Abstract
We have investigated whether the signaling protein phospholipase D is implicated in leukocyte cell motility. Treating differentiated HL-60 cells with small interfering RNAs (siRNAs), to deplete endogenous expression of the PLD1 isoform, led to an abolishment of basal chemokinesis that could not be rescued with chemoattractants ENA-78, FMLP, and IL-8. Transient overexpression of PLD1 increased both chemokinesis and chemotaxis toward IL-8 and FMLP but not toward ENA-78. Chemokinesis was not mediated by the enzymatic activity of PLD1, but the chemotactic response was, because a lipase-inactive mutant (PLD1-K830R) negated all chemokine-induced potentiating actions and because IL-8 and FMLP increased activity in vitro. Gene expression silencing of the other mammalian isoform, PLD2, also led to cell migration arrest, whereas ENA-78 selectively increased endogenous PLD2 activity and chemotaxis of HL-60 cells overexpressing a myc-pcDNA-PLD2 construct. Thus, PLD1 is differentially activated by CXCR-1, whereas CXCR-2 (and possibly CXCR-1) mediates PLD2 activation. Finally, immunofluorescence microscopy showed that both isoforms were associated with cell polarity and directionality concomitantly with adhesion and F-actin polymerization in response to IL-8. These data represent the first demonstration of the involvement of PLD and its enzymatic activity toward chemokines in the key physiologic process of leukocyte migration.
Introduction
Inflammation, wound repair, and angiogenesis have in common an initial physiologic event of cell migration or chemotaxis. Related pathologic processes, such as chronic inflammation, atherosclerosis, and cancer metastasis, are also heavily dependent on cell chemotaxis. In the case of normal leukocyte function, cell migration begins with the reorientation and alignment of the cells (polarization) in the direction of the inflammation site followed by the directional migration (chemotaxis) toward host- or pathogen-derived chemical stimuli (chemoattractants).
Known major neutrophil chemoattractants are the tripeptide FMLP, the lipids LTB4 and PAF, the activated complement protein C5a,1,2 and a group of cytokines collectively known as ELR+ CXC chemokines.3-6 These chemokines are characterized by the invariable presence of the Cys-X-Cys (CXC) consensus motif in the N-terminus of the protein, which is preceded by the amino acid sequence Glu-Leu-Arg (ELR). Classical neutrophil ELR+ CXC chemokines are IL-8 (CXCL8); ENA-78 (CXCL5); GROα, GROβ, and GRIγ; NAP-2; and GCP-2. They all induce cytosolic calcium changes, chemotaxis, and exocytosis3 and recruit neutrophils by binding and activating a specific class of receptors, called CXCR-2.7 Apart from binding to CXCR-2, IL-8 and GCP-2 also bind to another class of receptors, the CXCR-1.4,8
Neutrophil chemotaxis depends on PI3Kγ and Akt/PKB,9-11 whereas FMLP fails to induce cell directionality in PI3Kγ-deprived neutrophils.12 Pharmacologic studies, however, do not indicate an absolute requirement for PIP3.13,14 However, not all neutrophil or
Dictyostelium chemotaxis can be explained solely on the intracellular requirement for PI3K or the companion phosphatase PTEN. This leaves open the possibility of the participation of phosphoinositide-independent pathways.15 As further recognized by the same researchers, despite all the advances in this area, our understanding of the complexity of the signaling pathways that regulate cell migration might still be limited.
In attempting to fill this gap, we have investigated whether the signaling protein phospholipase D is implicated in leukocyte cell motility, and, if so, to what extent. Members of the PLD family (reviewed in Cockcroft,16 Exton,17 Gomez-Cambronero and Keire,18 Liscovitch et al,19 and Morris et al20 ) extend from prokaryotes to eukaryotes and have been associated with the biology of the cytoskeleton, which prompted us to formulate the hypothesis of a possible involvement of PLD in chemotaxis. Human PLDs are products of 2 genes, PLD1 and PLD2, and sufficient knowledge exists at the molecular level to analyze their signal transduction mechanisms of action.
We have taken advantage of a transfectable model of leukocytes: the human promyelocyte leukemia HL-60 cell line that can be induced to express the neutrophilic phenotype (dHL-60) with a demonstrated capability of response toward chemokines, as well as the normal cell counterpart (peripheral blood neutrophil) needed for validation assays. In this phagocyte dHL-60/neutrophil model, we have demonstrated for the first time a link between the PLD1 and PLD2 isoforms and cell chemotaxis.
Materials and methods
Materials
PC8, PA, anti-goat IgG (agarose beads), and TRITC were from Sigma (St Louis, MO); n-[1-3H]-butanol (5 Ci/mmol [18.5 × 1010 Bq/mmol]) was purchased from American Radiolabeled Chemicals (St Louis, MO); 1,2-dioleoyl-sn-glycero-3-phosphatidyl-butanol (PBut) lipid standard and cell-permeable 1,2-dioleoyl-sn-glycero-3-phosphate (di-oleoyl PA, or DO-PA) were from Avanti Polar Lipids (Alabaster, AL); polyclonal anti-PLD1 (F-12) and anti-PLD2 (N-20) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA); anti-HA and anti-myc antibodies were from Cell Signaling (Beverly, MA); electrophoresis chemicals, 96-well polymerase chain reaction (PCR) plates and iQ Supermix were from Bio-Rad Laboratories (Richmond, CA); phCMV2-HA vector (4.26 kb [kilobase]) was from Gene Therapy Systems (San Diego, CA); M-MLV reverse transcriptase, RNase inhibitor dNTPs, negative siRNA, and PLD1 (ex10) and PLD2 Silencer Pre-designed siRNAs were from Ambion (Austin, TX); SMARTs-election PLD1 siRNA (ex21) were from Dharmacon (La Fayette, CO); the RNeasy minikit and the Qiaquick gel purification kit were from Qiagen (Valencia, CA); PLD1/2 Taqman gene expression assays were from Applied Biosystems (Foster City, CA).
Plasmids and mutagenesis
A pcDNA3-PLD1b wild-type construct21 was modified as follows. The ORF of human PLD1b (RefSeq NM_002662) was first HA-tagged by direct subcloning into Xho and PspOM of a phCMV2-HA vector (Gene Therapy Systems, San Diego, CA), producing phCMV2-HA-PLD1b (hereafter named HA-PLD1 for simplicity). HA-PLD1 was mutagenized22 in the second HKD catalytic domain (as originally described in Chen et al23 ) to derive a lipase-inactive mutant (PLD1-K830R). The pcDNA3-myc-PLD2a wild-type construct (hereafter named myc-PLD2 for simplicity) has been characterized previously24 and was used in this study without modification.
Real-time (quantitative) RT-PCR (QRT-PCR)
Total RNA was isolated from dHL-60 cells with the RNeasy minikit. RNA concentrations were determined by fluorometric assay with Packard Fusion and Ribogreen RNA Quantitation Kit, and samples were normalized to 20 ng/μL RNA. Reverse transcription (RT) was performed with 210 ng RNA, 210 ng random hexamers, 500 μM dNTPs, 84 units RNase out and 210 units of M-MLV reverse transcriptase and incubated at 42°C for 55 minutes. Q-PCR reactions were run with 30 ng total input RNA (5 μL), 1 μL PLD1 gene expression assay (FAM-labeled), or 2 μL PLD2 gene expression assay (FAM-labeled) multiplexed with the housekeeping gene (β-tubulin) (Texas Red-labeled) with the final concentrations being 200 pmol and 400 pmol for the primers and probe, respectively. Sequences were as follows: PLD1 probe, FAM-5-ACG AAA AGC ACA ACA AGG AGT GAG G; PLD2 probe, FAM-5-CCT TGG CCC GGT GGT TTG TGA ATG G. β-Tubulin primers and probe were designed by our laboratory using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi), and the sequences were as follows: sense, 5′-AGT ACC CAG ACC GCA TCA TGA ACA-3′; antisense, 5′-ACA GGG CCT CGT TAT CAA TGG AGT-3′; probe, 5′-/Texas Red/TTC AGC GTC ATG CCC TCA CCC AAG GT/Black Hole Quencher 2/-3′. β-Tubulin primers and fluorescent probe were synthesized by Integrated DNA Technologies (Coralville, IA). Q-PCR conditions for the Bio-Rad i-cycler were 95°C for 3 minutes and then 50 cycles of the next 3 steps: 30 seconds at 95°C, 1 minute at 60°C, and then 1 minute at 72°C. The cycle threshold (Ct) values were arbitrarily chosen from the linear part of the PCR amplification curve where an increase in fluorescence can be detected 10 or more SEM above the background signal. ΔCt was calculated as ΔCt = Avg PLD Ct - Avg Housekeeping Ct and gene fold expression as 2-(ΔΔCt) = 2-(experimental Condition ΔCt - Control ΔCt).
Peripheral blood neutrophils, HL-60 cell differentiation, and plasmid transfection
Neutrophils were isolated from peripheral blood of volunteer donors, who signed an institutional review board-approved consent form. Promyelocytic leukemic HL-60 cells were grown at 37°C in a 5% CO2 incubator in Iscove Modified Eagle Media (IMEM) + 20% (vol/vol) heat-inactivated fetal bovine serum, 2 mM l-glutamine. Cell density was maintained between 0.1 and 1.0 × 106/mL. HL-60 cells were induced to differentiate (to dHL-60) with 1.75% (vol/vol) DMSO for 3 to 4 days to achieve the expression of the neutrophilic phenotype. Viability assays were routinely conducted with 0.4% trypan blue stain in cell preparations prior to all analyses and were greater than 95%. The assessment of cell differentiation was performed by flow cytometric analysis of surface expression of differentiation-related antigens as indicated in Lehman et al.25 One day after induction of differentiation, cells were transiently transfected with either HA-PLD1 or myc-PLD2 plasmid constructs using lipofectamine as the DNA carrier.23
PLD gene expression silencing and nucleofection
Two different sets of silencer siRNAs (at 100 nM) were used, one from Ambion (Silencer Pre-designed) that targets PLD1 on exon 10 (sense sequence, GGC AAA UGA AGA GAU UUU UTT) and the other from Dharmacon (La Fayette, CO) (SMARTselection) that targets exon 21 on PLD1 (sense sequence, UAA CUG AGC UUA UCU AUG UUU). siRNA targeting PLD2 (exon 15) was from Ambion (sense sequence, GGA CUA CAG CAA UCU UAU CTT). A negative control for siRNA was also from Ambion. Neg-siRNA#2 is a 19-bp scrambled sequence with 3′ dT overhangs (sequence not disclosed by Ambion) certified not to have significant homology to any known gene sequences from mouse, rat, or human and causes no significant changes in gene expression of transfected cells after 48 hours at the same concentration as the siRNA in test. The protocol for gene silencing involved first induction of differentiation of HL-60 cells with 1.75% DMSO on day 0. Then, on day 1, cells were transfected with siRNA. To initiate the transfection, siRNA was mixed with OptiMEM and incubated at room temperature for 5 minutes. siRNAs were added to an Amaxa (Gaithersburg, MD) electroporation cuvette with 2 × 106 HL-60 cells/100 μL in nucleofector solution V. Cells were zapped in program T-019 (as per the manufacturer's instructions) after which they were resuspended in 1 mL IMEM + 20% FBS + 1.75% DMSO (to allow cells to continue differentiation). Cells were immediately plated into 6-well plates and incubated at 37°C for 48 hours to allow for maximum gene expression silencing. Finally, on day 3, the cells were harvested and used for the experiments described herein.
PLD immunocomplex enzymatic activity
PLD enzymatic activity was quantified by an immunocomplex assay as reported previously26 with some modifications. Cells were resuspended in 0.2 mL diluted Triton X-100-based lysis buffer to preserve enzyme activity (5 mM HEPES, pH 7.8, 1 μM leupeptin, 768 nM aprotinin, and 0.5% Triton X-100) and sonicated briefly. Samples were immunoprecipitated overnight with either anti-PLD1 or anti-PLD2 antibodies, and the resulting immunocomplex beads were added to 1.5 mL Eppendorf tubes containing (final concentrations) 4.5 mM PC8 phospholipid, 75 mM HEPES, pH 7.8, and 1.6 μCi (59.2×103 μBq) [3H]-butanol in a liposome form. Samples were incubated for 20 minutes at 30°C with continuous shaking. Lipids were isolated, and [3H]PBut was separated out by thin-layer chromatography (TLC). Silica gel sections that migrated with PBut authentic standards were counted by scintillation spectrometry. Controls were run in parallel with all reagents except PC8, and counts were subtracted from test samples.
Immunofluorescence microscopy
HL-60 cells in suspension were attached to microscope slides by centrifugation (800g, 5 minutes) using a Shandon cytospin-4 centrifuge (Thermo Electron, Pittsburgh, PA). Neutrophils were allowed to adhere naturally to microscope slides placed on 6-well tissue culture plates and incubated with specific chemokines directly added into the chamber (upper-right corner) and with minimal disturbance to create a temporary gradient. Cells were then processed for immunofluorescence as in Gomez-Cambronero et al27 with 2 μg/mL of either anti-HA or anti-myc antibodies and with 0.5 μg/mL FITC-conjugated secondary antibody. For triple labeling, the dyes were TRITC-phalloidin, FITC-secondary Ab, and DAPI. Coverslips were mounted in Vectashield (Vector Laboratories, Burlingame, CA) and used for microscopy. For confocal microscopy, coverslips were examined on an Olympus Fluoview FX Laser scanning confocal microscope (Olympus, Melville, NY) with a 100×/1.35 objective lens. Images were acquired with Fluoview 5.0 software (Olympus) and analyzed by Image Pro Plus 5.1 software (Media Cybernetics, Silver Spring, MD); processing was performed with Adobe Photoshop 8.0 software (Adobe Systems, San Jose, CA). For epifluorescence microscopy, coverslips were examined on a Nikon Eclipse 50i microscope (Nikon, Tokyo, Japan) equipped with a Planfluor 100×/1.30 objective lens and a Spot 6 Insight camera (Diagnostic Instruments, Sterling Heights, MI). Images were acquired with MetaVue v. 6.1r0 (Molecular Devices, Downington, PA) and processed with CorelDraw 12 software (Corel, Ottawa, ON, Canada).
Cell migration assay
dHL-60 or neutrophils were resuspended at the density of 5 × 105 cells/mL or 5 × 106 cells/mL, respectively, in chemotaxis buffer (RPMI + 0.5% BSA).28 Two hundred micoliters were placed in the upper chambers (or inserts) of Transwell plates that are separated from the lower wells by a 6.5-mm diameter, 5-μm pore Nucleopore polycarbonate membrane (VWR International, West Chester, PA). For the study of chemotaxis, IL-8, ENA-78, or FMLP was added separately in 500 μL chemotaxis buffer to the lower wells of Transwell plates. For the study of random cell migration/chemokinesis, no chemoattractant (vehicle alone) was added to the bottom wells. In either case, the Transwell plates were incubated for 1 hour at 37°C under a 5% CO2 atmosphere. The number of cells that migrated to the lower wells was calculated by placing 10-μL aliquots on a hemocytometer and counting 4 fields in duplicate.
Directionality assay
For directionality assays, IL-8 was added at a precise location on a microscope slide of adherent neutrophils with a 200-μm micropipette to form a temporary gradient. Migration was allowed to occur for 10 minutes, and then slides were fixed and processed for immunostaining with anti-PLD antibody as indicated above, under “Immunofluorescence microscopy.” After this, fields of 23 to 26 cells lying within approximately 15 mm from the added cytokine were selected for microscope quantitation. Cells exhibiting polarization caps that were pointing within a 90° swath toward IL-8 were counted, and the percentage over total number of cells was calculated.
Statistical analysis
Data are presented as the mean ± SEM. The difference between means was assessed by the Single Factor Analysis of Variance (ANOVA) test. Probability of P less than .05 was considered to indicate a significant difference.
Results
PLD1 gene expression knock-down reduces chemokinesis and chemotaxis
To test whether PLD1 plays a functional role in leukocyte motility, double-stranded siRNAs were used to deplete PLD1 gene expression of dHL-60 cells (Figure 1). This was followed by an analysis of chemokinesis (cells displaying stochastic movement, or, experimentally, those cells that are found in the bottom well of the Transwell plates in the absence of any stimuli) and by an analysis of chemotaxis (cells moving directionally toward a stimulus that, experimentally, is placed in the bottom well of Transwell plates).
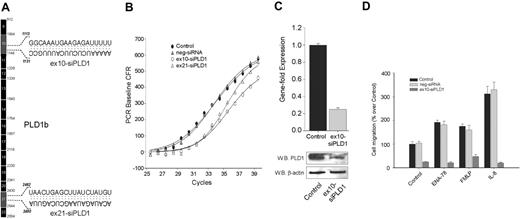
Two siRNAs, targeting exons 10 (ex10-siPLD1) and 21 (ex21-siPLD2) of PLD1b (Figure 1A) were initially used. Validation of siRNA-mediated knockdown in neutrophilic dHL-60 cells was performed by QRT-PCR (Figure 1B). A shift to the right in the sigmoidal curve with a ΔCt of 2.2 represents an approximate 78% gene expression knockdown by either ex10-siPLD1 or ex21-siPLD2. Because the effect was very similar, subsequent studies were carried out with only ex10-siPLD1. The silencing effect was isoform specific, because PLD1 siRNA duplex transfection did not affect the mRNA levels of PLD2 (data not shown), and a negative control siRNA showed no effect (Figure 1B). The level of endogenous PLD1 was found depleted with siRNA PLD1 by Western blot analysis (Figure 1C). Further, siPLD1 (but not negative control siRNA) knocked-down dHL-60 cells exhibited greater than 75% inhibition of random migration (Figure 1D) that paralleled the level of gene silencing, and that could not be rescued even in the presence of classical granulocyte chemoattractants, such as ENA-78, FMLP, and IL-8. This indicates that chemotaxis toward chemokines IL-8 and ENA-78 or phagocyte chemoattractant FMLP depends on the presence of an endogenous expression of PLD1.
Effect of PLD1 gene expression knock-down on cell migration. (A) cDNA map of PLD1b showing the exons to which 2 duplex siRNAs target. (B) PLD1-siRNAs and a negative control siRNA were transfected into dHL-60 cells, RNA was isolated and used to analyze gene expression by QRT-PCR using a FAM-labeled PLD1 probe multiplexed with a Texas-Red-labeled housekeeping gene. (C) Gene fold expression was calculated from ΔCt as indicated in “Materials and methods” (error bars are SEM with n = 3 in duplicate). Duplicate cell samples were used to generate cell lysates for the detection of protein expression by Western blotting. Blots were cut in half, the upper part (containing an ∼ 110-kDa region) was probed with anti-PLD1 antibodies, whereas a lower part (containing an ∼ 45-kDa region) was probed with anti-β-actin, to ascertain equivalent protein loading. (D) Analysis of chemokinesis/chemotaxis. Cells were resuspended in RPMI-based chemotaxis buffer at a 5 × 105 cells/mL density and placed on the upper or insert chambers of 6.5-mm Transwell plates. Appropriate bottom wells contained either buffer only, 30 nM ENA-78, 50 nM FMLP, or 12 nM IL-8; 100% represents 1.3 × 104 ± 0.15 cells/mL migrating to the bottom wells.
Effect of PLD1 gene expression knock-down on cell migration. (A) cDNA map of PLD1b showing the exons to which 2 duplex siRNAs target. (B) PLD1-siRNAs and a negative control siRNA were transfected into dHL-60 cells, RNA was isolated and used to analyze gene expression by QRT-PCR using a FAM-labeled PLD1 probe multiplexed with a Texas-Red-labeled housekeeping gene. (C) Gene fold expression was calculated from ΔCt as indicated in “Materials and methods” (error bars are SEM with n = 3 in duplicate). Duplicate cell samples were used to generate cell lysates for the detection of protein expression by Western blotting. Blots were cut in half, the upper part (containing an ∼ 110-kDa region) was probed with anti-PLD1 antibodies, whereas a lower part (containing an ∼ 45-kDa region) was probed with anti-β-actin, to ascertain equivalent protein loading. (D) Analysis of chemokinesis/chemotaxis. Cells were resuspended in RPMI-based chemotaxis buffer at a 5 × 105 cells/mL density and placed on the upper or insert chambers of 6.5-mm Transwell plates. Appropriate bottom wells contained either buffer only, 30 nM ENA-78, 50 nM FMLP, or 12 nM IL-8; 100% represents 1.3 × 104 ± 0.15 cells/mL migrating to the bottom wells.
Overexpression of PLD1 in HL-60 cells increases random cell migration and chemotaxis. (A) Undifferentiated, exponentially growing HL-60 cells (uHL-60) or DMSO-treated cells (dHL-60) were mock-transfected or transfected with WT HA-PLD1 (1 μg plasmid DNA), and cell migration was assayed on Transwell plates. The controls were cells transferred into Transwell plates immediately after (0 day) mock transfection. (B) Confocal immunofluorescence of transfected dHL-60 cells. The brightfield view is shown for mock-transfected cells, which had low fluorescence signal despite being incubated with the same anti-HA antibodies, as were the WT-transfected cells. Confocal epifluorescence micrographs represent single optical sections of a representative cell. (C) dHL-60 cells either mock- or WT HA-PLD1-transfected were assayed for their chemotactic responses; 100% represents 1.2 × 104 ± 0.1 cells/mL. The mean ± SEM from 3 independent experiments performed in duplicate are shown.
Overexpression of PLD1 in HL-60 cells increases random cell migration and chemotaxis. (A) Undifferentiated, exponentially growing HL-60 cells (uHL-60) or DMSO-treated cells (dHL-60) were mock-transfected or transfected with WT HA-PLD1 (1 μg plasmid DNA), and cell migration was assayed on Transwell plates. The controls were cells transferred into Transwell plates immediately after (0 day) mock transfection. (B) Confocal immunofluorescence of transfected dHL-60 cells. The brightfield view is shown for mock-transfected cells, which had low fluorescence signal despite being incubated with the same anti-HA antibodies, as were the WT-transfected cells. Confocal epifluorescence micrographs represent single optical sections of a representative cell. (C) dHL-60 cells either mock- or WT HA-PLD1-transfected were assayed for their chemotactic responses; 100% represents 1.2 × 104 ± 0.1 cells/mL. The mean ± SEM from 3 independent experiments performed in duplicate are shown.
Overexpression of PLD1 increases chemokinesis and chemotaxis
Transfection of either undifferentiated HL-60 (uHL-60) or differentiated, neutrophil-like cells (dHL-60) with a WT HA-PLD1 construct significantly increased chemokinesis in both cell populations in a time-dependent fashion, with 3 days being sufficient to show significant increase in cell function (Figure 2A). In uHL-60, the number of migrating cells increased approximately 2.1-fold under the influence of PLD1, whereas in dHL-60, the increase was approximately 2.4-fold over mock-transfected cells.
A further control for the transfection strategy of dHL-60 cells with HA-PLD1 involved immunofluorescence microscopy. Figure 2B indicates that fluorescent staining was diffusely present throughout the cytoplasm, as well as a concentrated ring around the cell nucleus. We next investigated the chemotactic response of these transfected cells. dHL-60 cells overexpressing PLD1, with the demonstrated enhancement of basal chemokinesis seen in Figure 2A, responded readily to chemoattractants (Figure 2C). FMLP and IL-8 were able to increase the already high response to 3.8- and 2.7-fold, respectively, whereas ENA-78 failed to do so. In conclusion, these data indicate that PLD1 overexpression potentiates basal cell migration and IL-8- and FMLP-induced chemotaxis.
On the mechanism of PLD action: the differential role of enzymatic activity
To ascertain the intracellular mechanism that PLD1 uses to cause the observed increase in cell migration, we investigated first whether its enzymatic activity was responsible for the observed effect. We generated in the laboratory a lipase-inactive mutant of PLD1 (K830R-PLD1) by site-directed mutagenesis. This mutant is known to exhibit a dominant-negative effect in control cells, as indicated before by other researchers.29,30 Figure 3A, inset shows that the K830R-PLD1 mutant did not alter the pattern of subcellular expression seen with the wild type of Figure 2B. QRT-PCR measurements indicate that K830R-PLD1 mRNAs were expressed at a similar level than that of wild type (Figure 3A). In both cases, a strong shift to the left of approximately 5.1 ΔCt (approximately 34-fold) was observed over mock-transfected cells. Figure 3B presents 2 important points. First, chemokinesis (control migration) is not affected by the PLD1 mutant. Thus, although chemokinesis is increased by PLD1 overexpression (Figure 2A), this effect is not mediated by the enzymatic activity of the enzyme. Second, the large IL-8- and FMLP-induced chemotactic responses were negated in cells transfected with PLD1-K830R. Therefore, basal chemokinesis is independent on enzymatic activity, and the chemokine-induced component is activity dependent.
Overexpression of either WT-PLD1 or K830R-PLD1 did not lead to an increase in chemotaxis in ENA-78-treated cells, as FMLP or IL-8 did, but it appeared to inhibit ENA-78-induced migration (Figure 3B). The reason for this could be that cell transfection might affect the level of protein expression of the chemokine receptors. Figure 3C shows that both CXCR1 and CXCR2 receptors are present in dHL-60 cells. However, PLD1 transfection seems to down-regulate the level of expression of CXCR2. This appears to be a specific effect, because neither overexpression of the other PLD isoform (PLD2) nor silencing of PLD1 (or PLD2) changes the level of expression of CXCR1 or CXCR2 receptors in dHL-60 cells.
Effect of a PLD enzyme-inactive mutant. (A) dHL-60 cells were mock-transfected or transfected with WT HA-PLD1 or with a lipase-inactive PLD1 mutant (K830R). Both wild-type and lipase-inactive mutants were expressed equally at the RNA level, as assayed by QRT-PCR. (Inset) Anti-HA confocal immunofluorescence showing the pattern of intracellular expression. (B) Chemotactic response of WT- or K830R-overexpressing dHL-60 cells toward the indicated chemoattractants. The mean ± SEM from 5 independent experiments performed in duplicate are shown; 100% represents 3.0 × 104 ± 0.4 cells/mL. (C) Chemokine receptor expression levels during HL-60 cell transfection. dHL-60 cells were transfected with HA-PLD1, myc-PLD2 plasmids, or siRNAs for PLD1 or PLD2. CXCR1 and CXCR2 were detected in Western blots with specific antibodies. A gel loading control is presented at the bottom of panel C, as protein staining (GelCode Blue; Pierce Biotechnology, Rockford, IL) in the region of β-actin.
Effect of a PLD enzyme-inactive mutant. (A) dHL-60 cells were mock-transfected or transfected with WT HA-PLD1 or with a lipase-inactive PLD1 mutant (K830R). Both wild-type and lipase-inactive mutants were expressed equally at the RNA level, as assayed by QRT-PCR. (Inset) Anti-HA confocal immunofluorescence showing the pattern of intracellular expression. (B) Chemotactic response of WT- or K830R-overexpressing dHL-60 cells toward the indicated chemoattractants. The mean ± SEM from 5 independent experiments performed in duplicate are shown; 100% represents 3.0 × 104 ± 0.4 cells/mL. (C) Chemokine receptor expression levels during HL-60 cell transfection. dHL-60 cells were transfected with HA-PLD1, myc-PLD2 plasmids, or siRNAs for PLD1 or PLD2. CXCR1 and CXCR2 were detected in Western blots with specific antibodies. A gel loading control is presented at the bottom of panel C, as protein staining (GelCode Blue; Pierce Biotechnology, Rockford, IL) in the region of β-actin.
To validate the leukemic HL-60 results of the role of PLD1 in cell migration presented in Figures 1 to 3 to a physiologic model, we measured PLD1 enzymatic activity in normal peripheral blood neutrophils. Figure 4A shows that there is a correlation between lipase activity and chemotaxis in those cells, whereby activity precedes chemotaxis in vivo for both IL-8 and FMLP. ENA-78 was unable to significantly activate PLD1, whereas it was capable of eliciting chemotaxis, as expected. As a negative control for the measurement of PLD1 activity, cell lysates were immunoprecipitated with an irrelevant antibody (anti-PLA2), which did not pull down any measurable PLD enzymatic activity (not shown).
An enzyme-chemotaxis correlation further extends to the treatment of neutrophils (Figure 4B) and dHL-60 cells (Figure 4C) with 3 commonly used PLD inhibitors: 4-(2-aminoethyl)-benzenesulfonyl fluoride (AEBSF),31,32 ethanol, and butanol. These alcohols are widely used in the scientific literature as an index of PLD presence/absence.33,34 In conclusion, known PLD inhibitors are also able to inhibit leukocyte chemotaxis in vivo, and a correlation between chemotaxis and PLD1 enzyme activity exists.
The PLD2 isoform also mediates cell migration: selective role in ENA-78 signaling
As presented in Figures 3 and 4, an enhanced activity of PLD1 mediates the chemotactic effects elicited by IL-8 and FMLP. However, this was not applicable to ENA-78, because this chemokine failed to enhance the activity of both endogenous or overexpressed PLD1. In attempting to understand what could mediate ENA-78 activation of chemotaxis that is not PLD1 related, we turned our attention to the other mammalian isoform, PLD2. A wild-type PLD2 construct (pcDNA3-myc-PLD2) was transfected into dHL-60 cells and cell migration was analyzed. Control experiments for this construct, mRNA and protein expression, as well as subcellular localization, are presented in Figure 5A-C. PLD2 transfection led to a 3-fold increase in basal chemokinesis (Figure 5D). The significant difference between IL-8- and ENA78-induced chemotaxis levels in this figure may be explained by the diminished expression level of CXCR1 by PLD transfection seen in Figure 3C, although IL-8 can still signal through CXCR2. At any rate, PLD2-transfected cells responded to chemoattractants, and, although all 3 chemokines induced chemotaxis, it was ENA-78 that showed the largest effect. This response to ENA-78 was in sharp contrast to the PLD1 situation described earlier. Silencing PLD2 gene expression with siRNA, however, led to a decrease in cell migration (Figure 5E) that could not be rescued by ENA-78, FMLP, or IL-8, further indicating the role of PLD2 in chemokine-induced cell migration.
Neutrophil PLD1 enzymatic activity is increased by chemoattractants. (A) Correlation activity/chemotaxis time wise. For thick-line tracings (activity), neutrophil suspensions (5 × 106 cells/mL) were incubated in the presence of the indicated chemoattractants. At the appropriate times, cells were lysed and immunoprecipitated with anti-PLD1 antibodies. Immunocomplex beads were used to assay the activity of the enzyme in vitro with [3H]-butanol and PC8 liposomes; 100% activity represents 1034 ± 76 dpm/mg protein. For thin-line tracings (chemotaxis), either IL-8, FMLP, or ENA-78 was added to the bottom wells of Transwell plates, and migrated cells were counted under the microscope at the times indicated; 100% represents 3.5 × 105 ± 0.25 cells/mL. (B) Correlation activity/chemotaxis in relation to PLD inhibitors. Neutrophils were preincubated with AEBSF, ethanol, or butanol and assayed for chemotaxis (□) or were used for IP and enzyme analysis (▪). (C) Confirmation of the inhibitory effect of PLD inhibitors on chemotaxis of dHL-60 cells. Viability at the end of the assays remained greater than 90%. Results indicated in the figure are the mean ± SEM from 4 independent experiments conducted in duplicate.
Neutrophil PLD1 enzymatic activity is increased by chemoattractants. (A) Correlation activity/chemotaxis time wise. For thick-line tracings (activity), neutrophil suspensions (5 × 106 cells/mL) were incubated in the presence of the indicated chemoattractants. At the appropriate times, cells were lysed and immunoprecipitated with anti-PLD1 antibodies. Immunocomplex beads were used to assay the activity of the enzyme in vitro with [3H]-butanol and PC8 liposomes; 100% activity represents 1034 ± 76 dpm/mg protein. For thin-line tracings (chemotaxis), either IL-8, FMLP, or ENA-78 was added to the bottom wells of Transwell plates, and migrated cells were counted under the microscope at the times indicated; 100% represents 3.5 × 105 ± 0.25 cells/mL. (B) Correlation activity/chemotaxis in relation to PLD inhibitors. Neutrophils were preincubated with AEBSF, ethanol, or butanol and assayed for chemotaxis (□) or were used for IP and enzyme analysis (▪). (C) Confirmation of the inhibitory effect of PLD inhibitors on chemotaxis of dHL-60 cells. Viability at the end of the assays remained greater than 90%. Results indicated in the figure are the mean ± SEM from 4 independent experiments conducted in duplicate.
To ascertain a possible role of PLD2 enzymatic activity in cell migration, we examined the fact that overexpression of this isoform leads to the presence of a high level of basal activity,35,36 as opposed to PLD1 which has a negligible level of basal activity.37,38 To mimic in cells the presence of PA (as the index of PLD activity) that transfection with PLD2 can create, dHL-60 cells were continuously incubated for 24 to 48 hours in culture with exogenous dioleoyl-phosphatidic acid (DO-PA) at different concentrations. Figure 5F shows that chemokinesis is enhanced with a continuous exposure of exogenous PA to cell cultures. The bell-shaped curve is characteristic of a cell migration response. Finally, to show that results obtained with a transformed cell line such as HL-60 pertain to a normal blood cell, experiments were conducted in human neutrophils freshly isolated from peripheral blood. Figure 5G indicates that ENA-78 increased the enzymatic activity of PLD2 to a greater extent than IL-8. These data paralleled the overexpression experiments and indicate that the observed chemotaxis induced by ENA-78 can be explained by the selective activation of PLD2 over PLD1.
Chemokines IL-8 and ENA-78 induce cell polarization and preferential directionality of PLD isoforms
We next wanted to investigate the effect of chemokines on cellular expression of endogenous PLD isoforms. For this, cells were adhered to 6-well tissue culture plates and were challenged with ENA-78 or IL-8, which were added at one corner of the well (as to create a temporary gradient). This was followed by in situ immunostaining and epifluorescence confocal microscopy, as presented in Figure 6A. Unstimulated adherent neutrophils showed a diffuse pattern of fluorescence staining. Stimulation with IL-8 caused a strong polarizing effect that manifested itself as the selective accumulation of PLD1 bright caps (seen as half-moons in the confocal micrographs) or PLD2 bright caps (horseshoe formations) at the edge of the cell. ENA-78 elicited the same pattern of polarization on PLD2 but failed to do so in PLD1 stainings. This selective response of ENA-78 activating only PLD2, while IL-8 activates both PLD1 and PLD2, paralleled that of chemotaxis and enzymatic activity as shown in previous figures. To further understand the polarization results, PLD, nuclear, and F-actin staining were performed simultaneously. Figure 6B and 6C shows that F-actin colocalizes with PLD1 and PLD2 in the polarization cups for all chemoattractants tested, except ENA-78, that again failed to induce a significant polarization in PLD1-stained cells.
Finally, Figure 7 represents a directionality analysis, indicating that most of the increase in polarization occurs within a 90° swath region of a slide-chamber toward the direction of a micropipette-added chemokine. Figure 7A indicates the layout of the experiment, and quantitation of the direction of polarization within this 90° swath region of the slide-chamber is presented in Figure 7B. The response of PLD2 is slightly greater (3.4-fold versus 1.9-fold) than that of PLD1 for IL-8. An example of PLD2 directional bright-cap orientation during chemotaxis is given in Figure 7C.
PLD2 also mediates cell migration. (A-C) Transfection controls. dHL-60 cells were transfected with pcDNA-myc-PLD2 for the length of time indicated. Cell samples were harvested, and whole lysates were taken for RNA isolation (A) for the detection of the overexpressed protein by immunoblotting with anti-PLD2 antibodies (B) (a gel loading control is presented at the bottom as the IgG heavy chain antibody), or for the subcellular detection of overexpressed protein by confocal immunofluorescence (C). (D) Cells overexpressing myc-PLD2 (or mock-transfected) were assayed for their ability to respond to chemotactic agents ENA-78, FMLP, and IL-8 (or buffer alone, Control). (E) PLD2 gene silencing. PLD2-duplex siRNA was nucleofected into HL-60 cells 1 day after differentiation induction with DMSO and challenged where appropriate with chemoattractants in Transwell. (Inset) Western blot of endogenous PLD2 and its silencing with siPLD2 RNA. (F) Effect of PA on chemokinesis. DO-PA was added at the indicated concentrations to dHL-60 cultures for 36 hours. Cells were then harvested and transferred to Transwell plates, and random cell migration was measured; 100% represents 1.1 × 104 ± 0.09 cells/mL. (G) PLD2 enzymatic activity. Neutrophil suspensions were incubated with the chemoattractants for the lengths of time indicated. Total cell lysates were immunoprecipitated with anti-PLD2 antibodies, and PLD activity was assayed using immunocomplex beads. Results are mean ± SEM from 4 independent experiments conducted in duplicate. *P < .05 for comparison of ENA-78 versus IL-8; 100% activity represents 2152 ± 160 dpm/mg protein.
PLD2 also mediates cell migration. (A-C) Transfection controls. dHL-60 cells were transfected with pcDNA-myc-PLD2 for the length of time indicated. Cell samples were harvested, and whole lysates were taken for RNA isolation (A) for the detection of the overexpressed protein by immunoblotting with anti-PLD2 antibodies (B) (a gel loading control is presented at the bottom as the IgG heavy chain antibody), or for the subcellular detection of overexpressed protein by confocal immunofluorescence (C). (D) Cells overexpressing myc-PLD2 (or mock-transfected) were assayed for their ability to respond to chemotactic agents ENA-78, FMLP, and IL-8 (or buffer alone, Control). (E) PLD2 gene silencing. PLD2-duplex siRNA was nucleofected into HL-60 cells 1 day after differentiation induction with DMSO and challenged where appropriate with chemoattractants in Transwell. (Inset) Western blot of endogenous PLD2 and its silencing with siPLD2 RNA. (F) Effect of PA on chemokinesis. DO-PA was added at the indicated concentrations to dHL-60 cultures for 36 hours. Cells were then harvested and transferred to Transwell plates, and random cell migration was measured; 100% represents 1.1 × 104 ± 0.09 cells/mL. (G) PLD2 enzymatic activity. Neutrophil suspensions were incubated with the chemoattractants for the lengths of time indicated. Total cell lysates were immunoprecipitated with anti-PLD2 antibodies, and PLD activity was assayed using immunocomplex beads. Results are mean ± SEM from 4 independent experiments conducted in duplicate. *P < .05 for comparison of ENA-78 versus IL-8; 100% activity represents 2152 ± 160 dpm/mg protein.
Chemoattractants induce strong cell polarization associated with PLD1 and PLD2 staining. (A) Single labeling. Neutrophils, at a density of 5 × 106 cells/mL, were adhered for 20 minutes to microscope slides in 6-well tissue culture plates. They were then incubated for 14 minutes at 37°C in 5% CO2 with chemokines, added to the upper part of the slide (more detail in Figure 7). After this, cells were fixed and immunostained with anti-PLD1 or anti-PLD2 antibodies. They were further treated with FITC-conjugated secondary antibodies, and finally observed by confocal fluorescence microscopy. (B-C) Triple labeling. Cells were treated as above but processed with anti-PLD1 (B) or anti-PLD2 antibodies (C), FITC-secondary Ab, TRITC-phalloidin, and DAPI, in sequential order, and were observed by epifluorescence microscopy.
Chemoattractants induce strong cell polarization associated with PLD1 and PLD2 staining. (A) Single labeling. Neutrophils, at a density of 5 × 106 cells/mL, were adhered for 20 minutes to microscope slides in 6-well tissue culture plates. They were then incubated for 14 minutes at 37°C in 5% CO2 with chemokines, added to the upper part of the slide (more detail in Figure 7). After this, cells were fixed and immunostained with anti-PLD1 or anti-PLD2 antibodies. They were further treated with FITC-conjugated secondary antibodies, and finally observed by confocal fluorescence microscopy. (B-C) Triple labeling. Cells were treated as above but processed with anti-PLD1 (B) or anti-PLD2 antibodies (C), FITC-secondary Ab, TRITC-phalloidin, and DAPI, in sequential order, and were observed by epifluorescence microscopy.
Discussion
The biologic importance of this study is that one of the most important leukocyte functionalities, cell migration, is enhanced because of PLD and, specifically, chemotaxis (but not chemokinesis) is being mediated by PA, the product of PLD's enzymatic activity. PLD appears to harness the ability of granulocytes (differentiated promyelocytic cells and normal peripheral neutrophils) to seek inflammatory cytokines, a prelude to microbial killing and tissue remodeling. We also show here that PLD is associated with cell polarity and F-actin polymerization in response to chemoattractants. Polarization (the asymmetric redistribution of cytoskeletal proteins, organelles, and signaling proteins in response to environmental clues) is the initial step in cell migration, concomitantly with adhesion. Cell adhesion to substrates or to other cells via ECM proteins serves to stabilize the polarization process in vivo.9
Studies in Drosophila and Xenopus embryos have uncovered key parts of the intracellular mechanism that might mediate polarization. Specifically, 2 Wnt-initiated pathways that mobilize Rho/Rac and Ca2+/PKC are known.11 Coincidentally, those very proteins modulate the PLD1 isoform. A role of PLD1 and PLD2 isoforms in macrophage phagocytosis39 has already been shown. At the early stages of cytoskeletal reorganization, strong polarization of PLD1 and PLD2 occurs, as evidenced in the immunofluorescence images of Figure 6. It seems plausible that PLD could be recruited along with other signaling molecules, such as PI3Kγ or Akt/PKB,40,41 known to spatially localize at the leading edge of the migrating cell.
Directionality analysis of PLD1 and PLD2. (A) Experimental setup. IL-8 was added at a precise location (red dot) with a 200-μm micropipette to a microscope slide of adherent neutrophils to form a temporary gradient prior to anti-PLD2 immunostaining. (B) Cells exhibiting polarization caps that were pointing within a 90° swath toward IL-8 were counted, and the percentage over total number of cells was calculated. Results are the mean ± SEM from 3 independent experiments conducted in duplicate. An example of one such field, observed by epifluorescence microscopy, is presented in panel C. The yellow lines (added with Adobe Illustrator software, v. 11.0; San Diego, CA) were defined as intersecting the main polarization cap (bright PLD2 immunostaining) of the cells in the direction of the chemokine.
Directionality analysis of PLD1 and PLD2. (A) Experimental setup. IL-8 was added at a precise location (red dot) with a 200-μm micropipette to a microscope slide of adherent neutrophils to form a temporary gradient prior to anti-PLD2 immunostaining. (B) Cells exhibiting polarization caps that were pointing within a 90° swath toward IL-8 were counted, and the percentage over total number of cells was calculated. Results are the mean ± SEM from 3 independent experiments conducted in duplicate. An example of one such field, observed by epifluorescence microscopy, is presented in panel C. The yellow lines (added with Adobe Illustrator software, v. 11.0; San Diego, CA) were defined as intersecting the main polarization cap (bright PLD2 immunostaining) of the cells in the direction of the chemokine.
At this point, we can only speculate as to whether PLD is a primary or a secondary regulator of cell migration. However, the role of microfilamentous cytoskeleton in neutrophil function and the role of PLD in cell motility and actin cytoskeleton reorganization are well-known facts10,29,42,43 and might indeed serve to confer PLD in a leading role. Further strengthening this point is that the product of PLD enzymatic action, PA, is known to be capable of modifying membrane architecture. PA can regulate several key steps in cytoskeletal reorganization and membrane trafficking.44,45 PLD is also involved in receptor endocytosis and/or recycling, which raises the possibility that chemokine receptors may be regulated in a similar way.46,47
Moreover, tubulin binding to PLD2 inhibits muscarinic receptor activation in COS-7 cells48 and PLD regulates Dictyostelium actin localization.49 Cytoskeletal dynamics controlled by Rho GTPase are needed for cell migration and other neutrophil functions.50 Rac GTPase controls chemotaxis in live neutrophils,51 and suppression of Rac2 activation concomitantly with ROS generation is mediated by the Rho family exchange factor (GEF) Vav-1.52 Additionally, Rac1 is essential for gradient detection and actin assembly through regulation of PI-3K and Akt in neutrophils.53 Our laboratory has uncovered recently that the ribosomal S6 protein kinase p70S6K is also involved in leukocyte chemotaxis.27 Because this signaling protein is regulated by the mammalian target for rapamycin (mTOR), itself a target of PA regulation,30 the interrelation between PLD-produced lipids and signaling kinases warrants further exploration.
In addition to stimulating chemotaxis, IL-8 (an ELR+ CXC chemokine) is also involved in neutrophil lipid metabolism. Sozzani et al54 demonstrated that IL-8 activates phospholipase D (PLD) and the respiratory burst, thus involving PA, the product of the PLD reaction, in neutrophil signaling. Furthermore, the formation of IL-8-induced PA is inhibited by pertussis toxin and tyrosine kinase inhibitors.55 During inflammation and phagocytosis, PA promotes degranulation.56 One of the questions that the present study has addressed is whether IL-8 activates chemotaxis through PLD. Results shown here for the first time indicate that PLD enzymatic activity mediates cell chemotaxis. However, as the study with a lipase-inactive mutant indicates, PLD1 activity does not affect chemokinesis (ie, basal or random cell migration).
Remarkably, the situation with ENA-78 was entirely different, because this chemokine failed to elicit any significant increase in PLD1 activity or polarization. In PLD1 overexpression, CXCR2 protein expression seems to be inhibited in dHL-60 cells (Figure 3C). However, even if this were not to occur, PLD1 would most likely still not lead to a significant response through ENA-78, based on 2 complementary lines of evidence: (1) when endogenous PLD1 enzymatic activity is measured in neutrophils (Figure 4A), ENA-78 elicits the smallest response of the 3 chemoattractants; and (2) polarization of endogenous PLD1 in neutrophils (Figure 6A-B) does not occur with ENA-78. At any rate, the lack of involvement of PLD1 mediating ENA-78 cell stimulation prompted us to study the other mammalian PLD isoform, PLD2. When the chemotactic response of dHL-60 cells toward ENA-78 was analyzed considering the PLD2 isoform, a more complete picture emerged. A potentiated response to ENA-78 was observed in HL-60 cells overexpressing myc-pcDNA-PLD2, and cell polarization of PLD2 occurred under direct stimulation of ENA-78. IL-8 and ENA-78 ELR+ CXC chemokines recruit neutrophils by binding and activating CXCR-2 receptors,7 whereas IL-8, but not ENA-78, binds to CXCR-1 receptors.4,8 Results in the present report indicate that PLD1 is differentially activated by CXCR-1, whereas CXCR-2 (and possibly CXCR-1) mediates PLD2 activation.
PLD-mediated chemotaxis in leukocytes should be viewed in light of the currently understood mechanisms for cellular motility. Cell migration is crucial during vital physiologic processes, such as inflammation and wound repair (the latter involving neovascularization), but it also plays a key role in pathophysiology. It would be most interesting to investigate whether PLD isoforms underlay circulatory pathologic situations such as chronic inflammation and cancer metastasis.
Prepublished online as Blood First Edition Paper, July 27, 2006; DOI 10.1182/blood-2006-02-005959.
Supported by National Institutes of Health grant HL056653 (J.G.-C.).
The authors declare no competing financial interests.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Kathleen Frondorf, Karen Henkels, and Tara Liao for excellent technical assistance; Dr Dan Halm for insights on the physiology of chemotaxis; and Dr Robert Fyffe for acquiring the confocal micrographs. The pcDNA-PLD1b wild-type construct was a generous gift from Dr S. H. Ryu (Pohang University of Science and Technology, Republic of Korea), and the pcDNA-PLD2 wild-type construct was a generous gift from Dr J. D. Lambeth (Emory University School of Medicine, Atlanta, GA) and Dr I. Lopez.



![Figure 4. Neutrophil PLD1 enzymatic activity is increased by chemoattractants. (A) Correlation activity/chemotaxis time wise. For thick-line tracings (activity), neutrophil suspensions (5 × 106 cells/mL) were incubated in the presence of the indicated chemoattractants. At the appropriate times, cells were lysed and immunoprecipitated with anti-PLD1 antibodies. Immunocomplex beads were used to assay the activity of the enzyme in vitro with [3H]-butanol and PC8 liposomes; 100% activity represents 1034 ± 76 dpm/mg protein. For thin-line tracings (chemotaxis), either IL-8, FMLP, or ENA-78 was added to the bottom wells of Transwell plates, and migrated cells were counted under the microscope at the times indicated; 100% represents 3.5 × 105 ± 0.25 cells/mL. (B) Correlation activity/chemotaxis in relation to PLD inhibitors. Neutrophils were preincubated with AEBSF, ethanol, or butanol and assayed for chemotaxis (□) or were used for IP and enzyme analysis (▪). (C) Confirmation of the inhibitory effect of PLD inhibitors on chemotaxis of dHL-60 cells. Viability at the end of the assays remained greater than 90%. Results indicated in the figure are the mean ± SEM from 4 independent experiments conducted in duplicate.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/10/10.1182_blood-2006-02-005959/4/m_zh80220603540004.jpeg?Expires=1765892437&Signature=yVrsnYyXYvE29qHZxmSjWDMGk-NJcZ8nYxV9ILfBEXMRe7BMRR2RkfnjxR8tnS8zfHnwV0eXRdSJu~nkCiU86xAOtEVcB4mJl2xfd~m6eUDj-yWnJ94vWYexeb8nlPThEfGe18LJvkMrc8mpHhOL~QB6l7lNcRLHja1dyM0nQIxSVLnmyHJ4Zd58OFcKAGRvsKfXCsepMjxrB1hpNftrsu5E8jgSOUfWrXcAuoPimQQ7ttjAAWBgnx-zVnyEBSFF5BnR4PP5KpG4upcpBB2jBJhsxNwcF3Z89FSy5RO9UH1jy07voSENtasHZz0ON5-J08bSnr1h-jakW3WopFNvgg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal