Abstract
Fanconi anemia (FA) is a genomic instability disorder, clinically characterized by congenital abnormalities, progressive bone marrow failure, and predisposition to malignancy. Cells derived from patients with FA display a marked sensitivity to DNA cross-linking agents, such as mitomycin C (MMC). This observation has led to the hypothesis that the proteins defective in FA are involved in the sensing or repair of interstrand cross-link lesions of the DNA. A nuclear complex consisting of a majority of the FA proteins plays a crucial role in this process and is required for the monoubiquitination of a downstream target, FANCD2. Two new FA genes, FANCB and FANCL, have recently been identified, and their discovery has allowed a more detailed study into the molecular architecture of the FA pathway. We demonstrate a direct interaction between FANCB and FANCL and that a complex of these proteins binds FANCA. The interaction between FANCA and FANCL is dependent on FANCB, FANCG, and FANCM, but independent of FANCC, FANCE, and FANCF. These findings provide a framework for the protein interactions that occur “upstream” in the FA pathway and suggest that besides the FA core complex different subcomplexes exist that may have specific functions other than the monoubiquitination of FANCD2.
Introduction
Multiple pathways are required by the cell to deal with endogenous and exogenous damage to the DNA and to maintain the genome's integrity. The inactivation of these pathways leads to an unstable genome, which increases the risk of tumorigenesis. Many of the genes involved in DNA repair and genomic stability are mutated in cancer predisposition syndromes such as XPA-G (xeroderma pigmentosum), NBS1 (Nijmegen breakage syndrome), and ATM (ataxia telangiectasia). The function of the gene products that are defective in the Fanconi anemia (FA) pathway, which is speculated to act in DNA repair, remains elusive.
FA is clinically characterized by congenital abnormalities, progressive bone marrow failure, and predisposition to malignancy, especially acute myeloid leukemia (AML) and squamous cell carcinoma (SCC). Cells derived from patients with FA are hypersensitive to DNA cross-linking agents, such as mitomycin C (MMC) and diexpoxybutane (DEB), which suggests that the FA pathway may be involved in the sensing and/or repair of interstrand cross-links. Cell fusion experiments have identified 12 different complementation groups, and 11 of their corresponding disease genes have been cloned: FANCA, FANCB, FANCC, FANCD1/BRCA2, FANCD2, FANCE, FANCF, FANCG, FANCJ/BRIP1, FANCL, and FANCM.1-14 Many of the FA proteins interact in a nuclear complex, the assembly of which is required for the monoubiquitination of a downstream target, FANCD2.15 This protein has been the subject of intense investigation in recent years, and several studies have revealed that the monoubiquitinated isoform of FANCD2 enters discrete nuclear foci where it colocalizes with multiple proteins involved in genomic stability including BRCA1, NBS1, and RAD51.15-17 Furthermore, it has been shown that this modification is cell-cycle regulated, occurring specifically during S phase.17 More recently, the nuclear FA core complex18 and FANCD219 have been found to associate with chromatin, which may indicate that the monoubiquitinated form of FANCD2 is involved in interstrand cross-link repair at the site of DNA damage. However, little is known about the sequence of events that leads to the monoubiquitination of this protein. What is clear is that an intact FA core complex is required for this modification to take place, but the molecular architecture of this complex is not fully defined. The recent identification of a core complex member with helicase and nuclease motifs (FANCM)13 and the observation that restoration of the FANCD2 monoubiquitination alone is not sufficient to complement the FA defect20 strongly suggest that the FA proteins might have other functions besides the monoubiquitination of FANCD2.
Considerable effort has been placed in investigating the relationship between the FA proteins with respect to their interactions, but until very recently all members of the FA core complex had not yet been discovered. Two new FA genes, FANCB and FANCL, have been identified, and their discovery has allowed a more detailed study into the molecular architecture of the FA pathway. FANCB and FANCL were found through their association with FANCA by complex purification experiments.3,12,21 FANCL is a RING finger protein probably acting as the E3 ubiquitin ligase of the FA pathway that monoubiquitinates FANCD2. The nuclear accumulation of FANCL seems to be regulated by FANCA and FANCB.12 In this study, we have sought to characterize the biochemical relationship of FANCB and FANCL with other members of the FA core complex. We show that FANCB and FANCL directly interact and that a complex of these proteins can bind FANCA. The interaction between FANCA and FANCL is abnormal in FA-B, -G, and -M cells, but normal in complementation groups C, E, and F. We propose a model in which the association between FANCA, FANCB, FANCG, FANCL, and FANCM represents the initial steps in the sequential assembly of the FA core complex. In each step, subcomplexes of FA proteins are formed that may have specific functions in the FA pathway. The monoubiquitination of FANCD2 is just one event in this whole cascade.
Materials and methods
Cell lines and culture conditions
Epstein-Barr virus (EBV)–transformed lymphoblasts were cultured in RPMI 1640 medium containing 10% fetal calf serum (FCS). Stably expressing lymphoblasts were generated by electroporation of pIRESneo constructs (Clontech, Palo Alto, CA) and selection on 600 μg/mL neomycin or by electroporation of pMEP4 constructs (Invitrogen, Carlsbad, CA) and selection on 100 μg/mL hygromycin. Human embryonic kidney 293 cells were maintained in D-MEM medium containing 10% FCS. 293 cells stably expressing FLAG-FANCB were generated by electroporation of a pIRESneo construct followed by selection on 600 μg/mL neomycin. A single clone was selected and grown out in order to establish a clonal cell line. Expression of FLAG-FANCB was confirmed by Western blotting. Human MCF-7 breast cancer cells were grown in D-MEM supplemented with 10% FCS and antibiotics. The MMC-induced growth inhibition assays were performed as previously described.22 All cells were grown in a humidified incubator at 37°C in 5% CO2.
Plasmids
Mammalian 2-hybrid constructs containing FANCA, FANCC, FANCD2, FANCE, FANCF, and FANCG have been previously described.23 Full-length FANCL was polymerase chain reaction (PCR) amplified with AccuPrime Pfx DNA polymerase (Invitrogen) using primers containing suitable restriction sites and ligated into the NdeI and EcoRI restriction sites of pGBKT7 (Clontech). To create a mammalian 2-hybrid construct encoding full-length FANCL, the 5′ region was PCR amplified using an internal primer and a primer that contained an EcoRI site and the product digested with EcoRI and an internal PstI. In addition, a PstI-SalI fragment was cut from pGBKT7-FANCL, and a 3-point ligation was performed into pVP16. FANCL fragments were PCR amplified using primers that contained EcoRI restriction sites at the indicated amino acid positions and ligated into pM to create an in-frame cDNA with the GAL4-BD. Mammalian 2-hybrid constructs containing FANCL point mutations were generated by PCR with oligonucleotides encoding the amino acid substitutions of the FANCL sequence. Full-length FANCB was PCR amplified with AccuPrime Pfx DNA polymerase (Invitrogen) using primers containing suitable restriction sites and ligated into the BamHI and NotI restriction sites of pM and pVP16. FANCA mutants were generated by using a QuickChange II Site Directed Mutagenesis Kit (Clontech) or by PCR. The full-length sequences, frame, and orientation were confirmed by sequencing, and the pathogenic character of these mutants was demonstrated by their inability to restore the MMC sensitivity of FA-A cells in an MMC growth inhibition test.
Subcellular fractionation, immunoprecipitations, and Western blotting
Nuclear and cytoplasmic fractionations were prepared from 2 × 107 lymphoblasts as previously described.24 Whole-cell extracts were prepared in lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, and 1% Triton X-100 supplemented with protease and phosphatase inhibitors). Lysates were subjected to immunoprecipitation with the indicated antibodies at 4°C overnight. The following day, immune complexes were captured with Protein A sepharose (Amersham Biosciences, Freiburg, Germany) for an additional 30 minutes. Immunoprecipitations using the FLAG epitope were performed using an anti–FLAG M2 affinity gel (Sigma, St Louis, MO). After extensive washing with lysis buffer, immunoprecipitated proteins were separated on 8% polyacrylamide gels or 8% to 16% Tris-glycine gels (Invitrogen) and transferred to Immobilon-P membrane (Millipore, Billerica, MA). Membranes were blocked and probed with the indicated antibodies as previously described.24
Mammalian 2-hybrid analysis
Mammalian 2- and 3-hybrid assays were performed as previously described.25 Transient expression of FANCL and FANCB constructs was confirmed by Western blotting with GAL4-BD–specific antisera (Clontech).
Immunofluorescence
Transfected MCF-7 cells were fixed with 3.7% formaldehyde in PBS for 30 minutes and permeabilized with 0.2% Triton in PBS for 10 minutes. Following a blocking step with 3% BSA (Sigma) in PBS for 1 hour, an anti–FLAG M2 monoclonal antibody (Stratagene, La Jolla, CA) was diluted 1:350 in blocking solution and applied for 1 hour. After washing with PBS, cells were incubated with an FITC-conjugated goat antimouse secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for 45 minutes. Finally, coverslips were mounted onto microscope slides with Vectashield (Vector Laboratories, Burlingame, CA). Hoechst 33342 (Sigma) was used to counterstain the cell nuclei. Slides were examined under UV light on an inverted Leica DMIRB/E fluorescence microscope (Leica, Heidelberg, Germany). Images (obtained using a 40×/0.75 numeric aperture oil objective) were collected using Leica Q500MC Quantimet software V01.01 (Leica, Cambridge, United Kingdom) and a Kappa CF 8/1 FMC camera (Kappa Opto-Electronics, Gleichen, Germany).
Results
FANCL is required for the assembly of the Fanconi anemia core complex
Purification of FANCA-associated proteins led to the identification of FANCL as a novel component of the FA core complex and the putative E3 ubiquitin ligase of the FA pathway.12 Initially we sought to confirm and extend this finding by examining FANCL coprecipitation with functionally active FLAG-tagged FA proteins from stably transfected FA lymphoblasts. FANCL copurified with FANCA-FLAG, which confirmed the original finding that FANCA and FANCL are together in a complex (Figure 1A, lane 2). Likewise, FANCF-FLAG (Figure 1B, lane 2) and FLAG-FANCC (Figure 1C, lane 2) were able to coprecipitate both FANCA and FANCL, providing further evidence that FANCL is an integral member of the FA core complex. We then decided to investigate what effect a lack of FANCL would have on the assembly of the complex. Whole-cell extracts were prepared from WT and FA lymphoblasts and subjected to immunoprecipitation using antibodies raised against different FA proteins. In WT cells, immunoprecipitation with an anti-FANCC antibody coprecipitates FANCA, FANCG, and weakly FANCF (Figure 1D, lane 1); however, in FA-L cells these interactions are disrupted (Figure 1D, lane 2). A direct Western blot on whole-cell extracts shows that the lack of interaction is not due to the absence of FANCL protein in FA-L cells (Figure 1E). Similarly, in WT cells an anti-FANCF immunoprecipitation coprecipitates FANCA and FANCG (Figure 1D, lane 4), whereas in FA-L cells these interactions are lost (Figure 1D, lane 5). Conversely, the interaction between FANCA and FANCG appeared to be retained in FA-L cells (Figure 1D, lane 8). These findings are consistent with FANCL being an important member of the FA core complex, its presence being required for the correct assembly of this multisubunit complex.
FANCL coimmunoprecipitates with FLAG-tagged FA proteins and is required for the assembly of the FA core complex. Lymphoblastoid cell lines stably expressing functionally active FLAG-tagged FANCA (A), FANCF (B), and FANCC (C) were generated. Whole-cell extracts of 10 million cells were immunoprecipitated with an α-FLAG affinity gel and precipitated proteins eluted by competition with a FLAG peptide. Untransfected cells (lane 1) were included as negative controls. Precipitated proteins were detected with α-FANCA (Rb89), α-FANCL,12 α-FANCF (Rb7), and α-FANCC (Rb39). In panel B, the bottom part of the blot was first probed for FANCL and then subsequently probed for FANCF, thus allowing detection of both proteins that run at a very similar molecular weight, 45 and 42 kDa, respectively. The asterisk in panels B and C mark a nonspecific polypeptide that cross-reacts with the FANCL antiserum. (D) Whole-cell extracts of 10 million WT, FA-L, and respective patient control lymphoblasts were immunoprecipitated using guinea pig antibodies against FANCC6-105, FANCF1-245, and FANCG480-622. Precipitated proteins were detected using rabbit antibodies against FANCA (Rb89), FANCG (Rb43), and FANCF (Rb7). The FANCG protein is indicated with an asterisk. (E) Direct FANCC Western blot on whole-cell extracts of 500 000 lymphoblastoid cells to show the total cellular FANCC levels.
FANCL coimmunoprecipitates with FLAG-tagged FA proteins and is required for the assembly of the FA core complex. Lymphoblastoid cell lines stably expressing functionally active FLAG-tagged FANCA (A), FANCF (B), and FANCC (C) were generated. Whole-cell extracts of 10 million cells were immunoprecipitated with an α-FLAG affinity gel and precipitated proteins eluted by competition with a FLAG peptide. Untransfected cells (lane 1) were included as negative controls. Precipitated proteins were detected with α-FANCA (Rb89), α-FANCL,12 α-FANCF (Rb7), and α-FANCC (Rb39). In panel B, the bottom part of the blot was first probed for FANCL and then subsequently probed for FANCF, thus allowing detection of both proteins that run at a very similar molecular weight, 45 and 42 kDa, respectively. The asterisk in panels B and C mark a nonspecific polypeptide that cross-reacts with the FANCL antiserum. (D) Whole-cell extracts of 10 million WT, FA-L, and respective patient control lymphoblasts were immunoprecipitated using guinea pig antibodies against FANCC6-105, FANCF1-245, and FANCG480-622. Precipitated proteins were detected using rabbit antibodies against FANCA (Rb89), FANCG (Rb43), and FANCF (Rb7). The FANCG protein is indicated with an asterisk. (E) Direct FANCC Western blot on whole-cell extracts of 500 000 lymphoblastoid cells to show the total cellular FANCC levels.
FANCL directly interacts with FANCB
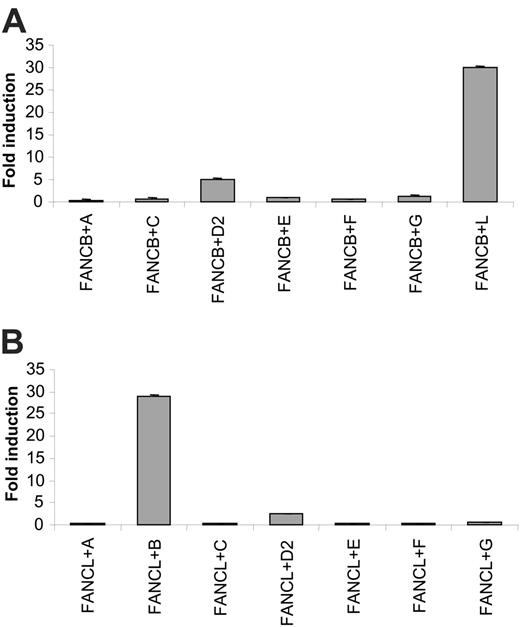
Because FANCL is an essential member of the FA core complex, we further investigated whether there was a direct binding between FANCL and other core complex members using the mammalian 2-hybrid assay. We tested FANCL for direct interactions with other FA proteins when FANCL was fused to the VP16 activation domain (AD). No luciferase reporter gene activation was detected when AD-FANCL was coexpressed with BD-FANCA, -FANCC, -FANCE, -FANCF, and -FANCG (Figure 2A). Weak reporter gene activation was detected when coexpressed with BD-FANCD2, compared with expression of FANCL alone (Figure 2A), but this was due to slight autoactivation of luciferase observed when FANCD2 was expressed by itself (data not shown). When AD-FANCL was coexpressed with BD-FANCB, an approximately 27-fold induction of reporter gene activation over controls was detected, indicating a direct interaction between this protein pair (Figure 2A). We then performed a similar experiment, this time testing FANCB for interaction with other FA proteins. We were unable to find any evidence for a direct interaction between FANCB and FANCA, FANCC, FANCD2, FANCE, FANCF, and FANCG, but we confirmed the direct interaction of FANCB with FANCL (Figure 2B).
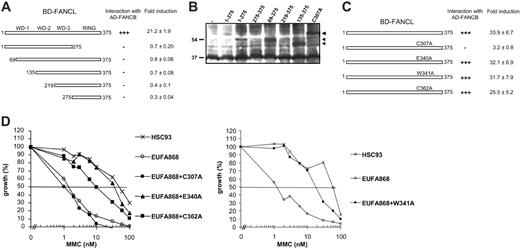
The FANCL sequence contains 3 putative WD40 repeat motifs and a C-terminal RING domain.12 Both of these sequences are thought to be involved in protein-protein interactions, and we investigated whether one or more of these domains were critical for the interaction of FANCB with FANCL. Mutant fragments have previously been used successfully to study interaction domains between FA proteins in yeast 2-hybrid analyses,26-28 and therefore we applied this technique in mammalian cells. A series of FANCL fragments was constructed and tested for interaction with full-length FANCB in the mammalian 2-hybrid assay. None of these fragments activated the luciferase reporter gene when cotransfected with FANCB (Figure 3A), although the majority of the fragments was expressed (Figure 3B). These results indicate that both the N-terminus and the RING domain of FANCL are needed for interaction with FANCB.
Direct interactions between FANCL and FANCB in the mammalian 2-hybrid assay. Protein pairs were cotransfected in 293 cells in the presence of a luciferase reporter construct to test for direct interactions. All experiments were performed in triplicate. (A) AD-FANCL was cotransfected with BD-FA proteins. Fold induction of luciferase expression is relative to FANCL alone. (B) AD-FANCB was cotransfected with BD-FA proteins. Fold induction of luciferase expression is relative to FANCB alone.
Direct interactions between FANCL and FANCB in the mammalian 2-hybrid assay. Protein pairs were cotransfected in 293 cells in the presence of a luciferase reporter construct to test for direct interactions. All experiments were performed in triplicate. (A) AD-FANCL was cotransfected with BD-FA proteins. Fold induction of luciferase expression is relative to FANCL alone. (B) AD-FANCB was cotransfected with BD-FA proteins. Fold induction of luciferase expression is relative to FANCB alone.
Characterizing the regions that are required for direct interaction between FANCB and FANCL. (A) Mammalian 2-hybrid assays coexpressing mutant fragments of BD-FANCL with full-length AD-FANCB. Fold induction of luciferase expression was measured relative to AD-FANCB alone. +++ indicates a strong activation of the luciferase reporter gene; –, no activation of the reporter gene. (B) FANCL constructs that failed to activate the luciferase reporter in the mammalian 2-hybrid experiments were transfected in 293 cells. Whole-cell extracts were immunoblotted with anti–GAL4-BD to show expression of the constructs. The fragments 219-375 and 275-375 are probably difficult to detect because of the presence of background bands. (C) BD-FANCL–containing point mutations of the RING domain were coexpressed with full-length AD-FANCB and fold induction was measured relative to AD-FANCB alone. +++ indicates a strong activation of the luciferase reporter gene; –, no activation of the reporter gene. (D) FANCL missense mutants were transfected in FA-L cell line EUFA868 and tested for their ability to restore the MMC hypersensitive phenotype of this cell line in an MMC growth inhibition test.
Characterizing the regions that are required for direct interaction between FANCB and FANCL. (A) Mammalian 2-hybrid assays coexpressing mutant fragments of BD-FANCL with full-length AD-FANCB. Fold induction of luciferase expression was measured relative to AD-FANCB alone. +++ indicates a strong activation of the luciferase reporter gene; –, no activation of the reporter gene. (B) FANCL constructs that failed to activate the luciferase reporter in the mammalian 2-hybrid experiments were transfected in 293 cells. Whole-cell extracts were immunoblotted with anti–GAL4-BD to show expression of the constructs. The fragments 219-375 and 275-375 are probably difficult to detect because of the presence of background bands. (C) BD-FANCL–containing point mutations of the RING domain were coexpressed with full-length AD-FANCB and fold induction was measured relative to AD-FANCB alone. +++ indicates a strong activation of the luciferase reporter gene; –, no activation of the reporter gene. (D) FANCL missense mutants were transfected in FA-L cell line EUFA868 and tested for their ability to restore the MMC hypersensitive phenotype of this cell line in an MMC growth inhibition test.
Our studies with mutant fragments might indicate that the conformation of the entire FANCL protein is important for its function. Therefore we decided to create point mutations in FANCL, anticipating a more subtle effect on the structure of FANCL. We created 4 different point mutations at conserved residues within the RING domain (C307A, E340A, W341A, and C362A) and used the mammalian 2-hybrid assay to test their ability to bind FANCB. We detected luciferase activation similar to wild-type levels when FANCL-E340A, FANCL-W341A, and FANCL-C362A were coexpressed with FANCB, indicating that these residues are not essential for this interaction (Figure 3C). In contrast, mutant C307A that has been previously described to disrupt the ability of FANCL to act as an ubiquitin ligase, affecting the interaction between FANCA and FANCL, and to inactivate the ability to correct MMC sensitivity of FA-L cells12 was not able to activate the reporter gene when coexpressed with FANCB. This implies that residue C307 is important for the interaction between FANCB and FANCL.
The 3 novel FANCL mutations presented in this study were also tested for their functionality in an MMC growth inhibition test. As shown in Figure 3D, the FANCL mutants E340A, W341A, and C362A were able to complement the MMC sensitivity of FA-L cells, indicating that these mutations do not interfere with the function of FANCL as suggested from their ability to interact with FANCB.
Interaction between FANCA and FANCL is dependent on other FA proteins
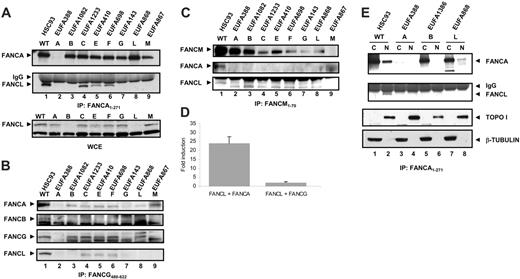
FANCL was initially identified through its association with FANCA, but the mammalian 2-hybrid results indicate that this interaction is probably indirect. This observation led us to ask whether the interaction between FANCA and FANCL was dependent on other FA core complex members. Interactions between members of the FA core complex are retained at close to wild-type levels in cells derived from complementation groups D1, D2, I, and J, therefore we chose to examine the FANCA interaction with FANCL in cells with core complex defects. In whole-cell extracts from WT cells, FANCL clearly coprecipitates with FANCA (Figure 4A, lane 1). This interaction is also detected in cell lines from complementation groups FA-C, FA-E, and FA-F, suggesting that FANCC, FANCE, and FANCF are not essential for the interaction between FANCA and FANCL (Figure 4A, lanes 4-6). In groups FA-B, FA-G, and FA-M, the interaction between FANCL and FANCA was undetectable (Figure 4A, lanes 3, 7, and 9), indicating that FANCB, FANCG, and FANCM either bridge this interaction or are required for the stability of this interaction. Direct Western blotting on whole-cell extracts showed that FANCL levels are strongly reduced in FA-B cells (Figure 4A), suggesting that the direct association between FANCB and FANCL stabilizes the FANCL protein and as such affects the binding between FANCA and FANCL. Likewise, FANCG and FANCM have been shown to stabilize FANCA12,13,24,29 and in that way may influence the interaction between FANCA and FANCL. Immunoprecipitation with an antibody against FANCG showed that FANCA and FANCL coprecipitate with FANCG in WT, FA-C, FA-E, and FA-F lymphoblasts (Figure 4B), indicating that FANCA, FANCG, and FANCL form a stable subcomplex independent of FANCC, FANCE, an FANCF. In contrast, FANCB was coprecipitated only with FANCG from WT cells (Figure 4B) and appears to be weakly associated with the FANCA/FANCG/FANCL subcomplex. Similarly, FANCM coprecipitated only FANCA and FANCL from WT cells (Figure 4C).
The fact that FANCL was coprecipitated with FANCA and FANCG, in a FANCB-dependent way, might suggest that this subset of FA proteins acts “upstream” in the FA pathway. Therefore, we sought to investigate whether there was a direct relationship between the FANCB/FANCL dimer and either FANCA or FANCG. We performed mammalian 3-hybrid assays using 293 cells that were stably expressing FLAG-FANCB. We cotransfected BD-FANCL with both AD-FANCA and AD-FANCG and assayed for luciferase activation. Coexpression of BD-FANCL with AD-FANCG in these cells did not activate the reporter gene over background levels; however, when BD-FANCL and AD-FANCA were expressed, there was a strong activation of the reporter gene almost 25-fold higher than controls (Figure 4D). Therefore we conclude that a complex of FANCB and FANCL is sufficient to interact with FANCA and that this interaction is stabilized by the presence of FANCG.
The interaction between FANCA and FANCL is dependent on other FA proteins and detected in the nucleus. (A) Whole-cell extracts of 10 million lymphoblastoid cells were immunoprecipitated with an antiserum against the N-terminus of FANCA (amino acid 1-271). After blotting, membranes were probed with α-FANCA antibody (Rb89) and α-FANCL antibody12 to show the interaction between FANCA and FANCL. A direct FANCL Western on whole-cell extracts (WCE) of 500 000 lymphoblastoid cells was performed to show the total cellular FANCL levels. (B) Whole-cell extracts of 10 million lymphoblastoid cells were immunoprecipitated with an antiserum against the C-terminus of FANCG (amino acid 480-622). After blotting, membranes were probed with α-FANCA antibody (Rb89), α-FANCB antibody,3 α-FANCG antibody (Rb43), and α-FANCL antibody12 to show the interaction between FANCA, FANCB, FANCG, and FANCL. (C) Whole-cell extracts of 10 million lymphoblastoid cells were immunoprecipitated with an antiserum against the N-terminus of FANCM (amino acid 1-70). After blotting, membranes were probed with α-FANCM antibody,13 α-FANCA antibody (Rb89), and α-FANCL antibody12 to show the interaction between FANCA, FANCL, and FANCM. (D) Mammalian 3-hybrid assay in 293 cells expressing FLAG-FANCB. Cells were cotransfected with the indicated protein pairs and assayed for luciferase activation. Fold induction relative to reporter gene activation when full-length FANCL is expressed alone. Error bars represent SD of 3 experiments. (E) Nuclear and cytoplasmic fractions of lymphoblasts cell lines were immunoprecipitated with an anti-FANCA antibody (as described for panel A). In addition, aliquots of protein lysate were probed with anti–beta tubulin and anti–TOPO I to check integrity of fractionation procedure.
The interaction between FANCA and FANCL is dependent on other FA proteins and detected in the nucleus. (A) Whole-cell extracts of 10 million lymphoblastoid cells were immunoprecipitated with an antiserum against the N-terminus of FANCA (amino acid 1-271). After blotting, membranes were probed with α-FANCA antibody (Rb89) and α-FANCL antibody12 to show the interaction between FANCA and FANCL. A direct FANCL Western on whole-cell extracts (WCE) of 500 000 lymphoblastoid cells was performed to show the total cellular FANCL levels. (B) Whole-cell extracts of 10 million lymphoblastoid cells were immunoprecipitated with an antiserum against the C-terminus of FANCG (amino acid 480-622). After blotting, membranes were probed with α-FANCA antibody (Rb89), α-FANCB antibody,3 α-FANCG antibody (Rb43), and α-FANCL antibody12 to show the interaction between FANCA, FANCB, FANCG, and FANCL. (C) Whole-cell extracts of 10 million lymphoblastoid cells were immunoprecipitated with an antiserum against the N-terminus of FANCM (amino acid 1-70). After blotting, membranes were probed with α-FANCM antibody,13 α-FANCA antibody (Rb89), and α-FANCL antibody12 to show the interaction between FANCA, FANCL, and FANCM. (D) Mammalian 3-hybrid assay in 293 cells expressing FLAG-FANCB. Cells were cotransfected with the indicated protein pairs and assayed for luciferase activation. Fold induction relative to reporter gene activation when full-length FANCL is expressed alone. Error bars represent SD of 3 experiments. (E) Nuclear and cytoplasmic fractions of lymphoblasts cell lines were immunoprecipitated with an anti-FANCA antibody (as described for panel A). In addition, aliquots of protein lysate were probed with anti–beta tubulin and anti–TOPO I to check integrity of fractionation procedure.
In order to find the cellular compartment in which FANCA and FANCL interact, we also performed FANCA immunoprecipitations on nuclear and cytoplasmic extracts of WT, FA-A, FA-B, and FA-L lymphoblastoid cell lines. The interaction between FANCA and FANCL is found only in the nuclear fraction of WT cells (Figure 4E, lane 2) and again not observed in FA-B cells (Figure 4E, lane 6). Furthermore, FANCA is almost exclusively precipitated from the cytoplasm of FA-B and FA-L cells (Figure 4E, lanes 5 and 7), indicating that the nuclear localization of FANCA depends on the interaction with FANCB and FANCL. Since in nuclear extract of FA-L cells some FANCA was found, FANCL seems less essential for the nuclear accumulation of FANCA.
Patient-derived missense mutations in FANCA disrupt the binding to FANCL
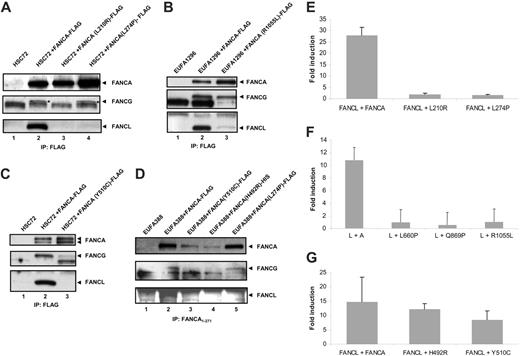
We further investigated the association between FANCA and FANCL using FANCA mutants. Patients with defects in FANCA exhibit many missense mutations, which can help to delineate how and where the core complex is formed. We tested a series of 5 patient-derived pathogenic missense mutations for their effect on binding FANCL using coimmunoprecipitation experiments: 3 novel mutations L210R, L274P, and Y510C, and the previously described mutations H492R and R1055L.30 FA-A lymphoblasts were stably transfected with constructs encoding the FANCA mutants with a FLAG-tag (or HIS-tag) at their C-terminus, and subsequent protein lysates were immunoprecipitated with anti-FLAG or anti-FANCA. The FANCA mutants L210R, L274P, and Y510C were unable to coprecipitate endogenous FANCL (Figure 5A, lanes 3-4, and Figure 5C, lane 3), while FANCL was coimmunoprecipitated with the FANCA mutant R1055L, albeit at strongly reduced levels compared with wild-type FANCA (Figure 5B, lane 3). A lack of interaction between FANCL and the FANCA mutants L274P and Y510C was again found when an antiserum against the N-terminus of FANCA was used and also mutant H492R was unable to interact with FANCL in this assay (Figure 5D, lanes 3-5). In addition, we examined the effect of these mutations on the association between FANCA and FANCL in the mammalian 3-hybrid assay and included the novel pathogenic mutations L660P and Q869P. The mutants L210R, L274P, L660P, Q869P, and R1055L were expressed (data not shown), but were unable to activate the reporter gene, indicating an inability of these proteins to interact directly with the FANCB/FANCL complex (Figure 5E-F). Surprisingly, the mutants H492R and Y510C were able to interact with FANCB/FANCL in the mammalian 3-hybrid assay (Figure 5G), indicating that these mutations do not affect the FANCB/FANCL binding site.
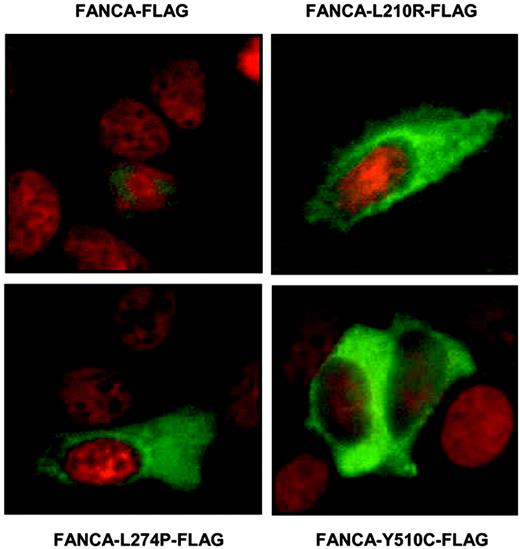
When judging the effect of the FANCA mutations on the FANCL association, there are 2 major differences between a mammalian 3-hybrid assay and an immunoprecipitation experiment that are important to consider. These differences involve the localization and coexpression of the proteins tested. In the mammalian 3-hybrid assay, the mutant proteins are forced to enter the nucleus by the SV40 large T-nuclear localization signal present in the mammalian 3-hybrid constructs, while in immunoprecipitation experiments the nuclear accumulation of the mutants depends on the properties of the mutant protein itself. This could create a problem when the natural interaction (as detected by immunoprecipitation experiments) occurs in the nuclear compartment. In this case, FANCA mutants in which the nuclear accumulation is affected, without changing the FANCB/FANCL interaction site, will score positive in the mammalian 3-hybrid assay and negative in the immunoprecipitation assay. Since we found these apparently conflicting results with FANCA mutants H492R and Y510C, and mutant H492R has been previously described as being expressed exclusively in the cytoplasm,30 we analyzed the subcellular localization of mutant Y510C and the other novel FLAG-tagged FANCA mutants in transiently transfected MCF-7 cells. Indirect immunofluorescence of FLAG-FANCA with an anti-FLAG antibody displayed a predominantly nuclear localization of the wild-type FANCA protein, while the FANCA mutants, L210R, L274P, and Y510C were all found to be retained in the cytoplasm (Figure 6). These results indicate that the mutants H492R and Y510C have lost their ability to interact with FANCL because of their failure to accumulate in the nucleus, rather than because they affect the FANCL interaction site. These results also strengthen the idea that FANCA interacts with FANCL in the nuclear compartment.
Interaction of FANCA mutants with the FANCB/FANCL complex and with FANCG. Stable cell lines were generated expressing FANCA protein containing patient-derived missense mutations and immunoprecipitation with an α-FLAG affinity gel (A-C), or an antiserum against the N-terminus of FANCA (D) was performed. Western blots were probed for FANCA (Rb89), FANCG (Rb43), and FANCL.12 The FANCG protein is indicated with an asterisk. (E-G) Mammalian 3-hybrid assay in 293 cells expressing FLAG-FANCB. Cells were cotransfected with the indicated protein pairs and assayed for luciferase activation. Fold induction relative to reporter gene activation when full-length FANCL is expressed alone. (E-G) Error bars represent SD of 3 experiments.
Interaction of FANCA mutants with the FANCB/FANCL complex and with FANCG. Stable cell lines were generated expressing FANCA protein containing patient-derived missense mutations and immunoprecipitation with an α-FLAG affinity gel (A-C), or an antiserum against the N-terminus of FANCA (D) was performed. Western blots were probed for FANCA (Rb89), FANCG (Rb43), and FANCL.12 The FANCG protein is indicated with an asterisk. (E-G) Mammalian 3-hybrid assay in 293 cells expressing FLAG-FANCB. Cells were cotransfected with the indicated protein pairs and assayed for luciferase activation. Fold induction relative to reporter gene activation when full-length FANCL is expressed alone. (E-G) Error bars represent SD of 3 experiments.
Another important difference between a mammalian 3-hybrid assay and an immunoprecipitation experiment is the fact that in a mammalian 3-hybrid assay only 3 FA proteins are overexpressed, whereas in an immunoprecipitation experiment all FA proteins are present at endogenous levels. These additional FA proteins are able to stabilize interactions in immunoprecipitation experiments. One important FA protein in this respect is FANCG, because no interaction between FANCA and FANCL was detected in cells that lack FANCG (Figure 4A). Therefore, we also investigated the coimmunoprecipitation of FANCG with the FANCA mutants. The mutant L210R is unable to bind FANCG (Figure 5A, lane 3), suggesting that this mutation interferes with the binding of FANCG to the N-terminus of FANCA. The mutants H492R (Figure 5D, lane 4) and Y510C (Figure 5C, lane 3, and Figure 5D, lane 3) showed a reduced interaction with FANCG compared with wild-type FANCA, whereas mutants L274P and R1055L bind FANCG like wild-type FANCA (Figure 5A, lane 4, Figure 5B, lane 3, and Figure 5D, lane 5). These results suggest that the R1055L mutant, which is able to accumulate in the nucleus,30 can coprecipitate FANCL despite a strongly reduced affinity (as measured in the mammalian 3-hybrid assay), because FANCG partially stabilizes this interaction. The mutation L274P might cause a change in the conformation of FANCA that affects both nuclear accumulation and interaction with FANCB/FANCL. The results of the different assays are summarized in Table 1.
Cytoplasmic expression of FLAG-tagged FANCA mutant proteins in transiently transfected MCF-7 cells by indirect immunofluorescence. The different FLAG-tagged FANCA mutant proteins are shown in green. The counterstaining with Hoechst is shown in red to visualize the nuclei.
Cytoplasmic expression of FLAG-tagged FANCA mutant proteins in transiently transfected MCF-7 cells by indirect immunofluorescence. The different FLAG-tagged FANCA mutant proteins are shown in green. The counterstaining with Hoechst is shown in red to visualize the nuclei.
Summary of the characteristics of different FANCA mutants
Mutant . | Interaction L* . | Interaction B/L† . | Interaction G* . | Localization . | Possible defect . |
|---|---|---|---|---|---|
| L210R | — | — | — | C | Interaction FANCG |
| L274P | — | — | + | C | Conformation |
| H492R | — | + | ± | C‡ | Phosphorylation |
| Y510C | — | + | ± | C | Phosphorylation |
| R1055L | ± | — | + | C + N‡ | Interaction B/L |
Mutant . | Interaction L* . | Interaction B/L† . | Interaction G* . | Localization . | Possible defect . |
|---|---|---|---|---|---|
| L210R | — | — | — | C | Interaction FANCG |
| L274P | — | — | + | C | Conformation |
| H492R | — | + | ± | C‡ | Phosphorylation |
| Y510C | — | + | ± | C | Phosphorylation |
| R1055L | ± | — | + | C + N‡ | Interaction B/L |
—indicates no interaction; C, predominantly cytoplasmic; +, normal interaction; ±, weak interaction; and C + N cytoplasmic and nuclear.
Immunoprecipitation.
Mammalian 3-hybrid assay.
Described by Adachi et al.30
Discussion
The upstream part of the Fanconi anemia pathway is currently known to involve the formation of a multisubunit protein complex, which is essential for the monoubiquitination of FANCD2. However, the sequence of events required to form this nuclear core complex has not been fully investigated. This is the first study that reveals a role for the recently identified FANCB and FANCL proteins in the assembly of this complex. We found that FANCB and FANCL directly interact and that a heterodimer of these proteins has the ability to bind FANCA, thus providing a direct link with the other FA proteins. FANCB and FANCL are essential for the nuclear localization of FANCA and for the assembly of the FA core complex. Furthermore, we show that the interaction between FANCA, FANCG, and FANCL is independent of FANCC, FANCE, and FANCF, suggesting the formation of different subcomplexes that may have specific functions in the FA pathway.
Despite the characterization of many interactions within the FA core complex, little is known about the order of assembly. Nuclear accumulation of the FA proteins is required for an active FA pathway; however, several FA proteins reside within the cytoplasm. Partial localization of a subset of FA proteins within the cytoplasmic compartment is probably representative of an initial stage of the FA pathway. Our previous findings, along with data presented in this study show that FANCA, FANCB, FANCG, and FANCL are all partially cytoplasmic. The requirement for one another with respect to protein stability and nuclear localization supports the hypothesis that this group of proteins forms a subcomplex in the FA pathway. When studying the interaction between FANCA, FANCG, and FANCL in multiple complementation groups, we found that binding is retained in cells derived from complementation groups FA-C, FA-E, and FA-F, which suggests that the proteins defective in these groups may act “downstream” in core complex formation. In addition, cells derived from FA-C, FA-E, and FA-F patients show that the nuclear accumulation of FANCA is not impaired,31 yet in FA-B and FA-L cells FANCA is almost exclusively cytoplasmic. In support of this downstream role, it has recently been shown that a dimer of FANCE and FANCC can directly interact with FANCF.25 These 3 FA proteins might represent another subcomplex that associates with FANCA, -B, -G, and -L after their accumulation in the nuclear compartment. Our observations shed new light on the FA pathway, because they suggest a sequential assembly of the FA core complex and may leave room for specific functions of these subcomplexes in the FA pathway. Of interest, the interaction between FANCA, FANCG, and FANCL is dependent on a novel FA protein (FANCM), which has a predominant nuclear localization.13 Cells that lack FANCM have markedly reduced FANCA and FANCG levels, while FANCL levels are modestly reduced.13 This suggests that the FA core complex assembles in the nucleus around the FANCM protein starting with the FANCA/FANCG complex. Immunoprecipitation experiments with FANCA13 and FANCM antiserum (this study) suggest that FANCM weakly associates with the FANCA/FANCG/FANCL subcomplex and needs the other core complex proteins for a stable association.
Studies have been performed that have gone some way to characterize the domains required for FA protein-protein interactions and nuclear accumulation. There is evidence that the region of FANCA that binds FANCG overlaps the bipartite NLS sequence26,28,32 ; therefore it is conceivable that this interaction blocks the NLS, thus retaining FANCA in the cytoplasm. All the patient-derived FANCA missense mutations described to date, except for mutant R1055L, have lost their ability to accumulate in the nucleus,30 but have retained their potential to interact with FANCG in coimmunoprecipitation experiments. This suggests that the FANCA mutants are unable to expose their NLS by displacing FANCG from the NLS. Possibly, a conformational change in FANCA has to alter the FANCG binding site and has to translocate FANCG to another region in the FANCA protein. A candidate region for FANCG binding is the C-terminus of the FANCA protein, which has been shown to be important for the nuclear accumulation of FANCA but does not contain a nuclear localization signal.33 This potential conformational change may be induced by phosphorylation of FANCA. The mutations H492R and Y510C are located in a highly conserved region of the FANCA protein and border a potential SH3 binding domain (PxxP). It is tempting to speculate that this region binds the cytoplasmic kinase that has been proposed to phosphorylate FANCA,34,35 which then may change the conformation of the FANCA protein and induce its nuclear translocation.
The observation that there is a strong direct interaction between FANCB and FANCL (Figure 2), and that they appear to be required for the stability of each other, is reminiscent of the interaction between FANCA and FANCG and between FANCC and FANCE. Certainly one role for FANCB appears to be the stabilization of FANCL. It seems that the overall conformation of the FANCL protein is important for the interaction with FANCB, since we were unable to show any interaction with deletion mutants. Also the disruption of the FANCL RING domain by changing the first cysteine (C307) of this motif into an alanine interfered with the ability to bind FANCB and restore the MMC hypersensitivity of FA-L cells. Of interest, mutagenesis of the last cysteine (C362) of the RING domain did not interfere with the binding of FANCB and only slightly affected the ability to complement FA-L cells. Possibly, a mutation in this amino acid may have a minimal affect on the RING domain, without affecting the total structure of the protein.
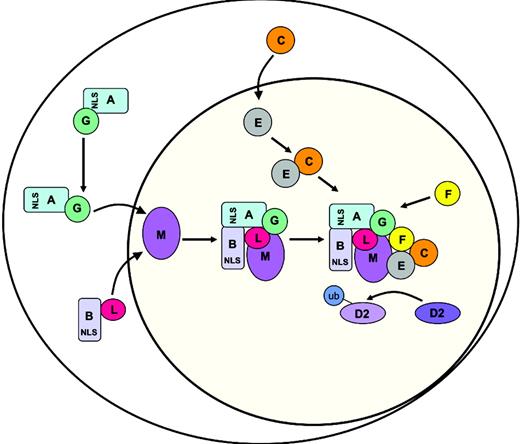
A model for the sequential assembly of the nuclear FA core complex. The model is explained in “Discussion.”
A model for the sequential assembly of the nuclear FA core complex. The model is explained in “Discussion.”
Combining the data we present here with previously reported findings, we propose our working model for the assembly of the FA core complex (Figure 7). Our results suggest a sequential assembly of the FANCA/FANCG and FANCB/FANCL subcomplexes around FANCM, because in cells that do not express the nuclear FANCM protein no interaction between FANCA, FANCG, and FANCL is found. These subcomplexes are probably imported to the nucleus independently, since the interaction between FANCA and FANCL is found only in nuclear fractions, and FANCA mutants that have lost their ability to accumulate in the nucleus are not able to interact with FANCL in coimmunoprecipitation experiments. The nuclear accumulation of the FANCA/FANCG subcomplex may be regulated by phosphorylation of FANCA, which could involve the highly conserved potential SH3 binding domain (PxxP) close to the mutations H592R and Y510C. In the absence of the FANCB/FANCL subcomplex, the FANCA/FANCG subcomplex is possibly exported from the nucleus through the nuclear export signals present in FANCA.36 In the presence of the FANCB/FANCL subcomplex, FANCA, FANCB, FANCG, and FANCL form a complex around FANCM in which FANCG stabilizes the interaction between FANCA and FANCL. FANCB and FANCM appear to be weakly associated with this subcomplex. We propose that FANCA and FANCB are important for regulating the nuclear accumulation of FANCL whereby it can associate with the core complex. Complex assembly tethers the pathways E3 ligase (FANCL) to its substrate (FANCD2) via its interaction with a subcomplex of FANCE and FANCC. The FANCF protein seems to stabilize the entire core complex by interaction with the FANCC/FANCE and FANCA/FANCG subcomplexes.25 The requirement for multiple proteins in this pathway seems indicative of a highly regulated and complex process for the handling and/or repair of interstrand cross-links. It remains to be seen whether these subcomplexes have a specific function in this process or even in separate processes.
Prepublished online as Blood First Edition Paper, May 23, 2006; DOI 10.1182/blood-2005-11-008151.
Supported by the Dutch Cancer Society, The Netherlands Organisation for Health Research and Development, the Intramural Research Program of the NIH, National Institute on Aging, and the Fanconi Anemia Research Foundation.
A.L.M. designed research, performed research, and wrote the paper; E.H.L., J.S., M.F., C.F., J.d.G., and M.A.R. performed research; R.J.S., A.R.M., and W.W. contributed vital new reagents; H.J. controlled data; and J.P.d.W. designed research, controlled data, and wrote the paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank K. J. Patel for the mammalian 2-hybrid constructs and Eric Blom for critically reading the paper.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal