, CA), and mouse anti-GAPDH (Acris Antibodies, Hiddenhausen, Germany). Human autoimmune serum recognizing Sp100 was obtained from the Institute of Rheumatology (Prague, Czech Republic) with the patients' consent and according to the guidelines of the local ethics committee.25 The secondary antibodies for immunofluorescence were as follows: donkey anti–human IgG conjugated with FITC, donkey anti–mouse IgG conjugated with Cy3 (both from Jackson ImmunoResearch Laboratories, West Grove, PA); and for immunoblotting: goat anti–human IgG (Sigma, St Louis, MO), anti–mouse IgG, and anti–rabbit IgG (Bio-Rad Laboratories, Hercules, CA), all conjugated with horseradish peroxidase.
Cell cultures and chemicals
HeLa, SaOS-2, HEK293T, NIH-3T3, and human skin fibroblasts (HSFs) were grown in Dulbecco modified Eagle medium, whereas H1299, K562, and Jurkat cells were grown in RPMI medium. Both media were supplemented with 10% fetal bovine serum (Gibco, Invitrogen, Paisley, United Kingdom). Cells were incubated in humidified atmosphere under 5% CO2 at 37°C. Exponentially growing cells were exposed to 1000 U/mL IFN-α(Interferonum α-2b; Schering-Plough, Kenilworth, NJ) and/or to inhibitors of HDAC: TSA, sodium butyrate (both from Sigma), MS-275, SAHA, and valproic acid (all from Alexis Biochemicals, Lausen, Switzerland) in doses and time intervals specified further in the “Results.” Mouse interferon-β (1000 U/mL for 24 hours; Calbiochem, San Diego, CA) was used in case of mouse NIH-3T3 cells.
Indirect immunofluorescence
Cells growing as a monolayer on glass coverslips were fixed and permeabilized in 4% formaldehyde/0.1% Triton X-100 for 20 minutes at room temperature (RT). Mowiol was used as a mounting medium. After washing with PBS, cells were incubated for 1 hour with primary antibody, washed in PBS/0.05% Tween-20, and then stained for 45 minutes with secondary antibody at RT. DNA was stained with DAPI (Sigma) or Sytox (Molecular Probes, Eugene, OR). Immunofluorescence was visualized using fluorescence microscope Olympus VANOX-S (Olympus Optical, Tokyo, Japan) or with confocal laser-scanning microscope Leica TCS SP (Leica, Wetzlar, Germany). Captured images were processed with Adobe Photoshop (Adobe Systems, San Jose, CA). The number of PML NBs per nucleus was estimated by QWin program (Leica, Wetzlar, Germany).
Immunoblotting
Harvested cells were lysed in SDS lysis buffer (62 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 100 mM DTT, and 0.01% bromphenol blue). Equal amounts of total protein (25 or 50 μg/lane) were loaded on sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels. Proteins were then electrotransferred to nitrocellulose membrane (Pall, Pensacola, FL) as described previously.26 For total protein estimation, gels were stained with Coomassie Brilliant Blue R-250, while the membranes were stained with Ponceau S. Membranes were blocked with 5% nonfat dried milk in PBS for 1 hour at RT. After 1-hour incubation with primary antibodies, membranes were treated with secondary antibodies conjugated with horseradish peroxidase for 1 hour and visualized by enhanced chemiluminescence (ECL, SuperSignal West Femto Chemiluminescence Kit; Pierce, Rockford, IL).
Cell fractionation
Cytosolic and nuclear fractions were prepared according to a modified protocol of Dignam et al.27 Briefly, cells were scraped into ice-cold PBS, collected by centrifugation (500g, 4°C, 5 minutes), resuspended in cytosolic lysis buffer containing 10 mM HEPES (pH 8.0), 1.5 mM MgCl2, 10 mM KCl, 10 mM iodoacetamide, 0.5 mM Pefabloc, 0.5 μg/mL pepstatin, and 1 μg/mL leupeptin, and stroked several times through injection needle (29 G). After incubation on ice (10 minutes), the nuclei were pelleted by centrifugation, and supernatant was removed and stored as cytosolic fraction. The nuclei were washed and resuspended in cytosolic lysis buffer. Both fractions were denatured in SDS sample buffer, boiled for 5 minutes, and shortly sonicated before loading onto the gel.
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was isolated with TriReagent (Sigma) and reverse transcribed with TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster City, CA) using random hexamers as primers. qRT-PCR was performed in ABI Prism 7000 (Applied Biosystems) using JumpStart SYBR Green PCR Master Mix (Sigma) with the following set of primers: PML: 5′-TGACCAGCATCTACTGCCG-3′ and 5′-AGCTCACTGTGGCTGCTGTC-3′; GAPDH: 5′-GTCGGAGTCAACGGATTTGG-3′ and 5′-AAAAGCAGCCCTGGTGAC C-3′; actin: 5′-AGGCACCAGGGCGTGAT-3′ and 5′-TCGCCCACATAGGAATCCTT-3′; and 18S rRNA: 5′-CGTTCAGCCACCCGAGATT-3′ and 5′-CGGACATCTAAGGGCATCACA-3′. Three control genes—actin, GAPDH, and 18S rRNA—were used for normalization. The ΔΔCt method was used for quantification.28
Chromatin immunoprecipitation (ChIP)
HeLa cells (about 5 × 106 cells for each ChIP) were treated with IFNα, TSA, or with both drugs simultaneously for 8 hours, then fixed with 1% formaldehyde for 10 minutes (reaction was stopped by 0.125 M glycine), harvested, and lysed in cell lysis buffer (5 mM PIPES, pH 8.0, 85 mM KCl, 0.5% NP-40) for 10 minutes on ice. All buffers were supplemented with protease inhibitors (1 mM Pefabloc, 1 μg/mL pepstatin, 10 μg/mL leupeptin). The nuclei were resuspended in nuclei lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 0.2% SDS) and sonicated 8 times for 30 seconds (30 W, Sanyo Soniprep 150; Sanyo Gallenkamp, Lougborough, Great Britain) on ice. Lysates were diluted and adjusted to contain all components of radioimmunoprecipitation assay (RIPA) buffer (final concentration: 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS). Lysates were precleared with G-Sepharose beads equilibrated in RIPA buffer containing 1 mg/mL BSA and 1 mg/mL sonicated salmon sperm. Immunoprecipitation with desired antibody was performed at 4°C overnight. The same total protein amount was used for each reaction. Immunocomplexes bound on beads were washed 2 times with RIPA, 4 times with LiCl buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 500 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate), and once with TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA). Protein-DNA complexes were eluted with 0.1 M NaHCO3 and 1% SDS, de–cross-linked in the presence of 200 mM NaCl for 5 hours at 65°C, and then treated with RNase A (fc 40 μg/mL, 20 minutes, 37°C) and proteinase K (fc 200 μg/mL, 3 hours, 50°C). DNA was phenol/chloroform extracted, precipitated, and PCR amplified. The following primers encompassing ISRE element in PML promoter20 were used: 5′-TAGAACCGCCCCCAGCTTCT-3′ and 5′-CCACCACAGCAAACCAACAAA-3′. The supernatant after the bead collection was saved as reaction input; DNA was isolated, and 1/16 000 of total input was amplified by PCR.
Results
IFNα-induced increase in the number of PML NBs is suppressed by TSA
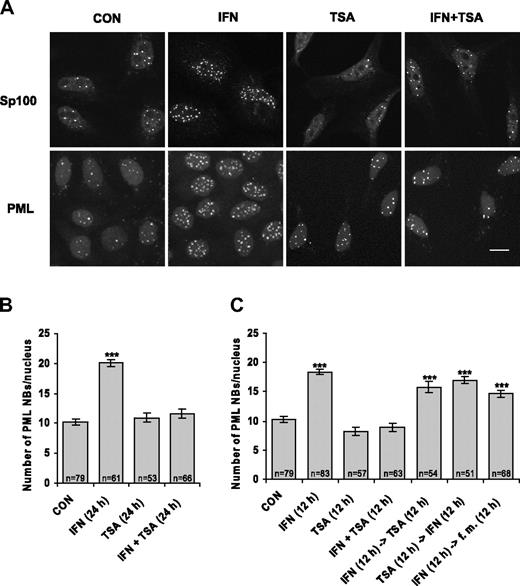
When examining the effect of protein acetylation on the redistribution of specific proteins between nuclear compartments, we found that HDAC inhibitor TSA negatively affects IFNα-dependent increase of specific nuclear compartment PML NBs. Since this finding is inconsistent with predominantly stimulatory action of TSA-mediated chromatin hyperacetylation on gene expression, we explored the effect of TSA on PML regulation in more detail. We followed the number of PML NBs in HeLa cells treated by IFNα, TSA, or combination of both drugs by indirect immunofluorescence using antibodies recognizing the 2 main structural components of PML NBs—PML and Sp100 protein. As expected, we observed at least 2-fold increase in PML NB number in HeLa cells stimulated by IFNα for 24 hours.18 However, this increase was completely suppressed when the cells were simultaneously treated with IFNα and TSA (see Figure 1A for immunofluorescence data and Figure 1B for quantification of PML NBs). The suppressive effect could be seen already after 12 hours of combined IFNα and TSA treatment (Figure 1C). Note that TSA alone did not noticeably influence the number of PML NBs in comparison with control untreated cells. When the cells were pretreated for 12 hours with IFNα and then exposed to TSA after IFNα removal, only an insignificant decrease in PML NB number was observed in comparison with cells treated with IFNα alone. The observation that once-induced PML NBs remained stable for at least 12 hours even in presence of TSA (note also last column in Figure 1C) suggests that TSA blocks only certain steps of PML expression and cannot cause PML NB dissociation. Conversely, when the cells were exposed to TSA (12 hours) prior to IFNα treatment (12 hours), the ability to increase PML NB number was still observed, which indicates that the effect of TSA on IFNα pathway is reversible (Figure 1C). Taken together, these data clearly show that PML NBs are not induced by IFNα in the presence of HDAC inhibitor TSA and suggest that activation of PML gene by IFNα pathway is impaired by inhibition of protein deacetylation.
IFNα-induced increase of PML NB number is suppressed by TSA. (A) Confocal images of HeLa cells untreated (CON) or treated with 1000 U/mL interferon-α (IFN), 500 ng/mL trichostatin A (TSA), or their combination (IFN + TSA) for 24 hours, immunostained with Sp100 or PML antibody; nuclei were costained with Sytox. Images were acquired by a Leica TCSSP confocal laser-scanning microscope equipped with a Leica HCX PL APO 63×/1.32 NA oil-immersion objective lens, and were processed by Leica Confocal Software version 2.5 (Leica Microsystems Heidelberg GmbH, Mannheim, Germany) and Adobe Photoshop 5.5 (Adobe Systems, San Jose, CA). Bar represents 10 μm. (B-C) Quantification of PML NBs. HeLa cells were treated with IFN, TSA, or IFN + TSA for the indicated time; were incubated first with one drug for a period of 12 hours and then incubated with the second drug after washing with PBS; or were maintained in the fresh medium (fm) for the next 12 hours as indicated for each column. The number of PML NBs per nucleus detected by PML antibody was counted on a total projection of confocal sections (1 section per 0.4 μm) through entire cell nucleus. n indicates the number of counted cells for each condition. Error bars represent standard error. Statistically significant differences to the control (untreated cells) are indicated by ***P < .001.
IFNα-induced increase of PML NB number is suppressed by TSA. (A) Confocal images of HeLa cells untreated (CON) or treated with 1000 U/mL interferon-α (IFN), 500 ng/mL trichostatin A (TSA), or their combination (IFN + TSA) for 24 hours, immunostained with Sp100 or PML antibody; nuclei were costained with Sytox. Images were acquired by a Leica TCSSP confocal laser-scanning microscope equipped with a Leica HCX PL APO 63×/1.32 NA oil-immersion objective lens, and were processed by Leica Confocal Software version 2.5 (Leica Microsystems Heidelberg GmbH, Mannheim, Germany) and Adobe Photoshop 5.5 (Adobe Systems, San Jose, CA). Bar represents 10 μm. (B-C) Quantification of PML NBs. HeLa cells were treated with IFN, TSA, or IFN + TSA for the indicated time; were incubated first with one drug for a period of 12 hours and then incubated with the second drug after washing with PBS; or were maintained in the fresh medium (fm) for the next 12 hours as indicated for each column. The number of PML NBs per nucleus detected by PML antibody was counted on a total projection of confocal sections (1 section per 0.4 μm) through entire cell nucleus. n indicates the number of counted cells for each condition. Error bars represent standard error. Statistically significant differences to the control (untreated cells) are indicated by ***P < .001.
TSA inhibits PML transcription induced by IFNα
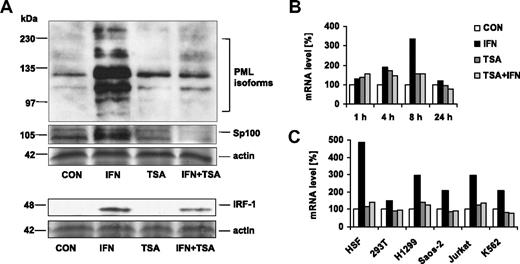
To clarify whether the suppressive effect of TSA on IFNα-induced increase of PML NB number is caused by the dispersion of PML and Sp100 proteins from bodies to nucleoplasm or by their decreased cell pool, we assessed cellular levels of both proteins by specific antibodies on immunoblots in a similar experimental setup as described in the previous paragraph. As shown on Figure 2A, TSA suppressed the IFNα-mediated induction of both proteins. For PML, identical results were obtained in human skin fibroblasts and HEK293 cells (not shown). In addition, PML was suppressed by TSA in mouse NIH-3T3 cells induced by IFNβ (not shown). Notably, IFNα-induced protein levels of IRF-1 were slightly decreased after TSA treatment in HeLa cells (Figure 2A). These findings suggested that the inhibitory effect of TSA on the expression of these IFNα-induced genes is mediated at the transcriptional level. For further confirmation, we measured specific PML mRNA levels by real-time RT-PCR. The primers were designed to cover the region of PML gene that is common to all PML isoforms (forward primer was designed over the junction of exons 2 and 3, and reverse primer was positioned in the exon 3; first 4 exons are shared by all PML isoforms). PML mRNA levels were assessed in HeLa cells treated with IFNα, TSA, or combination of both for 1, 4, 8, or 24 hours and normalized to GAPDH as endogenous control gene. As shown in Figure 2B, the level of PML mRNA increased more than 3 times in comparison with control cells, with a maximum after approximately 8 hours of IFNα treatment, and returned back to the original level 24 hours after the start of treatment. Consistently with previous findings, IFNα-induced up-regulation of PML mRNA was substantially blocked in the cells treated simultaneously with TSA (within the whole range of concentrations: 30, 60, 125, 250, 500 ng/mL; not shown). Next, we performed PML mRNA analyses also with the SaOS-2, HSF, HEK293T, H1299, K562, and Jurkat cell lines. Notably, we observed a similar suppressive effect of TSA on IFNα-mediated induction of PML mRNA in all human cell lines tested (Figure 2C). In summary, these findings show that TSA blocks IFNα-mediated transcription of PML gene and that the suppressive effect of TSA is independent of cellular origin. Moreover, the same effect seen in mouse NIH-3T3 cells suggests an operation of a similar mechanism controlling IFNα-induced expression of PML and other ISGs in different species.
TSA suppresses IFNα-induced PML transcription. (A) Western blot analysis of PML, Sp100, and IRF1 proteins in HeLa cells. Cell lysates (50 μg total protein) were loaded on the SDS gels. Proteins were detected by the indicated antibodies; Coomassie- or Ponceau S–stained bands of actin were used as a loading control. (B-C) Real-time RT-PCR quantification of PML mRNA. (B) HeLa cells were treated with 1000 U/mL interferon-α (IFN), 500 ng/mL trichostatin A (TSA), or both (IFN + TSA) for 1, 4, 8, and 24 hours. PML mRNA levels were normalized to GAPDH; the same results were obtained using other 2 control genes—actin and 18S rRNA (not shown). (C) Various cell lines were treated with IFN, TSA, or IFN + TSA for 8 hours, and PML mRNA was quantified by real-time RT-PCR relative to GAPDH.
TSA suppresses IFNα-induced PML transcription. (A) Western blot analysis of PML, Sp100, and IRF1 proteins in HeLa cells. Cell lysates (50 μg total protein) were loaded on the SDS gels. Proteins were detected by the indicated antibodies; Coomassie- or Ponceau S–stained bands of actin were used as a loading control. (B-C) Real-time RT-PCR quantification of PML mRNA. (B) HeLa cells were treated with 1000 U/mL interferon-α (IFN), 500 ng/mL trichostatin A (TSA), or both (IFN + TSA) for 1, 4, 8, and 24 hours. PML mRNA levels were normalized to GAPDH; the same results were obtained using other 2 control genes—actin and 18S rRNA (not shown). (C) Various cell lines were treated with IFN, TSA, or IFN + TSA for 8 hours, and PML mRNA was quantified by real-time RT-PCR relative to GAPDH.
Other histone deacetylase inhibitors suppress IFNα-induced PML up-regulation
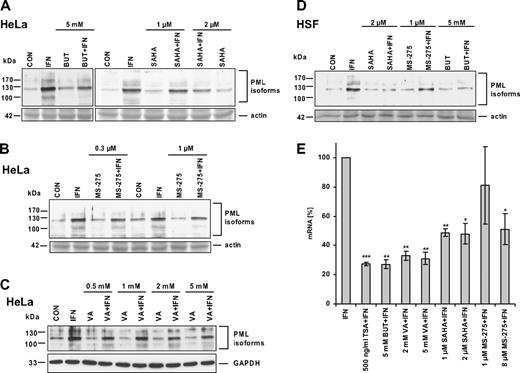
As the individual types of HDACs differ in their sensitivity to various inhibitors,12 we tested the effect of selected HDAC inhibitors on IFNα-induced PML expression. Sodium butyrate (BUT; 5 mM; 24 hours) blocked IFNα induction of PML protein in HeLa cells (Figure 3A) and in HSFs (Figure 3D). We were also interested if MS-275, SAHA, and valproic acid (VA), the HDAC inhibitors currently used in the early phases of clinical trials, have the same suppressive effect on IFNα-induced PML expression as TSA and BUT. MS-275 at a concentration of 0.3 μM (IC50 for HDAC129 ), 1 μM, or 8 μM (IC50 for HDAC329 ; not shown) showed only a slight inhibition of IFNα induction of PML protein in HeLa cells (Figure 3B) and HSFs (Figure 3D). A slight inhibition was also observed with SAHA at the 1-μM concentration (Figure 3A; only HeLa cells shown), whereas 2 μM and 10 μM (not shown) SAHA exhibited more pronounced suppression in both HeLa cells and HSFs (Figure 3A,D). Valproic acid, a suggested inhibitor of class I HDACs,30 had a mild suppressive effect on IFNα induction of PML protein level at concentration up to 5 mM (Figure 3C), that is, at the concentration reported to induce comparable levels of histone acetylation as 5 mM BUT or 100 nM TSA.30 Because we observed variable intensity of suppression of IFNα-induced PML protein levels by different HDAC inhibitors (especially VA and MS-275), we decided to confirm and compare the effects of all inhibitors at the level of PML mRNA. PML mRNA levels in HeLa cells treated with IFNα alone or in combination with particular HDAC inhibitor for 8 hours (ie, at the time interval when IFNα induction of PML mRNA is peaking; Figure 2B) were measured by real-time RT-PCR (Figure 3E). All tested inhibitors showed a significant suppression of PML mRNA in the following order: TSA > BUT/VA > SAHA > MS-275 (8 μM). With the exception of VA, these data correspond well with the suppressive effect of HDAC inhibitors observed at the protein level. In summary, all tested HDAC inhibitors showed, although to various extents, a significant suppressive effect on PML expression induced by IFNα.
HDAC inhibitors differ in their ability to suppress IFNα induction of PML. Western blot analysis of PML protein levels in HeLa cells (A-C) or HSFs (D) simultaneously treated for 24 hours with IFNα and one of the following HDAC inhibitors: BUT, VA, SAHA, or MS-275. The final concentrations of the inhibitors are indicated. Cell protein lysates (25 μg total protein) were loaded on 8% SDS gels (actin band of Ponceau S–stained membrane or GAPDH signal is shown to demonstrate equal loading). (E) Real-time RT-PCR quantification of HDAC inhibitors' suppression of IFNα-induced PML mRNA. HeLa cells were treated with 1000 U/mL IFNα alone or simultaneously with a particular HDAC inhibitor for 8 hours. PML mRNA levels were normalized to GAPDH. Statistically significant differences compared with the IFNα-treated cells are indicated by ***P < .001, **P < .005, or *P < .05; 3 independent experiments were evaluated. Error bars represent standard error.
HDAC inhibitors differ in their ability to suppress IFNα induction of PML. Western blot analysis of PML protein levels in HeLa cells (A-C) or HSFs (D) simultaneously treated for 24 hours with IFNα and one of the following HDAC inhibitors: BUT, VA, SAHA, or MS-275. The final concentrations of the inhibitors are indicated. Cell protein lysates (25 μg total protein) were loaded on 8% SDS gels (actin band of Ponceau S–stained membrane or GAPDH signal is shown to demonstrate equal loading). (E) Real-time RT-PCR quantification of HDAC inhibitors' suppression of IFNα-induced PML mRNA. HeLa cells were treated with 1000 U/mL IFNα alone or simultaneously with a particular HDAC inhibitor for 8 hours. PML mRNA levels were normalized to GAPDH. Statistically significant differences compared with the IFNα-treated cells are indicated by ***P < .001, **P < .005, or *P < .05; 3 independent experiments were evaluated. Error bars represent standard error.
TSA suppressive effect on IFNα stimulation of PML is not caused by impaired translocation of STAT2 to nucleus
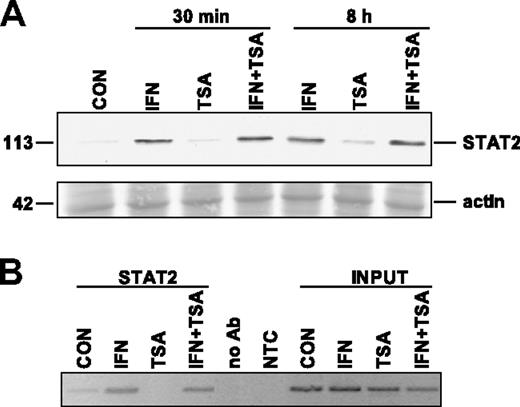
It has been shown previously that TSA suppresses the response to viral infection in mice and concurrently inhibits cytoplasmic-nuclear relocalization of signal transducer and activator of transcription 2 (STAT2) in virus-infected L929 cells.31 STAT2 is a component of IFN-stimulated gene factor 3 (ISGF3) complex, which is a transcription factor binding to ISRE elements in promoters of IFNα/β-induced genes. To test whether TSA has the same inhibitory effect on IFNα-induced translocation of STAT2 to the nucleus in our model, we performed immunoblot analysis of STAT2 localization in the nuclear fraction of HeLa cells treated with IFNα, TSA, or both. Consistently with the published data, we found that within as soon as 30 minutes after IFNα treatment, STAT2 appeared in the nuclear fraction (Figure 4A). STAT2 persisted in the nuclear fraction for at least 8 hours after IFNα treatment, when a peak of PML mRNA induction was detected by real-time RT-PCR. However, in the cells simultaneously treated by IFNα and TSA either for 30 minutes or for 8 hours, STAT2 signal in the nuclear fraction was similar to that of the cells treated by IFNα alone (Figure 4A). This indicates that in our model TSA did not block IFNα-induced translocation of STAT2 into the nucleus.
TSA does not affect IFNα-induced translocation of STAT2 to nucleus and STAT2 binding to PML promoter. (A) Western blot analysis of STAT2 in nuclei of HeLa cells treated with 1000 U/mL IFNα, 500 ng/mL TSA, or both simultaneously for either 30 minutes or 8 hours. Nuclear extracts (25 μg total protein) were loaded on 8% SDS gels (Ponceau S–stained band of actin is shown for equal loading). (B) Chromatin immunoprecipitation using STAT2 antibody and PCR amplification of PML promoter region containing ISRE element (located at +606 to +618 after the most 5′ major transcription start according to Stadler et al20 ). To exclude unspecific binding to G-Sepharose beads, we performed ChIP in the absence of the antibody (no Ab). The negative nontemplate control (NTC) reaction excluded unspecific PCR amplification. Cells were treated with the indicated drugs for 8 hours.
TSA does not affect IFNα-induced translocation of STAT2 to nucleus and STAT2 binding to PML promoter. (A) Western blot analysis of STAT2 in nuclei of HeLa cells treated with 1000 U/mL IFNα, 500 ng/mL TSA, or both simultaneously for either 30 minutes or 8 hours. Nuclear extracts (25 μg total protein) were loaded on 8% SDS gels (Ponceau S–stained band of actin is shown for equal loading). (B) Chromatin immunoprecipitation using STAT2 antibody and PCR amplification of PML promoter region containing ISRE element (located at +606 to +618 after the most 5′ major transcription start according to Stadler et al20 ). To exclude unspecific binding to G-Sepharose beads, we performed ChIP in the absence of the antibody (no Ab). The negative nontemplate control (NTC) reaction excluded unspecific PCR amplification. Cells were treated with the indicated drugs for 8 hours.
TSA treatment does not impair STAT2 binding to ISRE element of PML promoter
To investigate whether TSA affects the binding of ISGF3 complex to ISRE element of PML promoter in vivo, we performed chromatin immunoprecipitation of the PML promoter region containing ISRE element using STAT2 antibody in the control or in IFNα/TSA-treated HeLa cells. An increase of STAT2 bound to PML promoter in IFNα but also in IFNα + TSA–treated cells was detected repeatedly in independent experiments when compared with control or TSA-treated cells, where almost no signal was detected (Figure 4B). In summary, data from the cell fractionation and chromatin immunoprecipitation experiments suggest that in human cells TSA does not block translocation of STAT2 to the nucleus after IFNα stimulation and that TSA does not block STAT2 binding to PML promoter.
Discussion
Acetylation/deacetylation of histones is known to modulate chromatin organization and thus play a key role in the regulation of gene expression. Several specific HDAC inhibitors found to date represent not only valuable research tools, but also prospective drugs with a potential clinical use, especially in cancer therapy, since inhibitors of HDAC have been shown to cause differentiation, growth arrest, and/or apoptosis of cancer cells both in vitro and in vivo.12,32-34 Chromatin hyperacetylation prevailingly has a stimulatory effect on gene expression, when about 2% of total genes are influenced.35,36 Nevertheless, some genes, including a subset of IFN-controlled genes, are not stimulated but repressed. Genin et al31 found that TSA was able to suppress virus-induced expression of IRF-7 gene and IFNα-induced expression of ISG54, ISG56, ISG15, and IFI6-16 genes in murine L929 cells. At the same time, Nusinzon and Horvath37 reported a suppressive effect of TSA on IFNα and IFNγ induction of ISGs in several human cell lines, and Sakamoto et al38 demonstrated the suppressive effect of TSA on IFNβ induction of a similar set of ISGs in human foreskin fibroblasts. Our finding that TSA and other HDAC inhibitors block IFNα-mediated induction of PML NB components further supports and extends these reports. They indicate that many if not all IFN-inducible genes might be affected by HDAC inhibitors through a common mechanism operating during the propagation of interferon type I and type II signaling pathways. Although a direct deacetylation-regulated target has not yet been identified, Genin et al31 showed that TSA impairs STAT2 translocation into the nucleus in virus-infected murine cells and the formation and cytoplasmic-nuclear translocation of ISGF3, a trimeric complex of STAT1, STAT2, and IRF-9.39 This complex with a binding capacity to ISRE elements is formed on the cytoplasmic side of plasma membrane after the activation of IFNα/β receptors40 and then is translocated into the nucleus to transactivate ISGs. Our data show, however, that at least in HeLa cells the entry of STAT2 into the nucleus after IFNα stimulation was not blocked by TSA. Moreover, as proved by the ChIP analysis, STAT2 was bound to PML ISRE element to the same extent as in the cells treated by IFNα alone, even when PML expression was simultaneously suppressed by TSA. In addition, the transcription of IRF-1 gene, which is regulated by γ-interferon–activated factor (GAF)/α-interferon–activated factor (AAF) through GAS element after IFN stimulation, was also partially suppressed by TSA. This indicates that the effect is not mediated exclusively via ISGF3. Thus, our results are in accordance with the study of Nusinzon and Horvath showing no effect of pharmacological doses of TSA on STAT activation, heterodimerization, and subcellular trafficking.37 Their and our data also implicate that the effect of TSA is targeted to the downstream stages of IFN signaling pathways and that the effect of TSA on ISGF3 translocation previously observed in virus-infected murine cells is probably not present in IFN-treated human cells. These differences can be caused by different stimuli used; cellular response to virus infection is more complex than to IFNs themselves. In addition, transactivation domain (TAD) at carboxyl-terminus of murine and human STAT2 homologs interacts with a distinct though overlapping set of cellular proteins.41 It should be pointed out that the ISGF3 transcriptional signal is mediated exactly by STAT2 TAD42 through interactions with various cofactors including histone acetyltransferase CBP/p300.43 In summary, it is tempting to speculate that deacetylation of some other component of a transcription machinery is a necessary step for the full transcriptional activation of ISGs after IFN induction. Since the reports concerning the acetylation of individual members of STAT family are emerging,44-47 STAT proteins themselves might be the subjects of such regulatory event.
Several HDAC inhibitors are currently in clinical trials as anticancer agents (for reviews, see Marks et al12 and Kelly et al13 ). Synthetic derivates of hydroxamic acid such as SAHA48 exert anticancer activity at the well-tolerated doses. Growth inhibitory activity of HDAC inhibitors has been demonstrated for a large range of transformed cell types including solid tumor cell lines and hematopoietic transformed cell lines.34
The fact that several if not all genes controlled by interferons type I are affected by HDAC inhibitors can be of practical importance for cancer therapy as can be implied from known antiviral and anticancer effects of interferons (for a recent review on interferons in cancer therapy, see, for example, Borden et al49 ). Here, we report an adverse effect of TSA on the induction of PML by interferons. Our data show that though TSA does not decrease basal (ie, IFNα-noninduced) expression of PML gene, the IFNα-mediated activation of PML is strongly suppressed by TSA at the transcriptional level.
There are about 300 genes under the control of interferon type I as determined by DNA oligonucleotide microarray analysis.50 Many of them are transcription regulators such as members of the IRF family. These transcription activators or repressors act in positive (delayed interferon response) or negative feedback loops sharing partly the DNA binding elements of ISGs with factors of an early immediate IFN response. Apart of IRF-3 and IRF-7, IRF-1 is another possible target for further analysis of the suppressive effect of HDAC inhibitors on the interferon pathway, since this factor participates in a delayed interferon response and as such might be a potential transcription regulator of PML gene. This idea is supported by the fact that the maximal levels of IFNα-induced PML mRNA are observed 8 hours after interferon stimulus; the levels of IRF-1 protein are peaking within 2 and 4 hours after IFNα stimulation (Lehtonen et al51 and our observation). Since IRF-1 up-regulation by IFNα was partially inhibited by TSA and we also identified binding of IRF-1 to ISRE element of PML promoter by ChIP (J.V., unpublished observation, September 14, 2004), we can speculate that the inhibition of PML expression can be caused, at least partially, by suppression of IRF-1 cellular level.
Although HDAC inhibitors used in this study are thought to be nonselective or poorly selective inhibitors of all or most of class I (HDAC1, HDAC2, HDAC3, HDAC8, and HDAC11—newly classified into class IV) and class II (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10) enzymes,52 we observed different suppressive effects at equimolar doses of hydroxamates TSA and SAHA on IFNα-induced PML expression. This is in agreement with published data showing that SAHA is a weaker inhibitor of HDACs than TSA.53 In contrast to both hydroxamates, benzamidine-type inhibitor MS-275 was reported to have concentration-dependent effect on class I HDAC1 and HDAC3 and does not inhibit HDAC8 even at 2 order higher concentrations in one study,29 while an equal inhibition of both HDAC1 and HDAC3 was found in another study.54 Both studies show congruently that HDAC8 is not inhibited by MS-275 at the concentrations used in this study. Since TSA is inhibiting almost equally all of these 3 HDACs, one can speculate from differential suppressive effects of TSA and MS-275 that HDAC8 could be one of the HDAC candidates involved in PML gene expression induced by IFNα. Although we observed a variable suppressive effect of valproic acid in comparison with chemically related sodium butyrate at PML protein levels, the suppressive effect of VA detected by a more quantitative approach using PML mRNA estimation was comparable with that of sodium butyrate. This indicates that the effect of both drugs is mediated at the transcriptional level by a similar mechanism.
The discrimination of the specific HDAC involved in the regulation of interferon-stimulated genes would be the next step in our understanding of the transcriptional regulation of interferon-dependent genes. Regarding the importance of interferon system for the maintenance of organism homeostasis, this can also lead to design or to selection of antitumor HDAC inhibitors that would not perturb the interferon pathway.
Authorship
Contribution: J.V. performed research, analyzed data, and wrote the paper; Z.N. performed research and analyzed data; L.R. performed research and analyzed data; M.K. analyzed data; P.H. analyzed data; and Z.H. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Zdenĕk Hodný, Institute of Experimental Medicine, Vídeňská 1083, 142 20 Prague, Czech Republic; e-mail: hodny@biomed.cas.cz.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by Grant Agency of the Czech Republic (projects no. 304/03/1210 and no. DYN/04/E002), Grant Agency of the Academy of Sciences of the Czech Republic (project no. IAA500390501), and by the institutional grant no. AV0Z50390512.
The authors would like to thank Iva Jelinkova for a technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal