Abstract
Epigallocatechin-3-gallate (EGCG), a polyphenol extracted from green tea, is an antioxidant with chemopreventive and chemotherapeutic actions. Based on its ability to modulate growth factor-mediated cell proliferation, we evaluated its efficacy in multiple myeloma (MM). EGCG induced both dose- and time-dependent growth arrest and subsequent apoptotic cell death in MM cell lines including IL-6-dependent cells and primary patient cells, without significant effect on the growth of peripheral blood mononuclear cells (PBMCs) and normal fibroblasts. Treatment with EGCG also led to significant apoptosis in human myeloma cells grown as tumors in SCID mice. EGCG interacts with the 67-kDa laminin receptor 1 (LR1), which is significantly elevated in myeloma cell lines and patient samples relative to normal PBMCs. RNAi-mediated inhibition of LR1 resulted in abrogation of EGCG-induced apoptosis in myeloma cells, indicating that LR1 plays an important role in mediating EGCG activity in MM while sparing PBMCs. Evaluation of changes in gene expression profile indicates that EGCG treatment activates distinct pathways of growth arrest and apoptosis in MM cells by inducing the expression of death-associated protein kinase 2, the initiators and mediators of death receptor-dependent apoptosis (Fas ligand, Fas, and caspase 4), p53-like proteins (p73, p63), positive regulators of apoptosis and NF-κB activation (CARD10, CARD14), and cyclin-dependent kinase inhibitors (p16 and p18). Expression of related genes at the protein level were also confirmed by Western blot analysis. These data demonstrate potent and specific antimyeloma activity of EGCG and provide the rationale for its clinical evaluation.
Introduction
Tea leaves, derived from a shrub Camellia sinensis, contain high amounts of polyphenols or catechins. During the extraction process, the polyphenols in black tea are rendered inactive by fermentation; however, the extraction process for green tea involves only steaming and thus leaves the polyphenols active.1
In recent years, chemopreventive and chemotherapeutic effects of green tea have been reported in different malignancies.2-9 Because epigallocatechin-3-gallate (EGCG) is the most abundant and biologically active polyphenol with antioxidant activity in green tea, the majority of the mechanistic studies have focused on this compound. It selectively inhibits cell growth and induces apoptosis in cancer cells without adversely affecting normal cells.10 The antitumor effects of EGCG include inhibition of angiogenesis, modulation of growth factor-mediated proliferation, suppression of oxidative damage, induction of apoptosis, and cell-cycle arrest.8,11-14 Both epidemiologic studies on tea consumption15,16 and animal studies6,17 have shown that the polyphenols prevent the development of chemically induced cancer.
Cell growth inhibition and apoptosis-inducing effects of EGCG have been shown in several cancers.18,19 In prostate carcinoma cells (LNcaP), EGCG induces apoptosis by activation of p53 and p14ARF-mediated suppression of MDM2. In this study we have demonstrated that EGCG induces apoptotic cell death in multiple myeloma (MM) cells including IL-6-dependent cells and primary MM cells in vitro, while having no significant effect on growth of normal cells (peripheral blood mononuclear cells [PBMCs] and fibroblasts), and induces apoptosis and inhibition of growth in vivo in a murine model of human MM. Antimyeloma effects of EGCG are mediated through laminin receptor 1 (LR1), which is overexpressed on MM cells, and it activates multiple interrelated pathways of apoptosis and cell-cycle arrest.
Materials and methods
Chemicals
(-)-Epigallocatechin-3-gallate (EGCG) was purchased from Sigma-Aldrich (St Louis, MO) and dissolved in phosphate-buffered saline (PBS).
MM and normal cells
The MM cell line INA6 was kindly provided by Dr Renate Burger (University of Erlangen-Nuernberg, Erlangen, Germany) and the ARP cell line was kindly provided by Dr J. Epstein (University of Arkansas for Medical Sciences, Little Rock). Normal diploid fibroblasts (GM07675) were obtained from the American Type Culture Collection (Rockville, MD). ARP cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (HyClone, South Logan, UT), whereas INA6, an interleukin 6 (IL-6)-dependent cell line, was cultured in RPMI 1640 medium supplemented with 20% fetal bovine serum (HyClone) and 2.5 ng/mL recombinant human IL-6 (R&D Systems, Minneapolis, MN). Normal diploid fibroblasts (GM07675) were cultured in Dulbecco modified Eagle medium (DMEM; Sigma-Aldrich) containing 10% fetal bovine serum. All cell lines were maintained in a state of logarithmic growth at 37°C in humidified air with 5% CO2, as described previously.20-23 For RNA and protein analyses, cultures were harvested at the same final cell density (5 × 105/mL) and immediately processed.
Primary MM cells were isolated from bone marrow aspirate samples, obtained following informed consent, obtained in accordance with the Declaration of Helsinki, from patients with MM, by positive selection using anti-CD138 antibody-coated immunomagnetic beads and magnetic-assisted cell sorting (MACS), according to the manufacturer's instructions (Miltenyi Biotech, Auburn, CA). Purity of plasma cells (> 95%) was confirmed by monitoring cell-surface expression of CD38 and CD45.
Treatment and growth of cells
Cells (5 × 105) were plated in 100-mm dishes and treated with EGCG at various concentrations and live cell number was determined by trypan blue exclusion or by measuring 3H-thymidine incorporation on alternate days. For thymidine incorporation, 2 × 104 cells/well were incubated in 96-well culture plates with or without EGCG in triplicate. 3H-thymidine (0.5 μCi [0.0185 MBq]; NEN Life Science Products, Boston, MA) was then added to each well for the last 8 hours. Cells were harvested onto glass filters with an automatic cell harvester (Cambridge Technology, Cambridge, MA), and 3H-thymidine uptake was measured using a Micro-Beta Trilux counter (Wallac, Gaithersburg, MD).
siRNA and transfections
Nontargeting Cy3-labeled control siRNA and siRNA targeting LR1 (67 kDa) were purchased from Dharmacon Research (Lafayette, CO). siRNAs were transfected into MM cells using TransIT-TKO transfection reagent (Mirus, Madison, WI), as described by the manufacturer. Briefly, cells were plated at 2 × 105/mL in complete growth medium 24 hours prior to transfection and incubated overnight. Immediately prior to transfection, TransIT-TKO reagent was added dropwise to serum-free medium (RPMI 1640) and incubated at room temperature for 20 minutes. siRNA duplexes (100 nM) were added to diluted TransIT-TKO reagent, mixed, and incubated at room temperature for 20 minutes. siRNA-TKO complexes were then layered dropwise onto the cells and incubated as described.24,25
To monitor uptake of siRNA, cells transfected with Cy3-labeled control siRNA were incubated for 72 hours and Cy3 labeling was examined using an Olympus BX61 fluorescence microscope (Olympus America, Center Valley, PA) equipped with a Cy3 filter and a UPlanApo Olympus 20×/0.70 numeric aperture objective. Images were photographed using a SPOT RT color 2.2.1 digital camera (Diagnostic Instruments, Sterling Heights, MI), and were acquired using SPOT 3.4.2 image software (Diagnostic Instruments).
Animals
SCID mice (CB-17), obtained from Taconic (Germantown, NY), were maintained and monitored in the Farber Cancer Institute's Animal Research Facility. All animal studies were conducted according to protocols approved by the Institutional Animal Care and Use Committee. Animals were humanely killed when their tumors reached 2 cm in diameter or when paralysis or major compromise in their quality of life occurred.
Human MM xenograft murine model
CB-17 SCID mice were inoculated subcutaneously in the interscapular area with 2.5 × 106 OPM1 cells in 100 μL RPMI 1640 medium. Following appearance of palpable tumors, mice were injected intraperitoneally daily with PBS alone or EGCG (33 mg/kg). At the time of the animals' death, tumors were excised and cell-cycle profiles of tumor cells derived from control and EGCG-treated mice were analyzed using propidium iodide (PI) staining and flow cytometry. Briefly, cells (1 × 106) were washed with PBS, permeabilized by a 30-minute exposure to cold 70% ethanol at 4°C, washed with PBS, incubated with PI (5 μg/mL) in 500 mL PBS containing 10 μg/mL RNase for 30 minutes at room temperature, and analyzed for DNA content by Cytomics FC 500 Flow Cytometer (Beckman Coulter, Fullerton, CA).
Apoptosis assay
Apoptotic MM cells were detected using the annexin V-biotin apoptosis detection kit (Oncogene Research Products, San Diego, CA). Untreated or EGCG-treated myeloma cells (1 × 106 cells/mL) were mixed with annexin V-biotin and media-binding reagent and incubated in the dark for 15 minutes at room temperature. Cells were then centrifuged and medium was replaced with 1 × binding buffer (Oncogene Research Products) containing fluorescein isothiocyanate (FITC)-streptavidin (Amersham Life Sciences, Arlington Heights, IL). A portion of cell suspension (50 μL) was placed onto a glass slide, covered with a coverslip, and viewed immediately using a fluorescence microscope equipped with a FITC (green) filter. Imaging was conducted as described in “siRNA and transfections.” Two hundred cells, representing at least 5 distinct microscopic fields, were analyzed to assess the fraction of FITC-labeled cells for each sample.
Gene expression profile
Myeloma (INA6) cells, untreated or treated with 10 μM EGCG for 24 hours, were harvested and total RNA was isolated using an RNeasy kit (Qiagen, Valencia, CA) as described by the manufacturer. Total RNA (10-15 μg) was reverse-transcribed to get cDNA using the Superscript II reverse transcription kit (Invitrogen Life Technologies, Carlsbad, CA). cDNA was used in an in vitro transcription reaction to synthesize biotin-labeled cRNA using the Enzo RNA labeling kit (Enzo Diagnostics, Farmingdale, NY). Labeled cRNA was purified with the RNeasy mini-kit (Qiagen, Valencia, CA) and quantitated. Purified cRNA (15 μg) was hybridized to Human Genome U133 (HG-U133) GeneChip arrays (Affymetrix, Santa Clara, CA) according to the manufacturer's protocol. The HG-U133 set consists of 2 GeneChip arrays representing approximately 33 000 human genes. GeneChip arrays were scanned on a GeneArray scanner (Affymetrix).
Microarray data analysis
Normalization of arrays and calculation of expression values was performed using the DNA-chip analyzer (dChip) program.26,27 Arrays were normalized based on relative signal produced for an invariant subset of genes. This model-based method was used for probe selection and computing expression values.26,27 By pooling hybridization information across multiple arrays, it is possible to assess standard errors for the expression level indexes. This approach also allows automatic probe selection in the analysis stage to reduce errors due to cross-hybridization of probes and image contamination. We also used several high-level analysis functions in dChip for comparative analysis and hierarchic clustering.
Western blotting
Approximately 50 mg protein was suspended in Laemmli sample buffer (0.1 M Tris-HCl buffer, pH 6.8, 1% SDS, 0.05% β-mercaptoethanol, 10% glycerol, and 0.001% bromphenol blue), boiled for 2 minutes, and electrophoresed on 4% to 20% glycerol gradient SDS-polyacrylamide gel for 4 hours at 120 V. Gels were electroblotted onto Trans-Blot nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA) at 40 V for 3 hours in a Tris-glycine buffer system. Incubation with indicated antibodies was performed for 2 hours in PBS-Tween 20 (PBST) containing 1% BSA with constant rocking. Blots were washed with PBST and incubated in either anti-rabbit or anti-mouse horseradish peroxidase (HRP) conjugates for 2 hours in PBST containing 3% nonfat dry milk. After washing, specific
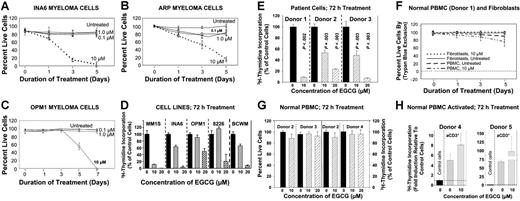
Effect of EGCG on cell survival. MM cells were cultured in the medium containing no EGCG or various concentrations of EGCG ranging from 0.1 to 10 μM. Cells were harvested at different time points as indicated and proliferative potential was assessed by trypan blue exclusion or 3H-thymidine labeling. The growth curves show the mean of 3 independent experiments, with SEM. (A) IL-6-dependent INA6 myeloma cells. (B) ARP myeloma cells. (C) OPM1 myeloma cells. (D) Myeloma cell lines MM1S, INA6, OPM1, 8226, and Waldenstrom cells (BCWM) were treated with 10 and 20 μM EGCG for 72 hours and proliferative potential was assessed by 3H-thymidine incorporation. (E) Three samples of CD138+ purified myeloma cells derived from patient bone marrow were treated with 10 and 20 μM EGCG for 72 hours and cell proliferation was evaluated by 3H-thymidine incorporation. (F) Effect of EGCG treatment in normal diploid fibroblasts and PBMCs from a healthy donor at days 1, 3, and 5. (G) Effect of EGCG on proliferation of PBMC from healthy donors is shown by trypan blue exclusion and 3H-thymidine incorporation, following 72 h treatment with 10 and 20 μM drug. (H) PBMCs from healthy donors (donors 4 and 5) were activated with anti-CD3 antibody, treated with 10 μM EGCG for 72 hours, and cell proliferation was evaluated by 3H-thymidine incorporation.
Effect of EGCG on cell survival. MM cells were cultured in the medium containing no EGCG or various concentrations of EGCG ranging from 0.1 to 10 μM. Cells were harvested at different time points as indicated and proliferative potential was assessed by trypan blue exclusion or 3H-thymidine labeling. The growth curves show the mean of 3 independent experiments, with SEM. (A) IL-6-dependent INA6 myeloma cells. (B) ARP myeloma cells. (C) OPM1 myeloma cells. (D) Myeloma cell lines MM1S, INA6, OPM1, 8226, and Waldenstrom cells (BCWM) were treated with 10 and 20 μM EGCG for 72 hours and proliferative potential was assessed by 3H-thymidine incorporation. (E) Three samples of CD138+ purified myeloma cells derived from patient bone marrow were treated with 10 and 20 μM EGCG for 72 hours and cell proliferation was evaluated by 3H-thymidine incorporation. (F) Effect of EGCG treatment in normal diploid fibroblasts and PBMCs from a healthy donor at days 1, 3, and 5. (G) Effect of EGCG on proliferation of PBMC from healthy donors is shown by trypan blue exclusion and 3H-thymidine incorporation, following 72 h treatment with 10 and 20 μM drug. (H) PBMCs from healthy donors (donors 4 and 5) were activated with anti-CD3 antibody, treated with 10 μM EGCG for 72 hours, and cell proliferation was evaluated by 3H-thymidine incorporation.
proteins were detected using an enhanced chemiluminescence, according to the instructions provided in the manual (Amersham Life Sciences).
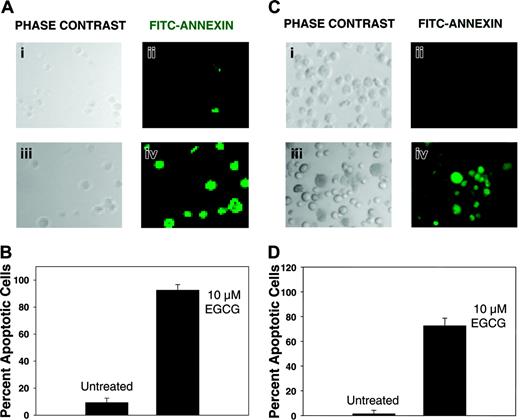
Apoptosis following EGCG treatment of myeloma cells. Myeloma cells were treated with 10 μM EGCG for 72 hours and analyzed for apoptosis using annexin V-biotin apoptosis detection kit. Cells were sequentially treated with annexin V-biotin and FITC-streptavidin. FITC-streptavidin-labeled apoptotic cells within the same microscopic field were viewed and photographed by phase contrast (PC) or by fluorescence emitted at 518 nm (FITC filter). Using the FITC filter, apoptotic cells appear bright green. (A) INA6 myeloma cells, untreated (i-ii) or treated with EGCG (iii-iv). (B) Bar graph shows percent apoptotic cells in control or EGCG-treated INA6 cells. (C) ARP myeloma cells, untreated (i-ii) or treated with EGCG (iii-iv). (D) Bar graph shows percent apoptotic cells in control or EGCG-treated ARP cells. (B, D) Error bars indicate SEM of percentage of apoptotic cells in 5 distinct microscopic fields.
Apoptosis following EGCG treatment of myeloma cells. Myeloma cells were treated with 10 μM EGCG for 72 hours and analyzed for apoptosis using annexin V-biotin apoptosis detection kit. Cells were sequentially treated with annexin V-biotin and FITC-streptavidin. FITC-streptavidin-labeled apoptotic cells within the same microscopic field were viewed and photographed by phase contrast (PC) or by fluorescence emitted at 518 nm (FITC filter). Using the FITC filter, apoptotic cells appear bright green. (A) INA6 myeloma cells, untreated (i-ii) or treated with EGCG (iii-iv). (B) Bar graph shows percent apoptotic cells in control or EGCG-treated INA6 cells. (C) ARP myeloma cells, untreated (i-ii) or treated with EGCG (iii-iv). (D) Bar graph shows percent apoptotic cells in control or EGCG-treated ARP cells. (B, D) Error bars indicate SEM of percentage of apoptotic cells in 5 distinct microscopic fields.
Results
EGCG induces inhibition of myeloma cell growth
INA6, ARP, and OPM1 MM cells were cultured in the presence or absence of EGCG at various concentrations and for variable lengths of time and viable cell number was determined as described. EGCG induced both time- and dose-dependent decline in survival of myeloma cells; at 10 μM concentration it induced 85% and over 55% cell death in INA6 and ARP myeloma cells, respectively, at day 3 and more than 99% cell death in INA6 and ARP cell lines at day 5 and OPM1 cells at day 7 (Figure 1A-C). In all myeloma cell lines tested and primary myeloma cells derived from 3 different patients, exposure to 10 or 20 μM EGCG led to a significant inhibition of cell proliferation as assessed by 3H-thymidine incorporation, within 72 hours treatment (Figure 1D-E). Importantly, the same concentrations of EGCG (10 or 20 μM) had no effect on survival of normal diploid fibroblasts and normal PBMCs from 4 healthy donors (Figure 1F-G). Normal PBMCs were treated with EGCG and cell proliferation was assessed by trypan blue exclusion or 3H-thymidine incorporation or both. As seen in Figure 1G, EGCG at 10 or 20 μM had no effect on cell proliferation following 72 hours of treatment. To further confirm the lack of effect on normal cells, normal PBMCs were activated with anti-CD3 antibody and treated with EGCG. As indicated by 3H-thymidine incorporation, exposure to 10 μM EGCG did not have inhibitory effect on proliferation of PBMCs. These data confirm that EGCG, at concentrations used, specifically inhibits the proliferation of myeloma cells while having no significant effect on normal cells.
EGCG induces apoptotic cell death
Myeloma cells (INA6 and ARP) were treated with EGCG (10 μM) and analyzed for apoptotic cell death. Both untreated or EGCG-treated myeloma cells were sequentially treated with annexin V-biotin and FITC-streptavidin and apoptotic cells were evaluated by a fluorescence microscope. Approximately 200, representing at least 5 distinct microscopic fields, were analyzed to assess the fraction of annexin V+ cells for each sample. Following a 3-day exposure to EGCG, 92% ± 8% INA6 cells and 73% ± 6% of ARP cells were annexin V+, whereas only 8% ± 2% and less than 2% of untreated INA6 and ARP cells, respectively, were annexin V+ (Figure 2), indicating that EGCG induces apoptosis in myeloma cells.
EGCG mediates its activity via LR1
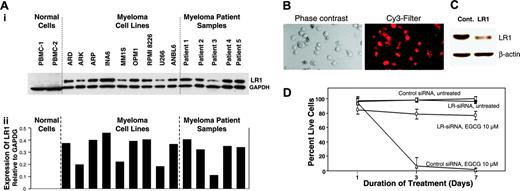
EGCG has been reported to confer its effects through its interaction with LR1 (67 kDa).28 We therefore evaluated the protein levels of LR1 in myeloma cell lines and patient samples using Western blot analysis. As seen in Figure 3Ai and quantitation following normalization with α-tubulin levels (Figure 3Aii), a 10-fold or greater increase in the protein levels of LR1 in all myeloma cell lines and patient samples is observed.
Next, to confirm the role of LR1 in EGCG-mediated growth inhibition of MM cells, we transfected INA6 myeloma cells with Cy3-labeled nontargeting control siRNA or siRNA directed against LR1. Uptake of siRNA was confirmed by fluorescence microscopy (Figure 3B), and reduction of LR1 protein level was confirmed by Western blot analysis (Figure 3C). Transfected cells were treated on the next day with EGCG (10 μM) and cell viability was measured on alternate days for 7 days. As seen in Figure 3D, EGCG had no significant effect on the growth of INA6 cells transfected with LR1-specific siRNA, whereas more than 98% of cells transfected with control siRNAs died within 3 days following exposure to EGCG.
EGCG induces apoptosis in myeloma cells in vivo
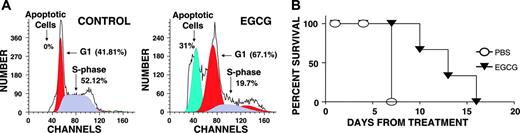
To evaluate in vivo activity of EGCG on MM cells, OPM1 MM cells were injected subcutaneously in CB17/ICr-SCID mice, and following appearance of palpable tumors, mice were given intraperitoneal injections of PBS alone or EGCG dissolved in PBS. As the tumors reached more than 2 cm in size, the mice were humanely killed, the tumors were excised, and the cell-cycle profile of MM cells was analyzed using PI staining and flow cytometry. Percentage of apoptotic cells in tumors derived from 3 control mice remained less than 1%, whereas the fraction of apoptotic cells in EGCG-treated mice ranged from 32% to 39%, indicating significant (P < .004) in vivo antimyeloma activity.
Consistent with these data, the survival of EGCG-treated mice was also prolonged relative to control mice (Figure 4B).
EGCG activates multiple proapoptotic pathways
To identify the molecular mechanisms of EGCG-induced apoptosis, we analyzed change in gene expression profile of INA6 cells following exposure to 10 μM EGCG for 24 hours, using HG-U133A GeneChip array (Affymetrix), as reported previously.20,21,29,30 Reproducibility of expression change was confirmed by correlation coefficients (0.96-0.99) of independently conducted experiments.
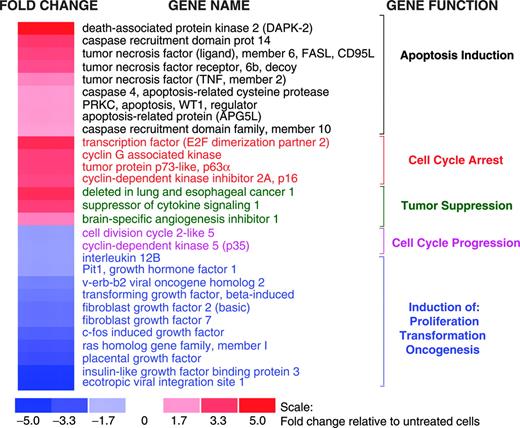
Exposure of myeloma cells to EGCG led to up-regulation of major regulatory genes involved in apoptosis and cell cycle arrest as well as down-regulation of genes implicated in oncogenic transformation (Figure 5).
Role of LR1 in EGCG-induced myeloma cell death. (A) Elevated levels of LR1 (67 kDa) protein in myeloma cells. (Ai) Protein levels of LR1 were evaluated in 2 samples of normal PBMCs, 9 myeloma cell lines, and 5 myeloma patient samples, using a monoclonal antibody specific for LR; (ii) bar graph shows expression of LR1 protein, normalized to GAPDH levels, in normal and myeloma cells. (B-C) INA6 myeloma cells were cotransfected with Cy3-labeled nontargeting control (cont) and laminin receptor 1 (LR1) siRNAs. (B) Transfected cells were incubated for 72 hours and uptake of siRNA was monitored by a fluorescence microscope equipped with a Cy3 filter. (C) INA6 cells were transfected as described for panel B and LR1 protein level was determined by Western blot analysis. (D) EGCG induced myeloma cell death. INA6 myeloma cells were transfected with control siRNAs or siRNAs directed against LR1, and 24 hours later treated with 10 μM EGCG. Cell viability was determined on alternate days. Error bars indicate SEM of 3 independent experiments.
Role of LR1 in EGCG-induced myeloma cell death. (A) Elevated levels of LR1 (67 kDa) protein in myeloma cells. (Ai) Protein levels of LR1 were evaluated in 2 samples of normal PBMCs, 9 myeloma cell lines, and 5 myeloma patient samples, using a monoclonal antibody specific for LR; (ii) bar graph shows expression of LR1 protein, normalized to GAPDH levels, in normal and myeloma cells. (B-C) INA6 myeloma cells were cotransfected with Cy3-labeled nontargeting control (cont) and laminin receptor 1 (LR1) siRNAs. (B) Transfected cells were incubated for 72 hours and uptake of siRNA was monitored by a fluorescence microscope equipped with a Cy3 filter. (C) INA6 cells were transfected as described for panel B and LR1 protein level was determined by Western blot analysis. (D) EGCG induced myeloma cell death. INA6 myeloma cells were transfected with control siRNAs or siRNAs directed against LR1, and 24 hours later treated with 10 μM EGCG. Cell viability was determined on alternate days. Error bars indicate SEM of 3 independent experiments.
Effect of EGCG on proliferation of myeloma cells in vivo. CB-17 SCID mice were inoculated subcutaneously in the interscapular area with 5 × 106 OPM1 myeloma cells. Following appearance of tumors, the mice were treated intraperitoneally with PBS alone or EGCG 33 mg/kg/d. When mice were humanely killed, tumors were excised and analyzed for apoptosis by flow cytometry. (A) Cell-cycle profiles of tumor cells derived from control and EGCG-treated mice. (B) Survival curve of control and EGCG-treated mice.
Effect of EGCG on proliferation of myeloma cells in vivo. CB-17 SCID mice were inoculated subcutaneously in the interscapular area with 5 × 106 OPM1 myeloma cells. Following appearance of tumors, the mice were treated intraperitoneally with PBS alone or EGCG 33 mg/kg/d. When mice were humanely killed, tumors were excised and analyzed for apoptosis by flow cytometry. (A) Cell-cycle profiles of tumor cells derived from control and EGCG-treated mice. (B) Survival curve of control and EGCG-treated mice.
EGCG activated multiple pathways associated with growth arrest by inducing the expression of: (1) death-associated protein kinase 2 (DAPK2), a multifunctional protein kinase implicated in apoptotic pathways mediated by death receptors, p19/p53, and modulation of cytoskeleton; (2) initiators and mediators of death receptor-mediated apoptosis including Fas, Fas ligand, and caspase 4; (3) p63, a p53-like protein involved in induction of apoptosis; (4) caspase recruitment domain proteins (CARD10 and CARD14) associated with induction of apoptosis via activation of BCL10 and NF-κB; and (5) cyclin-dependent kinase inhibitors, p16 and p18 (Figure 5), which induce cell-cycle arrest by inhibiting phosphorylation of retinoblastoma (RB).
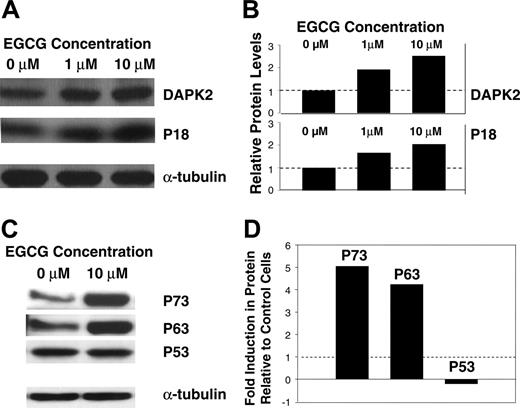
For selected genes, we have further confirmed the observed changes in gene expression profile at protein levels. Myeloma cells were treated with EGCG at 10 μM for 24 hours and the cell lysates were resolved on a gradient SDS-polyacrylamide gel, electroblotted, and probed with specific antibodies. Consistent with gene expression data, the exposure of MM cells to EGCG was associated with elevated protein levels of DAPK2, p18, and p63 (Figure 6A-D). Both the gene expression (not shown) and Western blot (Figure 6C-D) analyses indicated no change in level of p53 following exposure to EGCG. However, the Western blot analysis indicated a 6-fold increase in p73 protein (Figure 6C-D). Overall these data confirm the gene expression and protein changes and provide the molecular basis for observed growth arrest and apoptosis following exposure of myeloma cells to EGCG.
Effect of EGCG on gene expression in myeloma cells. Gene expression profile was analyzed in untreated or EGCG-treated (10 μM for 24 hours) MM cells using HG-U133A gene arrays (Affymetrix). Fold change in the expression in EGCG-treated cells relative to expression in untreated INA6 cells is shown by the intensity of red (induction) or blue (suppression) colors.
Effect of EGCG on gene expression in myeloma cells. Gene expression profile was analyzed in untreated or EGCG-treated (10 μM for 24 hours) MM cells using HG-U133A gene arrays (Affymetrix). Fold change in the expression in EGCG-treated cells relative to expression in untreated INA6 cells is shown by the intensity of red (induction) or blue (suppression) colors.
Discussion
Here, we demonstrate that EGCG, an antioxidant from green tea, induces growth arrest and apoptosis in MM cells while having no significant effect on normal PBMCs as well as fibroblasts. Anticancer effects of EGCG have been demonstrated in vitro in several malignancies including human lung, cervical, colon, and oral squamous carcinoma cells with effective IC50 values ranging from 22 to 200 μM.14,18,31 However, this is the first report demonstrating its activity in hematologic malignancy and elucidating the molecular mechanisms of EGCG-induced apoptosis in cancer cells, specifically in MM. Exposure of myeloma cell lines and primary patient cells to 10 to 20 μM EGCG led to apoptotic cell death within 5 to 7 days. Our data suggest higher susceptibility of MM cells to EGCG, which may provide a higher therapeutic index. Importantly EGCG at 10 to 20 μM had no significant effect on survival or proliferation of normal diploid fibroblasts and normal PBMCs.
The effect of EGCG on protein expression in INA6 myeloma cells. Equal amounts of protein were fractionated on SDS-polyacrylamide gels and electroblotted onto nitrocellulose membranes. The membranes were sequentially treated with primary antibodies and HRP-conjugated secondary antibodies, and the proteins were detected using an enhanced chemiluminescence. The same blots were then stripped and incubated with a monoclonal antibody for α-tubulin. Signal intensity of each band was quantitated and the amount of each protein was normalized to that of α-tubulin. (A) Expression of DAPK2 and p18 proteins in INA6 cells, untreated or treated with 1 μM and 10 μM EGCG for 24 hours. (B) Bar graph shows relative expression of DAPK2 and p18 proteins, following normalization with corresponding α-tubulin levels. (C) Expression of p53 family of proteins (p53, p63, p73) in INA6 cells, untreated or treated with 10 μM EGCG for 24 hours. (D) Bar graph shows fold induction of p53 family members, following normalization with corresponding α-tubulin levels.
The effect of EGCG on protein expression in INA6 myeloma cells. Equal amounts of protein were fractionated on SDS-polyacrylamide gels and electroblotted onto nitrocellulose membranes. The membranes were sequentially treated with primary antibodies and HRP-conjugated secondary antibodies, and the proteins were detected using an enhanced chemiluminescence. The same blots were then stripped and incubated with a monoclonal antibody for α-tubulin. Signal intensity of each band was quantitated and the amount of each protein was normalized to that of α-tubulin. (A) Expression of DAPK2 and p18 proteins in INA6 cells, untreated or treated with 1 μM and 10 μM EGCG for 24 hours. (B) Bar graph shows relative expression of DAPK2 and p18 proteins, following normalization with corresponding α-tubulin levels. (C) Expression of p53 family of proteins (p53, p63, p73) in INA6 cells, untreated or treated with 10 μM EGCG for 24 hours. (D) Bar graph shows fold induction of p53 family members, following normalization with corresponding α-tubulin levels.
To further confirm the lack of its effect on normal cells, we treated normal PBMCs activated with anti-CD3 antibody with EGCG at 10 μM (Figure 1H) and 20 μM (data not shown) and observed no effect on 3H-thymidine incorporation. These data further confirm that EGCG specifically inhibits the proliferation of myeloma cells while having no effect on normal cells at a concentration that can be achieved in vivo, as demonstrated in a rat model in which plasma concentration of 96.0 μM was achieved without reported toxicity.32
We have also confirmed in vivo activity of EGCG in MM. Administration of EGCG at 33 mg/kg/d led to induction of apoptosis in MM cells and prolongation of survival in SCID mice bearing subcutaneous MM tumors. The survival of EGCG-treated mice was also significantly increased (P < .05), demonstrating an antimyeloma activity in vivo.
We also demonstrate that the antimyeloma effects of EGCG are mediated through a 67-kDa LR1, a cell-surface receptor implicated in the interaction of myeloma cells with basement membrane and subsequent infiltration/migration of these cells in surrounding tissue.33 As reported here, LR1 knock-down cells are not susceptible to EGCG-induced myeloma cell death. The majority of myeloma cell lines and patient samples have elevated levels of LR1 protein. Gene expression profiling also showed up-regulated transcript levels of LR1 and its pseudogene in primary patient myeloma cells compared to normal plasma cells (data not shown). Extremely low expression of LR1 in normal cells and overexpression in myeloma cells provides the molecular explanation for specific activity of EGCG on MM cells with minimum effect on normal cells and may also provide the basis on which to expect a higher pharmacologic index for this agent in clinical practice.
The gene expression profile following EGCG treatment showed the most prominent induction of death-associated protein kinase 2 (DAPK2). DAPK2, a member of calcium/calmodulin-dependent serine/threonine kinases,34 is implicated in multiple apoptotic pathways. Apoptosis initiated by TNF-α, activated Fas, and IFN-γ is mediated by DAPK,35 which functions as a key regulatory step between the formation of death-inducing signaling complex (DISC) and activation of caspases.35,36 DAPK can also induce apoptosis by p19ARF-p53 pathway37,38 as well as by phosphorylation of myosin light chain, which leads to abnormal cytokinesis.36
EGCG treatment was also associated with elevated transcript and protein levels of p73 and p63, the members of the p53 family with ability to induce apoptosis in a p53-like manner.39,40 Because p53 is frequently mutated in cancers, the induction of p53-like proteins (p73 and p63) by EGCG provides an important alternate mechanism of cell growth arrest in the absence of p53. These proteins are not only implicated in the induction of genes involved in apoptosis but also regulate genes involved in cell-cycle arrest and DNA repair.
EGCG also induced the expression of tumor necrosis factor ligand (member 6; FASL) and tumor necrosis factor receptor (member 6b; FAS), implicated in the death receptor-dependent apoptosis. The interaction between the ligand and its death receptor leads to the recruitment of DISC that subsequently initiates a cascade of events leading to activation of initiator and effector caspases. An in vitro evaluation in liver cancer has confirmed the increased protein levels of FASL following EGCG treatment.41 There is also evidence that EGCG may directly bind and activate FAS leading to induction of apoptosis.42
Both gene expression and protein data also indicate that EGCG treatment is associated with the induction of cyclin-dependent kinase (CDK) inhibitors p16 and p18. Inhibitors of CDKs can induce cell-cycle arrest by preventing RB phosphorylation and E2F1 release.
In conclusion, these studies demonstrate that EGCG is a potent suppressor of MM cell growth with specificity provided by its interaction with LR1, a cell-surface receptor implicated in the interaction of myeloma cells with basement membrane. It leads to induction of multiple interrelated pathways implicated in growth arrest, providing a concerted activity leading to MM cell death both in vitro and in vivo. These data, therefore, indicate that a natural product with antioxidant properties from green tea has a specific activity against MM, making it an ideal compound for therapy and possible chemoprevention of this disease.
Prepublished online as Blood First Edition Paper, June 29, 2006; DOI 10.1182/blood-2006-05-022814.
Supported in part by National Institutes of Health (NIH) grant DK031092, a Merit Review Award from the Research Service Veterans Health Care (VHA) (R.K.G.), NIH-P50-100007 Developmental Research Award (M.A.S.), a Merit Review Award from the Research Service, a Merit Review Award from Epidemiology Service VHA (N.C.M.), and NIH-P050-100007 and NIH-PO1-78378 (N.C.M. and K.C.A.). N.C.M. is a Leukemia Society Scholar in Translational Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal