Abstract
The identification of an acquired mutation of JAK2 in patients with myeloproliferative disorders has raised questions about the relationship between mutation-positive and mutation-negative subtypes, timing of the JAK2 mutation, and molecular mechanisms of disease progression. Here we demonstrate that patients with V617F- essential thrombocythemia do not commonly progress to become V617F+. Consistent with the concept of distinct pathogenetic mechanisms, we show that patients with and without the JAK2 mutation have different patterns of cytogenetic abnormality, with virtually all patients carrying the 20q deletion or trisomy 9 being V617F+. We also investigated the existence of a “pre-JAK2” phase by comparing the proportion of clonally derived granulocytes, estimated from X-chromosome inactivation patterns (XCIPs), with the proportion of V617F+ granulocytes. Our results demonstrate that inherent XCIP variability between granulocytes and T cells produces a systematically biased pattern of results that may be misinterpreted as evidence for an excess of clonally derived granulocytes, an observation that limits the utility of XCIP analysis in this context. Lastly, we studied 4 patients with V617F+ myeloproliferative disorders who subsequently developed acute myeloid leukemia. In 3 patients the leukemic cells were V617F-, suggesting that in these patients the leukemia arose in a V617F- cell.
Introduction
An acquired mutation in JAK2 has been described in nearly all patients with the myeloproliferative disorder (MPD) polycythemia vera (PV), and half those with essential thrombocythemia (ET) and idiopathic myelofibrosis (IMF).1-5 The V617F mutation arises in a multipotent progenitor, 1 is associated with erythropoietin-independent growth of erythroid progenitors,1 confers constitutive activity on the kinase, with enhanced downstream signaling,2-4,6 and was sufficient for the development of erythrocytosis in a murine retroviral model.2 The V617F mutation was not found in an extensive survey of nonhematologic cancers and lymphoid malignancies7 and is relatively uncommon in myeloid malignancies other than the classic MPD.8-10
Analysis of prospective data from the PT-1 study11 has demonstrated that JAK2 status distinguishes 2 biologically distinct subtypes of ET, with the V617F+ subgroup exhibiting many laboratory and clinical similarities to PV.12 Similar observations have been reported by other groups.13,14 These results suggest that V617F+ ET and PV form a biologic continuum, with the degree of erythrocytosis determined by physiologic or genetic modifiers.12
The relationship of V617F- ET to V617F+ ET is unclear. It has been suggested that V617F- ET may represent an earlier phase of the same disease, with subsequent acquisition of the JAK2 mutation.4 In support of this concept it has been reported that patients with V617F+ MPD had a longer duration of disease compared to those lacking the mutation.4 In addition, the existence of rare families in whom multiple members have an MPD, which may be negative or positive for the V617F mutation, has been used to suggest the existence of a pre-JAK2 mutation.15 However, a prospective study of ET did not confirm a longer duration of disease in V617F+ patients and found no difference between mutation-positive and -negative patients in the frequency of features of advanced disease, such as splenomegaly, abnormal cytogenetics, and transformation rates.12 These considerations suggest an alternative model in which JAK2-positive and -negative ET represent pathogenetically distinct diseases.12
The MPDs are associated with several cytogenetic abnormalities, including trisomy 9, trisomy 8, and deletion of 20q and 13q.16 The rates of cytogenetic abnormalities have been reported to be comparable in V617F+ and V617F- MPDs,1,12,17 although the small number of patients preclude definitive conclusions. Such cytogenetic abnormalities may provide clues to the as yet unknown pathogenesis of V617F- MPDs and may illuminate the potential role of cooperating mutations in progression of V617F+ disease.
Both PV and ET may terminate in an accelerated phase, with patients experiencing falling blood counts, myelofibrotic transformation, and progressive systemic symptoms, as well as frank acute leukemic transformation. However, neither is an inevitable consequence of the MPD,18 and acquisition of additional mutations seems likely to contribute to the evolution of these more aggressive stages. It is not known what role the JAK2 mutation plays in transformation to acute myeloid leukemia (AML). The V617F mutation is rare in de novo AML,7,9,10 although fusion genes involving JAK2 are associated with acute leukemia.19
Here we have investigated the relationship of V617F- ET to V617F+ ET, the existence of a “pre-JAK2” phase of V617F+ MPDs, and the molecular mechanisms associated with disease progression.
Patients, materials, and methods
Patients and samples
Granulocytes were prepared by centrifugation of whole blood through a Ficoll density gradient, and T cells were isolated from the mononuclear-cell layer by anti-CD2 magnetic beads, as described.20,21 Mean purity was greater than 95% for granulocytes and 91% for T cells. For samples from ET patients after AML transformation, archival bone marrow or peripheral-blood smears were scraped at the thickest part of the film and DNA isolated as previously described.22 Methods for sequencing, allele-specific polymerase chain reaction (PCR; 3-primer method) for the JAK2 V617F mutation and microsatellite genotyping have been described previously.1 Patients for the clonality study were women, younger than age 75 years, with a JAK2+ MPD selected to minimize the chances of including patients with significant homozygosity for V617F. All patients gave written informed consent for the research. The research was approved by Multi-Region Ethics Committee (Fulbourn, United Kingdom) for Causes of Clonal Haematological Disorders project, which permits the use of biologic samples and clinical data from patients with clonal hematologic disorders, and was carried out in accordance with the principles of the Declaration of Helsinki.
HUMARA and JAK2 quantitation
For quantitation of the JAK2 V617F mutation, fluorescently labeled primers were used in a modification of the 4-primer protocol published by Jones et al.8 The forward wild-type (WT) inner primer was labeled with FAM and the reverse mutation-specific inner primer with HEX, and the PCR products were run on an ABI Prism 3700 machine (Applied Biosystems, Foster City, CA). A dilution series of mutant and Wt DNA was run with each reaction and used to estimate the proportion of V617F alleles in each patient's DNA, as described in “Statistical analysis.” To calculate the proportion of V617F granulocytes from the V617F/WT allelic ratio, we assume that JAK2+ granulocytes are heterozygous for the mutation. Because a heterozygous cell contributes 1 WT and 1 mutant allele to the DNA sample, the estimated proportion of JAK2+ granulocytes is double the V617F/WT allelic ratio. The presence of both heterozygous and homozygous cells would lead to overestimation of the proportion of JAK2+ granulocytes. We have minimized the impact of this by excluding patients with known 9p loss of heterozygosity (LOH), trisomy 9, or V617F homozygosity (6 patients). We selected any ET patients (in whom homozygosity is rare) and focused on PV patients with low levels of JAK2 positivity, defined as mutation detectable by allele-specific PCR alone (3 patients), mutant peak less than 25% total height by sequencing (5 patients), or mutant peak height 25% to 50% (2 patients).
The human androgen receptor assay (HUMARA) was performed using fluorescently labeled primers after overnight digestion with HhaI.23 The proportion of clonal granulocytes was estimated from the HUMARA using well-established formulas.21,24 This corrects for uneven X-chromosome inactivation patterns (XCIPs) through concomitant assessment of granulocyte and T-cell DNA and PCR amplification bias through the use of undigested DNA. The assumptions implicit in the formula are that there is no T-cell involvement in the abnormal clone (supported by several papers on the JAK2 mutation1,2,4,25-27 ) and that T cells and the polyclonal fraction of granulocytes have the same patterns of X-chromosome inactivation. All assays were performed in duplicate.
Cytogenetics
The cytogenetics analyses used data pooled from several sources, including the prospective ET trials (599 patients with cytogenetics),11,12 the European Myelofibrosis Network (71 patients),17 the Addenbrooke MPD clinic (89 patients),1 and previous collaborative studies of 20q and 13q deletions in MPDs.21 Because 20q deletions detected on G-banded cytogenetics are often unreliable,21 all putative cases of 20q deletion were confirmed by testing of microsatellite markers in the common deleted region.
Statistical analysis
The quantitation of V617F alleles used the logarithm of the ratio of peak areas for the mutant and WT peaks. A 3-parameter logistic curve was fitted to log(peak area ratio) versus V617F proportion from the dilution series. Parameter values were estimated by nonlinear least squares regression and point estimates of V617F proportion were calculated for patients. The measurement standard error for JAK2 and HUMARAs was estimated by variance components models.
For the Monte Carlo simulations28 of estimated proportions of clonal granulocytes under the null hypothesis of no mutation preceding JAK2 V617F, the data of Gale et al were used.29 The XCIPs of paired T-cell and granulocyte samples were used to estimate the variability of granulocyte XCIPs for young (< 50 years) and elderly (> 75 years) women. A logit transform (defined as logit (p) = log(p/(1 - p)) was taken of granulocyte and T-cell inactivation proportions, and as expected, these were linearly associated with slope of 1 and normally distributed residuals. The age-specific standard deviation of the difference between granulocyte and T-cell XCIPs was used in the simulations (0.44 for women < 50 years, 1.25 for women > 75 years, and an interpolated estimate of 0.75 for women 50-75 years).
To perform the simulations,28 the logit of a given proportion of active Xm in T cells was used as the mean of a normal distribution for the logit of granulocyte XCIPs, with standard deviation as detailed. The JAK2 mutation was assumed to arise in a cell with an active maternal X chromosome with probability p, where p was the proportion of active maternal X chromosomes in granulocytes simulated from the previous step. For a given proportion of JAK2+ cells, the observed XCIPs in the mixed clonal and polyclonal granulocyte population could be calculated. The estimated proportion of clonal granulocytes could be calculated using the formulas described previously21,24 :
when RT ≤ RG,, where GC is the proportion of clonal granulocytes, and where RT and RG are the Xm/Xp allele ratios for T cells and granulocytes, respectively. For each level of T-cell skewing, standard deviation and JAK2+ proportion, 50 000 simulated cases were used to generate the medians and 95% confidence intervals shown in Figure 2A. The simulations were used to assess whether the pattern we observed provided evidence against the null hypothesis of no pre-JAK2 phase. Under this hypothesis, the centile of the observed proportion of clonal granulocytes compared to the simulated population is uniformly distributed on [0,1], with mean 0.5. The mean of the centiles for the 20 patients was the test statistic. Power calculations show that a sample size of 20 would reject the null hypothesis with 80% power if, on average, each patient fell on the upper third (ie, ≥ 67%) of the simulated population. This would equate to a 15% population of clonal JAK2- granulocytes in patients with a 30% population of JAK2+ granulocytes, for example. Therefore, this sample size is adequate to detect even modest populations of V617F- clonal granulocytes if they are present in most patients.
Hypothesis tests for whether there was any difference in proportions of patients positive or negative for V617F with a given cytogenetic abnormality were based on estimates of the frequency of V617F positivity in large cohorts of the 3 MPD subtypes using appropriately sensitive detection methods, namely, 97% for PV1 (pPV), 53% for ET12 (pET) and 55% for IMF17 (pIMF). Assuming the number of patients with a given cytogenetic abnormality is fixed as nPV, nET, and nIMF, then the probability that exactly x patients, x = 0,1,2,... nPV + nET + nIMF, will be V617F+ in the whole cohort is given by:
To calculate a 2-sided p value, values for P(x) were summed over 0,...,x and nPV + nET + nIMF - x,..., nPV + nET + nIMF.
Results
Patients with V617F- ET do not subsequently become V617F+
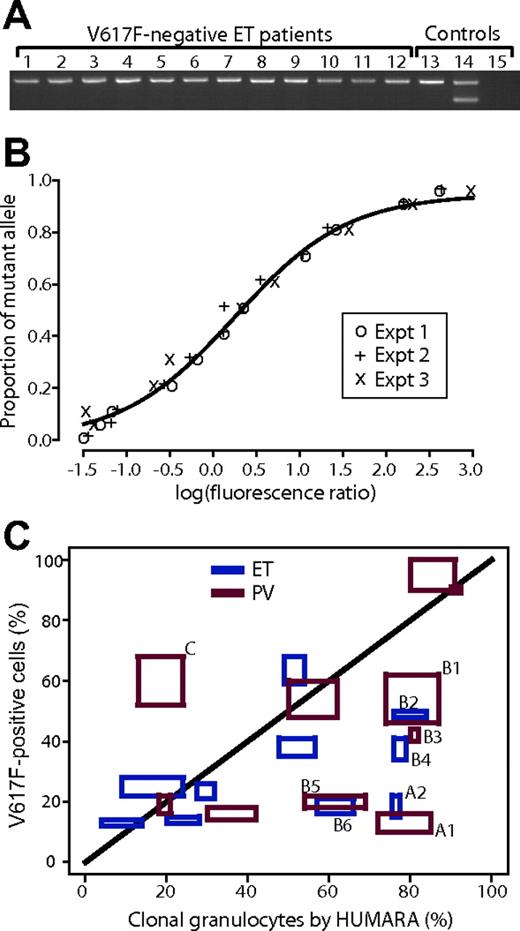
To investigate the possibility that V617F- ET progresses into V617F+ ET, we studied prospectively collected, paired DNA samples from 50 patients with V617F- ET enrolled in the high-risk PT-1 trial11 and the on-going low- and intermediate-risk trials. Allele-specific PCR1 was used to detect the mutation in DNA from unfractionated blood collected at trial entry and again after a median follow-up of 77 months (range, 61-90 months). None of the 50 patients showed evolution from V617F- to V617F+ ET (Figure 1A), suggesting that if it occurs at all, it is a rare phenomenon. These data suggest that V617F+ and V617F- thrombocytosis are distinct disorders, rather than temporally linked phases of the same disease.
Comparison of XCIPs with proportion of V617F+ granulocytes
The identification of V617F- and V617F+ ET as separate disorders does not exclude the possibility that the V617F mutation may require one or more prior mutations to cause an overt MPD phenotype. This scenario suggests that it might be possible to detect a pre-JAK2 phase by comparing the proportion of clonally derived granulocytes (using XCIPs) with the proportion of V617F+ granulocytes. We therefore assessed granulocyte and T-cell DNA from 20 informative women under the age of 75 who had a V617F+ MPD (10 with ET and 10 PV). To calculate the proportion of V617F+ cells from the V617F/WT ratio, we assume that granulocytes are heterozygous for the V617F mutation. Because 30% of PV patients have evidence of 9p LOH and V617F homozygosity,1-4,30 we excluded patients with known 9p LOH, trisomy 9, or a mutant peak greater than 50% of total peak height by sequencing. In addition, we studied 10 patients with ET (in whom V617F homozygosity is rare) and focused on PV patients with a low burden of disease. HUMARA was used to calculate the proportion of clonal granulocytes21 and a 4-primer allele-specific PCR method8 to quantify the proportion of V617F+ cells. The method for quantifying the V617F/WT ratio was reproducible and accurate, with little evidence for interexperiment variability (Figure 1B), as was the case for HUMARA (Figure 1C). The estimated standard errors of measurement for the HUMARA and JAK2 assays were 6.7% and 5.5%, respectively.
Relationship of V617F-ET to V617F+ ET and comparison of proportion of V617F+ granulocytes to proportion of clonal granulocytes estimated by HUMARA. (A) Allele-specific PCR for V617F mutation (3-primer method) performed on peripheral-blood leukocyte samples obtained at follow-up of 12 ET patients who were V617F- a median 77 months earlier. Lanes 1-12, follow-up ET samples; lane 13, healthy individual; lane 14, patient with V617F+ ET; lane 15, water. (B) Standard curve for quantitation of proportion of V617F+ alleles using 4-primer allele-specific PCR. Three separate experiments are shown, and the line is the nonlinear least squares fit of a 3-parameter logistic curve. The x-axis shows the logarithm of the ratio of mutant allele peak area to WT peak area and the y-axis is the known proportion of mutant allele in a dilution series of known homozygous V617F DNA into normal DNA. Expt indicates experiment. (C) Estimated proportion of clonal granulocytes by XCIPs (x-axis) and proportion of V617F+ granulocytes (y-axis) in 20 women with V617F+ MPDs (10 ET, blue; 10 PV, red). Each assay was performed in duplicate and the boxes for each patient represent the 2 values for each measurement for both HUMARAand V617F to demonstrate the interassay variability. For discussion of patients marked A, B, and C, see “Comparison of XCIPs with proportion of V617F+ granulocytes” in the text.
Relationship of V617F-ET to V617F+ ET and comparison of proportion of V617F+ granulocytes to proportion of clonal granulocytes estimated by HUMARA. (A) Allele-specific PCR for V617F mutation (3-primer method) performed on peripheral-blood leukocyte samples obtained at follow-up of 12 ET patients who were V617F- a median 77 months earlier. Lanes 1-12, follow-up ET samples; lane 13, healthy individual; lane 14, patient with V617F+ ET; lane 15, water. (B) Standard curve for quantitation of proportion of V617F+ alleles using 4-primer allele-specific PCR. Three separate experiments are shown, and the line is the nonlinear least squares fit of a 3-parameter logistic curve. The x-axis shows the logarithm of the ratio of mutant allele peak area to WT peak area and the y-axis is the known proportion of mutant allele in a dilution series of known homozygous V617F DNA into normal DNA. Expt indicates experiment. (C) Estimated proportion of clonal granulocytes by XCIPs (x-axis) and proportion of V617F+ granulocytes (y-axis) in 20 women with V617F+ MPDs (10 ET, blue; 10 PV, red). Each assay was performed in duplicate and the boxes for each patient represent the 2 values for each measurement for both HUMARAand V617F to demonstrate the interassay variability. For discussion of patients marked A, B, and C, see “Comparison of XCIPs with proportion of V617F+ granulocytes” in the text.
Comparison between the proportions of clonal and V617F+ granulocytes showed 2 main points. First, the distribution of PV patients was similar to ET patients, suggesting that the assumption of heterozygosity had not substantially affected the pattern of results for PV patients (Figure 1C). Second, some patients appeared to have more clonal granulocytes than V617F+ cells. Two patients (marked with A1 and A2 in the figure) had clonal granulocytes estimated by XCIP analysis to be 70% to 85%, but V617F+ cells in the range 10% to 20%. These patients were 70 and 74 years old at the time of sampling, and there has been nothing unusual about the presentation or subsequent course of their MPDs. Six other patients, marked with B1-6 in the figure, had clonal granulocytes estimated by HUMARA that exceeded the proportion of V617F+ cells, although the degree of discrepancy was less marked than for patients A1 and A2.
Clonality studies: Monte Carlo modeling demonstrates unexpected variability and systematic bias
These data might be taken to suggest the existence of a population of clonally derived granulocytes that are V617F-, and therefore used to support the concept of a pre-JAK2 phase of MPD development. However, the calculation of the proportion of clonally derived granulocytes using X-chromosome inactivation methods is based on a critical assumption, namely, that the pattern of X-chromosome inactivation in normal granulocytes is the same as that in T cells. This assumption underlies the estimation of the proportion of clonal granulocytes by any method of X-linked inactivation patterns, whether based on DNA or RNA, but, as has been shown in previous studies of healthy women, the actual proportion of granulocytes with a given X chromosome inactivated is distributed around the value for T cells, with the variability of the distribution increasing with age.24,29 We reasoned that this variability may have a significant impact on the accuracy of the calculated proportion of clonal granulocytes.
To assess whether discrepancy between granulocyte and T-cell XCIPs could explain why we found apparently clonal V617F- granulocytes in some MPD patients, we undertook Monte Carlo simulations28 based on the null hypothesis of no pre-JAK2 phase. Using given values for the proportion of V617F+ granulocytes (the predefined “true” clonal proportion) and the T-cell XCIP together with the variance of granulocyte XCIP around the value for T cells calculated from healthy women,29 we modeled the resultant granulocyte XCIPs. These granulocyte XCIPs were then used to calculate the proportion of clonal granulocytes that would be obtained by XCIP analysis, thus allowing comparison of this estimated proportion with the predefined “true” proportion of clonal granulocytes.
Several observations emerged. First, the 95% confidence intervals for the population distribution of clonal granulocytes estimated from XCIPs were wide. As expected, this was more marked for older patients (Figure 2A). To see why discrepancies between granulocyte and T-cell XCIPs have such a dramatic effect, consider a patient in whom 50% of T cells but 75% of granulocytes carry an active maternal X chromosomes (Xm; Figure 2B). In this patient, if a JAK2 mutation arises in a cell with Xm active and results in 20% V617F+ granulocytes (the “true” clonal population), the total percentage of granulocytes with Xm active is calculated as 80% (ie, the 20% V617F+ cells plus 0.75 × the remaining polyclonal granulocytes). The Xm/Xp ratio in granulocytes will thus be 80:20% but in T cells it will be 50:50%, and so the estimated proportion of clonally derived granulocytes using the standard formula21,24 will be 60% (ie, 3 times higher than the true value of 20%). Second, the variability is particularly evident when levels of V617F are low and tends to zero as the proportion of V617F+ cells approaches 100%. This is because when the V617F+ population is small, even small discrepancies between T cells and granulocytes in XCIPs will have a disproportionately large impact on the estimated proportion of clonally derived granulocytes. Third, there is a systematic bias in the distribution of estimated proportion of clonal granulocytes, evident as a shift to the right compared to the true proportion of clonal granulocytes (dotted line in Figure 2A). To see why this occurs, consider again the example in Figure 2B. Here, the T-cell XCIP is balanced, but the granulocytes (and presumably their progenitors) are skewed 75:25%. If the JAK2 mutation occurs in a progenitor with Xm active, the proportion of clonal granulocytes will tend to be overestimated, whereas if it arises in a cell with Xp active, an underestimate will result. However, because 3 times as many myeloid cells carry an active Xm, the JAK2 mutation is 3 times more likely to strike a cell with an active Xm, and this will produce a systematic bias toward overestimating the proportion of clonal granulocytes.
Thus, not only does the problem of discrepancy between T-cell and granulocyte XCIPs cause significant variability in the estimated proportion of clonal granulocytes, but the estimates are actually biased toward concluding that there is evidence of a population of clonal V617F- granulocytes. Comparison between the confidence intervals in Figure 2A and the observed patterns in our patients suggests that the observed discrepancies are no more than would be expected under the null hypothesis, with a P value of 0.17. The 2 patients marked A in Figure 1C may well represent benign, age-related skewing, rather than a pre-JAK2 phase of disease. Our data therefore demonstrate that considerable caution must be exercised when using XCIP analysis to quantify the proportion of clonal granulocytes.
V617F+ and V617F- MPDs are associated with distinct patterns of cytogenetic abnormalities
The concept that V617F+ and V617F- ET may represent pathogenetically distinct disorders raises the possibility that they may be associated with different patterns of cytogenetic abnormality. Previous studies assessing the relationship of cytogenetic abnormalities to JAK2 status have not shown any significant associations,1,12,17 although numbers have generally been too small to make definitive comment. We therefore assessed the JAK2 V617F mutation status of 68 patients who had one of the common cytogenetic abnormalities described in MPDs (Table 1). There were significant differences in the pattern of cytogenetic abnormalities by V617F status (P < .001, Fisher exact test), suggesting that the abnormalities are not distributed evenly between V617F+ and V617F- patients.
Cytogenetic abnormalities in V617F+ and V617F– patients with MPDs
. | V617F+ . | V617F– . | P . |
|---|---|---|---|
| Del(20q) total | 28 | 1 | < .001 |
| PV | 12 | 0 | |
| ET | 5 | 0 | |
| IMF | 10 | 1 | |
| MPD/MDS | 1 | 0 | |
| Del(13q) total | 3 | 9 | .17 |
| ET | 1 | 0 | |
| IMF | 2 | 9 | |
| Translocations total | 2 | 8 | .12 |
| ET | 0 | 4 | |
| IMF | 2 | 4 | |
| Trisomy 9 | 10 | 0 | .01 |
| PV | 3 | 0 | |
| ET | 3 | 0 | |
| IMF | 4 | 0 | |
| Trisomy 8 | 6 | 1 | .23 |
| PV | 2 | 0 | |
| ET | 2 | 0 | |
| IMF | 2 | 1 |
. | V617F+ . | V617F– . | P . |
|---|---|---|---|
| Del(20q) total | 28 | 1 | < .001 |
| PV | 12 | 0 | |
| ET | 5 | 0 | |
| IMF | 10 | 1 | |
| MPD/MDS | 1 | 0 | |
| Del(13q) total | 3 | 9 | .17 |
| ET | 1 | 0 | |
| IMF | 2 | 9 | |
| Translocations total | 2 | 8 | .12 |
| ET | 0 | 4 | |
| IMF | 2 | 4 | |
| Trisomy 9 | 10 | 0 | .01 |
| PV | 3 | 0 | |
| ET | 3 | 0 | |
| IMF | 4 | 0 | |
| Trisomy 8 | 6 | 1 | .23 |
| PV | 2 | 0 | |
| ET | 2 | 0 | |
| IMF | 2 | 1 |
Intriguingly, of the 29 patients with a 20q deletion, 28 were V617F+ (P < .001). In all of these patients, microsatellite PCR confirmed the presence of 20q deletion (data not shown). The patient who was V617F- with 20q deletion is a 72-year old woman with a typical presentation of IMF, characterized by pancytopenia, leukoerythroblastic blood film, and bone marrow fibrosis. V617F positivity was also significantly associated with trisomy 9, with all 10 patients having the mutation (P = .01). Although the differences were not statistically significant, greater numbers of patients with translocations and deletions of chromosome 13q were V617F-, suggesting that these abnormalities may contribute to the development of V617F- MPDs. These distinct associations of cytogenetic abnormalities with V617F+ and V617F- MPDs confirm the concept that the 2 subtypes are distinct entities, and also reinforce the likely role of cooperating mutations in the pathogenesis of both disorders.
V617F- AML in patients with a preceding V617F+ MPD
To study the role of the JAK2 mutation in leukemic transformation, we identified 4 patients with V617F+ MPDs (2 ET, 2 PV) who subsequently transformed to AML. Patient PV1 was aged 48 years at diagnosis of PV, and received 32P, busulfan, and hydroxyurea before transformation to AML 25 years later. Interestingly, cytogenetics on 2 occasions before transformation to AML showed a del(20q) and a balanced t(1;9)(q21;q24) translocation, but cytogenetics after transformation to AML showed a complex karyotype, which did not include either the del(20q) or the t(1;9) translocation. Patient PV2 was aged 74 at diagnosis of PV and had the disease 12 years before AML transformation, during which time she received only hydroxyurea. Cytogenetics were normal at diagnosis of PV and not performed when the AML developed. The 2 ET patients, ET1 and ET2, were aged 75 and 70 years at diagnosis of ET and transformed to AML 26 and 44 months after diagnosis, respectively. Both had normal cytogenetics at diagnosis of ET, but ET1 had a 5q deletion at the time of AML transformation, whereas ET2 still had normal cytogenetics. Both had received only hydroxyurea before developing AML.
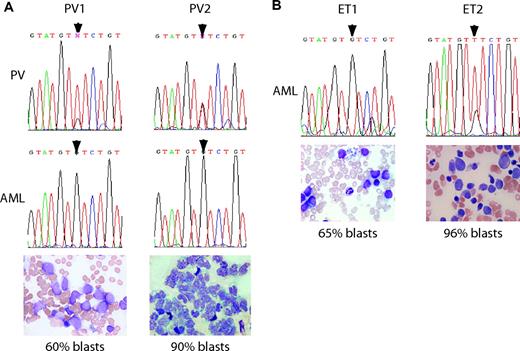
Paired samples before and after AML transformation were available for all 4 patients and were used to assess JAK2 V617F mutation status (Figure 3). A panel of microsatellite markers on chromosome 20 was used to confirm that the paired DNA samples for each patient did indeed have the same genotype (data not shown). For patient PV1, granulocytes taken during the PV phase showed a predominantly mutant sequencing trace, but DNA from the AML bone marrow sample was WT (Figure 3A). This finding is consistent with the cytogenetic evidence for the appearance of a distinct clone at the time of transformation. Patient PV2 also had a mixed sequence trace in granulocytes but subsequently a WT trace in AML peripheral-blood blasts (Figure 3A). For the ET patients, V617F positivity was demonstrated in pre-transformation samples using allele-specific PCR (data not shown). Only the WT allele was detected in the AML sample of patient ET1, whereas the V617F allele predominated in the sample from patient ET2 (Figure 3B).
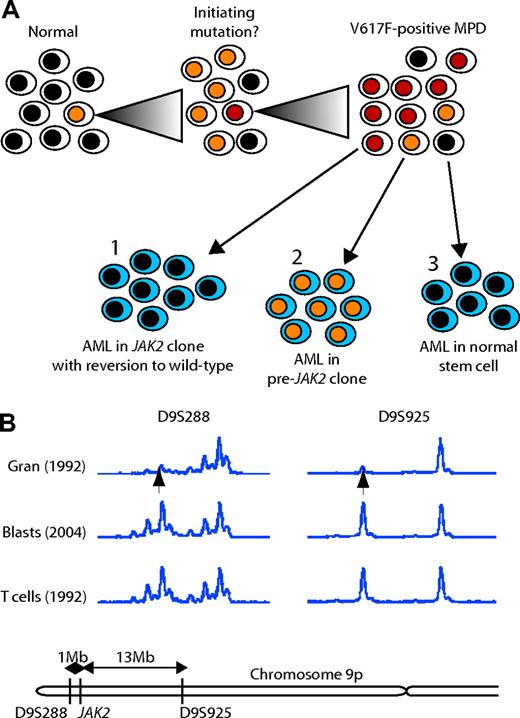
Thus, in 3 of 4 patients with a V617F+ MPD who transformed to AML, the leukemic cells were V617F-. There are 3 possible models to explain this unexpected finding (Figure 4A). First, the mutations responsible for transformation to AML may have developed in a V617F+ clone that subsequently reverted to V617F negativity. Loss of the V617F mutation may provide a growth advantage to leukemic cells if, for example, the V617F mutation is associated with a strong differentiation signal. Second, if mutation of JAK2 is not the initiating event in some MPDs, the AML may result from transformation within the pre-JAK2 clone. Third, AML may result from transformation of a normal stem cell, perhaps as a consequence of mutagenic therapy.
Evidence for the first model (reversion) was sought using cytogenetic analysis and microsatellite PCR. No deletion of chromosome 9p was detected by G-banding in the 3 patients for whom metaphases were available at the time of their AML. Microsatellite PCR was performed on samples from patients PV1 (data not shown) and PV2 (Figure 4B). In the PV phase of the disease, both patients exhibited LOH for 9p markers. By contrast, the AML samples from both patients showed retention of heterozygosity for markers on 9p, thus excluding mitotic recombination as a mechanism for reversion to WT JAK2. Unfortunately, the 2 women (PV2 and ET1) were uninformative at the androgen receptor locus, preventing us from testing whether model 3 was operative. Our results therefore demonstrate that model 1 (reversion) is unlikely and suggest that, in patients with a V617F+ MPD, AML can arise in a V617F- cell.
V617F- leukemic transformation developed in 3 of 4 patients with a preceding V617F+ MPD. (A) Paired sequence traces on 2 patients with PV from the PV phase (top panel) and leukemic phase (middle panel) of disease showing reversion of mutated nucleotide (arrowheads) to WT. Bottom panels are smears from samples used for sequencing (bone marrow aspirate for PV1 and peripheral-blood mononuclear cells for PV2). (B) Sequence traces from bone marrow samples taken after leukemic transformation in 2 patients with ET (top panel). Samples from before AML transformation showed both patients to be V617F+ by allele-specific PCR (data not shown). Both samples had high percentage of blast cells in their bone marrow aspirates (bottom panels). Romanowsky-stained slides were imaged on an Olympus BX51 microscope (Olympus, Center Valley, PA) equipped with a 40×/0.85 oil-immersion objective lens. A Pixera Penguin 600CL camera (Pixera, Los Gatos, CA) was used to capture images, and Viewfinder 3.0.1 (Pixera) was used to acquire them.
V617F- leukemic transformation developed in 3 of 4 patients with a preceding V617F+ MPD. (A) Paired sequence traces on 2 patients with PV from the PV phase (top panel) and leukemic phase (middle panel) of disease showing reversion of mutated nucleotide (arrowheads) to WT. Bottom panels are smears from samples used for sequencing (bone marrow aspirate for PV1 and peripheral-blood mononuclear cells for PV2). (B) Sequence traces from bone marrow samples taken after leukemic transformation in 2 patients with ET (top panel). Samples from before AML transformation showed both patients to be V617F+ by allele-specific PCR (data not shown). Both samples had high percentage of blast cells in their bone marrow aspirates (bottom panels). Romanowsky-stained slides were imaged on an Olympus BX51 microscope (Olympus, Center Valley, PA) equipped with a 40×/0.85 oil-immersion objective lens. A Pixera Penguin 600CL camera (Pixera, Los Gatos, CA) was used to capture images, and Viewfinder 3.0.1 (Pixera) was used to acquire them.
Discussion
We have addressed several issues raised by the discovery of the JAK2 V617F mutation, including the relationship between V617F- and V617F+ subtypes of ET, the timing of the JAK2 mutation, and the molecular mechanisms associated with leukemia transformation.
We have previously demonstrated that the V617F+ and V617F- subtypes of ET exhibit multiple laboratory and clinical differences and have suggested that the 2 subtypes represent pathogenetically distinct diseases rather than sequential phases of the same disease.12 We now provide direct evidence for this model. Comparison of paired samples from 50 patients with V617F- ET showed no evidence that they subsequently acquire a JAK2 V617F mutation. In addition, we demonstrate that V617F+ and V617F- MPD patients exhibit very different patterns of cytogenetic abnormalities. In particular, the demonstration that 28 of 29 patients with a 20q deletion were V617F+ provides striking evidence for cooperation between the JAK2 mutation and gene(s) located on the long arm of chromosome 20.
Kralovics and colleagues have proposed a “sequential disorder” model in which V617F- MPDs represent an earlier phase of V617F+ disease.4 Support for this model came from 2 lines of evidence. First, they found that V617F+ patients had a longer duration of disease.4 The discrepancy between this result and data from the PT-1 trial12 may reflect methodologic differences. Kralovics and colleagues used sequence analysis that is now recognized to be less sensitive than allele-specific PCR.12 Because the proportion of V617F+ granulocytes increases with time,31 sequence analysis will preferentially detect patients relatively late in the course of their disease, whereas patients in the early stage may be falsely labeled as V617F-. This bias may produce an apparent difference in the duration of the disease between the 2 subtypes of ET. The second line of evidence is that familial MPD pedigrees have both V617F+ and V617F- members. However, it is not clear whether the genetic lesion that predisposes to an MPD in such families contributes to the pathogenesis of sporadic MPDs.
Our results highlight several issues that limit the ability of XCIP analysis to demonstrate a pre-JAK2 clone of V617F- cells in V617F+ patients. First, several groups have demonstrated small differences in XCIPs from T cells and granulocytes in healthy subjects.24,29,32,33 We show here that these differences result in wide confidence limits for the calculated proportion of V617F- cells that are apparently clonally derived. Importantly, this variability is systematically biased toward overestimating the proportion of clonally derived granulocytes. These problems result from inherent differences in granulocyte and T-cell XCIPs (due to selection for X-linked polymorphisms that affect stem-cell behavior24,34 ) and therefore apply to both DNA and RNA methods of XCIP analysis. Second, when estimating the proportion of clonal cells, it is assumed that T cells are not involved in the malignant clone. In support of this, T cells lack the JAK2 mutation1,2,4,25-27 and, in the majority of patients, have balanced X-inactivation patterns.35,36 However, T-cell involvement by a pre-JAK2 clone in a minority of patients cannot be excluded. The third important assumption is that granulocytes are heterozygous. A mixed population of homozygous, heterozygous, and WT cells would lead to overestimation of the proportion of JAK2+ granulocytes. In this report, we have minimized the impact of this by studying patients with ET (in whom homozygosity is rare), by excluding PV patients with a homozygous sequence trace, and by selecting patients with a low V617F/WT ratio. Nevertheless, such a phenomenon may explain patient C in Figure 1C and remains a further caveat to the use of XCIPs to identify a pre-JAK2 phase.
Models for the development of V617F- AML from a V617F+ MPD. (A) Three possible models are shown, in which V617F- AML could develop from (1) a V617F+ cell that subsequently reverts to V617F-; (2) a cell that had some other initiating mutation prior to JAK2; or (3) a normal stem cell. (B) Microsatellite analysis on peripheral-blood granulocytes and T cells from the PV phase of disease (taken in 1992) and blasts from leukemic transformation (2004) for 2 markers on chromosome 9p either side of JAK2 from patient PV2.
Models for the development of V617F- AML from a V617F+ MPD. (A) Three possible models are shown, in which V617F- AML could develop from (1) a V617F+ cell that subsequently reverts to V617F-; (2) a cell that had some other initiating mutation prior to JAK2; or (3) a normal stem cell. (B) Microsatellite analysis on peripheral-blood granulocytes and T cells from the PV phase of disease (taken in 1992) and blasts from leukemic transformation (2004) for 2 markers on chromosome 9p either side of JAK2 from patient PV2.
These problems inherent in comparisons between JAK2 quantitation and XCIPs make it difficult to draw meaningful conclusions about the existence of a pre-JAK2 clone. However, it is important to emphasize that these considerations do not exclude a pre-JAK2 phase of disease in V617F+ patients. Indeed, this remains a potential explanation for our striking observation that several patients with a V617F+ MPD transformed to a V617F- AML. An analogous sequence of events is highly unusual in other hematologic malignancies. For example, the blast transformation of chronic myeloid leukemia is BCR-ABL+ and the cytogenetic abnormalities of AML evolving from preceding myelodysplasia include those of the original myelodysplastic clone.37 Our observations in V617F+ MPDs extend the findings of Jelinek et al,10 who found that the prevalence of the JAK2 mutation was lower in samples from patients with AML transformed from antecedent PV and IMF than in their cohort of PV and IMF patients without transformation.
There are 3 possible models for the development of a V617F- AML in a patient with V617F+ MPD (Figure 4A). First, leukemic transformation may have occurred in a V617F+ cell that subsequently became V617F-. It is possible that the JAK2 V617F mutation provides a strong differentiating signal to hematopoietic progenitors in which case leukemic blasts that lose this signal may gain a growth advantage. However, this model seems unlikely because, in patient PV1, the leukemic cells did not have either the 20q deletion or the t(1;9) translocation that were present in the PV phase. Furthermore, we were unable to find any evidence for mitotic recombination or deletion of the originally mutated JAK2 gene in either patient PV1 or PV2 (Figure 4B). We cannot exclude the possibility that point mutation could have caused reversion to WT JAK2, but overall there seems little evidence in favor of this model.
Another possibility is that the leukemia could have developed from a normal stem cell, not part of the original clone. This could explain patient PV1, because he received known mutagens (32P and busulphan). However, the other patients received only hydroxyurea. Although there is controversy over the leukemogenicity of hydroxyurea in MPDs, extensive experience with the drug in sickle cell anemia suggests that hydroxyurea does not commonly induce leukemic transformation of normal stem cells.38 It is therefore difficult to explain why patients PV2 and ET1 would develop leukemia in a normal stem cell following therapy with hydroxyurea alone.
The third model to explain the development of V617F- AML is that, in these patients, the JAK2 mutation followed an initial pre-JAK2 phase of their disease, which resulted from an unidentified initial mutation. Such an initiating mutation could either confer a growth or survival advantage in cooperation with the JAK2 and the AML mutations or, alternatively, could promote genomic instability, thus predisposing the target cell to the acquisition of further mutations. This model raises the question of why the AML did not develop in a stem cell with both the initiating mutation and the V617F mutation, because with increasing time the V617F+ cells come to predominate. One possibility is that the V617F mutation selects against proliferation of immature myeloblasts, perhaps by providing a strong differentiation signal, although it then becomes difficult to explain both patient ET2 who actually evolved toward a homozygous V617F pattern in her leukemic cells, and rare occurrences of V617F+ de novo AML.7,9,10
Our results argue against the first model to explain the absence of the JAK2 mutation in the leukemic phase of V617F+ MPDs and demonstrate that distinguishing between models 2 and 3 will be difficult. Importantly, confirmation of model 3 will require identification of the putative initiating mutation since indirect methods based on XCIP analysis are too sensitive to departures from the assumption of equal XCIPs in T cells and granulocytes.
Prepublished online as Blood First Edition Paper, July 27, 2006; DOI 10.1182/blood-2005-12-013748.
Supported by grants from the Leukaemia Research Fund, United Kingdom.
The authors declare no competing financial interests.
P.J.C. performed HUMARA, JAK2 quantitation, allele-specific PCR, and statistical analyses; E.J.B. performed the microsatellite PCR, AML slide scraping and analysis, and cytogenetic data collation; P.A.B. extracted and analyzed the DNA from the V617F- ET follow-up samples and L.M.S. genotyped the initial ET samples; A.J.B. and B.J.P.H. collected samples and clinical information on the Cambridge cohort of 20q and 13q deletions, respectively; W.N.E. analyzed the bone marrow aspirates and trephines; R.K., T.S.L., S.G., M-C. Le B-K., M.G., J.T.R., B.Y.C., and C.N.H. provided clinical information, cytogenetic analyses, and JAK2 status on patients with recurrent cytogenetic abnormalities; and A.R.G. coordinated and directed the research. All authors have reviewed and contributed to the manuscript.
P.J.C. and E.J.B. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We would like to thank Kath Andrews and Clare Bryant for their assistance with the cytogenetics analyses. We would also like to thank the Addenbrooke's Haematological Disorders Sample Bank for assistance with sample processing.

![Figure 2. Comparison of XCIPs with proportion of V617F+ granulocytes. (A) Median and 95% confidence intervals produced by Monte Carlo simulations of estimated clonal granulocytes for a given proportion of V617F+ cells. Two estimates for the variability of granulocytes around T-cell proportion were used, based on published age-specific data in healthy women.29 The dashed line shows the “true” line under the null hypothesis that V617F is the initiating mutation, that is, that the proportion of clonal granulocytes is the same as the proportion of V617F+ cells. The individual point estimates from the 20 women studied (Figure 1C) are shown. (B) Worked examples demonstrating the effects of assuming equal T-cell and granulocyte X-inactivation patterns on the estimated proportion of clonal granulocytes from (i) a patient in whom 50% of T cells and granulocytes carry active Xm and (ii) a patient in whom 50% of T cells and 75% of granulocytes carry active Xm. The estimated proportion of clonally derived granulocytes from XCIP analysis was derived from (ii) using the standard formula21,24 as follows: with similar calculations for (i). Note that if the V617F mutation occurs in an Xm cell, the unequal XCIPs in T cells and granulocytes result in an apparent excess of clonally derived cells. GC indicates proportion of clonally derived granulocytes; Xm, cell with active maternal X chromosome; Gr, granulocytes; T, T cells. \batchmode \documentclass[fleqn,10pt,legalpaper]{article} \usepackage{amssymb} \usepackage{amsfonts} \usepackage{amsmath} \pagestyle{empty} \begin{document} \[\ G_{C}=\frac{R_{G}-R_{T}}{R_{G}+1}=\frac{80{/}20-50{/}50}{1+80{/}20}=60\%,\ \] \end{document}](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/10/10.1182_blood-2005-12-013748/4/m_zh80220603730002.jpeg?Expires=1769085938&Signature=GGNMBlrZCoPeGAYJl7HNlmlqHkXWBMpvBUUGQwU2V9GF-LIPKhwwl3JOFFFBZirs~mRuNK4mY3k26jc8rVdLW-rm03r9ZVA4cB1TgUdx7XPukpES4jj1nHJj-8OMIcXsrYmI8wvjWey3XVEMee4ddazBy4FFNizrsOO0o0MMEew-Gs0HtgqxmlPW6UUKomdOIDPUe0li42ZK68WCrFDdsLc-FclPr2mFO2BDxOjkYR5LwQ2GRmvYnIyNCxgRBScNA~KfDHBVcyDo~JV4fmjA7nc3qTnnEmRFOU9ChcMB~-RMgZAVLdHMfLVCqNw2n4F2kZ9s7veEckJLYbd97HIdjg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal