Abstract
Basophil numbers are typically elevated in chronic myeloid leukemia (CML) and increase during disease progression. Histamine is an essential mediator and marker of basophils and is highly up-regulated in CML. We examined the biochemical basis of histamine synthesis in CML cells. The CML-specific oncoprotein BCR/ABL was found to promote expression of histidine decarboxylase (HDC) and synthesis of histamine in Ba/F3 cells. Moreover, the BCR/ABL tyrosine kinase inhibitors imatinib (STI571) and nilotinib (AMN107) decreased histamine levels and HDC mRNA expression in BCR/ABL-transformed Ba/F3 cells, in the CML-derived basophil cell line KU812, and in primary CML cells. Synthesis of histamine was found to be restricted to the basophil compartment of the CML clone and to depend on signaling through the PI3-kinase pathway. CML cells also expressed histamine receptors (HRs), including HR-1, HR-2, HR-4, and histamine-binding CYP450 isoenzymes which also serve as targets of HR antagonists. The HR-1 antagonists loratadine and terfenadine, which bind to CYP450, were found to counteract proliferation of CML cells, whereas no growth inhibition was observed with the HR-1 antagonist fexofenadine which is not targeted or metabolized by CYP450. Moreover, DPPE, an inhibitor of histamine-binding CYP450 isoenzymes, produced growth inhibition in CML cells. Together, these data show that BCR/ABL promotes histamine production in CML cells and that certain HR-targeting drugs exert antileukemic effects on CML cells.
Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative disease in which leukemic cells exhibit the reciprocal translocation t(9;22) and the BCR/ABL oncogene.1-3 The resulting oncoprotein, BCR/ABL, displays constitutive tyrosine kinase activity and activates a number of signaling pathways that contribute to abnormal growth and survival of leukemic cells.4-6 Clinically, CML can be divided into 3 phases: the chronic phase, the accelerated phase, and the blast phase of CML, which resembles an acute leukemia.7-10
A number of prognostic factors correlate with the phase of CML and are indicative of a poor survival. Among them are the blast count, basophilia, cytopenia, and additional chromosomal defects indicating clonal evolution.7-13 A strong independent prognostic variable in CML is basophilia.11-16 Thus, basophils are almost invariably elevated in CML, and basophil numbers typically increase during disease progression. Therefore, basophilia is an accepted criterion of disease acceleration in CML.7-10
Histamine is an essential mediator and marker of basophils and their progenitors and is highly up-regulated in CML.17-20 Studies performed on basophil progenitors in healthy subjects and in patients with CML suggest that production of histamine is an early event in basophil development.21-23 Thus, even the most immature basophil progenitor cells express histamine.22,23 Therefore, histamine has been considered as a useful marker to determine basophil lineage involvement.17-20 Other studies have shown that histamine is a potential regulator of growth and differentiation of myeloid progenitor cells.24-27
The key enzyme involved in histamine generation is histidine decarboxylase (HDC).28 This enzyme is expressed in basophil-committed progenitors as well as in mature basophils.21,23 Interleukin-3 (IL-3), a major differentiation factor for basophils, has been reported to induce the expression of HDC and thus synthesis of histamine in myeloid progenitors.21-23 By contrast, in CML, the biochemical basis of synthesis and expression of histamine in (basophil) progenitors remains unknown. In fact, although basophils in CML possess the BCR/ABL oncogene,29,30 it has not been clarified whether (increased) histamine synthesis and (enhanced) basophil differentiation are directly induced by this oncogene, by IL-3, or by other factors.
The aims of the current study were to clarify whether the disease-associated oncoprotein BCR/ABL, which uses signaling pathways similar to the IL-3 receptor, is involved in the regulation of synthesis of histamine in CML cells, and to define the role of histamine and of histamine receptors (HRs) in the growth of leukemic cells.
Materials and methods
Monoclonal antibodies and other reagents
The PE-labeled monoclonal antibody (mAb) 97A6 (CD203c)31,32 was purchased from Immunotech (Marseille, France), a polyclonal anti-HDC antibody was from Progen (Heidelberg, Germany), biotinylated horse anti-mouse IgG and goat anti-rabbit IgG avidin-biotin-peroxidase complex, as well as streptavidin-biotin-peroxidase complex from Vector Laboratories (Burlingame, CA), and goat anti-rabbit IgG from Biocarta (San Diego, CA). The basophil-specific mAb 2D733 was a kind gift from Dr L. B. Schwartz (Virginia Commonwealth University, Richmond). The Protoscript First Strand cDNA Synthesis kit was purchased from New England Biolabs (Beverly, MA); RPMI 1640 medium and fetal calf serum (FCS) from PAALaboratories (Pasching,Austria); α-fluoromethylhistidine (α-FMH), histamine, and histamine receptor (HR) antagonists (HR-1: loratadine, terfenadine, fexofenadine; HR-2: famotidine, cimetidine, ranitidine) from Sigma (St Louis, MO); PD98059, LY294002, and rapamycin were from Calbiochem (San Diego, CA); and recombinant human granulocyte macrophage colony-stimulating factor (GM-CSF) was from PeproTech (Rocky Hill, NJ). N,N-diethyl-2-(4-(phenylmethyl)phenoxy)-ethanamine (DPPE)34,35 was a kind gift from Dr V. Laszlo (Semmelweis University, Budapest, Hungary). Imatinib (STI571) and nilotinib (AMN107)36 were kindly provided by Dr P. W. Manley and Dr D. Fabbro, Novartis Pharma AG (Basel, Switzerland).
Cell lines and culture conditions
Ton.B210 and Ton.B210-X cells are interleukin-3 (IL-3)-dependent Ba/F3-derived cell lines, in which BCR/ABL can conditionally be induced through addition of doxycycline 1 μg/mL.37,38 Both cell lines were grown in RPMI 1640 medium with 10% FCS and 10% WEHI-3B-conditioned medium (source of murine IL-3) at 37°C and 5% CO2. For starvation, cells were cultured in the absence of IL-3 for up to 24 hours. The BCR/ABL+ cell lines K562 (multilineage) and KU812 (basophil committed39 ), as well as the BCR/ABL-negative cell lines HL60, KG1a, A431, and lung fibroblasts (LUFs), were maintained in RPMI 1640 medium and 10% FCS.
Isolation and culture of primary cells
Primary leukemic cells were obtained from 20 patients with untreated chronic-phase CML and 6 with accelerated-phase CML (with basophilia) after obtaining approval from the Institutional Review Board of the Medical University of Vienna (Vienna, Austria). Normal bone marrow (BM) cells were obtained from 6 patients with lymphoma (routine staging) without BM involvement. In accordance with the Declaration of Helsinki, informed consent was obtained in each case prior to blood donation or BM puncture. BM mononuclear cells (BMMCs) and peripheral-blood mononuclear cells (PBMCs) were isolated by centrifugation using Ficoll. Isolated cells were recovered and cultured in RPMI 1640 medium containing 10% FCS with or without GM-CSF (100 ng/mL). In 2 patients with CML and marked basophilia and 1 without basophilia, PBMCs were separated into 2 fractions by cell sorting, namely a CD203c-positive fraction (> 98% basophils), and a CD203c-negative fraction (< 1% basophils) using the PE-labeled CD203c mAb 97A6.31 In one patient with accelerated-phase CML, CD34+ BMMCs were purified by cell sorting using a mAb against CD34 (BD Pharmingen, San Diego, CA). Cell sorting was performed on a fluorescence-activated cell sorter (FACS)-Vantage (Becton Dickinson, San Jose, CA). After sorting, cell viability was more than 95% in all experiments.
Immunohistochemistry (IHC) and immunocytochemistry (ICC)
In 3 patients with chronic-phase CML and 3 with accelerated-phase CML, expression of HDC was analyzed by IHC on serial sections (2 μm) prepared from paraffin-embedded formalin-fixed BM specimens. Immunohistochemistry was performed using the indirect immunoperoxidase-staining technique, an anti-HDC antibody (work dilution, 1:500) and the basophil-specific mAb 2D7 (work dilution, 1:500) as described.40-43 For ICC, cells (primary CML cells, cell lines) were spun on cytospin slides and stained with the anti-HDC antibody or 2D7 antibody as described.40-43 Acquisition of figures was performed by an Olympus DP11 camera connected to an Olympus microscope equipped with 40×/0.85 (Figure 1A, B) and 100×/1.35 (Figure 1C, D) UPlan-Apo objective lenses (Olympus, Hamburg, Germany). Images were acquired with Adobe Photoshop CS2 software version 9.0 (Adobe Systems, San Jose, CA) software and processed with PowerPoint software (Microsoft, Redmond, WA).
Western blotting
K562 cells and KU812 cells were cultured in control medium, imatinib (1 μM), AMN107 (1 μM), loratadine (5 μM), or terfenadine (5 μM) at 37°C for 4 hours. Thereafter, cells were recovered and subjected to Western blot analysis. Western blotting was performed as described44 using the antiphosphoprotein mAb 4G10 (1:1000) (Upstate Biotechnology, Lake Placid, NY) and a rabbit-anti-human β-actin antibody (1:1000) (Sigma).
Treatment of cells with compounds and signal transduction inhibitors
Primary CML cells and cell lines (KU812 cells, Ton.B210-X cells) were incubated with the PI3-kinase inhibitor LY294002 (20 μM), MEK (MAP-kinase/ERK-kinase) inhibitor PD98059 (50 μM), the mTOR-inhibitor rapamycin (20 nM) (all from Calbiochem), imatinib (60 nM to 1 μM) (Novartis Pharma AG), AMN10736 (10 pM to 1 μM) (Novartis Pharma AG), or control medium in 10% FCS at 37°C for up to 24 hours. In a separate set of experiments, K562 cells and KU812 cells were incubated with various concentrations (10 pM to 100 mM) of histamine, HR antagonists (HR-1: loratadine, terfenadine, fexofenadine, each 100 fM to 1 mM; HR-2: famotidine, cimetidine, ranitidine, each 100 fM to 1 mM), or with combinations of HR antagonists and BCR/ABL-targeting drugs (imatinib, AMN107). In other experiments, DPPE34,35 (1 nM to 1 mM), or α-FMH45 (1 nM to 100 μM) were applied.
Northern blot analysis and RT-PCR
Total RNA was isolated from primary CML cells or cell lines using Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Northern blotting was performed as described.38 In brief, 15 μg total RNA was size-fractionated on 1.0% formaldehyde-agarose gels and transferred to nylon membranes (Hybond-N; Amersham, Aylesbury, United Kingdom).46 Hybridization was performed with 32P-labeled cDNAs specific for HDC or β-actin (see below). Labeling was performed using the Megaprime-kit (Amersham). Bound radioactivity was visualized by exposure to Biomax-MS film (Kodak, Rochester, NY) at -80°C using intensifying screens (Kodak). mRNA expression levels were quantified by densitometry using E.A.S.Y.-Win32 software (Herolab, Wiesloch, Germany).
Primers for reverse transcriptase-polymerase chain reaction (RT-PCR) and generation of Northern blot probes were as follows: murine β-actin, 5′-GACGGCCAGGTCATCACTAT-3′ (forward) and 5′-AGGGAGACCAAAGCCTTC AT-3′ (reverse); human β-actin, 5′-ATGGATGATGATATCGCCGCG-3′ (forward) and 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGGGCC-3′ (reverse); murine HDC, 5′-ATGATGGAGCCCTGTGAATACCG-3′ (forward) and 5′-CCAGAGTTGGCATGTCGGAGGTAG-3′ (reverse); human HDC, 5′-ATGATGGAGCCTGAGGAGTACAG-3′ (forward) and 5′-CCTGAGTTGGCATGCCTGAGGTAG-3′ (reverse); human HR1, 5′-AGGGAGTGAGCCATAACTGGTGGC-3′ (forward) and 5′-CAGATAGTGCTCAGGACCACCACCAG-3′ (reverse); human HR2, 5′-GGACTGAGCCGTAGAGTCCCAGGATG-3′ (forward) and 5′-GCCAGCAACGGTGATGAGGATG-3′ (reverse); human HR3, 5′-CCAGAACAACTTCTTCCTGCTCAACC-3′ (forward) and 5′-CCACTACCAGCCACAGCTTGCAG-3′ (reverse); human HR4, 5′-CATCCCTCACACGCTGTTCGAATG-3′ (forward) and 5′-CGGCCACCATCAGAGTAACAATCTTC-3′ (reverse). Primers specific for CYP450 isoenzymes were identical to those described by Nagai et al.47 RT-PCR reactions were performed using the Protoscript First Strand cDNA Synthesis kit (New England Biolabs) using 1 μg total RNA in a 50-μL reaction volume with primer pairs mentioned earlier in this section. PCR conditions were as follows: initial denaturation at 94°C for 40 seconds, annealing at 59 to 67°C for 40 seconds, polymerization at 72°C for 40 seconds (35 cycles), and terminal extension at 72°C for 10 minutes. The products were resolved in 1% agarose gels containing 0.5 μg/mL ethidium bromide.
For real-time PCR, total RNA (1 μg) was reverse transcribed into cDNA with random hexamers (1 μg/μg total RNA). The reaction mixture was incubated at 37°C for 45 minutes followed by 15 minutes at 45°C and 20 minutes at 70°C. All real-time PCRs were performed using the iTaq SYBR Green kit (BioRad, Hercules, CA). The following primer pairs were used: murine HDC, 5′-ACTCCAAATGTGCAGCCTGGATACC-3′ (forward) and 5′-GGCTAGATGCCCACGTGAATCCTAA-3′ (reverse); murine β-actin, 5′-GTGGGCCGCTCTAGGCACCAA-3′ (forward) and 5′-CTCTTTGATGTCACGCACGATTTC-3′ (reverse). Using the ABI-Prism 7700 sequence detection system (PE Applied Biosystems, Warrington, United Kingdom), PCR cycling conditions were as follows: initial denaturation at 95°C for 10 minutes, followed by 40 cycles at 94°C for 30 seconds, 60°C for 15 seconds, and 72°C for 30 seconds and a 10-minute terminal incubation at 72°C. Expression of target genes were measured in triplicates and were normalized to β-actin levels.
Measurement of histamine in lysates and supernatants of leukemic cells
Histamine levels were quantified in lysates and/or supernatants of primary CML cells, KU812 cells, Ton.B210 cells, or Ton.B210-X cells. KU812 cells were cultured in the presence or absence of imatinib (0.1-1 μM) or AMN107 (0.1-1 μM) for up to 14 days before being analyzed. Ton.B210 and Ton.B210-X cells were kept in control medium with or without IL-3 or were exposed to doxycycline (1 μg/mL) in the absence or presence of IL-3 for up to 24 hours. In control experiments, nontransformed Ba/F3 cells (lacking BCR/ABL) were incubated with doxycycline for 24 hours. In separate experiments, KU812 and Ton.B210-X cells (after exposure to doxycycline) were incubated with imatinib (1 μM), AMN107 (0.1 μM), PD98059 (50 μM), LY294002 (20 μM), or rapamycin (20 nM) for up to 24 hours. After incubation, total (cellular + extracellular) histamine was measured in cell suspensions. To show factor-independent synthesis and release of histamine in primary cells, histamine levels were measured in supernatants and lysates of cultured mononuclear cells (up to 24 days) in 7 patients with CML (chronic phase, n = 4; accelerated phase, n = 3) and in 3 donors with normal bone marrow. Cellular histamine levels were measured by a commercial radioimmunoassay (Immunotech, Marseille, France) as described.39
Determination of cell viability and 3H-thymidine incorporation
Cell viability was determined by trypan blue exclusion. The percentage of apoptotic cells was quantified on Wright-Giemsa-stained cytospin preparations.48 To investigate growth inhibitory effects, K562 and KU812 cells were cultured in 96-well microtiter plates (5 × 104 cells/well) in the absence or presence of various concentrations of imatinib, AMN107, histamine, HR antagonists, α-FMH, or DPPE at 37°C for 48 hours. In a separate set of experiments, K562 and KU812 cells were incubated with combinations (in fixed ratio according to IC50) of BCR/ABL inhibitors (imatinib, AMN107) and HR antagonists loratadine and terfenadine at 37°C for 48 hours. In select experiments, primary CML (progenitor) cells were cultured in the presence of BCR/ABL inhibitors, control medium, or HR-1 antagonists. After incubation, 1 μCi (0.037 MBq) 3H-thymidine was added to cells in each well. Twelve hours later, cells were harvested on filter membranes (Packard Bioscience, Meriden, CT). Filters were air-dried, and bound radioactivity was measured in a β-counter (Top-Count NXT; Packard Bioscience). All experiments were performed in triplicates.
Statistical evaluation of data
To determine the level of significances in differences in growth rates in leukemic cells, standard statistical tests, including the paired Student t test, were applied. Results were considered to be significantly different when the P value was less than .05. To determine cooperative antileukemic effects between HR antagonists and tyrosine kinase inhibitors, combination index values were calculated using a commercially available software program (Calcusyn; Biosoft, Ferguson, MO).
Results
Expression of histidine decarboxylase (HDC) and histamine in CML cells
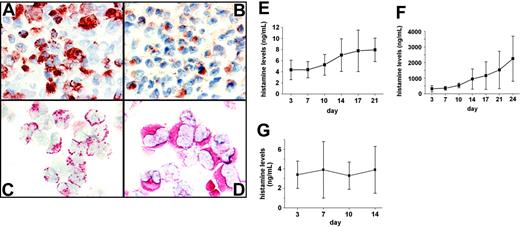
As assessed by IHC and ICC, primary CML cells were found to express HDC in all patients. Figure 1A and B show adjacent BM sections from a patient with CML, stained with the basophil-specific antibody 2D7 (1A) and an antibody against HDC (1B). These experiments revealed that the percentage and distribution of HDC-positive cells in the BM match those of 2D7-positive cells, suggesting that HDC is expressed in basophils. Expression of HDC was also demonstrable in enriched CML basophils by ICC (Figure 1C-D). Expression of HDC in CML cells was found to be associated with constitutive production and secretion of histamine (Figure 1E-F). Whereas the levels of (secreted) histamine in supernatants of cultured CML cells were relatively low in patients with chronic-phase CML (Figure 1E), histamine levels were high and were found to accumulate extensively over time in supernatants of cultured cells in accelerated-phase CML (Figure 1F). In normal BMMCs, histamine concentrations measured in supernatants remained at baseline levels (Figure 1G).
Immunohistochemical and immunocytochemical detection of HDC in CML cells. (A-B) Immunohistochemical analysis of bone marrow (BM) cells for expression of histidine decarboxylase (HDC) in a patient with accelerated-phase CML. Adjacent BM sections were stained with the basophil-specific antibody 2D7 (A) and an antibody against HDC (B). Original magnification, ×100. (C-D) Immunocytochemistry of primary CML cells obtained from the same patient. Slides were stained with the basophil-specific antibody 2D7 (C) and an antibody against HDC (D). Original magnification, ×40. (E-G) Measurement of histamine in cell-free supernatants of leukemic cells obtained from patients with chronic-phase CML (n = 4) (E), from patients with accelerated-phase CML (n = 3) (F), and in normal BMMCs (n = 3) (G). Isolated CML cells were cultured in RPMI 1640 medium and 10% FCS for various time periods as indicated. Concentrations of histamine in cell-free supernatants were determined by radioimmunoassay (RIA). Results represent the mean ± SD of 3 or 4 independent experiments.
Immunohistochemical and immunocytochemical detection of HDC in CML cells. (A-B) Immunohistochemical analysis of bone marrow (BM) cells for expression of histidine decarboxylase (HDC) in a patient with accelerated-phase CML. Adjacent BM sections were stained with the basophil-specific antibody 2D7 (A) and an antibody against HDC (B). Original magnification, ×100. (C-D) Immunocytochemistry of primary CML cells obtained from the same patient. Slides were stained with the basophil-specific antibody 2D7 (C) and an antibody against HDC (D). Original magnification, ×40. (E-G) Measurement of histamine in cell-free supernatants of leukemic cells obtained from patients with chronic-phase CML (n = 4) (E), from patients with accelerated-phase CML (n = 3) (F), and in normal BMMCs (n = 3) (G). Isolated CML cells were cultured in RPMI 1640 medium and 10% FCS for various time periods as indicated. Concentrations of histamine in cell-free supernatants were determined by radioimmunoassay (RIA). Results represent the mean ± SD of 3 or 4 independent experiments.
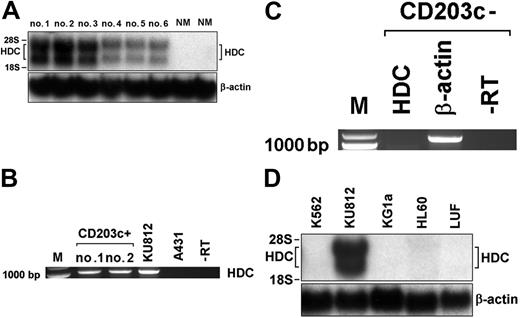
Detection of HDC mRNA expression in primary CML cells by Northern blotting and RT-PCR. (A) Northern blot analysis of HDC mRNA expression in PBMCs obtained from 3 patients with accelerated-phase CML (nos. 1-3), 3 with chronic-phase CML (no. 4-6), and 2 donors with normal bone marrow (NM). Total RNA was isolated and subjected to Northern blot analysis using an HDC-specific cDNA probe. β-actin served as loading control. (B) RT-PCR analysis of HDC mRNA expression in purified CD203c+-sorted basophils (purity > 98%) obtained from 2 patients (nos. 1, 2) with accelerated-phase CML, and in KU812 cells. (C) RT-PCR analysis of basophil-depleted (CD203c-negative) CML cells using primers specific for HDC or β-actin. The RT omission controls (-RT) are also shown. (D) Northern blot analysis of expression of HDC mRNA in various myeloid cell lines (K562, KU812, KG1a, HL60) and human lung fibroblasts (LUFs) using cDNA probes specific for HDC and β-actin (loading control).
Detection of HDC mRNA expression in primary CML cells by Northern blotting and RT-PCR. (A) Northern blot analysis of HDC mRNA expression in PBMCs obtained from 3 patients with accelerated-phase CML (nos. 1-3), 3 with chronic-phase CML (no. 4-6), and 2 donors with normal bone marrow (NM). Total RNA was isolated and subjected to Northern blot analysis using an HDC-specific cDNA probe. β-actin served as loading control. (B) RT-PCR analysis of HDC mRNA expression in purified CD203c+-sorted basophils (purity > 98%) obtained from 2 patients (nos. 1, 2) with accelerated-phase CML, and in KU812 cells. (C) RT-PCR analysis of basophil-depleted (CD203c-negative) CML cells using primers specific for HDC or β-actin. The RT omission controls (-RT) are also shown. (D) Northern blot analysis of expression of HDC mRNA in various myeloid cell lines (K562, KU812, KG1a, HL60) and human lung fibroblasts (LUFs) using cDNA probes specific for HDC and β-actin (loading control).
Subsequently, we analyzed expression of HDC mRNA in CML cells. As assessed by Northern blotting, primary CML cells were found to express HDC mRNA in a constitutive manner in all patients (Figure 2). As visible in Figure 2A, higher levels of HDC mRNA transcripts were found in patients with accelerated-phase CML with basophilia compared with chronic-phase CML or normal BM (densitometry results, AP: 100% ± 26% versus CP: 19% ± 14%; P < .05). To confirm expression of HDC in basophils in CML, basophils were sorted from the peripheral blood of 2 patients with accelerated-phase CML using the basophil-specific CD203c-antibody 97A6.31 As assessed by RT-PCR, isolated CD203c-positive cells (basophils) expressed substantial amounts of HDC mRNA (Figure 2B), whereas CD203c-negative (basophil-depleted) cells did not express substantial amounts of HDC mRNA (Figure 2C). Finally, we examined expression of HDC mRNA in various myeloid cell lines. As visible in Figure 2D, the CML-derived basophil cell line KU812 was found to express substantial amounts of HDC mRNA (both transcripts49 ), whereas the nonbasophil cell line K562 did not express significant amounts of HDC mRNA (Figure 2D). Corresponding data were obtained when cellular histamine levels were measured by RIA. In particular, KU812 cells expressed relatively high concentrations of histamine (0.01-0.1 pg/cell), whereas K562 cells expressed significantly lower histamine levels (100-1000 times less compared with KU812). Together, these data show that expression of HDC and synthesis of histamine in CML cells depend on the cellular background (basophil differentiation capacity) of the clone.
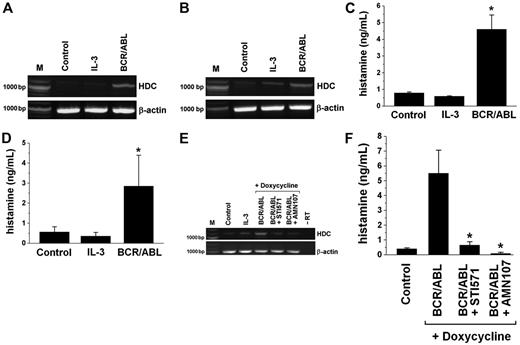
Detection of HDC and histamine levels in Ba/F3 cells inducibly expressing the BCR/ABL oncoprotein. (A-B) RT-PCR analysis of HDC mRNA expression in Ton.B210 cells (A) and Ton.B210-X cells (B) cultured in the absence of IL-3 (Control), in the presence of IL-3, or in the presence of doxycycline (1 μg/mL) to induce BCR/ABL for 24 hours. RT-PCR was performed using primers specific for HDC or β-actin (loading control). (C-D) Histamine levels in lysates of Ton.B210 cells (C) and Ton.B210-X cells (D) cultured in the absence of IL-3 (Control), in the presence of IL-3, or in the presence of doxycycline (1 μg/mL) to express BCR/ABL for 24 hours. Histamine was quantified by RIA. Results represent the mean ± SD from 3 independent experiments. *P < .05. (E) RT-PCR analysis of HDC mRNA expression in Ton.B210-X cells cultured in the absence of IL-3 (Control), in the presence of IL-3, or in the presence of doxycycline (BCR/ABL) for 24 hours. BCR/ABL-expressing cells were incubated with control medium, imatinib, 1 μM (BCR/ABL + STI571), or AMN107, 100 nM (BCR/ABL + AMN107) for another 24 hours. RT-PCR was performed using primers specific for HDC or β-actin (loading control). The RT omission control (-RT) is also shown. (F) Histamine levels (by RIA) in Ton.B210-X cells cultured in the absence of IL-3 (Control) or in the presence of doxycycline with or without inhibitory drugs (STI571, 1 μM; AMN107, 100 nM) for 24 hours. Results represent the mean ± SD from 3 independent experiments. *P < .05.
Detection of HDC and histamine levels in Ba/F3 cells inducibly expressing the BCR/ABL oncoprotein. (A-B) RT-PCR analysis of HDC mRNA expression in Ton.B210 cells (A) and Ton.B210-X cells (B) cultured in the absence of IL-3 (Control), in the presence of IL-3, or in the presence of doxycycline (1 μg/mL) to induce BCR/ABL for 24 hours. RT-PCR was performed using primers specific for HDC or β-actin (loading control). (C-D) Histamine levels in lysates of Ton.B210 cells (C) and Ton.B210-X cells (D) cultured in the absence of IL-3 (Control), in the presence of IL-3, or in the presence of doxycycline (1 μg/mL) to express BCR/ABL for 24 hours. Histamine was quantified by RIA. Results represent the mean ± SD from 3 independent experiments. *P < .05. (E) RT-PCR analysis of HDC mRNA expression in Ton.B210-X cells cultured in the absence of IL-3 (Control), in the presence of IL-3, or in the presence of doxycycline (BCR/ABL) for 24 hours. BCR/ABL-expressing cells were incubated with control medium, imatinib, 1 μM (BCR/ABL + STI571), or AMN107, 100 nM (BCR/ABL + AMN107) for another 24 hours. RT-PCR was performed using primers specific for HDC or β-actin (loading control). The RT omission control (-RT) is also shown. (F) Histamine levels (by RIA) in Ton.B210-X cells cultured in the absence of IL-3 (Control) or in the presence of doxycycline with or without inhibitory drugs (STI571, 1 μM; AMN107, 100 nM) for 24 hours. Results represent the mean ± SD from 3 independent experiments. *P < .05.
BCR/ABL induces expression of HDC and synthesis of histamine in Ba/F3 cells
We next asked whether BCR/ABL contributes to the synthesis of histamine in leukemic cells. As shown in Figure 3, BCR/ABL induced the production of histamine in Ton.B210 cells and Ton.B210-X cells. In fact, exposure of these cells to doxycycline (to induce BCR/ABL) was followed by an increase in the expression of HDC mRNA (Figure 3A-B) and a substantial increase in histamine (Figure 3C-D). The increase in HDC mRNA in Ton.B210-X cells could be confirmed by real-time PCR (3.0 ± 1.2-fold increase compared with control). Doxycycline per se failed to induce expression of HDC mRNA or synthesis of histamine in Ba/F3 cells lacking BCR/ABL (not shown). The histamine-inducing effects of BCR/ABL in Ton.B210 and Ton.B210-X cells were seen in the presence as well as in the absence of IL-3 (Table 1). Confirming the data of Dy et al,50 IL-3 per se (without BCR/ABL) showed no significant effect on histamine production in Ba/F3 cells (Figure 3; Table 1).
Effects of IL-3 and BCR/ABL on histamine levels in Ton.B210-X cells
Culture condition, h 1-24 . | Culture condition, h 25-48 . | Cellular histamine levels, ng/mL . |
|---|---|---|
| Control medium | Control medium | 0.22 ± 0.13 |
| IL-3 (WEHI-CM) | IL-3 (WEHI-CM) | 0.30 ± 0.06 |
| IL-3 (WEHI-CM) | Control medium | 0.28 ± 0.14 |
| BCR/ABL, control medium without IL-3 | BCR/ABL, control medium without IL-3 | 3.74 ± 2.00* |
| IL-3 + BCR/ABL | IL-3 + BCR/ABL | 4.45 ± 2.01* |
| IL-3 + BCR/ABL | BCR/ABL, control medium without IL-3 | 4.32 ± 1.71* |
Culture condition, h 1-24 . | Culture condition, h 25-48 . | Cellular histamine levels, ng/mL . |
|---|---|---|
| Control medium | Control medium | 0.22 ± 0.13 |
| IL-3 (WEHI-CM) | IL-3 (WEHI-CM) | 0.30 ± 0.06 |
| IL-3 (WEHI-CM) | Control medium | 0.28 ± 0.14 |
| BCR/ABL, control medium without IL-3 | BCR/ABL, control medium without IL-3 | 3.74 ± 2.00* |
| IL-3 + BCR/ABL | IL-3 + BCR/ABL | 4.45 ± 2.01* |
| IL-3 + BCR/ABL | BCR/ABL, control medium without IL-3 | 4.32 ± 1.71* |
Ton.B210-X cells were kept in control medium (RPMI 1640 + 10% FCS), in medium containing WEHI-conditioned medium (WEHI-CM) as source of murine IL-3, and/or in the presence of doxycycline to induce expression of BCR/ABL, as indicated. After incubation, cells were harvested and subjected to measurement of cellular histamine levels by RIA. No significant differences in cell numbers were found after 48 hours when comparing different culture conditions. Results represent the mean ± SD of 4 independent experiments
P < .05 compared with control medium
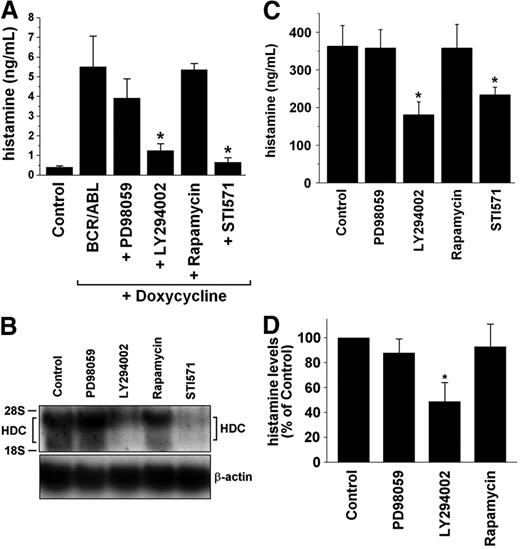
Effects of targeting of BCR/ABL in leukemic cells by imatinib and nilotinib (AMN107)
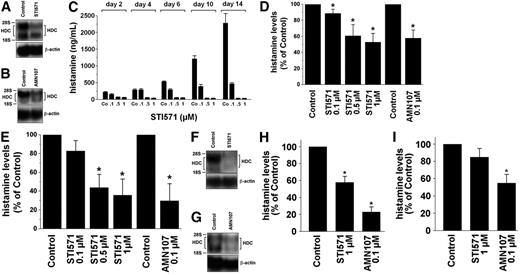
To confirm that BCR/ABL is responsible for histamine generation, specific inhibitors of BCR/ABL (ie, imatinib [STI571] and nilotinib [AMN107])36 were applied. Both agents were found to counteract BCR/ABL-induced expression of HDC mRNA (Figure 3E) and BCR/ABL-induced synthesis of histamine (Figure 3F) in Ton.B210-X cells. We next examined KU812 cells and primary CML cells. As shown in Figure 4A and B, exposure of KU812 cells to imatinib (1 μM) resulted in a significant decrease in HDC mRNA expression (densitometry of transcript levels, 48% ± 26% of control, n = 3, P < .05). Corresponding results were obtained with AMN107, 100 nM (44% ± 17%, n = 3, P < .05). Imatinib and AMN107 also decreased the levels of histamine in KU812 cells (Figure 4C-E). Figure 4C shows the dose- and time-dependent effects of imatinib on expression of cellular histamine levels in KU812 cells. Consistent with its greater potency as a BCR/ABL inhibitor,36 AMN107 decreased the levels of histamine in KU812 cells at lower concentrations compared with imatinib (Figure 4D-E). Thus, targeting of BCR/ABL by specific pharmacologic inhibitors in KU812 cells is associated with reduced histamine production. Corresponding results were obtained with primary CML cells. In fact, imatinib and AMN107 were found to decrease HDC mRNA levels in primary CML cells (HDC mRNA transcript levels compared with control: imatinib, 1 μM, 34% ± 47%; AMN107, 100 nM, 34% ± 65%; P < .05; Figure 4F-G). As shown in Figure 4H and I, the imatinib- and AMN107-induced decrease in expression of HDC mRNA in CML cells was also associated with a decrease in cellular histamine levels. Figure 4I shows the effects of imatinib and AMN107 on expression of histamine in leukemic cells in patients with accelerated-phase CML.
Effects of imatinib and AMN107 on histamine levels and HDC mRNA expression in CML cells. (A-B) KU812 cells were incubated in control medium (Control) or imatinib (STI571; 1 μM) for 8 hours (A), or in control medium (Control) or AMN107 (0.1 μM) for 8 hours (B). Thereafter, total RNA was isolated and subjected to Northern blot analysis using an HDC-specific cDNA probe. The β-actin loading control is also shown. (C) Cellular histamine levels in KU812 cells cultured in control medium (Control) or various concentrations of imatinib (STI571; 0.1, 0.5, or 1 μM) for 2, 4, 6, 10, or 14 days (C). (D-E) Histamine levels in KU812 cells cultured in control medium (Control), imatinib (STI571; 0.1-1 μM) or AMN107 (0.1 μM) for 24 hours (D) or 48 hours (E). Cellular histamine levels were determined by RIA. Results represent the mean ± SD from 3 independent experiments. *P < .05. (F-G) Peripheral-blood mononuclear cells (PBMCs) were isolated from a patient with chronic-phase CML (F) and a patient with imatinib-resistant accelerated-phase CML (G). Cells were incubated in control medium (Control) or imatinib (STI571; 1 μM) (F), or in control medium (Control) or AMN107 (0.1 μM) (G), for 14 hours. Thereafter, total RNA was isolated. Northern blotting was performed with an HDC-specific cDNA probe and a β-actin probe (loading control). (H-I) Histamine levels in CML cells obtained from patients with chronic-phase CML (n = 5) (H) and with imatinib-resistant CML in accelerated-phase (n = 4) (I). Isolated PBMCs were incubated in control medium (Control) or imatinib (STI571; 1 μM), or in control medium (Control) or AMN107 (0.1 μM), for 24 hours. Thereafter, cells were lysed in distilled water, and the content of histamine was quantified by RIA. Results represent the mean ± SD from 5 (H) or 4 (I) independent experiments, respectively. *P < .05 compared with controls.
Effects of imatinib and AMN107 on histamine levels and HDC mRNA expression in CML cells. (A-B) KU812 cells were incubated in control medium (Control) or imatinib (STI571; 1 μM) for 8 hours (A), or in control medium (Control) or AMN107 (0.1 μM) for 8 hours (B). Thereafter, total RNA was isolated and subjected to Northern blot analysis using an HDC-specific cDNA probe. The β-actin loading control is also shown. (C) Cellular histamine levels in KU812 cells cultured in control medium (Control) or various concentrations of imatinib (STI571; 0.1, 0.5, or 1 μM) for 2, 4, 6, 10, or 14 days (C). (D-E) Histamine levels in KU812 cells cultured in control medium (Control), imatinib (STI571; 0.1-1 μM) or AMN107 (0.1 μM) for 24 hours (D) or 48 hours (E). Cellular histamine levels were determined by RIA. Results represent the mean ± SD from 3 independent experiments. *P < .05. (F-G) Peripheral-blood mononuclear cells (PBMCs) were isolated from a patient with chronic-phase CML (F) and a patient with imatinib-resistant accelerated-phase CML (G). Cells were incubated in control medium (Control) or imatinib (STI571; 1 μM) (F), or in control medium (Control) or AMN107 (0.1 μM) (G), for 14 hours. Thereafter, total RNA was isolated. Northern blotting was performed with an HDC-specific cDNA probe and a β-actin probe (loading control). (H-I) Histamine levels in CML cells obtained from patients with chronic-phase CML (n = 5) (H) and with imatinib-resistant CML in accelerated-phase (n = 4) (I). Isolated PBMCs were incubated in control medium (Control) or imatinib (STI571; 1 μM), or in control medium (Control) or AMN107 (0.1 μM), for 24 hours. Thereafter, cells were lysed in distilled water, and the content of histamine was quantified by RIA. Results represent the mean ± SD from 5 (H) or 4 (I) independent experiments, respectively. *P < .05 compared with controls.
Effects of signal transduction inhibitors on histamine levels and HDC expression in Ba/F3 cells and CML cells. (A) Ton.B210-X cells were cultured in the absence (Control) or in the presence of doxycycline, 1 μg/mL (to express BCR/ABL) for 24 hours. BCR/ABL-expressing cells were incubated in control medium (BCR/ABL) or in the presence of inhibitors (PD98059, 50 μM; LY294002, 20 μM; rapamycin, 20 nM; imatinib, 1 μM) for another 24 hours. Thereafter, cells were harvested, and the levels of histamine were determined by RIA. Results represent the mean ± SD from 3 independent experiments. The asterisk indicates significant inhibition compared with control = BCR/ABL (P < .05). (B) KU812 cells were incubated with control medium (Control) or various signal transduction inhibitors (same type and dose as in panel A) for 14 hours. Thereafter, total RNA was isolated and subjected to Northern blot analysis. HDC mRNA expression was analyzed using an HDC-specific cDNA probe. β-actin served as a loading control. (C-D) Histamine levels (determined by RIA) in KU812 cells (C) and in PBMCs obtained from patients with CML (chronic phase, n = 5; accelerated phase, n = 4) (D). KU812 cells and isolated primary PBMCs were incubated with control medium (Control), PD98059 (50 μM), LY294002 (20 μM), rapamycin (20 nM), or imatinib (STI571; 1 μM) for 24 hours. Then cells were lysed in distilled water and subjected to RIA. Results represent the mean ± SD from 3 (C) or 9 (D) independent experiments, respectively. *P < .05 compared with controls.
Effects of signal transduction inhibitors on histamine levels and HDC expression in Ba/F3 cells and CML cells. (A) Ton.B210-X cells were cultured in the absence (Control) or in the presence of doxycycline, 1 μg/mL (to express BCR/ABL) for 24 hours. BCR/ABL-expressing cells were incubated in control medium (BCR/ABL) or in the presence of inhibitors (PD98059, 50 μM; LY294002, 20 μM; rapamycin, 20 nM; imatinib, 1 μM) for another 24 hours. Thereafter, cells were harvested, and the levels of histamine were determined by RIA. Results represent the mean ± SD from 3 independent experiments. The asterisk indicates significant inhibition compared with control = BCR/ABL (P < .05). (B) KU812 cells were incubated with control medium (Control) or various signal transduction inhibitors (same type and dose as in panel A) for 14 hours. Thereafter, total RNA was isolated and subjected to Northern blot analysis. HDC mRNA expression was analyzed using an HDC-specific cDNA probe. β-actin served as a loading control. (C-D) Histamine levels (determined by RIA) in KU812 cells (C) and in PBMCs obtained from patients with CML (chronic phase, n = 5; accelerated phase, n = 4) (D). KU812 cells and isolated primary PBMCs were incubated with control medium (Control), PD98059 (50 μM), LY294002 (20 μM), rapamycin (20 nM), or imatinib (STI571; 1 μM) for 24 hours. Then cells were lysed in distilled water and subjected to RIA. Results represent the mean ± SD from 3 (C) or 9 (D) independent experiments, respectively. *P < .05 compared with controls.
Role of PI3-kinase in BCR/ABL-induced generation of histamine
In a next step, we examined signaling pathways underlying BCR/ABL-dependent expression of HDC and synthesis of histamine in leukemic cells. As shown in Figure 5A, the PI3-kinase inhibitor LY294002 counteracted BCR/ABL-induced synthesis of histamine in Ton.B210-X cells, whereas the MEK-inhibitor PD98059 and rapamycin showed no effects. The same result was obtained with KU812 cells. In fact, LY294002 (24% ± 12%, P < .05) but not PD98059 (91% ± 13%) or rapamycin (82% ± 19%) inhibited the expression of HDC mRNA transcript levels assessed by densitometry (n = 3). A representative experiment is shown in Figure 5B. LY294002 was also found to inhibit the synthesis of histamine in KU812 cells (Figure 5C). Moreover, we were able to show that LY294002 counteracts histamine production in primary CML cells (Figure 5D). These data suggest that the BCR/ABL-induced production of histamine in leukemic cells is mediated through a PI3-kinase-dependent signaling pathway.
CML cells express various receptors for histamine
Several studies suggest that histamine promotes growth of hematopoietic progenitors through autocrine mechanisms involving transmembrane or intracellular histamine receptors.24-27,51,52 We therefore examined expression of histamine-binding sites in KU812 cells and K562 cells as well as in primary CML cells and normal BMMCs. As assessed by RT-PCR, K562 cells and KU812 cells were found to express transcripts for HR-1, HR-2, and HR-4, whereas HR-3 was not detectable in these cells (Table 2). An almost identical profile of HR was found in primary CML cells (Table 3). In addition, we were able to show that highly enriched CD34+ CML progenitor cells display transcripts for HR-1, HR-2, and HR-4 (Table 3). Because histamine may act as an intracellular regulator, we were also interested in examining the expression of intracellular HRs. An important family of intracellular HRs is the CYP450 isoenzymes.52 As shown in Table 2, K562 cells and KU812 cells were found to express transcripts for several CYP450 isoforms, including CYP-2D6 and CYP-1A1, both of which have been implicated in the binding of histamine via their heme moiety, and in the recognition and metabolization of various HR antagonists.52
Expression of cytochrome P450 (CYP450) and histamine receptor (HR) mRNA in K562 cells and KU812 cells
Gene product . | Product size, bp . | Expression of mRNA . | . | |
|---|---|---|---|---|
| . | . | K562 cells . | KU812 cells . | |
| CYP450 1A1 | 146 | + | + | |
| CYP450 1A2 | 226 | – | – | |
| CYP450 1B1 | 360 | – | – | |
| CYP450 2A6 | 589 | + | + | |
| CYP450 2A7 | 774 | + | + | |
| CYP450 2A13 | 320 | – | – | |
| CYP450 2C9 | 357 | – | – | |
| CYP450 2D6 | 419 | + | + | |
| CYP450 2E1 | 246 | + | – | |
| CYP450 3A4 | 353 | + | – | |
| CYP450 3A5 | 432 | + | + | |
| CYP450 3A7 | 447 | – | – | |
| HR-1 | 150 | + | + | |
| HR-2 | 128 | + | + | |
| HR-3 | 140 | – | – | |
| HR-4 | 200 | + | + | |
Gene product . | Product size, bp . | Expression of mRNA . | . | |
|---|---|---|---|---|
| . | . | K562 cells . | KU812 cells . | |
| CYP450 1A1 | 146 | + | + | |
| CYP450 1A2 | 226 | – | – | |
| CYP450 1B1 | 360 | – | – | |
| CYP450 2A6 | 589 | + | + | |
| CYP450 2A7 | 774 | + | + | |
| CYP450 2A13 | 320 | – | – | |
| CYP450 2C9 | 357 | – | – | |
| CYP450 2D6 | 419 | + | + | |
| CYP450 2E1 | 246 | + | – | |
| CYP450 3A4 | 353 | + | – | |
| CYP450 3A5 | 432 | + | + | |
| CYP450 3A7 | 447 | – | – | |
| HR-1 | 150 | + | + | |
| HR-2 | 128 | + | + | |
| HR-3 | 140 | – | – | |
| HR-4 | 200 | + | + | |
Expression of transcripts specific for cytochrome P450 (CYP450) isoenzymes and histamine receptors (HR) type 1-4 in K562 cells and KU812 cells was analyzed by RT-PCR using primers specific for CYP450 isoenzymes47 and histamine receptors
Expression of histamine receptor (HR) mRNA in CML cells
. | . | Expression of mRNA . | . | . | ||
|---|---|---|---|---|---|---|
| Gene product . | Product size, bp . | Primary CML cells . | CD34+ CML cells . | Normal BMMCs . | ||
| HR-1 | 150 | +/– | + | +/– | ||
| HR-2 | 128 | + | + | + | ||
| HR-3 | 140 | – | – | – | ||
| HR-4 | 200 | + | + | + | ||
. | . | Expression of mRNA . | . | . | ||
|---|---|---|---|---|---|---|
| Gene product . | Product size, bp . | Primary CML cells . | CD34+ CML cells . | Normal BMMCs . | ||
| HR-1 | 150 | +/– | + | +/– | ||
| HR-2 | 128 | + | + | + | ||
| HR-3 | 140 | – | – | – | ||
| HR-4 | 200 | + | + | + | ||
Expression of transcripts specific for histamine receptors (HRs) type 1-4 in leukemic cells was analyzed by RT-PCR using primers specific for histamine receptors.
BMMC indicates bone marrow mononuclear cells; +/–, not detectable in all patients examined
Effects of HR-targeting drugs on growth of CML cells
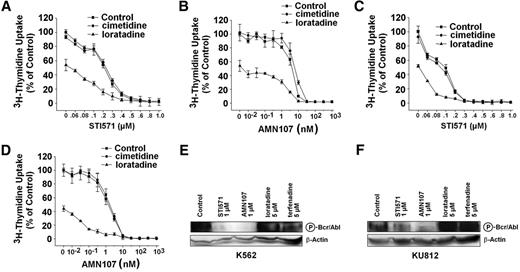
In a next step, we asked whether pharmacologic HR antagonists can inhibit the growth of CML cells. In these experiments, we found that the HR-1 antagonists loratadine and terfenadine, which also bind to various CYP450 isoenzymes, inhibit the growth of K562 and KU812 cells in a dose-dependent manner (Figure 6A-B). Loratadine and terfenadine were also found to inhibit the growth of primary CML cells dose dependently (Figure 6C-D), without differences observed in cells kept with or without GM-CSF (not shown). By contrast, the HR-1 antagonist fexofenadine, which does not bind to CYP450 (but represents the CYP450-metabolization product of terfenadine53 ) did not inhibit the growth of CML cells (Figure 6A-B). Moreover, DPPE, a compound that inhibits the binding of intracellular histamine as well as of various HR antagonists to CYP450,34,35 was found to inhibit proliferation of K562 cells and KU812 cells in a dose-dependent manner (Figure 6E). To show that the effects of HR-1 antagonists also include CML progenitors, we repeated some of the experiments using highly enriched CD34+ cells obtained from a patient with accelerated-phase CML. In these experiments, loratadine and terfenadine were found to inhibit growth of CD34+ CML cells in a dose-dependent manner (Figure 6F). We then asked whether HR-2 antagonists exert antileukemic effects. However, we were unable to substantiate growth-inhibitory effects of HR-2 antagonists (famotidine, cimetidine, ranitidine) on CML cells (not shown). Next, we asked whether histamine per se would modulate the growth of CML cells. However, under the experimental conditions and dose range applied, histamine (10 pM to 100 mM) failed to promote growth of CML cells. In addition, we were unable to see growth-inhibitory effects of the HDC inhibitor α-FMH (1 nM to 100 μM) in CML cells (not shown).
Cooperative antiproliferative effects of BCR/ABL inhibitors and HR antagonists on growth of CML cells
Because loratadine and terfenadine are used in clinical practice and are found to be potent inhibitors of growth of CML cells, we also asked whether these HR antagonists would cooperate with imatinib or AMN107 in producing growth inhibition in CML cells. As shown in Figure 7, both HR antagonists, loratadine and terfenadine, were found to augment the growth-inhibitory effects of imatinib and AMN107 in K562 cells (Figure 7A-B) and KU812 cells (Figure 7C-D). These drug effects were additive rather than synergistic in nature as determined by calculating combination index values by Calcusyn software (not shown). To further examine the potential mechanism of action of loratadine and terfenadine on CML cells, Western blot experiments using the antiphospho mAb 4G10 were performed. However, whereas imatinib and AMN107 were found to counteract BCR/ABL kinase activity, no effects were seen with loratadine or terfenadine (Figure 7E-F).
Effects of histamine receptor antagonists and intracellular histamine antagonists on growth of CML cells. (A-B) K562 cells (A) and KU812 cells (B) were incubated in control medium (Co) or in medium containing various concentrations of the HR-1 antagonists loratadine (▪), terfenadine (•), or fexofenadine (▴) at 37°C and 5% CO2 for 48 hours. After incubation, 1 μCi (0.037 MBq) 3H-thymidine was added. Twelve hours later, cells were harvested, and bound radioactivity was measured in a β-counter. Results are expressed as the percentage of Control (Co) and represent the mean ± SD of 3 independent experiments. (C) Primary CML cells obtained from 7 patients (chronic phase, n = 5; accelerated phase, n = 2) were incubated without (Co) or with various concentrations of loratadine as indicated (37°C, 5% CO2) for 48 hours. (D) Primary CML cells obtained from 6 patients (chronic phase, n = 5; accelerated phase, n = 1) were incubated without (Co) or with various concentrations of terfenadine for 48 hours. After incubation, 3H-thymidine uptake was measured. Results are expressed as the percentage of control (Co) and represent the mean ± SD of 7 (C) or 6 (D) independent experiments, respectively. (E) K562 cells and KU812 cells were incubated without (Co) or with various concentrations of N, N-diethyl-2-[4-(phenylmethyl)phenoxy]-ethanamine (DPPE) for 48 hours. After incubation, 3H-thymidine uptake was measured. Results are expressed as the percentage of control (Co) and represent the mean ± SD of 3 independent experiments. (F) Primary CD34+ CML progenitor cells were incubated in control medium (Co) or in medium containing various concentrations of loratadine (•)or terfenadine (▪) at 37°C and 5% CO2 for 48 hours. After incubation, 1 μCi (0.037 MBq) 3H-thymidine was added. Twelve hours later, cells were harvested, and bound radioactivity was measured in a β-counter. Results are expressed as the percentage of control (Co) and represent the mean ± SD of triplicates.
Effects of histamine receptor antagonists and intracellular histamine antagonists on growth of CML cells. (A-B) K562 cells (A) and KU812 cells (B) were incubated in control medium (Co) or in medium containing various concentrations of the HR-1 antagonists loratadine (▪), terfenadine (•), or fexofenadine (▴) at 37°C and 5% CO2 for 48 hours. After incubation, 1 μCi (0.037 MBq) 3H-thymidine was added. Twelve hours later, cells were harvested, and bound radioactivity was measured in a β-counter. Results are expressed as the percentage of Control (Co) and represent the mean ± SD of 3 independent experiments. (C) Primary CML cells obtained from 7 patients (chronic phase, n = 5; accelerated phase, n = 2) were incubated without (Co) or with various concentrations of loratadine as indicated (37°C, 5% CO2) for 48 hours. (D) Primary CML cells obtained from 6 patients (chronic phase, n = 5; accelerated phase, n = 1) were incubated without (Co) or with various concentrations of terfenadine for 48 hours. After incubation, 3H-thymidine uptake was measured. Results are expressed as the percentage of control (Co) and represent the mean ± SD of 7 (C) or 6 (D) independent experiments, respectively. (E) K562 cells and KU812 cells were incubated without (Co) or with various concentrations of N, N-diethyl-2-[4-(phenylmethyl)phenoxy]-ethanamine (DPPE) for 48 hours. After incubation, 3H-thymidine uptake was measured. Results are expressed as the percentage of control (Co) and represent the mean ± SD of 3 independent experiments. (F) Primary CD34+ CML progenitor cells were incubated in control medium (Co) or in medium containing various concentrations of loratadine (•)or terfenadine (▪) at 37°C and 5% CO2 for 48 hours. After incubation, 1 μCi (0.037 MBq) 3H-thymidine was added. Twelve hours later, cells were harvested, and bound radioactivity was measured in a β-counter. Results are expressed as the percentage of control (Co) and represent the mean ± SD of triplicates.
Cooperative growth-inhibitory effects of BCR/ABL inhibitors and HR antagonists on growth of CML cells. (A-D) K562 cells (A-B) and KU812 cells (C-D) were incubated with various concentrations of imatinib (STI571) (A,C) or AMN107 (B,D) in the absence (Control, ▪) or presence of cimetidine, 10 μM(•) or loratadine, 10 μM(▴)at 37°C for 48 hours. Thereafter, 3H-thymidine uptake was measured. Results are expressed as the percentage of control (Control) and represent the mean ± SD of triplicates. (E-F) K562 and KU812 cells were incubated in control medium (Control), imatinib (STI571), AMN107, loratadine, or terfenadine for 4 hours. Thereafter, cells were isolated and subjected to Western blot analysis using the antiphosphoprotein antibody 4G10 to detect the phosphorylated BCR/ABL oncoprotein (P-Bcr/Abl) (210 kDa) (top). β-actin served as loading control (bottom).
Cooperative growth-inhibitory effects of BCR/ABL inhibitors and HR antagonists on growth of CML cells. (A-D) K562 cells (A-B) and KU812 cells (C-D) were incubated with various concentrations of imatinib (STI571) (A,C) or AMN107 (B,D) in the absence (Control, ▪) or presence of cimetidine, 10 μM(•) or loratadine, 10 μM(▴)at 37°C for 48 hours. Thereafter, 3H-thymidine uptake was measured. Results are expressed as the percentage of control (Control) and represent the mean ± SD of triplicates. (E-F) K562 and KU812 cells were incubated in control medium (Control), imatinib (STI571), AMN107, loratadine, or terfenadine for 4 hours. Thereafter, cells were isolated and subjected to Western blot analysis using the antiphosphoprotein antibody 4G10 to detect the phosphorylated BCR/ABL oncoprotein (P-Bcr/Abl) (210 kDa) (top). β-actin served as loading control (bottom).
Discussion
Histamine is a key mediator of basophils and a potential growth regulator of myeloid cells.24-27 Whereas it is well established that histamine production in normal progenitors is regulated by cytokines such as IL-3,21-23 little is known about the regulation of production of histamine in neoplastic cells. The results of our study show that the CML-related oncoprotein BCR/ABL induces synthesis of histamine in leukemic progenitors. The BCR/ABL-induced synthesis of histamine was found to involve the PI3-kinase-signaling pathway. In addition, we found that the BCR/ABL tyrosine kinase inhibitors imatinib and nilotinib (AMN107) counteract histamine production in CML cells. To our knowledge this is the first report describing that a leukemia-specific oncogene is involved in the regulation of production of histamine in neoplastic cells.
HDC is the key enzyme involved in histamine generation.28 To directly show the effect of BCR/ABL on expression of HDC mRNA and synthesis of histamine, Ba/F3 cells with doxycycline-inducible expression of BCR/ABL (Ton.B210 and Ton.B210-X cells)38 were used. Using these cells, we were able to show that BCR/ABL induces the expression of HDC and the synthesis of histamine. In line with this observation, the BCR/ABL-targeting drugs imatinib and AMN107 were found to counteract BCR/ABL-induced expression of HDC and synthesis of histamine in these cells. Moreover, both drugs were found to counteract histamine expression in primary CML cells and in the CML cell line KU812. In imatinib-resistant CML cells, the histamine-reducing effects were observed with AMN107 but not with imatinib. These data are in line with the notion that AMN107 is an effective antileukemic agent in imatinib-resistant CML.36
Because IL-3 is a well-defined inductor of histamine-synthesis21-23 and may act as autocrine growth regulator in CML cells,54,55 it was important to compare histamine-inducing effects of IL-3 and BCR/ABL in Ba/F3 cells. However, IL-3 did not exert major effects on histamine synthesis in Ton.B210 cells or Ton.B210-X cells in the absence of BCR/ABL, confirming the data of Dy et al.50 Moreover, IL-3 did not significantly augment the BCR/ABL-induced production of histamine. Most importantly, we were able to show that BCR/ABL induces expression of HDC and synthesis of histamine in the absence of exogenous IL-3. This was an unexpected result because IL-3 receptor-dependent and BCR/ABL-dependent signaling pathways exhibit a significant overlap.4-6 The most likely explanation might be that Ba/F3 cells are extremely immature precommitted cells which may not produce histamine in response to IL-3 in the same way as more mature (myeloid-differentiated) cells.
Thus, apart from BCR/ABL, the cellular background may be an important determinant controlling the expression of HDC and production of histamine in leukemic cells. This notion was further supported by the observation that basophil-enriched populations of leukemic cells in our patients with CML expressed significantly higher levels of HDC mRNA and higher amounts of histamine than leukemic cells with lower numbers of basophils or basophil-depleted cell fractions. Thus, our data clearly show that histamine and HDC are expressed primarily in the basophil lineage compartment of the malignant CML clone. Moreover, significant levels of HDC mRNA and histamine were detectable in the basophil-committed CML cell line KU812 but not in the uncommitted cell line K562, which produces only trace amounts of histamine. All in all, both factors, namely the cellular background and the specific oncoprotein, BCR/ABL, may contribute to histamine generation in leukemic cells in CML.
A number of different signaling pathways underlie BCR/ABL-dependent growth and survival of leukemic cells in CML.4-10 The 2 most prominent pathways are the PI3-kinase pathway and the Ras/Raf/MEK/ERK pathway.4-10 Our results show that inhibition of the PI3-kinase by LY294002 is associated with reduced expression of HDC and reduced synthesis of histamine, whereas no effects were observed with PD98059. These data suggest that the PI3-kinase pathway, but not the MAP-kinase pathway, is involved in BCR/ABL-dependent synthesis of histamine in CML cells.
Several studies suggest that histamine is not only a cellular marker and mediator of basophils but also a potential growth regulator of neoplastic cells.24-27 We therefore were interested to learn whether histamine, derived from BCR/ABL-transformed cells, plays a role in neoplastic-cell growth in CML. To address this issue, we first examined whether CML cells express receptors for histamine. During the past few decades, a number of surface and intracellular receptors for histamine have been identified.52,56 In a first step, we were able to show that primary CML cells and the CML cell lines K562 and KU812 display HR-1, HR-2, and HR-4 mRNA. Another interesting observation was that the HR-1 antagonists loratadine and terfenadine inhibit growth of CML cells, including primary CML cells. An unexpected finding was that these drug effects were not only seen in KU812 cells and unfractionated CML cells but also in highly enriched CD34+ CML progenitors and K562 cells, which display only trace amounts of histamine. Moreover, when added as an exogenous substance, histamine did not promote in vitro growth of CML cells, and the same negative result was obtained with α-FMH, an HDC inhibitor. In addition, not all HR-1 antagonists tested in this study were found to counteract growth of CML cells. Together, these data suggest that concerning growth inhibition, the HR-1 does not serve as a critical target in CML cells.
These observations prompted us to search for further targets of loratadine and terfenadine in CML cells. One important family of intracellular histamine receptors, which can also interact with HR antagonists, are CYP450 isoenzymes.52 In the present study, we found that K562 and KU812 cells express transcripts for several different CYP450 isoforms, including CYP-2D6 and CYP-1A1, both of which have been implicated in the binding of histamine and in the recognition of HR antagonists.52 The hypothesis that CYP450 isoenzymes may serve as targets of loratadine and terfenadine was supported by the observation that fexofenadine, an HR-1 antagonist that is not targeted (metabolized) by CYP450 (but represents the CYP450 metabolization product of terfenadine53 ) did not counteract growth of CML cells. Moreover, DPPE, an arylalkylamine analog of tamoxifen that blocks the binding of histamine and of various HR antagonists to CYP450 isoenzymes34,35,52 was found to inhibit growth of K562 and KU812 cells. All in all, these data suggest that certain CYP450 isoenzymes may play a role as targets of loratadine and terfenadine in CML cells.
A considerable problem in the treatment of advanced-phase CML is resistance against imatinib or other BCR/ABL kinase inhibitors.5,6,10 To overcome resistance, a number of pharmacologic strategies can be envisaged. One approach is to combine targeted drugs with each other. We were therefore interested to learn whether HR-1 antagonists (loratadine and terfenadine) and BCR/ABL inhibitors (imatinib, AMN107) exert cooperative antiproliferative effects on CML cells. In respective experiments we were indeed able to show that these drugs cooperate with each other in producing growth inhibition, although the effects were found to be additive rather than synergistic in nature.
In summary, our data show that production of histamine in CML cells is determined by the cellular background and is driven by the disease-specific oncoprotein BCR/ABL. Our data also show that incubation of CML cells with loratadine and terfenadine is associated with inhibition of growth of leukemic cells and with increased sensitivity against BCR/ABL inhibitors. Whether these compounds can also augment the effects of imatinib and AMN107 in vivo in patients with CML remains to be determined in clinical trials.
Prepublished online as Blood First Edition Paper, July 18, 2006; DOI 10.1182/blood-2005-12-028456.
Supported by the Fonds zur Förderung der Wissenschaftlichen Forschung in Österreich (FWF) (grant P-16412), by the FWF Charlotte Bühler (grant H-185) (H.A.), by the Austrian Ministry for Education, Science, and Culture (grant G2 200.142/1-VI/1/2006), and by the Center for Molecular Medicine of the Austrian Academy of Sciences (CeMM 20030) (S.D. and W.F.P.).
The authors declare no competing financial interests.
K.J.A. performed the laboratory experiments and wrote the paper; M.M. provided vital new analytic tools and performed PCR experiments; C.S. provided logistics and patients; A.V. performed Northern blot and PCR experiments; M.-T.K. and H.A. contributed immunocytochemical and immunohistochemical staining experiments; K.V.G., S.D., and W.F.P. contributed by performing Western blots and laboratory experiments on cell growth and proliferation; M.B. and H.E. performed real-time PCR experiments; K.S. and S.F. contributed flow cytometry and cell-sorting experiments; A.F. provided essential reagents and logistic support; and P.V. contributed by designing the study, establishing the work plan, providing logistics, and by approving data and the final version of the manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Hans Semper for skillful technical assistance.

![Figure 6. Effects of histamine receptor antagonists and intracellular histamine antagonists on growth of CML cells. (A-B) K562 cells (A) and KU812 cells (B) were incubated in control medium (Co) or in medium containing various concentrations of the HR-1 antagonists loratadine (▪), terfenadine (•), or fexofenadine (▴) at 37°C and 5% CO2 for 48 hours. After incubation, 1 μCi (0.037 MBq) 3H-thymidine was added. Twelve hours later, cells were harvested, and bound radioactivity was measured in a β-counter. Results are expressed as the percentage of Control (Co) and represent the mean ± SD of 3 independent experiments. (C) Primary CML cells obtained from 7 patients (chronic phase, n = 5; accelerated phase, n = 2) were incubated without (Co) or with various concentrations of loratadine as indicated (37°C, 5% CO2) for 48 hours. (D) Primary CML cells obtained from 6 patients (chronic phase, n = 5; accelerated phase, n = 1) were incubated without (Co) or with various concentrations of terfenadine for 48 hours. After incubation, 3H-thymidine uptake was measured. Results are expressed as the percentage of control (Co) and represent the mean ± SD of 7 (C) or 6 (D) independent experiments, respectively. (E) K562 cells and KU812 cells were incubated without (Co) or with various concentrations of N, N-diethyl-2-[4-(phenylmethyl)phenoxy]-ethanamine (DPPE) for 48 hours. After incubation, 3H-thymidine uptake was measured. Results are expressed as the percentage of control (Co) and represent the mean ± SD of 3 independent experiments. (F) Primary CD34+ CML progenitor cells were incubated in control medium (Co) or in medium containing various concentrations of loratadine (•)or terfenadine (▪) at 37°C and 5% CO2 for 48 hours. After incubation, 1 μCi (0.037 MBq) 3H-thymidine was added. Twelve hours later, cells were harvested, and bound radioactivity was measured in a β-counter. Results are expressed as the percentage of control (Co) and represent the mean ± SD of triplicates.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/10/10.1182_blood-2005-12-028456/4/m_zh80220603670006.jpeg?Expires=1769168026&Signature=dxEtWEvtjOwb-uW~zwUxjRa853eINCw3l-MLncgNFvNDSONWLo76lWAJ-m7c826fb-~tTkD95g5JzN~9FUFpVPhaeheAhHHi1sH0Jud6jtr57YmF7LOyktgAB-f5uCKH-21f2nCFtDk9pGfW9f~ZT0S690Q3TQz9BSg1k0hVDMBHru3ZQJCV6-njOYSiJeLZYKlEt2yJA8MLunmvuI0kvXQSRkdKTKRILGM0LPBOwZIEwWWvlOSmCuCqVk3zsm1mf151vSbXauQ8aflBu01h4BwaQu2DHyVnaiDf3l-aIl3UrWST-SvdNJo-DBoAl-U~LI-9cKRouDGrf2WYgKTL1w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal