Abstract
Hepcidin is a key iron-regulatory hormone produced by the liver. Inappropriately low hepcidin levels cause iron overload, while increased hepcidin expression plays an important role in the anemia of inflammation (AI) by restricting intestinal iron absorption and macrophage iron release. Its expression is modulated in response to body iron stores, hypoxia, and inflammatory and infectious stimuli involving at least in part cytokines secreted by macrophages. In this study we established and characterized IL6-mediated hepcidin activation in the human liver cell line Huh7. We show that the proximal 165 bp of the hepcidin promoter is critical for hepcidin activation in response to exogenously administered IL6 or to conditioned medium from the monocyte/macrophage cell line THP-1. Importantly, we show that hepcidin activation by these stimuli requires a STAT3 binding motif located at position –64/–72 of the promoter. The same STAT binding site is also required for high basal-level hepcidin mRNA expression under control culture conditions, and siRNA-mediated RNA knockdown of STAT3 strongly reduces hepcidin mRNA expression. These results identify a missing link in the acute-phase activation of hepcidin and establish STAT3 as a key effector of baseline hepcidin expression and during inflammatory conditions.
Introduction
The iron-regulatory hormone and hepatic acute-phase protein hepcidin is strongly implicated in the anemia of inflammation (AI), a common clinical disorder that affects patients with acute and chronic infections, trauma, inflammatory disorders, and malignancies. The disease is characterized by hypoferremia, low serum iron-binding capacity, and normal to elevated ferritin levels.1 Several cytokines that participate in the pathogenesis of AI also modulate iron metabolism. For example, IL1 and TNFα induce hypoferremia in mice, and IL6 increases hepatic ferritin synthesis and transferrin uptake in rats.2-5 Importantly, hypoferremia is induced within a few hours after injection of hepcidin into mice.6 Hepcidin binds to the iron export protein ferroportin (also known as SLC11A3, IREG1, and MTP) and triggers its degradation to decrease iron egress from duodenal enterocytes, macrophages, and hepatocytes, thus contributing to the hypoferremia that is a hallmark of AI.7 Hepcidin may hence be considered to be a principal iron regulatory hormone, a key mediator of AI, and a bridge between innate immunity and iron metabolism.
IL6 induces hepcidin expression both in cell culture and in vivo.8,9 Conditioned medium (CM) from LPS-treated macrophages activates hepcidin mRNA expression in hepatocytes, a response that can be blocked by anti-IL6 antibodies.8,10 However, additional factors seem to regulate hepcidin expression, because mice with a targeted disruption of the IL6 gene (IL6−/−) still activate hepcidin expression in response to endotoxin injection.11 Interestingly, the treatment of primary mouse hepatocytes with IL-1α and IL1β strongly increases hepcidin mRNA expression.10 Moreover, a previous report suggests that mice lacking the protein HFE fail to mount an appropriate hepcidin response following LPS injection despite a normal increase in IL6 expression,12 although other investigators have reached different conclusions using different experimental systems.11 Taken together, these results reflect the complexity of hepcidin regulation by inflammatory stimuli.
HFE, transferrin receptor 2 (TfR2), and hemojuvelin (HJ), 3 genes mutated in a group of frequent iron overload disorders called hereditary hemochromatosis (HH), control appropriate hepcidin expression.13-20 Recent work demonstrates that HJ can function as a bone morphogenetic protein (BMP) coreceptor that mediates BMP signaling.21 BMP positively regulates hepcidin expression at the transcriptional level, a response that is enhanced by HJ expression.21 HJ-deficient mice show reduced levels of phosphorylated Smad 1/5/8, suggesting that BMP-dependent Smad activation is important for hepcidin activation in the mouse.21 Further work supports a role for the TGFβ pathway and SMAD4 in the transcriptional activation of hepcidin in response to iron overload or IL6.22 The hepcidin response to TGFβ was abrogated in SMAD4-deficient hepatocytes,22 and mice with a liver-specific SMAD4 deficiency develop iron overload.22 Hepcidin expression is further affected by the CCAAT/enhancer-binding protein (C/EBP), because mice with a liver-specific disruption of C/EBPα show decreased hepcidin mRNA levels and periportal hepatic iron overload.23
Little information is presently available about promoter elements and transcription factors that control hepcidin expression. To address this issue, we established a cell-based assay system to investigate the cis-acting elements and trans-acting factors for basal hepcidin expression and in response to inflammatory stimuli. We show that a STAT binding motif at position –64/–72 of the hepcidin promoter and STAT-3 are critical for the control of baseline hepcidin mRNA expression and under inflammatory conditions.
Materials and methods
Cell culture
Human hepatocarcinoma Huh7 and monocyte/macrophage THP-1 cell lines were cultured in Dulbecco modified Eagle medium (DMEM; high glucose; Invitrogen, Carlsbad, CA). Medium was supplemented with 10% heat-inactivated low-endotoxin fetal bovine serum (FBS; Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin. All cultures were maintained at 37°C under 5% CO2. Cellfree conditioned medium (CM) from THP-1 cells growing at an initial cell confluence of 70% was collected 12 hours, 24 hours, 48 hours, and 72 hours after plating of the cells and used undiluted to treat Huh7 cells. Human recombinant IL6 (Roche, Basel, Switzerland) was used at final concentrations of 1 ng/mL.
RNA isolation
Total RNA was isolated using the Qiagen RNAeasy kit according to the manufacturer's instruction (QIAGEN, Hilden, Germany). The concentration and purity of the RNA was determined by OD260/280 reading. The quality of the RNA was assessed by gel electrophoresis and ethidium bromide staining.
Reverse transcription and quantitative real-time PCR analysis
Two micrograms of total RNA was reverse transcribed using 10 μM each of dCTP, dGTP, dATP, and dTTP, 100 ng random primers, 1 × first-strand buffer (Gibco BRL, Carlsbad, CA), 0.01 M DTT, and 200 units of SuperScript II reverse transcriptase (Gibco BRL) in a 20 μL reaction for 90 minutes at 42°C. Real-time polymerase chain reaction (PCR) was performed using the ABI Prism 7500 Applied Biosystems (Applera Deutschland, Darmstadt, Germany). Amplification reactions were carried out in 25 μL volume using SYBR Green I dye and the following amplification conditions: 50°C for 2 minutes and 95°C for 10 minutes (95°C, 15 seconds; 60°C, 15 seconds) for 45 cycles. Primers were designed to specifically amplify 123 bp of hepcidin cDNA (forward 5′-CTCTGTTTTCCCACAACAGAC-3′, reverse 5′-TAGGGGAAGTGGGTGTCTC-3′); 113 bp of GAPDH cDNA (forward 5′-CATGAGAAGTATGACAACAGCCT-3′, reverse 5′-AGTCCTTCCACGATACCAAAGT-3′); 152 bp of STAT3 cDNA (forward 5′-CATATGCGGCCAGCAAAGAA-3′, reverse 5′-ATACCTGCTCTGAAGAAACT-3′); and 127 bp of C/EBPα cDNA (forward 5′-AAGAACAGCAACGAGTACCGG-3′, reverse 5′-CATTGTCACTGGTCAGCTCCA-3′).
The mRNA/cDNA abundance of each gene was calculated relative to the expression of a housekeeping gene, GAPDH (glyceraldehyde-3-phosphate-dehydrogenase).
Promoter analysis
The –942-bp and –165-bp nucleotide fragments of the 5′-flanking genomic region plus the 5′ untranslated region (UTR) of the human hepcidin gene were obtained from human genomic DNA by PCR amplifications using the forward primers 5′-GGCTCGAGGTACTCATCGGACTGTAGAT-3′ and 5′-GGCTCGAGTGAACACACCTCTGCCGGCTGA-3′, respectively, and reverse primer 5′-GGAAGCTTCGTGCCGTCTGTCTGGCTGT-3′; the incorporated XhoI and HindIII sites are underlined. PCR fragments were digested with XhoI/HindIII and inserted in the promoterless luciferase reporter vector pGL3-Basic (Promega, Madison, WI). The –385 bp fragment was obtained by XhoI/Bsu36I digestion of the –942 bp construct.
AP1 (position –242/–233) and STAT (position –69/–72) binding sites were deleted by site-directed mutagenesis (Invitrogen) using the following primer pairs: 5′-ACTTTTTTCCCGATCAGCAGTGATGGGGAAA-3′ and 5′-GTTTTCCCCCTTGAAAAAAGGGCTAGTCGTC-3′; and 5′-GCCTTTTCGGCGCCACCACCTGGAAATGAGA-3′ and 5′-GGTGGTGGCGCCGAAAAGGCGGGAGAGATA-3′, respectively.
Details of the constructs are available upon request. All constructs were confirmed by DNA sequencing. Search for putative transcription factor binding sites was performed using MatInspector24 (Genomatrix Software, München, Germany) and TFsearch version 1.3 (Computational Biology Research Center, National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan).
Cell transfection and luciferase assay
Huh7 cells were seeded at 30% confluency in 35 mm–diameter dishes and grown overnight. A total of 500 ng of pGL3 hepcidin promoter vectors was cotransfected with a control plasmid containing the Renilla gene under the control of the CMV promoter (details of the control plasmid are available upon request). Transfections were performed using lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Twenty-four hours after transfection the cells were harvested or treated with the indicated stimuli. At the indicated time points cells were lysed in passive lysis buffer (Promega), and cellular extracts were analyzed for luciferase activity using the Dual-Luciferase-Reporter assay system (Promega) and a Centro LB 960 luminometer (Berthold Technologies, Bad Wildbad, Germany).
siRNA-mediated knockdown of C/EBPα and STAT3
Huh7 cells were seeded at 30% confluency in DMEM supplemented with 10% low-endotoxin FBS without antibiotics. After 24 hours, cells were transfected using Oligofectamine Reagent (Invitrogen) and 100 nM siRNA directed against C/EBPα (Dharmacon, Lafayette, CO) or STAT3 (Dharmacon). After a further 24 hours a second round of transfection was performed, and the cells were harvested 24 hours later. As a control, siRNA directed against luciferase (Dharmacon) was transfected. The efficiency of the knockdown was analyzed at the mRNA level by quantitative real-time PCR.
Statistics
Results were expressed as mean plus or minus SD. The Student t test was used for estimation of statistical significance.
Results
IL6-dependent hepcidin activation is mediated by a STAT binding motif at position –64/–72 of the hepcidin promoter
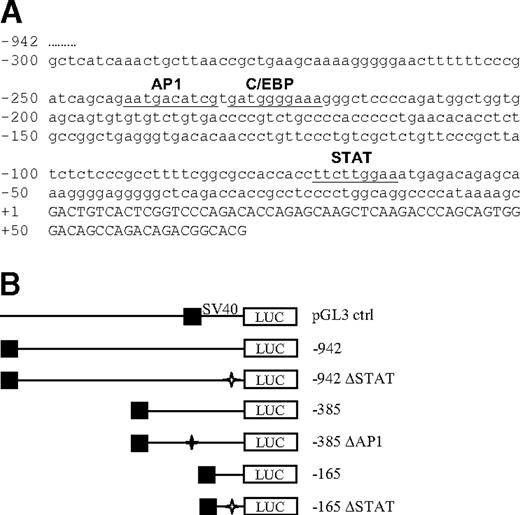
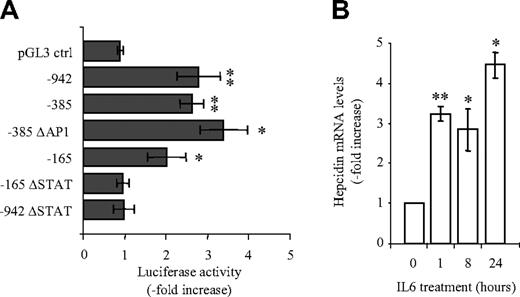
To investigate how the inflammatory cytokine IL6 activates hepcidin mRNA expression, we generated luciferase reporter vectors containing the 942 bp upstream of the transcription start site of the human hepcidin promoter or different truncations/mutations thereof (Figure 1) Promoter truncations were designed such that putative transcription factor binding sites, as defined by bioinformatic analyses (eg, an AP1 binding motif at position –242/–233 or a STAT binding motif at position –64/–72), were contained within the promoter fragments analyzed. The luciferase reporter constructs were transfected into the human hepatocyte cell line Huh7. Huh7 cells were selected for their high level of endogenous hepcidin mRNA expression compared with other hepatocyte cell lines (eg, HepG2, Hepa1-6, AML12) that we tested (data not shown). Hepcidin mRNA is also expressed in the human and mouse liver.25 Transfected Huh7 cells were treated with IL6 (1 ng/mL), and luciferase activity was measured 24 hours later. Luciferase activity increased 2- to 3-fold in response to IL6 treatment when Huh7 cells were transfected with the –942 bp, –385 bp, and -165 bp hepcidin promoter constructs (Figure 2A). By comparison, luciferase activity remained almost unaltered when expressed from the pGL3 control vector (Figure 2A). Importantly, endogenous hepcidin mRNA expression as an internal control also increased approximately 3-fold upon treatment with 1 ng/mL IL6 (Figure 2B). These results suggest that the –165 promoter fragment harbors the essential element(s) for IL-6 induction.
Luciferase reporter vectors containing the human hepcidin promoter. (A) Nucleotide sequence of the 5′-flanking region of human hepcidin. The sequence of the 300 bp of the transcription start site of the human hepcidin promoter is indicated in small letters. The 5′ UTR of the hepcidin gene is indicated in capital letters starting at position +1. Underlined sequences indicate transcription factor binding motifs. The AP1 and STAT binding motifs are phylogenetically conserved (100% identity of the core sequences) in the human, mouse, and rat. The transcription factor binding motifs were identified by MatInspector24 and TFsearch version 1.3. (B) Luciferase (firefly) reporter vectors. The –942 construct contains the longest promoter region analyzed in this study; the –385 construct contains the AP1, C/EBPα, and STAT binding motifs; in the –385 ΔAP1 construct the AP1 binding motif is deleted; the –165 construct contains the STAT binding motif whereas the AP1 and C/EBPα binding motifs are deleted; in the –165 ΔSTAT construct the STAT binding motif is deleted; in the –942 ΔSTAT construct the STAT binding motif is deleted, while the AP1 and C/EBPα binding motifs are preserved; SV40-driven pGL3 vector expressing luciferase (Renilla) was used as a control. Asterisks indicate the deletion of transcription factor binding motifs; ▪, the 5′ end of the promoter region.
Luciferase reporter vectors containing the human hepcidin promoter. (A) Nucleotide sequence of the 5′-flanking region of human hepcidin. The sequence of the 300 bp of the transcription start site of the human hepcidin promoter is indicated in small letters. The 5′ UTR of the hepcidin gene is indicated in capital letters starting at position +1. Underlined sequences indicate transcription factor binding motifs. The AP1 and STAT binding motifs are phylogenetically conserved (100% identity of the core sequences) in the human, mouse, and rat. The transcription factor binding motifs were identified by MatInspector24 and TFsearch version 1.3. (B) Luciferase (firefly) reporter vectors. The –942 construct contains the longest promoter region analyzed in this study; the –385 construct contains the AP1, C/EBPα, and STAT binding motifs; in the –385 ΔAP1 construct the AP1 binding motif is deleted; the –165 construct contains the STAT binding motif whereas the AP1 and C/EBPα binding motifs are deleted; in the –165 ΔSTAT construct the STAT binding motif is deleted; in the –942 ΔSTAT construct the STAT binding motif is deleted, while the AP1 and C/EBPα binding motifs are preserved; SV40-driven pGL3 vector expressing luciferase (Renilla) was used as a control. Asterisks indicate the deletion of transcription factor binding motifs; ▪, the 5′ end of the promoter region.
IL6-dependent hepcidin activation is mediated by STAT binding motif at position –64/–72 of the hepcidin promoter. (A) Luciferase reporter assays. Huh7 cells were transfected with luciferase reporter vectors and 24 hours later treated with IL6 (1 ng/mL; 24 hours). Transfections were performed in triplicates, and results are presented as a fold change ± SD of firefly/Renilla (F/R). (B) Hepcidin mRNA expression. Huh7 cells were treated with IL6 (1 ng/mL), and endogenous hepcidin mRNA expression was analyzed by quantitative real-time PCR. Data are normalized to mRNA expression of a housekeeping gene, GAPDH, and shown as fold change in comparison with untreated cells. Significant expression changes are marked by an asterisk, whereby * represents P < .05 and ** represents P < .005.
IL6-dependent hepcidin activation is mediated by STAT binding motif at position –64/–72 of the hepcidin promoter. (A) Luciferase reporter assays. Huh7 cells were transfected with luciferase reporter vectors and 24 hours later treated with IL6 (1 ng/mL; 24 hours). Transfections were performed in triplicates, and results are presented as a fold change ± SD of firefly/Renilla (F/R). (B) Hepcidin mRNA expression. Huh7 cells were treated with IL6 (1 ng/mL), and endogenous hepcidin mRNA expression was analyzed by quantitative real-time PCR. Data are normalized to mRNA expression of a housekeeping gene, GAPDH, and shown as fold change in comparison with untreated cells. Significant expression changes are marked by an asterisk, whereby * represents P < .05 and ** represents P < .005.
Interestingly, bioinformatic analysis24 identified a highly conserved, putative STAT binding motif within the first 165 bp of the hepcidin promoter at position –64/–72. The deletion of this STAT binding motif in the context of both the complete –942 bp or the shortened –165 bp construct completely abolished the IL6-dependent increase in luciferase activity (Figure 2A). By contrast, the deletion of the AP1 binding motif in the –385 bp construct did not affect IL6 activation. These data show that a functional STAT binding motif at position –69/–72 within the hepcidin promoter mediates the hepcidin response to exogenously administered IL6.
STAT3 mediates hepcidin expression
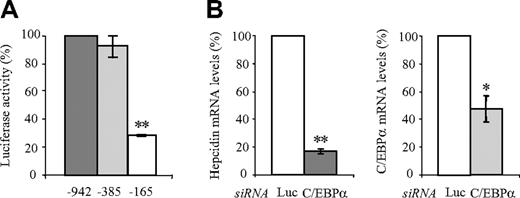
A previous report identified a putative C/EBPα binding site at position –231/–222 of the hepcidin promoter and suggested that C/EBPα is required for high-level hepcidin mRNA expression both in Huh7 cells and in mice.23 Here we show that a truncation of the hepcidin promoter to –385 bp does not affect the luciferase readout in unstimulated control Huh7 cells. A further truncation to -165 bp, however, reduces luciferase activity more than 3-fold after transfection (Figure 3A) Because the C/EBPα binding site is located within this deleted promoter region, we assessed the effect of siRNA-mediated knockdown of C/EBPα on endogenous hepcidin expression in Huh7 cells (Figure 3B). As expected, the knockdown of C/EBPα decreased hepcidin mRNA expression, confirming previous reports (Figure 3B).
C/EBPα mediates a high level of basal hepcidin mRNA expression. (A) Luciferase reporter assays. Huh7 cells were transfected with luciferase reporter vectors, and luciferase activity was measured after 72 hours. Transfections were performed in triplicates, and results are presented as fold change ± SD of firefly/Renilla (F/R). The –165 bp promoter construct (lacking the C/EBPα binding motif) shows more than 3-fold reduced luciferase activity compared with the –942 bp and –385 constructs. Luciferase activity of the –942 bp construct was set to 100%. (B) siRNA-mediated knockdown of C/EBPα. Hepcidin mRNA expression in Huh7 cells was assayed after transfection with specific siRNA directed against C/EBPα or luciferase (Luc siRNA) as a control. Hepcidin and C/EBPα mRNA expression was analyzed by quantitative real-time PCR, and data were normalized to mRNA expression of a housekeeping gene, GAPDH. Data are presented as fold change whereby Huh7 cells transfected with the luciferase control siRNA were set to 100%. Significant expression changes are marked by an asterisk, whereby * represents P < .05 and ** represents P < .005.
C/EBPα mediates a high level of basal hepcidin mRNA expression. (A) Luciferase reporter assays. Huh7 cells were transfected with luciferase reporter vectors, and luciferase activity was measured after 72 hours. Transfections were performed in triplicates, and results are presented as fold change ± SD of firefly/Renilla (F/R). The –165 bp promoter construct (lacking the C/EBPα binding motif) shows more than 3-fold reduced luciferase activity compared with the –942 bp and –385 constructs. Luciferase activity of the –942 bp construct was set to 100%. (B) siRNA-mediated knockdown of C/EBPα. Hepcidin mRNA expression in Huh7 cells was assayed after transfection with specific siRNA directed against C/EBPα or luciferase (Luc siRNA) as a control. Hepcidin and C/EBPα mRNA expression was analyzed by quantitative real-time PCR, and data were normalized to mRNA expression of a housekeeping gene, GAPDH. Data are presented as fold change whereby Huh7 cells transfected with the luciferase control siRNA were set to 100%. Significant expression changes are marked by an asterisk, whereby * represents P < .05 and ** represents P < .005.
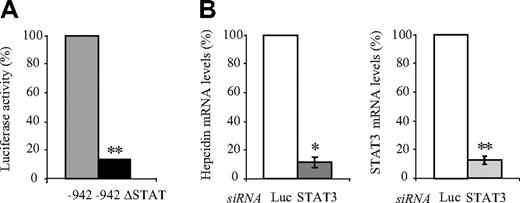
To investigate whether the STAT binding motif at position –64/–72, which mediates the IL6 activation of the hepcidin promoter (Figure 2A), is also required to maintain high-level luciferase expression in untreated Huh7 cells, we analyzed the –942 ΔSTAT construct. Interestingly, we observed a 9.3-fold reduction of luciferase activity compared with the 942 bp construct, suggesting a role for this putative STAT binding site in maintaining high-level hepcidin mRNA expression (Figure 4A)
STAT3 mediates hepcidin expression. (A) Luciferase reporter assays. Huh7 cells were transfected with the –942 bp or the –942 bp ΔSTAT luciferase reporter vectors, and luciferase activity was measured after 48 hours. Transfections were performed in triplicates, and results are presented as a fold change ± SD of firefly/Renilla (F/R). (B) siRNA-mediated knockdown of STAT3. Hepcidin mRNA expression in Huh7 cells was assayed after transfection with specific siRNAs directed against STAT3 or luciferase (Luc siRNA) as a control. Hepcidin and STAT3 mRNA expression was analyzed by quantitative real-time PCR and data normalized to mRNA expression of a housekeeping gene, Gapdh. Data are presented as fold change, whereby Huh7 cells transfected with the luciferase control siRNA were set to 100%. STAT3 mRNA expression was reduced to 12%. Significant expression changes are marked by an asterisk, whereby * represents P < .05 and ** represents P < .005.
STAT3 mediates hepcidin expression. (A) Luciferase reporter assays. Huh7 cells were transfected with the –942 bp or the –942 bp ΔSTAT luciferase reporter vectors, and luciferase activity was measured after 48 hours. Transfections were performed in triplicates, and results are presented as a fold change ± SD of firefly/Renilla (F/R). (B) siRNA-mediated knockdown of STAT3. Hepcidin mRNA expression in Huh7 cells was assayed after transfection with specific siRNAs directed against STAT3 or luciferase (Luc siRNA) as a control. Hepcidin and STAT3 mRNA expression was analyzed by quantitative real-time PCR and data normalized to mRNA expression of a housekeeping gene, Gapdh. Data are presented as fold change, whereby Huh7 cells transfected with the luciferase control siRNA were set to 100%. STAT3 mRNA expression was reduced to 12%. Significant expression changes are marked by an asterisk, whereby * represents P < .05 and ** represents P < .005.
To determine whether the STAT3 transcription factor that is predicted to interact with this binding motif controls hepcidin mRNA expression, we performed siRNA-mediated knockdown of STAT3. A knockdown of STAT3 mRNA levels to 90% caused a 9-fold decrease in endogenous hepcidin mRNA expression as measured by quantitative real-time PCR (Figure 4B).
Taken together, these results confirm the role of C/EBPα in promoting hepcidin expression. They identify STAT3 as a critical transcription factor for the basal expression of hepcidin mRNA under control culture conditions as well as for the IL6-mediated inflammatory response via the STAT binding motif located at position –64/–72 of the hepcidin promoter.
Hepcidin activation by conditioned medium from macrophages is also mediated by the –64/–72 STAT binding motif
In whole organisms, cytokine release by one cell type (eg, macrophages) can trigger changes in gene expression in other cell types (eg, hepatocytes). Previous reports demonstrated that medium supernatant from cultured macrophages or macrophage cell lines can activate hepcidin mRNA expression in hepatocytes.8,9
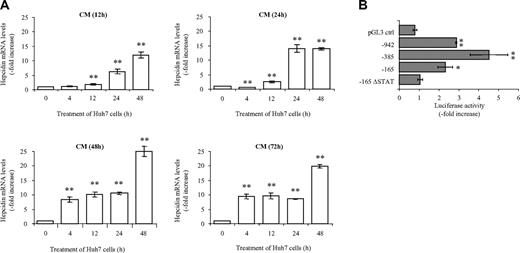
We therefore investigated hepcidin mRNA expression following treatment of Huh7 cells with cellfree conditioned medium (CM) from a monocyte/macrophage cell line, THP-1. Hepcidin mRNA expression strongly increases under these conditions. The time course of hepcidin activation depends on the time that THP-1 cells were cultured to generate the CM (12 hours, 24 hours, 48 hours, or 72 hours). CM from THP-1 cells cultured for 12 hours and 24 hours (CM(12h) and CM(24h), respectively; Figure 5A) activates hepcidin mRNA expression 6.3- and 13.9-fold, respectively, 24 hours after the addition of CM to Huh7 cells (Figure 5A). Interestingly, CM(48h) and CM(72h) causes a fast (4 hours or less) 8- to 9-fold increase in hepcidin mRNA levels (Figure 5A). These data indicate that the longer THP-1 cells are maintained in culture (eg, 48 hours or 72 hours) before collection of the CM, the stronger and earlier is the increase in hepcidin mRNA expression in Huh7 cells.
Cell-free conditioned medium (CM)–dependent hepcidin activation. (A) Conditioned medium (CM) from THP-1 cells was collected after 12, 24, 48, and 72 hours (CM(12h), CM(24h), CM(48h), CM(72h), respectively) in culture. The undiluted CM was used to stimulate Huh7 cells for the indicated time periods. Endogenous hepcidin mRNA expression in Huh7 cells was analyzed by quantitative real-time PCR, and data were normalized to mRNA expression of a housekeeping gene, GAPDH. Data are presented as fold change ± SD. (B) Increased luciferase activity after treatment with CM(48h) depends on a functional STAT binding site. Huh7 cells were transfected with the luciferase reporter vectors. Twenty-four hours after transfection cells were treated with CM(48h), and luciferase activity was assessed 24 hours later. Transfections were performed in triplicates, and results are presented as fold change ± SD of firefly/Renilla (F/R). Significant expression changes are marked by an asterisk, whereby * represents P < .05 and ** represents P < .005.
Cell-free conditioned medium (CM)–dependent hepcidin activation. (A) Conditioned medium (CM) from THP-1 cells was collected after 12, 24, 48, and 72 hours (CM(12h), CM(24h), CM(48h), CM(72h), respectively) in culture. The undiluted CM was used to stimulate Huh7 cells for the indicated time periods. Endogenous hepcidin mRNA expression in Huh7 cells was analyzed by quantitative real-time PCR, and data were normalized to mRNA expression of a housekeeping gene, GAPDH. Data are presented as fold change ± SD. (B) Increased luciferase activity after treatment with CM(48h) depends on a functional STAT binding site. Huh7 cells were transfected with the luciferase reporter vectors. Twenty-four hours after transfection cells were treated with CM(48h), and luciferase activity was assessed 24 hours later. Transfections were performed in triplicates, and results are presented as fold change ± SD of firefly/Renilla (F/R). Significant expression changes are marked by an asterisk, whereby * represents P < .05 and ** represents P < .005.
To identify promoter element(s) responsible for the CM-mediated induction, we transfected Huh7 cells with the luciferase reporter constructs and added CM(48h) 24 hours after transfection for a further 24 hours. Luciferase activity increases 2.9-, 4.5-, and 2.3-fold if luciferase is expressed from vectors containing 942 bp, –385 bp, and –165 bp hepcidin promoter fragments, respectively (Figure 5B). This shows that the hepcidin response (approximately 10-fold after addition of CM(48h) for 24 hours) is at least partially mediated via the hepcidin promoter. This induction is retained in the absence of the AP1 binding site in the –385 bp construct (data not shown), indicating that the AP1 site does not contribute to this activation. However, the deletion of the putative STAT binding site in the –165 bp hepcidin promoter construct completely abrogates the responsiveness of the promoter construct to the CM from macrophages, demonstrating that the STAT binding motif is critically important for the transcriptional activation of the hepcidin promoter in response to both IL6 and CM.
Discussion
Hepcidin is a key regulatory hormone that plays a central role in frequent clinical disorders like hereditary hemochromatosis (HH) and the anemia of inflammation (AI). Hepcidin expression increases in response to iron overload and inflammatory stimuli to reduce duodenal iron absorption and to increase iron retention in the reticuloendothelial system (RES). In the setting of AI, these responses result in hypoferremia, a common hallmark of this disease.26 On the contrary, inappropriately low hepcidin expression in the iron overload disorder HH causes increased iron absorption and iron accumulation in the liver and other parenchymal organs.
To understand the control of hepcidin expression, we investigated the regulatory, cis-acting elements and trans-acting factors in the hepcidin promoter required for baseline hepcidin mRNA expression and in response to inflammatory stimuli. We established experimental conditions in which hepcidin mRNA expression in Huh7 cells is activated by treatment with IL6 or following administration of conditioned medium from the macrophage/monocyte THP-1 cell line. Using luciferase reporter vectors, we show that the –942 bp hepcidin promoter is activated by IL6. Only the immediate 165 bp of the promoter suffice to retain IL6 activation (Figure 2A). Importantly, the deletion of a highly conserved STAT binding motif within these 165 bp or the –942 bp fragment at position –64/–72 completely abrogates the IL6 effect (Figure 2A). Transcriptional inflammatory responses via IL6 are classically mediated via IL6 binding to the IL6 receptor (p130) to activate the JAK kinases, which in turn switch on latent transcription factors—the signal transducers and activators of transcription (STAT) proteins.27 Our data show that hepcidin activation via IL6 follows this classic pathway and identifies the critical regulatory element. Together with the STAT3 ablation experiments (Figure 4B), our data unravel a missing link in the acute-phase activation of hepcidin in which IL6 activates the transcription of hepcidin via the STAT binding motif at position –64/–72 of the hepcidin promoter.
In addition to exogenously added IL6, CM from THP-1 cells also activates luciferase activity via the same STAT binding motif (Figure 5B). It is therefore possible that IL6 secreted from the THP-1 cells or IL6 endogenously produced in Huh7 cells in response to cytokines contained within the CM mediates hepcidin activation by CM. Notably, hepcidin activation is accelerated and more pronounced when CM from THP-1 cells maintained in culture for 48 hours or 72 hours is used, compared with CM(12h) and CM(24h). Thus, such differences in cytokine composition seem to alter the kinetics of hepcidin activation (Figure 5A).
Surprisingly, the STAT binding site at position –64/–72 of the hepcidin promoter controls not only its IL6-dependent transcriptional activation but also transcriptional activity under control culture conditions (Figure 4A). Knockdown of STAT3 by RNAi significantly reduces endogenous hepcidin mRNA expression in Huh7 cells (Figure 4B), suggesting that STAT3 controls hepcidin transcription both in the absence and presence of experimental inflammatory stimuli. It is possible, however, that agents contained in the control culture medium stimulate a low-level “inflammatory response,” although we use high-purity, low-endotoxin culture media and FBS. Nonetheless, the finding of STAT3-mediated hepcidin transcription even in the absence of experimental inflammatory stimuli implies that stimuli that mediate hepatic STAT3 activation may also enhance hepcidin mRNA expression.
A previous report suggested that C/EBPα also controls hepcidin transcription.23 In Huh7 cells, the deletion of the putative C/EBPα binding site at position –231/–222 from our reporter constructs reduced luciferase expression (Figure 3A), and siRNA-mediated knock-down of C/EBPα mRNA significantly diminished endogenous hepcidin mRNA expression (Figure 3B), confirming and extending these earlier data. In contrast to STAT3, however, C/EBPα does not seem to be involved in the activation of the hepcidin promoter in response to inflammatory stimuli (Figures 2A and 5B). Whether and how STAT3 and C/EBPα cooperate to maintain high-level hepcidin transcription requires further experimentation.
HH is a group of frequent iron overload disorders caused by mutations in the HFE, TfR2, or HJ genes.28 Interestingly, each of these gene defects results in inappropriately low expression of hepcidin, triggering increased intestinal iron absorption and hepatic iron overload. Signaling pathways that regulate gene expression through transcription factors like C/EBPα and STAT3 may thus be of interest in the context of HH. Of note are several reports that describe links between STAT and TGFβ/SMAD signaling.29-31 While such a link up to now was not described in the liver, SMAD proteins are implicated in the transcriptional control of hepcidin in HH21,22 as well as in the response to iron overload or IL6.22 Thus, the investigation of this link may warrant further study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: M.V.V.F. and M.V.S. designed research, performed research, and analyzed data; R.K. and J.S. performed research and collected data; and M.W.H. and M.U.M. designed research and wrote the paper.
M.V.V.F., M.V.S., M.W.H., and M.U.M. contributed equally to this study.
Acknowledgments
This work was partially supported by a long-term fellowship from Trieste University (M.V.V.F.). We thank Vladimir Benes and members of European Molecular Biology Laboratory (EMBL) Genomics Core Facility for support with quantitative real-time PCR.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal