Abstract
Iron homeostasis is maintained through meticulous regulation of circulating hepcidin levels. Hepcidin levels that are inappropriately low or high result in iron overload or iron deficiency, respectively. Although hypoxia, erythroid demand, iron, and inflammation are all known to influence hepcidin expression, the mechanisms responsible are not well defined. In this report we show that the inflammatory cytokine interleukin-6 (IL-6) directly regulates hepcidin through induction and subsequent promoter binding of signal transducer and activator of transcription 3 (STAT3). STAT3 is necessary and sufficient for the IL-6 responsiveness of the hepcidin promoter. Our findings provide a mechanism by which hepcidin can be regulated by inflammation or, in the absence of inflammatory stimuli, by alternative mechanisms leading to STAT3 activation.
Introduction
The anemia of inflammation is an acquired condition that affects patients with a variety of disorders, including infection, arthritis, inflammatory bowel disease, trauma, organ failure, and cancer. At times, it can be severe enough to require transfusion. Even when it does not, it causes a measurable decrease in quality of life and general well-being. Hepcidin is a central mediator of the anemia of inflammation. It is a circulating hormone that is induced by inflammation, which blocks the release of iron from macrophages and interrupts intestinal iron absorption.1,2 Accordingly, mice overexpressing hepcidin develop severe iron deficiency anemia.3
The first direct evidence supporting a role for hepcidin in the anemia of inflammation came from studies of patients with glycogen storage disease (GSD) type 1a. GSD1a is an autosomal recessive disorder caused by mutations in the glucose-6-phosphatase gene, resulting in an inability to maintain glucose homeostasis.4 A subset of the adult GSD1a patient population develops large hepatic adenomas and anemia with features characteristic of the anemia of inflammation.5,6 Although the cause of these adenomas is unknown, the anemia is believed to result from inappropriate overexpression of hepcidin in adenoma tissue.5 High levels of hepcidin are likely to cause the anemia since, upon removal of adenomas, iron homeostasis returns to normal.7
To date, studies of hepcidin regulation have given little insight into the aberrant expression of hepcidin in GSD1a patient adenomas. However, it has become clear that hepcidin is also induced in the anemia of inflammation resulting from other causes. Induction can be attributed, at least in part, to the inflammatory cytokine interleukin-6 (IL-6). IL-6 treatment stimulates hepcidin expression in isolated hepatocytes and in hepatocyte-like cell lines.1 Administration of IL-6 to human subjects stimulates increased hepcidin production and results in low serum iron (hypoferremia) in vivo.8 Mice lacking IL-6 fail to induce hepcidin and do not become hypoferremic after treatment with endotoxin.8 Taken together, these observations leave little doubt that IL-6 links inflammation to hepcidin production. However, the mechanism by which IL-6 induces hepcidin expression is not fully understood.
IL-6 signaling is a major regulator of the acute-phase response in hepatocytes. Upon an inflammatory stimulus, IL-6 is released and binds to a complex of the IL-6 receptor α and gp130.9 The IL-6 ligand-receptor interaction results in the activation of Janus kinases (JAKs) that phosphorylate signal transducers and activators of transcription (STAT) proteins, predominantly STAT3.10 Upon phosphorylation at tyrosine residue 705, STAT3 translocates into the nucleus where it regulates transcription of many target genes.11
The goals of this study were to determine whether IL-6 acts directly to up-regulate hepcidin expression and to elucidate the downstream mechanism of IL-6–mediated hepcidin induction. We have identified an IL-6 responsive element in the putative hepcidin promoter. We demonstrate that IL-6 regulates hepcidin expression through direct binding of STAT3 to the promoter. Finally, we show that STAT3 is necessary and sufficient to confer IL-6 responsiveness in a luciferase reporter assay. These observations not only illuminate IL-6 regulation of hepcidin, but also suggest that, even in the absence of elevated cytokine levels, aberrations in hepatic STAT3 regulation could lead to increased hepcidin and anemia.
Materials and methods
Promoter mapping
We mapped the 5′ end of the human hepcidin transcript using 5′ rapid amplification of cDNA ends (RACE) (Invitrogen, Carlsbad, CA). According to the manufacturer's specifications, we performed first-strand cDNA synthesis using 1 μg human total liver RNA (Clontech Laboratories, Mountain View, CA) and the hepcidin-specific primer 5′TGGGGCAGCAGGAATAAATA. We used tailed cDNA as a template for a subsequent polymerase chain reaction (PCR) using hepcidin-specific reverse primer 5′CAGGGCAGGTAGGTTCTACG. PCR products were used as templates for nested PCR using hepcidin-specific reverse primer 5′CTACGTCTTGCAGCACATCC, and resulting products were fractionated on a 2% agarose gel. We excised bands and extracted the DNA using the QIAGEN gel extraction kit (QIAGEN, Valencia, CA). DNA sequences were determined by the Molecular Genetics Core Facility of Children's Hospital, Boston, MA.
cDNA subcloning
We subcloned the hepcidin 0.6 kb minimal promoter from a PCR fragment amplified with the primers 5′CTTACGCGTGCTAGCTGAGGGCACTGATAGGGGTA and 5′ATCGCAGATCTCGAGTCCTGTTCTGCTGTGCAGTC using the 1.3 kb human hepcidin upstream flanking region as a template (genomic clone provided by H. Lin, Massachusetts General Hospital, Boston, MA). We subcloned the 1.3 kb and 0.6 kb fragments into the pGL3-Basic vector (Promega, Madison, WI) upstream of the firefly luciferase reporter gene. We introduced mutations into the hepcidin 0.6 kb promoter using site-directed mutagenesis (Invitrogen).
Cell culture and transfections
We cultured HepG2/2.2.1 (American Type Culture Collection [ATCC] Manassas, VA) cells at 37°C and 5% CO2 in a 1:1 mixture of Dulbecco modified Eagle medium and Ham F12 medium (Gibco, Grand Island, NY) supplemented with 2.5 mM l-glutamine (Sigma-Aldrich, St Louis, MO), 15 mM HEPES (Sigma-Aldrich), 0.5 mM sodium pyruvate (Sigma-Aldrich), 1200 mg/L sodium bicarbonate (Sigma-Aldrich), 10% Premium Fetal Bovine Serum (Cambrex Life Sciences, Walkersville, MD), and antibiotics (Sigma-Aldrich). We performed plasmid transfections using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Real-time quantitative PCR
We passaged HepG2/2.2.1 cells 1:5 and treated them with 20 ng/mL IL-6 (R&D Systems, Minneapolis, MN) or 10 μg/mL cycloheximide (Sigma-Aldrich). We isolated total RNA using the RNeasy Mini kit (QIAGEN) according to the manufacturer's instructions. We performed reverse transcription using the first-strand synthesis method (Invitrogen) with 1 μg total RNA treated with DNase (Roche, Basel, Switzerland). To quantify cDNA we performed real-time quantitative PCR using a SYBR green reaction mix (Bio-Rad Laboratories, Hercules, CA) with the following primers for hepcidin 5′ CTGCAACCCCAGGACAGAG and 5′GGAATAAATAAGGAAGGGAGGGG or β-actin, 5′AGGATGCAGAAGGAGATCACTG and 5′GGGTGTAACGCAACTAAGTCATAG, as an internal normalization control, according to the manufacturer's recommendations. Reactions were carried out in a Bio-Rad iCycler.
Luciferase reporter assay
We transiently transfected HepG2/2.2.1 cells with 2.5 μg of one of the following luciferase reporter constructs: promoterless pGL3-basic (negative control; Promega), IL-6 responsive M67-luc (positive control; kind gift from David Frank, Dana-Farber Cancer Institute, Boston, MA), hepcidin 1.3 kb-luc or hepcidin 0.6 kb-luc. All transfections were done in combination with 0.25 μg pRL-TK Renilla luciferase plasmid (Promega) as a normalization control. We also co-transfected each of these plasmids with or without 2.5 μg of one of the following STAT3 mutant constructs: constitutively active STAT3-C, dominant-negative STAT3 Y705F, short hairpin–mediated RNA interference construct pRS-STAT3, or pRS vector control. All STAT3 mutant and RNA interference constructs were provided by David Frank. We serum-starved cells 48 hours after transfection in Opti-Mem (Invitrogen) for 6 hours and treated with 20 ng/mL IL-6 (R&D Systems) for 12 to 16 hours. Following treatment, we lysed cells and measured luciferase activity using the Promega Dual Luciferase Reporter Assay according to the manufacturer's instructions. Experiments were performed in triplicate wells. We calculated relative luciferase activity as the ratio of firefly (reporter) to Renilla (normalization control) activity.
Chromatin immunoprecipitation assay
We performed chromatin immunoprecipitation (ChIP) using a commercially available kit (Upstate Biotechnology, Lake Placid, NY) according to the manufacturer's instructions. Specifically, we treated 5 × 107 HepG2/2.2.1 cells with or without 20 ng/mL IL-6 for 3 hours. We cross-linked chromatin-protein complexes by treating cells with 1% formaldehyde for 10 minutes at 37°C. Cells were washed with phosphate-buffered saline (PBS) and lysed with lysis buffer supplied by the kit, supplemented with protease inhibitor pellets (Roche). We then sonicated cell lysates (20 seconds followed by 4 minutes on ice, cycle repeated 3 times) to shear chromatin into 200- to 1000-base pair fragments. Samples (treated or untreated) were separated into 3 equal parts and diluted 10-fold. One percent of each sample was saved and designated input chromatin, the amount of chromatin present before immunoprecipitation. We incubated each sample with salmon sperm DNA/protein A agarose–50% slurry for 30 minutes at 4°C with agitation. We incubated precleared supernatant overnight at 4°C while agitating in the presence of no antibody, 1:100 anti-STAT3, or 1:100 anti-STAT1 (Cell Signaling Technology, Danvers, MA). The following day we added salmon sperm DNA/protein A agarose slurry to the antibody/DNA mixture for one hour at 4°C with rotation to collect the antibody/histone complex. We pelleted conjugated beads by gentle centrifugation and removed the supernatant fraction containing unbound chromatin. We washed the protein A agarose/antibody/chromatin complexes for 5 minutes with rotation 5 times with wash buffers provided by the manufacturer. At this point, we divided each sample into 2 equal portions. One portion was diluted with sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) running buffer for Western blot analysis. The other portion was incubated twice with 250 μL fresh elution buffer (1% SDS, 0.1 M NaHCO3) for 15 minutes at room temperature to elute protein A agarose/antibody/chromatin complexes. We combined the eluates (500 μL total) and treated them as well as input samples with 5 M NaCl and warmed to 65°C for 4 hours to reverse cross-links, and then treated with 0.5 M EDTA, 1 M Tris-HCl, pH 6.5 and 10 mg/mL Proteinase K for 1 hour at 45°C to digest proteins. We recovered DNA by phenol/chloroform extraction and ethanol precipitation. To determine the relative abundance of immunoprecipitated chromatin, we performed PCR using a fraction of input or output samples as template with primers flanking an approximately 150 bp region containing the putative STAT3 binding site (5′ GAGGGTGACACAACCCTGTT and 5′ ACCGAGTGACAGTCGCTTTT) or an approximately 200 bp region upstream that does not contain a putative STAT3 binding site (negative control 5′ TCTCTGCCTTCAGTGCCTTT and 5′CACTTCTGCACCAACTCAGC). We resolved PCR products on a 2% agarose gel. We estimated the volume of each band by densitometry and determined the ratio of output to input PCR product.
Statistical analysis
We performed a 2-tailed Student t test and considered P values of less than .05 as statistically significant. Error bars represent standard deviation of the mean unless otherwise indicated.
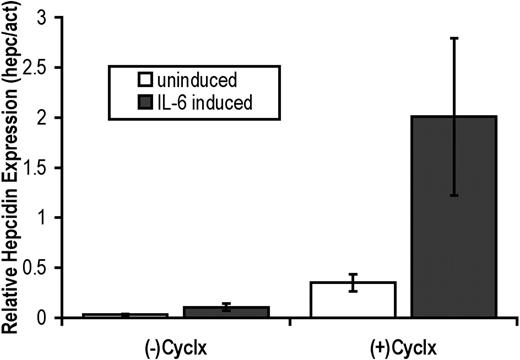
IL-6 induces hepcidin expression directly. Hep G2/2.2.1 cells were maintained without induction (□) or were induced with 20 ng/mL IL-6 () for 8 hours in the absence or presence of 10 μg/mL cycloheximide (Cyclx). Following treatment, total RNA was isolated and real-time quantification of hepcidin mRNA transcripts was performed in triplicate using a 2-step RT-PCR. The ratio of hepcidin (hepc) to β-actin (act, internal control) PCR products was used to determine the relative levels of hepcidin expression. Error bars represent standard deviation of the mean.
IL-6 induces hepcidin expression directly. Hep G2/2.2.1 cells were maintained without induction (□) or were induced with 20 ng/mL IL-6 () for 8 hours in the absence or presence of 10 μg/mL cycloheximide (Cyclx). Following treatment, total RNA was isolated and real-time quantification of hepcidin mRNA transcripts was performed in triplicate using a 2-step RT-PCR. The ratio of hepcidin (hepc) to β-actin (act, internal control) PCR products was used to determine the relative levels of hepcidin expression. Error bars represent standard deviation of the mean.
Results
IL-6 induces hepcidin expression directly
Treatment of human subjects, mice, primary hepatocytes, and hepatocyte cell lines with IL-6 induces hepcidin expression.1,8 However, IL-6 induces the transcription of many hepatic genes during the acute-phase response, including genes involved in metabolism, protein synthesis, and transcriptional regulation.12,13 To assess whether the induction of hepcidin by IL-6 is indirect (requiring protein synthesis) or direct, HepG2/2.2.1 cells were treated with cycloheximide, a potent translation inhibitor, in the presence or absence of IL-6 (Figure 1). Using real-time PCR to obtain quantitative results, we found that IL-6 treatment induced hepcidin expression 3.5-fold compared with untreated control cells in the absence of cycloheximide. In the presence of cycloheximide, IL-6 induced hepcidin expression by 5.7-fold above control cells treated with cycloheximide alone. We conclude that hepcidin induction by IL-6 does not require protein synthesis and is therefore direct.
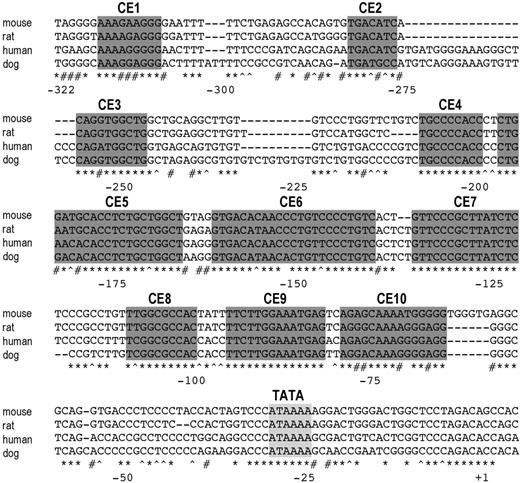
The 5′ flanking region of the hepcidin gene contains conserved elements. The transcription start site of human hepcidin was determined by 5′ RACE (+1). A putative TATA box (light gray shading) and 10 conserved elements (CEs; dark gray shading) were identified by aligning 5′ flanking sequences of the hepcidin coding region from mouse, rat, human, and dog. Identical bases in all 4 species (*), conserved purines (#), and conserved pyrimidines (^) are indicated.
The 5′ flanking region of the hepcidin gene contains conserved elements. The transcription start site of human hepcidin was determined by 5′ RACE (+1). A putative TATA box (light gray shading) and 10 conserved elements (CEs; dark gray shading) were identified by aligning 5′ flanking sequences of the hepcidin coding region from mouse, rat, human, and dog. Identical bases in all 4 species (*), conserved purines (#), and conserved pyrimidines (^) are indicated.
The 5′ flanking region of the hepcidin gene contains conserved elements
Although there was one previous report describing transcriptional regulation of hepcidin expression,14 the structure of the hepcidin mRNA had not been fully characterized. We performed 5′ RACE to map the 5′ end of the human hepcidin transcript and determine its transcriptional start site (Figure 2). Making the assumption that the transcriptional start sites would be similar or identical in other mammalian species, we aligned the 5′ flanking regions upstream of the human, dog, rat, and mouse hepcidin genes. Blocks of homologous sequence, likely to correspond to transcription factor binding sites, were concentrated within a 325-bp region, suggesting that this segment contains the probable hepcidin promoter (Figure 2). There was no obvious homology further upstream. Small regions with significant homology among species were denoted conserved elements (CEs) 1-10. Sequences of these elements and the mutations introduced into them are listed in Table 1.
Conserved element mutant summary
CE . | Sequence . | Mutation . |
|---|---|---|
| 1 | AAAAGGGGG | AAAAGGGcc |
| 2 | TGACATC | acACATC |
| 3 | CAGATGGCTG | gtGATGGCTG |
| 4 | TGCCCCACC | TGCCaaACC |
| 6 | GTGACACAACCCTGTTCCCTGTC | GacACACAACCCTGTTCCCTGTC |
| 7 | GTTCCCGCTTATCTC | GTTaaCGCTTATCTC |
| 9 | TTCTTGGAAATGAG | ggaTTGGAAATGAG |
| 10 | AGAGCAAAGGGGAGG | AGAGCAAAGccaAGG |
CE . | Sequence . | Mutation . |
|---|---|---|
| 1 | AAAAGGGGG | AAAAGGGcc |
| 2 | TGACATC | acACATC |
| 3 | CAGATGGCTG | gtGATGGCTG |
| 4 | TGCCCCACC | TGCCaaACC |
| 6 | GTGACACAACCCTGTTCCCTGTC | GacACACAACCCTGTTCCCTGTC |
| 7 | GTTCCCGCTTATCTC | GTTaaCGCTTATCTC |
| 9 | TTCTTGGAAATGAG | ggaTTGGAAATGAG |
| 10 | AGAGCAAAGGGGAGG | AGAGCAAAGccaAGG |
Wild-type sequences for conserved elements in the hepcidin promoter are listed. To disrupt putative transcription factor binding sites, mutations in conserved elements (lower-case letters) were introduced individually into the hepcidin 0.6 kb-luc (mini) plasmid by site-directed mutagenesis.
Conserved element 9 (CE9) mediates IL-6 responsiveness
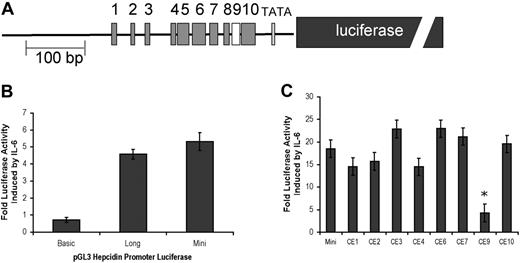
We subcloned the 1.3-kb (long) or the 0.6-kb (mini) fragment of human hepcidin 5′ flanking sequence upstream of a luciferase reporter gene in a pGL3 expression vector to assess IL-6 responsiveness of the hepcidin promoter (Figure 3A). As compared with untreated controls and a negative control vector that does not contain the hepcidin-flanking region (basic), both hepcidin long and mini were induced by IL-6 treatment (Figure 3B). A similar fold induction above untreated controls was seen with both promoter fragments, suggesting that the IL-6 responsive element is retained within the hepcidin mini construct.
Conserved element 9 mediates IL-6 responsiveness. (A) A diagram of hepcidin mini shows conserved elements (CEs) 1-10 and the luciferase reporter gene (dark gray box). (B) Hep G2/2.2.1 cells were transiently transfected with control plasmid-expressing Renilla luciferase along with one of the following firefly luciferase reporter genes: a 1.3-kb (long) or 0.6-kb (mini) fragment of the 5′ flanking region of the hepcidin gene or a promoterless negative control (basic). At 48 hours after transfection, cells were serum starved for 6 hours and induced with 20 ng/mL IL-6. After 16 hours, firefly luciferase activity (normalized to Renilla luciferase as a transfection control) was measured and compared with activity from uninduced cell lysates. (C) Mutations in hepcidin mini were introduced into CEs 1-4, 6-9, and 10 by site-directed mutagenesis. Each mutant promoter–firefly luciferase plasmid was transiently transfected into HepG2/2.2.1 cells and induced with IL-6 as described in “Materials and methods.” Fold luciferase activity above uninduced controls was compared with that of hepcidin mini. Error bars represent standard error. *P < .001.
Conserved element 9 mediates IL-6 responsiveness. (A) A diagram of hepcidin mini shows conserved elements (CEs) 1-10 and the luciferase reporter gene (dark gray box). (B) Hep G2/2.2.1 cells were transiently transfected with control plasmid-expressing Renilla luciferase along with one of the following firefly luciferase reporter genes: a 1.3-kb (long) or 0.6-kb (mini) fragment of the 5′ flanking region of the hepcidin gene or a promoterless negative control (basic). At 48 hours after transfection, cells were serum starved for 6 hours and induced with 20 ng/mL IL-6. After 16 hours, firefly luciferase activity (normalized to Renilla luciferase as a transfection control) was measured and compared with activity from uninduced cell lysates. (C) Mutations in hepcidin mini were introduced into CEs 1-4, 6-9, and 10 by site-directed mutagenesis. Each mutant promoter–firefly luciferase plasmid was transiently transfected into HepG2/2.2.1 cells and induced with IL-6 as described in “Materials and methods.” Fold luciferase activity above uninduced controls was compared with that of hepcidin mini. Error bars represent standard error. *P < .001.
We compared fold induction by IL-6 treatment among promoters carrying independent mutations in most of the identified conserved elements (Figure 3C). Notably, while most mutants showed slight differences in fold induction compared with the wild-type hepcidin mini; mutant CE9 retained only one-fourth the hepcidin induction activity. No other mutant showed statistically significant differences in fold luciferase activity in response to IL-6. These results suggest that CE9 plays a critical role in transducing the IL-6 signal to up-regulate hepcidin expression.
STAT3 binds to the hepcidin promoter
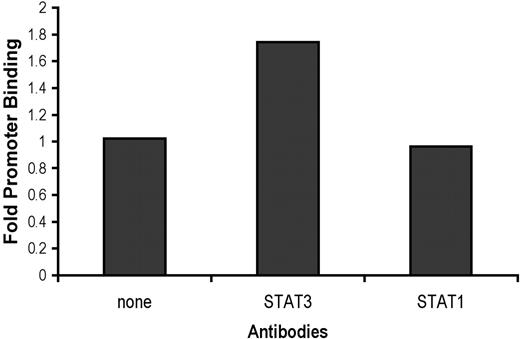
Based on results from searches using 3 different transcription factor binding site identification programs (TESS, http://www.cbil.upenn.edu/cgi-bin/tess/tess?RQ=WELCOME, TFSEARCH http://www.cbrc.jp/research/db/TFSEARCH.html, and MATCH, http://www.gene-regulation.com/pub/programs.html), CE9 was predicted to contain a putative binding site for STAT proteins. The consensus sequence is disrupted by the TTC to GGA mutation that we introduced into CE9. Receptor binding of IL-6 can activate both STAT1 and STAT3.15 To determine which of these STAT proteins binds directly to the hepcidin promoter downstream of IL-6, we performed chromatin immunoprecipitation assays. We found that immunoprecipitation with anti-STAT3, but not with anti-STAT1, enriched recovery of a specific PCR product containing CE9 in IL-6–treated samples as compared with untreated samples or controls without antibody (Figure 4). When negative control primers were used to amplify a region upstream of the hepcidin promoter that does not contain a STAT consensus site there were no differences between immunoprecipitates recovered with anti-STAT3 versus no antibody (data not shown).
STAT3 binds the hepcidin promoter. HepG2/2.2.1 cells were treated with IL-6 (20 ng/mL) or left untreated. After cells were fixed, we performed chromatin immunoprecipitation with no antibody, anti-STAT3, or anti-STAT1 and used PCR to amplify a 150 bp fragment containing the consensus STAT binding site of the hepcidin promoter. We fractionated PCR products and determined relative product amounts normalized to input chromatin. Fold promoter binding was calculated as the ratio of IL-6–treated to untreated output/input volume ratios of PCR products.
STAT3 binds the hepcidin promoter. HepG2/2.2.1 cells were treated with IL-6 (20 ng/mL) or left untreated. After cells were fixed, we performed chromatin immunoprecipitation with no antibody, anti-STAT3, or anti-STAT1 and used PCR to amplify a 150 bp fragment containing the consensus STAT binding site of the hepcidin promoter. We fractionated PCR products and determined relative product amounts normalized to input chromatin. Fold promoter binding was calculated as the ratio of IL-6–treated to untreated output/input volume ratios of PCR products.
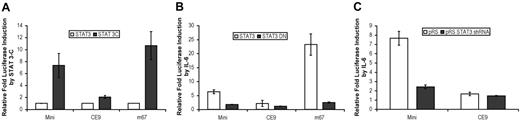
STAT3 is necessary and sufficient to activate hepcidin expression
To assess whether activated STAT3 alone can stimulate the putative hepcidin promoter in the absence of IL-6 treatment, we co-transfected HepG2/2.2.1 cells with luciferase reporter constructs and a constitutively active form of STAT3 (STAT3-C).16 While STAT3-C has no effect on the promoterless negative control (data not shown), it activated luciferase transcription through an IL-6 responsive positive control promoter, m67, which contains STAT3 binding elements (Figure 5A).17 Similarly, STAT3-C induced hepcidin mini, but not the CE9 mutant.
To assess the requirement of functional STAT3 for hepcidin induction, we co-transfected HepG2/2.2.1 cells with luciferase reporter constructs and a dominant-negative STAT3 mutant (STAT3-DN).18 STAT3-DN significantly inhibited the ability of IL-6 to induce both the positive control m67 and the hepcidin mini promoters, reducing their IL-6 responsiveness to levels comparable that of the CE9 mutant (Figure 5B).
As a complementary approach, we decreased STAT3 expression by co-transfection with a short hairpin RNA to induce STAT3-specific RNA interference.19 While vector control (pRS) expression had no effect on promoter responsiveness, short hairpin–mediated RNA interference of STAT3 (pRS-STAT3) significantly reduced promoter activity of both control and hepcidin promoters (Figure 5C).
Taken together, these results demonstrate that constitutively active STAT3 is sufficient to activate the hepcidin promoter in the absence of IL-6 stimulus and that this activation requires a wild-type CE9 binding site. Moreover, functional STAT3 is required for IL-6 induction of the 0.6 kb hepcidin promoter.
STAT3 is necessary and sufficient to activate the hepcidin promoter downstream of IL-6. HepG2/2.2.1 cells were transiently transfected with a firefly luciferase reporter gene under the control of hepcidin mini, the CE9 mutant or the IL-6 responsive m67 promoter in conjunction with a Renilla luciferase vector with or without constructs expressing mutant STAT3 proteins or siRNA. (A) Constitutively active STAT3 mutant STAT3-C (2.5 μg cDNA). (B) Dominant-negative STAT3 mutant STAT3-DN (2.5 μg cDNA). (C) 2.5 μg STAT3 short hairpin siRNA or vector control. Cells in panels B and C were treated for 12 to 16 hours with 20 ng/mL IL-6. Cell lysates were analyzed for luciferase activity 48 hours after transfection. Relative luciferase activity was calculated as the ratio of firefly to Renilla luciferase activity and is expressed as a multiple of the activity of unstimulated cells transfected with reporter alone. Error bars represent standard deviation of the mean.
STAT3 is necessary and sufficient to activate the hepcidin promoter downstream of IL-6. HepG2/2.2.1 cells were transiently transfected with a firefly luciferase reporter gene under the control of hepcidin mini, the CE9 mutant or the IL-6 responsive m67 promoter in conjunction with a Renilla luciferase vector with or without constructs expressing mutant STAT3 proteins or siRNA. (A) Constitutively active STAT3 mutant STAT3-C (2.5 μg cDNA). (B) Dominant-negative STAT3 mutant STAT3-DN (2.5 μg cDNA). (C) 2.5 μg STAT3 short hairpin siRNA or vector control. Cells in panels B and C were treated for 12 to 16 hours with 20 ng/mL IL-6. Cell lysates were analyzed for luciferase activity 48 hours after transfection. Relative luciferase activity was calculated as the ratio of firefly to Renilla luciferase activity and is expressed as a multiple of the activity of unstimulated cells transfected with reporter alone. Error bars represent standard deviation of the mean.
Discussion
Hepcidin is a master regulator of systemic iron homeostasis and a critical mediator of the anemia of inflammation. Until recently, little was understood about the regulation of its expression. We and others have shown that a bone morphogenetic protein/Smad signaling cascade is important for basal regulation of hepcidin transcription.20,21 That pathway did not, however, account for the induction of hepcidin expression in inflammation. Here we demonstrate the importance of a parallel IL-6/STAT signaling pathway involved in the hepcidin response to inflammation.
IL-6 has many activities, particularly in hepatocytes. We showed that IL-6 induction of hepcidin does not require new protein synthesis. Interestingly, treatment of cells with IL-6 in the presence of cycloheximide enhanced IL-6 responsiveness. This suggests the existence of a translation-dependent mechanism that negatively regulates hepcidin transcription. Since basal hepcidin levels increase upon cycloheximide treatment in the absence of IL-6, we infer that this negative regulator is active in HepG2.2.1 cells and is not dependent on IL-6 treatment. We speculate that this negative regulatory mechanism contributes to the relatively low levels of hepcidin expression observed in HepG2.2.1 cells and may explain their failure to induce hepcidin expression in response to treatment with iron or diferric transferrin (D.M.W. and N.C.A., unpublished data, July 17, 2002, and March 11, 2005).
IL-6 can activate at least 3 signaling pathways.22 To hone in on the pathway responsible for IL-6 induction of hepcidin, we first defined and analyzed the hepcidin promoter. We defined the putative hepcidin promoter by mapping the transcriptional start site and analyzing conserved sequences in 4 mammalian species. We showed that a 0.6-kb fragment of upstream flanking sequence, containing the putative promoter, conferred IL-6 responsiveness on a luciferase reporter gene. This was abrogated by mutations in a highly conserved element (CE9) that matched a consensus binding site for STAT proteins. We used several independent methods to show that IL-6 responsiveness was mediated by STAT3.
We have established that hepcidin is directly induced by IL-6 signaling through STAT3 activation and subsequent hepcidin promoter binding. Although IL-6 is known to induce hepcidin expression in inflammatory conditions, STAT3 can be activated by a plethora of other cytokines and growth factors,23 suggesting that hepcidin expression may respond to other mediators. In fact, cytokine levels do not explain elevated hepcidin expression in GSD1a patient adenomas.5 Studies of liver-specific STAT3 knockout mice show that STAT3 is a necessary regulator of glucose-6-phosphatase and other genes involved in gluconeogenesis.24 Therefore, it would not be surprising for STAT3 to be activated in GSD1a patients. Activated STAT3 can also promote tumorigenesis. We speculate that anemia, via hepcidin regulation, and adenoma formation, via antiapoptotic gene regulation25 in GSD1a patients may result from aberrant STAT3 activity in response to a compensatory signal that attempts to increase glucose production. Thus, dysregulation of STAT3 or an upstream mediator may explain clinical observations of anemia resembling the anemia of inflammation in patients with a GSD1a as well as in a variety of malignancies, including multiple myeloma and neuroblastoma.
The many signals that regulate hepcidin expression must be integrated to maintain normal iron homeostasis. Inappropriately low levels of hepcidin expression observed in patients and animals with genetic hemochromatosis suggest that genes involved in the pathogenesis of the disease regulate hepcidin expression. We previously studied the induction of hepcidin in Hfe–/– mice, lacking a protein that is mutated in most patients with genetic hemochromatosis. We reported that Hfe–/– mice failed to induce hepcidin expression appropriately in response to treatment with lipopolysaccharide, an inflammatory stimulus.26 Reports from other groups have not agreed with our findings.27 In subsequent experiments we have shown that primary hepatocytes isolated from Hfe–/– mice do induce hepcidin expression in response to IL-6 treatment, but that both basal and induced levels of hepcidin expression are lower than levels in control hepatocytes from wild-type mice (D.M.W. and N.C.A., unpublished data, February 11, 2004).
These results are consistent with the IL-6 regulatory mechanism that we describe here. While HFE has no known role in IL-6 signal transduction, it need not be directly involved to modulate the hepcidin response. We speculate that HFE modulates hepcidin expression through a parallel pathway, and that HFE must be present for a full hepcidin response to inflammation. In that sense, HFE is permissive for the induction of hepcidin in inflammation, but not obligatory. In support of this idea, others have recently shown that impairment of hepcidin expression through inactivation of hepatic SMAD4 abrogates IL-6 induction of hepcidin.21
Two pathways, BMP/SMAD signaling and IL-6/STAT3 signaling, are now known to be important for transcriptional regulation of hepcidin. The next challenge will be to determine how these pathways, or others, transduce the physiologic signals (body iron stores, erythroid drive, hypoxia) that are known to regulate hepcidin production and iron homeostasis. Molecular understanding will allow investigation of crosstalk between and hierarchy among the pathways. A detailed knowledge of the signaling pathways that regulate hepcidin should inform new pharmacologic approaches to modify iron homeostasis through hepcidin modulation, offering new treatments for the anemia of inflammation and for iron overload disorders.
Prepublished online as Blood First Edition Paper, July 11, 2006; DOI 10.1182/blood-2006-06-027631.
Supported by R01 DK053813 (N.C.A.) and T32 HL07623 (D.M.W.) from the National Institutes of Health and a research grant from the Roche Foundation for Anemia Research (N.C.A.).
D.M.W. and N.C.A. designed the experiments and wrote the paper. D.M.W. performed the experiments.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Jodie Babitt, Herbert Lin, Seth Frietz, Angela Ross, Cindy Roy, Sarah Walker, and David Frank for their kind sharing of reagents, technical advice, and support on the experiments in this manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal