Abstract
Erythroid progenitors differentiate in erythroblastic islands, bone marrow niches composed of erythroblasts surrounding a central macrophage. Evidence suggests that within islands adhesive interactions regulate erythropoiesis and apoptosis. We are exploring whether erythroid intercellular adhesion molecule 4 (ICAM-4), an immunoglobulin superfamily member, participates in island formation. Earlier, we identified αV integrins as ICAM-4 counterreceptors. Because macrophages express αV, ICAM-4 potentially mediates island attachments. To test this, we generated ICAM-4 knock-out mice and developed quantitative, live cell techniques for harvesting intact islands and for re-forming islands in vitro. We observed a 47% decrease in islands reconstituted from ICAM-4 null marrow compared to wild-type marrow. We also found a striking decrease in islands formed in vivo in knock-out mice. Further, peptides that block ICAM-4/αV adhesion produced a 53% to 57% decrease in reconstituted islands, strongly suggesting that ICAM-4 binding to macrophage αV functions in island integrity. Importantly, we documented that αV integrin is expressed in macrophages isolated from erythroblastic islands. Collectively, these data provide convincing evidence that ICAM-4 is critical in erythroblastic island formation via ICAM-4/αV adhesion and also demonstrate that the novel experimental strategies we developed will be valuable in exploring molecular mechanisms of erythroblastic island formation and their functional role in regulating erythropoiesis.
Introduction
Erythroid progenitors proliferate, differentiate, and enucleate within specialized bone marrow niches, termed erythroblastic islands.1-4 These structural units are composed of developing erythroblasts surrounding a central macrophage. It is apparent from ultrastructural studies that extensive cell-cell interactions, both erythroblast-macrophage, as well as erythroblast-erythroblast, occur within these 3-dimensional structures. However, little is known regarding either the molecular nature or functional role of the specific adhesive interactions. We are exploring the potential function of erythroid ICAM-4, a recently characterized member of the immunoglobulin superfamily, in erythroblastic island formation. ICAM-4 expression is limited to erythroid and placental tissue5 but, to date, there is no information on its role in erythropoiesis. We earlier identified α4β1 and αV family integrins as ICAM-4–binding partners.6 Because macrophages express αV and erythroblasts exhibit α4β1, ICAM-4 is an attractive candidate for mediating erythroblast-erythroblast interactions via ICAM-4/α4β1 binding and regulating adhesion of erythroblasts to central macrophages via ICAM-4/αV binding.
ICAM-4, which carries the Lansteiner Wiener (LW) blood group antigen system, has strong sequence homology with other members of the ICAM protein superfamily.7,8 It is composed of 2 extracellular immunoglobulin-like domains, an N-terminal I set and a membrane proximal I2 set, and a single membrane-spanning domain.8,9 ICAM-4 is detected early during terminal differentiation, concordant with surface expression of glycophorin A and RhGP.10 Hence, the timing of ICAM-4 expression during erythropoiesis is consistent with a functional role in erythroblastic islands.
To elucidate the structural basis of αV integrin–ICAM-4 interaction we earlier performed targeted mutagenesis of ICAM-4 surface-exposed amino acid residues, using a molecular model of ICAM-4 derived from the crystal structure of closely related ICAM-2.11 Using adhesion assays with cells that bind ICAM-4 via αVB1 and αVB5, we identified a patch or “footprint” that mediates adhesion to αV integrins composed of 2 series of residues on the N-terminal extracellular domain: F18, W19, V20 and R92, A94, T95, S96, R97. In the protein structure these 8 residues are close to one another, suggesting that this region is crucial for ICAM-4 attachment to αV integrins. We also tested synthetic peptides composed of sequences of ICAM-4 shown to be involved in adhesion to αV integrins and found that they inhibited cell binding, providing independent support for the role of the proposed footprint in αV integrin binding.11
To explore whether ICAM-4 participates in erythroblastic island formation, we generated ICAM-4 null homozygous mice and studied whether islands were perturbed. For these investigations we established quantitative and reproducible live cell techniques for harvesting intact islands from mouse bone marrow or re-forming islands in vitro from single cell suspensions of mouse marrow. Applying these methods, we observed a striking decrease in the number of islands formed in vivo or in vitro by ICAM-4 null erythroblasts. Collectively, the results of this phenotypic analysis provide convincing evidence for ICAM-4 in erythroblastic island formation. Further, we determined that synthetic peptides that block ICAM-4/αV adhesion caused a marked concentration-dependent decrease in islands reconstituted from single cell suspensions of wild-type mouse marrow, thereby identifying erythroblast ICAM-4 binding to macrophage αV integrins as functionally important attachments. We postulate that this newly identified erythroblast integrin counterreceptor may be crucial not only for adhesive integrity of island structure but also for initiating intracellular signaling essential for normal erythroid terminal differentiation.
Materials and methods
Generation of mice lacking ICAM-4
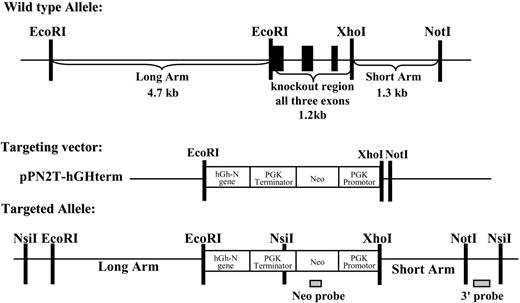
For targeted gene deletion, we designed a construct to delete 1.2 kb of ICAM4 genomic sequence encoding exons 1-3, which encompasses the entire protein coding region12 (Figure 1). To construct the targeting vector we ligated into the pPN2T-hGHterm vector13,14 a 4.7-kb “long arm” of gDNA from the 5′ end of the region to be deleted using EcoRI restriction site and a 1.3-kb “short arm” from the 3′ deletion boundary using XhoI and NotI restriction sites. The 2 arms flank a PGKneo cassette modified with polyA-signal/terminator from the human growth hormone N-gene generating a 1-kb targeting vector (Figure 1). After analysis with sequencing and restriction enzyme digestions to check for fidelity and correct orientation, the targeting vector was provided to Xenogen Biosciences (Cranberry, NJ) who generated the ICAM-4 null mice. Two embryonic stem (ES) cell lines with correct targeting were obtained. Injection of these lines into C57BL/6J (B6) blastocysts produced a number of chimeric males, ranging from 40% to 80% chimerism. The chimeric mice were mated to wild-type B6 mice to generate heterozygous ICAM-4 knock-out mice; breeding of heterozygous mice produced viable homozygous ICAM-4 null mice. Mice were maintained on a hybrid B6,129 background for all experiments.
Southern blot analysis
Neomycin-resistant colonies were screened by Southern blot analysis with a 0.5-kb 3′ probe derived from sequence just downstream of the “short arm.” This probe hybridized to a 12.8-kb NsiI fragment in normal mouse gDNA and a 5.2-kb NsiI fragment in the targeted variant, respectively.
Polymerase chain reaction analysis
gDNA was prepared from 3.0- to 4.0-mm tail samples with Dneasy tissue kit (Qiagen, Valencia, CA) and used to genotype progeny by polymerase chain reaction (PCR). To distinguish between wild-type, heterozygote, and ICAM-4 null mice, a multiplex PCR genotyping assay was developed using primers for the Icam4 gene and the neo gene. Icam4 primers generated a 528-bp fragment, whereas the neo primers produced a 381-bp fragment. Icam4 forward primer located in exon 1: 5′-CAGCAAGAGTGGATGCAAAGTCC-3′; reverse primer located in exon 2: 5′-CCAGGATCACCACCAAGAATC-3′. Neo forward primer: 5′-TTGTCACTGAAGCGGGAAGG-3′; reverse primer: 5′-CACAGTCGATGAATCCAGAAAAGC-3′. PCR was set up using PuReTaq Ready-To-Go PCR beads (GE Healthcare, Piscataway, PA) with 0.5 μM of each primer in a reaction volume of 100 μL. PCR conditions were 32 cycles at 94°C for 30 seconds, 64°C for 30 seconds, and 72°C for 60 seconds.
Western blot analysis
Blood was collected from ICAM-4 null and wild-type mouse tails in potassium EDTA-treated microtubes and from an anonymous, existing normal human blood sample complying with the United Kingdom National Blood Service Policy under the Nuffield Council on Bioethics and the Medical Research Council operational and ethical guidelines. Red cell membranes were prepared by washing red cells 3 times in 15 volumes of 10 mM phosphate-buffered saline (PBS), pH 7.4, followed by 3 washes in 15 volumes of lysis buffer (Na2HPO4 6.8 mM, NaH2PO4 2.25 mM, pH 8.0). Membrane proteins (30 μg/lane) were separated on a nonreducing 10% SDS-polyacrylamide gel, then transferred onto PVDF membrane (Hybond-P; Amersham, Bucks, United Kingdom) using a semidry electroblotter. After blocking for 1 hour in PBS, 0.1% Tween-20, 5% nonfat dry milk, blots were probed overnight at 4°C with polyclonal rabbit anti–mouse ICAM-4 (Pickwell Labs, Amsterdam, The Netherlands. Antigen: mouse ICAM-412 ) at 1:200 or anti–human ICAM-4 (BS5615 ; a gift from Dr H. Sonneborn, Biotest, Dreieich, Germany) at 50 μg/mL, washed, and then incubated with either anti–rabbit (1:100) or anti–mouse (1:1000) IgGs coupled to horseradish peroxidase (DakoCytomation, Ely, Cambridgeshire, United Kingdom). After several washes, blots were developed using the 3,3′-diaminobenzidine (DAB) method (Sigma-Aldrich, St Louis, MO).
Reconstitution of erythroblastic islands
Bone marrow was gently flushed from both tibias and femurs of 3- to 5-month-old adult mice by inserting the end of bones into flexible tubing attached to a 1-mL syringe. Extracted marrow was resuspended in 3 mL Iscove modified Dulbecco medium (IMDM; Invitrogen, Carlsbad, CA) containing 3.5% sodium citrate (BD PharMingen, San Diego, CA) and 20% fetal calf serum (FCS; Invitrogen) by pipetting 20 times with a Pasteur pipette; bone and tissue fragments were removed by passage through a 70-μm cell strainer. A single cell suspension was obtained by reaspirating cells through a 26-gauge needle 5 times and the cells viewed and counted using bright-field microscopy. We normally obtain about 90 million nucleated cells/mouse. Cells were aliquoted (1 × 106/tube), incubated on ice for 15 minutes in activation buffer (IMDM, 3.5% sodium citrate, 20% FCS, 2 mM Mn+2, 2 mM EGTA), in the presence or absence of specific synthetic peptides, and subsequently incubated for 2 hours at room temperature with erythroid-specific (Ter119-PE, eBiosciences, San Diego, CA), macrophage-specific (F4/80-FITC, eBiosciences), and DNA (Hoechst 33342, Sigma-Aldrich) probes. Then 1 × 105 cells were transferred into 8-well chambered cover glasses containing 400 μL IMDM and 20% FCS and allowed to settle for 15 minutes. The timing was carefully controlled throughout the experiments so that each aliquot of cells was analyzed after the same amount of time from introduction into Mn+2-containing buffer. Labeled live cell samples were analyzed by a blinded observer at room temperature using conventional fluorescence microscopy on a Nikon TE2000 (Nikon Instruments, Melville, NY) with a 10×/0.5 NA S Fluor S objective equipped with a Q-imaging RetigaEX CCD camera (QIMAGING, Burnaby, BC, Canada). The number of islands contained in 20 random fields covering 12% of the chamber was counted per concentration of peptide. Images were acquired/processed by Image Pro 4.5 (MediaCybernetics, Silver Spring, MD) and Adobe Photoshop (Adobe Systems, San Jose, CA).
Bone marrow was obtained from MacGreen mice16 for establishing the assay and for peptide inhibition studies or from B6,129 ICAM-4 null and wild-type littermates. Macrophages from MacGreen mice express macrophage colony-stimulating factor (M-CSF) receptor-green fluorescent protein transgene, thereby providing a useful macrophage identifier. In experiments in which MacGreen mice were used, F4/80-FITC was not used.
Harvesting intact erythroblastic islands
To harvest erythroblastic islands formed in vivo, bone marrow was gently flushed from both tibias and femurs of 3- to 5-month-old adult B6,129 ICAM-4 null and wild-type littermates by inserting the end of bones into flexible tubing attached to a 1-mL syringe. Extracted marrow was gently resuspended in 5-mL IMDM containing 3.5% sodium citrate and 20% FCS by pipetting 20 times with a Pasteur pipette; bone and tissue fragments were removed by passage through a 70-μm cell separator. To obtain a cell count, a single cell suspension was prepared from a small aliquot, as described (see “Reconstitution of erythroblastic islands”), the cell count determined, and then a volume of the filtered, island-containing suspension equivalent to the volume containing 1 × 106 cells was diluted to a final volume of 200 μL, labeled, as described, with erythroid-, macrophage-, and DNA-specific probes and incubated undisturbed for 2 hours at room temperature. Then 20 μL labeled cells were transferred into 8-well chambered cover glasses containing 400 μL IMDM, 20% FCS and analyzed by a blinded observer at room temperature using conventional fluorescence microscopy on a Nikon TE2000 with a 10 ×/0.5 NA S Fluor S objective equipped with a Q-imaging RetigaEX CCD camera. The number of islands contained in 20 random fields covering 12% of the chamber was counted per experiment. Images were acquired/processed by Image Pro 4.5 and Adobe Photoshop.
Deletion of Icam4 gene. Targeting strategy for homologous recombination in ES cells. Restriction map of wild-type Icam4 allele (top), targeting vector (middle), and targeted allele (bottom). In the wild-type Icam4 allele the black boxes represent the 3 exons. In the targeting vector, hGh-N indicates human growth hormone-N gene; PGK, phosphoglycerate kinase; Neo, bacterial neomycin resistance gene.
Deletion of Icam4 gene. Targeting strategy for homologous recombination in ES cells. Restriction map of wild-type Icam4 allele (top), targeting vector (middle), and targeted allele (bottom). In the wild-type Icam4 allele the black boxes represent the 3 exons. In the targeting vector, hGh-N indicates human growth hormone-N gene; PGK, phosphoglycerate kinase; Neo, bacterial neomycin resistance gene.
Peptide inhibition studies
Synthetic peptides V(16)PFWVRMS (FWV) and T(91)RWATSRI (ATSR) were reconstituted with activation buffer to a concentration of 5 mM, vortexed for 30 minutes, ultracentrifuged at 313 000g for 30 minutes at 4°C, and peptide concentrations determined on centrifuged supernatants. FWV and ATSR peptides (0.5-3.0 mM) were then used in erythroblastic island reconstitution assays.
Central macrophage αV phenotyping
To obtain central macrophages from erythroblastic islands for αV integrin phenotyping, intact erythroblastic islands were harvested as described and islands separated from single cells by velocity sedimentation on a 4-mL discontinuous gradient of 0.5% bovine serum albumin (BSA; Chemicon, Temecula, CA), 20% FBS-IMDM overlaying 1% BSA, and 20% FBS-IMDM at 4°C. After 1.0 hour, fractions were collected and those fractions highly enriched for islands and depleted of single cells were pooled. The islands were then dispersed into a single cell suspension by aspirating through a 25-gauge needle and erythroid cells removed from the single cell suspension using Ter119 microbeads (Miltenyi Biotec, Auburn, CA). Central macrophages were settled onto cover glass for 4 hours, fixed with 4% paraformaldehyde for 10 minutes, washed 6 times with 1% BSA-PBS, and then blocked overnight at 4°C with 10 μg/mL SeroBloc (Serotec, Raleigh, NC), 20 μg/mL goat IgG (Jackson ImmunoResearch, West Grove, PA), and 10% normal goat serum (Jackson ImmunoResearch). Following blocking, cells were stained with 25 μg/mL monoclonal anti–mouse CD51, which recognizes mouse αV integrin (Biolegend, San Diego, CA), and F4/80 for 1 hour at room temperature. After washing 6 times with 1% BSA-PBS, cells were stained with rhodamine red-X–conjugated F(ab′)2 fragment goat anti–rat IgG (Jackson ImmunoResearch) and Hoechst DNA dye for 1 hour at room temperature and washed 6 times with 1% BSA-PBS. Cells on cover glasses were mounted onto slides with Vectashield mounting medium (Vector Labs, Burlingame, CA) and analyzed using conventional fluorescence microscopy on a Nikon TE2000 with a 60 ×/1.40 NA plan Apochromat oil objective equipped with a Q-imaging RetigaEX CCD camera. Images were acquired/processed by Image Pro 4.5 and Adobe Photoshop.
Measurements of red blood cell parameters
Whole blood (∼20 μL) was drawn from the retro-orbital sinus into EDTA-coated microhematocrit tubes, mixed with RPMI 1640 media (Invitrogen), and adjusted to 200 μL final volume with PBS containing a final concentration of 5% BSA. Blood counts and erythrocyte indices were determined using an automated hematology analyzer (ADVIA 120; Bayer Diagnostics, Tarrytown, NY) and adjusted for the dilution factor. Each mouse was analyzed on at least 3 different occasions. Data were collected on 9 adult knock-out (> 3 months of age) and 7 age-matched littermate controls to determine mean values and SDs for the various parameters.
Results
Generation of knock-out mice
To study the physiologic role of ICAM-4 in erythroblastic islands we targeted ICAM4 for germline deletion in mice. The mouse erythrocyte ICAM4 gene of 1.2 kb is composed of 3 exons (Figure 1).12 Exons 1-3, encompassing the entire protein-coding domain, were targeted for replacement with the vector pPN2T-hGHterm, containing a PGKneomycin resistance cassette modified with polyA-signal/terminator from the human growth hormone N-gene (Figure 1). As summarized in “Materials and methods,” ES cells derived from inbred strain 129 were transfected and neomycin-resistant colonies screened by Southern blot analysis. Two ES cell lines with correct targeting were obtained. Injection into B6 blastocysts produced a number of chimeric males, ranging from 40% to 80% chimerism. Chimeric mice were mated to wild-type B6 mice to generate heterozygous Icam4 knock-out mice and breeding of heterozygous mice produced viable homozygous Icam4 null mice. Correct targeting of Icam4 in heterozygous and homozygous mice was documented by Southern blot analysis (Figure 2A). Deletion of Icam4 DNA and protein was further confirmed by PCR analysis of tail gDNA and Western blotting of erythrocyte membranes. Using a multiplex PCR genotyping assay using primers for the Icam4 and neo genes, Icam4 primers generated a 528-bp fragment and neo primers produced a 381-bp fragment (Figure 2B). This assay allowed us to readily distinguish between wild-type, heterozygous, and Icam4 null mice, thereby enabling determination of the genotype of each mouse that was used experimentally. When Western blots of equivalent amounts of erythrocyte membranes from wild-type and knock-out mice were probed with antibody recognizing mouse ICAM-4, a band of appropriate size for ICAM-4 was present in wild-type membranes and lacking in knock-out membranes (Figure 2C). No reactivity was observed using preimmune rabbit control serum (data not shown). As a positive control, human erythrocyte membranes were probed with BS56, a well-characterized antibody against an epitope on the LW blood group active extracellular region of ICAM-415 (Figure 2C). The immunoreactive band observed in human membranes migrated at a similar molecular weight as the band observed in wild-type mouse erythrocyte membranes. Together, these data confirm the targeted deletion of the ICAM4 gene.
Targeted disruption of Icam4. (A) Southern blot analysis of NsiI-digested DNA derived from tail vein samples of offspring from heterozygous mating pair. Blot probed with 3′ probe (shown in Figure 1) depicts homozygous animals containing only a 5.2-kb band derived from the targeted allele and the neomycin cassette (lanes 1 and 2). Heterozygote possesses both the 5.2-kb band and the endogenous DNA migrating at 12.8 kb (lane 3). Wild-type animal contains only the endogenous 12.8-kb band (lane 4). (B) PCR analysis of tail gDNA. Primers binding to Icam4 exon 1 and exon 2 amplified a 528-bp fragment; primers binding to the Neo gene amplified a 381-bp fragment. Molecular weight markers (lane 1); gDNA from wild-type mouse generated a 528-bp fragment (lane 2); gDNA from heterozygote generated 528-bp and 381-bp fragments (lane 3); gDNA from homozygous mouse generated a 381-bp fragment (lane 4). (C) Western blot analysis of erythrocyte membranes. Equivalent amounts of erythrocyte membranes from wild-type and knock-out mice probed with antibody recognizing mouse ICAM-4 produced a band of appropriate size for ICAM-4 in wild-type membranes, which was absent in knock-out membranes. As a positive control, human erythrocyte membranes were probed with BS56, a well-characterized antibody to ICAM-4 and produced an immunoreactive band migrating at a similar molecular weight as the band observed in wild-type mouse erythrocyte membranes.
Targeted disruption of Icam4. (A) Southern blot analysis of NsiI-digested DNA derived from tail vein samples of offspring from heterozygous mating pair. Blot probed with 3′ probe (shown in Figure 1) depicts homozygous animals containing only a 5.2-kb band derived from the targeted allele and the neomycin cassette (lanes 1 and 2). Heterozygote possesses both the 5.2-kb band and the endogenous DNA migrating at 12.8 kb (lane 3). Wild-type animal contains only the endogenous 12.8-kb band (lane 4). (B) PCR analysis of tail gDNA. Primers binding to Icam4 exon 1 and exon 2 amplified a 528-bp fragment; primers binding to the Neo gene amplified a 381-bp fragment. Molecular weight markers (lane 1); gDNA from wild-type mouse generated a 528-bp fragment (lane 2); gDNA from heterozygote generated 528-bp and 381-bp fragments (lane 3); gDNA from homozygous mouse generated a 381-bp fragment (lane 4). (C) Western blot analysis of erythrocyte membranes. Equivalent amounts of erythrocyte membranes from wild-type and knock-out mice probed with antibody recognizing mouse ICAM-4 produced a band of appropriate size for ICAM-4 in wild-type membranes, which was absent in knock-out membranes. As a positive control, human erythrocyte membranes were probed with BS56, a well-characterized antibody to ICAM-4 and produced an immunoreactive band migrating at a similar molecular weight as the band observed in wild-type mouse erythrocyte membranes.
Reconstituted erythroblastic islands
To begin to test whether ICAM-4 has a functional role in erythroblastic islands, we developed a quantitative live cell assay for re-forming islands from single cell suspensions of freshly harvested mouse bone marrow. Adults 3 to 5 months of age were used and all females were virgins. A single cell suspension was prepared, then cells were incubated for carefully controlled times in media containing manganese. We determined that we could identify islands and their cellular components by 3-color immunofluorescence microscopy using fluoresceinated erythroid-specific Ter119 antibody,17 macrophage-specific F4/80 antibody,18,19 or macrophage marker M-CSF receptor GFP transgene expression16 and a DNA probe (Figure 3A-C). We observed that the number of cells per island varied as did their stage of differentiation, consistent with observations by others of erythroblastic islands formed in vivo.20-24 Because surface expression of glycophorin A increases during terminal differentiation, the intensity of Ter119 staining served as an effective indicator of erythroblast stage. A faint blush of Ter119 fluorescence was present in early erythroblasts and increasing degrees of staining were observed in progressively more differentiated cells. We also found that the fluorescence intensity of Ter119 label varied among erythroblasts in an individual island, indicating that islands were composed of erythroblasts at various stages of differentiation. Young, multilobulated reticulocytes were present in many islands, again consistent with prior descriptions of erythroblastic islands formed in vivo. To determine the amount of variation in total number of islands that re-formed from a single cell suspension of 1 × 105 cells, we counted islands containing 6 or more erythroblasts in experiments on 10 different MacGreen mice. Total islands were counted at the beginning (ie, end of the incubation period) and conclusion of each experiment. We found that the number of reconstituted islands in control mice was highly reproducible (918 ± 148) and did not vary substantially from experiment to experiment using different mice (Figure 3D).
Erythroblastic island formation is decreased in ICAM-4 null mice
To test for functional ICAM-4–mediated adhesion in erythroblastic islands, we analyzed ICAM-4 knock-out mice, comparing the capacity of single cell suspensions from ICAM-4 null and wild-type bone marrow to form erythroblastic islands in vitro. We observed a marked decrease in the percentage of islands formed from bone marrow of ICAM-4 null mice compared to wild-type littermates. Strikingly, we found a 47% decrease in the total number of islands formed from 1 × 105 bone marrow cells from ICAM-4 null mice compared to wild-type littermates. Control wild-type cells re-formed 953 ± 141 islands, whereas ICAM-4 null cells re-formed 504 ± 88 islands (Figure 4A).
Erythroblastic islands formed in vivo
In addition to analyzing erythroblastic islands reconstituted in vitro, we wanted to develop the capability to study islands that had been formed in vivo. Hence, we performed experiments to establish methodology for reproducibly harvesting intact erythroblastic islands from mouse bone marrow. We determined that by gently extracting marrow and removing bone and tissue fragments we were able to retain erythroblastic island structures. As with the reconstituted islands, we identified islands formed in vivo and their cellular components by 3-color immunofluorescence microscopy using fluoresceinated erythroid-specific Ter119 antibody, macrophage-specific F4/80 antibody, and a DNA probe. We found that the number of harvested islands in normal mice was very reproducible and equaled 898 ± 246 in volume equivalent to 1 × 105 bone marrow cells (Figure 4B).
Reconstituted erythroblastic islands. Bright field (A) and immunofluorescent standard (B) and confocal (C) micrographs of typical erythroblastic islands formed from single cell suspensions of MacGreen mouse bone marrow. Immunofluorescent micrographs of islands show cells stained for erythroid-specific marker GPA (Ter119; red), macrophage marker M-CSF receptor GFP transgene expression (green), and DNA (Hoechst 33342; blue). In the confocal image some of the cells appear blurred because they are not in the plane of focus. However, macrophage staining is apparent in various regions of the island. Reticulocytes, arrowheads; macrophage, arrows; bars represent 10 μm. (D) Histogram shows number of erythroblastic islands formed from 1 × 105 single cells; n = 10. Results are shown as mean ± SD.
Reconstituted erythroblastic islands. Bright field (A) and immunofluorescent standard (B) and confocal (C) micrographs of typical erythroblastic islands formed from single cell suspensions of MacGreen mouse bone marrow. Immunofluorescent micrographs of islands show cells stained for erythroid-specific marker GPA (Ter119; red), macrophage marker M-CSF receptor GFP transgene expression (green), and DNA (Hoechst 33342; blue). In the confocal image some of the cells appear blurred because they are not in the plane of focus. However, macrophage staining is apparent in various regions of the island. Reticulocytes, arrowheads; macrophage, arrows; bars represent 10 μm. (D) Histogram shows number of erythroblastic islands formed from 1 × 105 single cells; n = 10. Results are shown as mean ± SD.
Erythroblastic islands from ICAM-4 null and wild-type mouse bone marrow. (A) Islands reconstituted from B6,129 ICAM-4 null and wild-type mouse bone marrow cells. Histogram of number of erythroblastic islands formed from 1 × 105 single cells obtained from wild-type (n = 10) and ICAM-4 null (n = 10). *P < .001 when compared to islands formed from wild-type marrow. (B) Erythroblastic islands formed in vivo in ICAM-4 null and wild-type mice. Histogram of number of erythroblastic islands obtained from wild type (n = 6) and ICAM-4 null (n = 6). *P < .001 when compared to islands formed from wild-type marrow. Results are shown as mean ± SD.
Erythroblastic islands from ICAM-4 null and wild-type mouse bone marrow. (A) Islands reconstituted from B6,129 ICAM-4 null and wild-type mouse bone marrow cells. Histogram of number of erythroblastic islands formed from 1 × 105 single cells obtained from wild-type (n = 10) and ICAM-4 null (n = 10). *P < .001 when compared to islands formed from wild-type marrow. (B) Erythroblastic islands formed in vivo in ICAM-4 null and wild-type mice. Histogram of number of erythroblastic islands obtained from wild type (n = 6) and ICAM-4 null (n = 6). *P < .001 when compared to islands formed from wild-type marrow. Results are shown as mean ± SD.
Erythroblastic islands formed in vivo are decreased in ICAM-4 null mice
To determine the ability of ICAM-4 null erythroblasts to form islands in vivo, we collected and quantitated intact islands from freshly harvested mouse bone marrow. Similar to the in vitro data, we found a marked decrease in the percentage of islands from bone marrow of ICAM-4 null mice compared to wild-type littermates. We observed a 64% decrease in the total number of islands from ICAM-4 null mice compared to wild-type littermates. Control wild-type cells re-formed 898 ± 246 islands, whereas ICAM-4 null cells re-formed 327 ± 97 islands (Figure 4B). Taken together, the results of this phenotypic analysis provide convincing evidence that ICAM-4 is critical in erythroblastic island formation.
Steady-state ICAM-4 null mice have normal hematocrit, hemoglobin, and red cell indices
To determine the characteristics of circulating erythrocytes in ICAM-4 null mice during steady-state erythropoiesis, we obtained complete blood counts and red cell indices using an automated hematology analyzer. Data were collected on 9 adult knock-out mice and 7 age-matched littermate controls. We found that red cell count and hemoglobin, hematocrit, and reticulocyte values, as well as red cell indices, were normal in ICAM-4 null mice (Table 1). Hence, in nonstressed erythropoiesis the observed decrease in erythroblastic islands does not result in anemia or any detectable abnormal phenotype of circulating erythrocytes.
Measurements of red blood cell parameters
. | RBC count, × 1012/L . | HGB level, g/dL . | HCT, % . | Reticulocytes, % . | MCV, fL . | MCH level, pg . | CHCM, g/dL . | RDW, % . |
|---|---|---|---|---|---|---|---|---|
| Wild type | 10.48 ± 0.59 | 15.83 ± 0.45 | 48.07 ± 2.49 | 2.54 ± 0.13 | 45.81 ± 1.39 | 15.14 ± 0.71 | 313.5 ± 4.7 | 13.24 ± 0.55 |
| Knock-out | 10.43 ± 0.53 | 15.68 ± 0.49 | 48.10 ± 2.71 | 2.52 ± 0.31 | 46.10 ± 0.88 | 15.14 ± 0.63 | 312.1 ± 5.7 | 13.70 ± 0.80 |
. | RBC count, × 1012/L . | HGB level, g/dL . | HCT, % . | Reticulocytes, % . | MCV, fL . | MCH level, pg . | CHCM, g/dL . | RDW, % . |
|---|---|---|---|---|---|---|---|---|
| Wild type | 10.48 ± 0.59 | 15.83 ± 0.45 | 48.07 ± 2.49 | 2.54 ± 0.13 | 45.81 ± 1.39 | 15.14 ± 0.71 | 313.5 ± 4.7 | 13.24 ± 0.55 |
| Knock-out | 10.43 ± 0.53 | 15.68 ± 0.49 | 48.10 ± 2.71 | 2.52 ± 0.31 | 46.10 ± 0.88 | 15.14 ± 0.63 | 312.1 ± 5.7 | 13.70 ± 0.80 |
Data were collected on 9 adult knock-out mice and 7 age-matched littermate controls. Values are mean ± SD.
RBC indicates red blood cell; HGB, hemoglobin; HCT, hematocrit; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; CHCM, cell hemoglobin concentration mean; RDW, red cell distribution width.
Erythroblastic island formation is inhibited by peptides that block adhesion of ICAM-4 to αV integrins
We next tested the effects on erythroblastic island formation of 2 synthetic peptides that we have previously shown block ICAM-4/αV adhesion.11 Peptides V(16)PFWVRMS (FWV) and T(91)RWATSRI (ATSR) correspond to sequences within the αV-binding region located on the A and G strands of ICAM-4 domain 1, respectively (Figure 5A). A single cell suspension was prepared from freshly harvested normal mouse marrow and then cells were incubated for carefully controlled times in media containing manganese and then in media containing various peptides or media alone. Both peptides caused a marked, concentration-dependent decrease in the percentage of islands formed. ATSR at 0 to 1.0 mM, 1.0 to 2.0 mM, and 2.0 to 3.0 mM inhibited island formation 17%, 28%, and 53%, respectively (Figure 5B), whereas 0 to 1.0 mM, 1.0 to 2.0 mM, and 2.0 to 3.0 mM FWV inhibited island formation 20%, 37%, and 57%, respectively (Figure 5C). These data strongly suggest that erythroblast ICAM-4 binding to macrophage αV integrins is critical for erythroblastic island formation.
Central macrophages isolated from erythroblastic islands express the αV integrin subunit
Macrophages vary phenotypically and although macrophages from various tissues have been shown to express the αV integrin subunit CD51, it was crucial to confirm its presence on the surface of erythroblastic island central macrophages. Therefore, we performed αV integrin phenotyping on central macrophages isolated from erythroblastic islands. For these investigations, intact islands were separated from single cells by velocity sedimentation, the islands were disrupted, and then erythroblasts removed from the single cell suspension using Ter119 microbeads. The remaining cells were analyzed by immunofluorescence microscopy using macrophage-specific probe F4/80 and anti-CD51. In double-label experiments we observed colocalization of the 2 markers (Figure 6), clearly showing that central macrophages isolated from erythroblastic islands express the αV integrin subunit.
Discussion
Terminally differentiating erythroblasts express a variety of cell adhesion molecules on their surfaces.10,25-34 These proteins mediate interactions between erythroblasts and stromal cells and between erythroblasts and extracellular matrix components, such as fibronectin and laminin. ICAM-4 is a cell adhesion protein with a narrow tissue distribution, its expression limited to erythroid and possibly placental tissues.5 Prior to the current investigations the function of ICAM-4 during erythropoiesis was unknown.
As a definitive exploration of whether ICAM-4 mediated adhesive interactions function in erythroblastic island integrity, we generated ICAM-4 null mice and quantitated these marrow substructures. In novel island reconstitution experiments we observed a 47% decrease in the total number of islands formed from bone marrow cells from ICAM-4 null mice compared to wild-type littermates. We felt that it was crucial to also obtain data on in vivo island formation in these null mice. To achieve this objective, we developed techniques for harvesting and analyzing intact erythroblastic islands that were both reproducible and quantitative. Applying these novel methods, we found a 64% decrease in islands harvested from ICAM-4 null mice compared to wild-type littermates. This striking decrease in islands formed both in vivo and in vitro by ICAM-4 null erythroblasts clearly shows that ICAM-4 protein is critical in erythroblastic island formation.
Erythroblastic island formation in the presence and absence of peptides blocking adhesion of ICAM-4 to αV integrins. Model of extracellular domain of ICAM-4 is shown revealing its solvent exposed surface in 3 orientations rotated 120° to each other. The region of ICAM-4 involved in adhesion to αV integrins is shown in yellow and green. Yellow designates area of FVW peptide sequence and green depicts location of ATSR peptide sequence (A). Histogram of percentage of islands formed in the presence of 0.5 to 1.0 mM FWV peptide (n = 11), 1.0 to 2.0 mM FWV peptide (n = 16), and 2.0 to 3.0 mM FWV peptide (n = 8) compared to islands formed in media alone. *P = .02, **P < .001, ***P < .001 when compared to islands formed in media alone (B). Histogram of percentage of islands formed in the presence of 0.5 to 1.0 mM ATSR peptide (n = 3), 1.0 to 2.0 mM ATSR peptide (n = 8), and 2.0 to 3.0 mM ATSR peptide (n = 4) compared to islands formed in media alone. *P = .1, **P = .004, ***P < .001 when compared to islands formed in media alone (C). Results are shown as mean ± SD.
Erythroblastic island formation in the presence and absence of peptides blocking adhesion of ICAM-4 to αV integrins. Model of extracellular domain of ICAM-4 is shown revealing its solvent exposed surface in 3 orientations rotated 120° to each other. The region of ICAM-4 involved in adhesion to αV integrins is shown in yellow and green. Yellow designates area of FVW peptide sequence and green depicts location of ATSR peptide sequence (A). Histogram of percentage of islands formed in the presence of 0.5 to 1.0 mM FWV peptide (n = 11), 1.0 to 2.0 mM FWV peptide (n = 16), and 2.0 to 3.0 mM FWV peptide (n = 8) compared to islands formed in media alone. *P = .02, **P < .001, ***P < .001 when compared to islands formed in media alone (B). Histogram of percentage of islands formed in the presence of 0.5 to 1.0 mM ATSR peptide (n = 3), 1.0 to 2.0 mM ATSR peptide (n = 8), and 2.0 to 3.0 mM ATSR peptide (n = 4) compared to islands formed in media alone. *P = .1, **P = .004, ***P < .001 when compared to islands formed in media alone (C). Results are shown as mean ± SD.
A major finding of the current study is that adhesive interactions between erythroblast ICAM-4 and its αV integrin counterreceptor on central macrophages is critical for erythroblastic island integrity. An important aspect of the present study is our unequivocal demonstration by live-cell microscopy that central macrophages of native bone marrow erythroblastic islands indeed express αV integrin. We consider this finding crucial for making a definitive conclusion regarding a role for macrophage αV integrin in erythroblastic island formation, in view of the marked heterogeneity of macrophage phenotypes. Using the quantitative and reproducible live cell technique that we developed for re-forming islands in vitro, we observed that synthetic peptides ATSR and FWV, which block ICAM-4/αV adhesion,11 caused a marked concentration-dependent decrease in the percentage of islands reconstituted from bone marrow single cell suspensions. ATSR and FWV inhibited island formation 53% and 57%, respectively, at the highest peptide concentrations tested. Significant similarity in the inhibiting effects of the 2 peptides was also observed at lower peptide concentrations. Our findings that 2 different peptides, each composed of amino acid residues within the αV integrin-binding region on ICAM-4, blocked island reconstitution to similar degrees, strongly argues for the importance of ICAM-4/αV attachments in 3-dimensional erythroblastic islands.
Expression of F4/80 and αV integrin on central macrophages. Central macrophages isolated from erythroblastic islands were analyzed by immunofluorescence microscopy in double antibody label experiments using macrophage-specific probe antibody F4/80 (green), anti-CD51 (red), and DNA (Hoechst 33342; blue). Two representative cells are shown, one in the top 3 panels and the other in the bottom 3 panels. Merged image signals showed colocalization (yellow) of the 2 antibody markers indicating that central macrophages isolated from erythroblastic islands express the αV integrin subunit. Bar represents 10 μm.
Expression of F4/80 and αV integrin on central macrophages. Central macrophages isolated from erythroblastic islands were analyzed by immunofluorescence microscopy in double antibody label experiments using macrophage-specific probe antibody F4/80 (green), anti-CD51 (red), and DNA (Hoechst 33342; blue). Two representative cells are shown, one in the top 3 panels and the other in the bottom 3 panels. Merged image signals showed colocalization (yellow) of the 2 antibody markers indicating that central macrophages isolated from erythroblastic islands express the αV integrin subunit. Bar represents 10 μm.
Earlier we reported marked similarities between mouse and human ICAM-4 protein with 68% overall identity.12 Critical cysteine residues and other key residues within the 2 extracellular IgSF domains are conserved, suggesting that these disulfide-bonded domains are similarly folded in human and murine proteins and may have analogous functional properties. In support of this we have determined that the αV integrin-binding properties of ICAM-4 are conserved across species.12 These data strongly suggest that our current findings regarding the adhesive role of ICAM-4 in mouse erythroblastic island integrity may be equally pertinent to human erythropoiesis.
We were interested to discover that, during basal state erythropoiesis, ICAM-4 null mice have normal hematocrit, hemoglobin, and red cell indices. We speculate that in stressed erythropoiesis the observed decrease in erythroblastic islands may result in anemia or a blunted reticulocyte response. This scenario would mirror that reported for mice lacking 2 important transcription factors, Stat5a and Stat5b. In the nonstressed state, Stat5 mutant mice have normal numbers of red cells and hematocrit and hemoglobin levels.35 However, Stat5a–/–5b–/– embryos are severely anemic, demonstrating that Stat5 is essential for the high rate of erythroid proliferation during fetal development.36 Additionally, Stat5a–/–5b–/– adult mice have a blunted reticulocyte response to stress.37 In future studies we plan to explore ICAM-4 null embryonic erythropoiesis and the effect of stress on adult circulating red cells to determine whether ICAM-4 is essential for high rates of proliferation.
Growing evidence supports the concept of erythroblastic islands as microenvironmental niches within bone marrow where cell-cell attachments, in concert with cytokines, are crucial for terminal erythroid differentiation and regulation of apoptosis. To date, only a few receptor-counterreceptor interactions have been described, but the data regarding their impact on erythropoiesis are tantalizing. Previous studies identified a transmembrane protein, erythroblast macrophage protein (Emp) present in both erythroblasts and macrophages, that appears to mediate erythroblast-erythroblast and erythroblast-macrophage attachments via homophilic binding.3,38 In erythroblasts cultured in the presence of anti-Emp or in the absence of macrophages, a marked decrease in erythroid cell proliferation, maturation, and enucleation is observed, accompanied by increased apoptosis.38 Another identified attachment within erythroblastic islands occurs between erythroblast α4β1 integrin and its counterreceptor, VCAM-1, in central macrophages.39 Island integrity is perturbed by antibodies to either VCAM-1 or α4β1. Additionally, anti-α4β1 induces mobilization of progenitor cells to peripheral blood40 and studies in α4 conditional knock-out mice demonstrate an impaired response to erythropoietic stress.41 Finally, a recent report appears to demonstrate the importance of intercellular signaling between erythroblasts in regulating GATA-1 activity.42 We postulate that the various linkages within erythroblastic islands are dynamic during erythroid development and that the signaling pathways stimulated by these attachments could influence differentiation and enucleation.
Interestingly, Fas/Fas ligand–related regulation of apoptosis also appears to occur within erythroblastic islands.43 Orthochromatic erythroblasts expressing Fas ligand demonstrate a Fas-based cytotoxicity against immature erythroblasts expressing Fas, which is abolished by high levels of erythropoietin. Fas-independent regulation of apoptosis may also occur within islands. Bone marrow macrophages secrete soluble receptor-binding cancer antigen expressed in SiSo cells (RCAS1) that permeabilizes mitochondrial membranes and activates caspase-8 and caspase-3 in immature erythroblasts that express RASC1 receptor.44 In sum, these findings delineate potential mechanisms for negative regulatory feedback between mature and immature erythroblasts.
In earlier investigations we discovered a novel secreted isoform of mouse ICAM-4, termed ICAM-4S.12 We found that ICAM-4S mRNA is up-regulated late in terminal differentiation, suggesting a regulatory role in late erythropoiesis. Secreted ICAM-4S may compete with cellular ICAM-4 for integrin counterreceptors, thereby interfering with adhesion between membrane ICAM-4 and its binding partners. This potential repressive function of ICAM-4S could enable young reticulocytes to detach from erythroblastic islands in preparation for their egress into the peripheral circulation.
We postulate that the novel receptor-counterreceptor interaction between erythroblast ICAM-4 and macrophage αV integrin identified in the current report may be important not only for adhesive integrity of erythroblastic island structures but also for initiating intracellular signaling essential for normal erythroid terminal differentiation. In addition, the novel quantitative island reconstitution assays we developed are likely to be extremely valuable in furthering our understanding of the molecular basis for cell-cell interactions in erythroblastic islands and in delineating the functional sequelae of these interactions.
Prepublished online as Blood First Edition Paper, May 11, 2006; DOI 10.1182/blood-2006-03-006759.
Supported in part by National Institutes of Health grants DK56267 and DK32094; by the National Health Service Research and Development Directorate, United Kingdom; and by the Director, Office of Health and Environment Research Division, US Department of Energy, under contract DE-AC03-76SF00098.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Dr H. Sonneborn (Biotest, Dreieich, Germany) for the gift of antibody BS56 and acknowledge Xenogen Biosciences (Cranberry, NJ) for generation of the ICAM-4 null mice. The MacGreen mice are owned by IMBcom and the University of Queensland, Australia and are provided as a service to the research community. We are very grateful to Dr Luanne Peters for providing the pPN2T-hGHterm vector and for helpful discussions.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal