Abdominal-type HoxA genes in combination with Meis1 are well-documented on-cogenes in various leukemias but it is unclear how they exert their transforming function. Here we used a system of conditional transformation by an inducible mixed lineage leukemia-eleven-nineteen leukemia (MLL-ENL) oncoprotein to overexpress Hoxa9 and Meis1 in primary hematopoietic cells. Arrays identified c-Myb and a c-Myb target (Gstm1) among the genes with the strongest response to Hoxa9/Meis1. c-Myb overexpression was verified by Northern blot and quantitative reverse transcription-polymerase chain reaction (RT-PCR). Also MLL-ENL activated c-Myb through up-regulation of Hoxa9 and Meis1. Consequently, short-term suppression of c-Myb by small inhibitory RNA (siRNA) efficiently inhibited transformation by MLL-ENL but did not impair transformation by transcription factor E2A-hepatic leukemia factor (E2A-HLF). The anti c-Myb siRNA effect was abrogated by coexpression of a c-Myb derivative with a mutated siRNA target site. The introduction of a dominant-negative c-Myb mutant had a similar but weaker effect on MLL-ENL-mediated transformation. Hematopoietic precursors from mice homozygous for a hypo-morphic c-Myb allele were more severely affected and could be transformed neither by MLL-ENL nor by E2A-HLF. Ectopic expression of c-Myb induced a differentiation block but c-Myb alone was not transforming in a replating assay similar to Hoxa9/Meis1. These results suggest that c-Myb is essential but not sufficient for Hoxa9/Meis1 mediated transformation. (Blood. 2006;108:297-304)
Introduction
The transcription factors encoded by the clustered Hox homeobox genes are key regulators of normal and malignant hematopoiesis. In particular, the abdominal-type HoxA genes and several members of the HoxB cluster are crucial for hematopoietic stem cell (HSC) development and blood-cell maturation.1,2 These genes are highly expressed in progenitor cells and become down-regulated during differentiation.3,4 Genetic ablation of Hoxb4 and also Hoxa9 in animals causes defects in the proliferative and repopulation ability of HSCs.5-7 For effective gene regulation Hox proteins form heterodimers and heterotrimers with “3-amino-acid loop extension” (TALE) homeobox-proteins of the Pbx and Meis families.8-12 It is therefore not surprising that an elevated expression of Meis and HoxA genes has been causally linked to a wide variety of hematologic malignancies. Several lines of evidence point to a central and important role of Hox/Meis during leukemogenesis. HOXA gene overexpression is a feature found in many human acute myeloid leukemias and high HOXA9 levels are associated with a dismal prognosis.13-15 In some T-cell acute lymphocytic leukemias (T-ALLs) a recurrent inversion inv(7)(p15q34) juxtaposes the HOXA and T-cell receptor beta (TCRB) loci, causing a dramatic deregulation of HOX genes across the whole locus.16,17 In the BXH2 leukemia mouse model Hoxa9, Hoxa7 and Meis1 (Meis is an acronym for mouse ecotropic integration site) are frequently coactivated by retroviral insertion,18 and transplanted HSCs transduced with retroviruses coding for Hoxa7/9 and Meis1 rapidly initiate acute leukemia in recipient animals.19-21 HOXA9 is fused to nucleoporin 98 (NUP98-HOXA9) in leukemias bearing a translocation t(7;11)(p15;p15).22,23 Finally, an over-abundance of HOX/MEIS transcripts is characteristic for several leukemias with defined genetic abnormalities like CALM-AF10 containing T-ALL24 and for almost all cases of mixed-lineage leukemia (MLL) harboring MLL fusion proteins.25-29 For the latter class we have shown that Hoxa9 and Meis1 are direct downstream targets of the prototypical MLL fusion protein MLL-ENL.30,31 Ectopic overexpression of Hoxa9 in combination with Meis1 was sufficient to recapitulate almost all aspects of MLL-ENL-mediated transformation, underscoring the crucial function of these 2 genes in MLL fusion protein controlled leukemogenesis. In contrast, neither Hoxa9 nor Meis1 on its own was able to replace MLL-ENL function.31 The absolute requirement for a cooperation between these 2 genes to achieve efficient transformation was also reflected by animal models. Mice that received transplants of HSCs expressing Hoxa9 alone succumbed to leukemia only rarely and with a long latency. Coexpression of Hoxa9 and Meis1, however, led to a fully penetrant and rapid-onset leukemia.20,32,33
Despite this overwhelming body of evidence for a participation of Hox and Meis proteins in the etiology of blood cancers, little is known about the underlying molecular mechanism. Two studies have been published that tried to identify HOXA9 and/or MEIS1 subordinate genes in hematopoietic cells. Dorsam and coworkers transiently transfected 3 established human leukemia lines with HOXA9 expression constructs and analyzed the transcriptome on cDNA arrays.34 Wang et al compared the RNA inventory of cell lines immortalized by Hoxa9 before and after subsequent transduction with Meis1-coding retroviruses.33 Unfortunately, there is very little overlap between these studies, indicating a strong context-dependent action of Hox/Meis. Here we wanted to study Hox/Meis controlled genes in a defined primary cell population to avoid any unwanted bias due to unknown mutations in established cell lines. For this purpose we used a conditional transformation system developed in our laboratory.31 Primary hematopoietic precursors can be reversibly transformed by transduction with a tamoxifen-inducible derivative of MLL-ENL (MLL-ENL-ERtm). In the presence of 4-hydroxy tamoxifene (4-OHT) these cells proliferate in culture and can be further transduced with any gene of interest. After withdrawal of 4-OHT the cells revert and the effects of the additional gene(s) can be studied in a homogeneous near-normal environment that would otherwise be hardly accessible.
With this method we were able to identify genes that are controlled by the cooperation of Hoxa9 with Meis1. Surprisingly, c-Myb, the cellular homolog of the “myeloblastosis leukemia virus oncogene,” and a known c-Myb target gene were among the genes that showed the most pronounced up-regulation in response to a combination of Hoxa9 and Meis1. Although clearly oncogenic, and despite many years of research, c-Myb has remained largely an “orphan” that could not yet specifically be associated with any known oncogenic pathway (for reviews see Weston35 and Ganter and Lipsick36 ). c-Myb levels are elevated in many leukemias and c-Myb has a critical function in HSC development as evidenced by hematopoietic defects in c-Myb-deficient animals.37,38 Nevertheless, c-Myb is not involved in any recurrent genetic abnormality, nor is it a common target for activating mutations. Here we provide the first evidence that c-Myb might be a crucial transmitter of oncogenic signals from Hox/Meis and upstream Hox-regulators to cellular growth control.
Materials and methods
Plasmid construction
The cDNAs for mouse Hoxa9, Meis1, and c-Myb were amplified by polymerase chain reaction (PCR) with epitope tag primers, and the corresponding amplification products were cloned into murine stem-cell virus (pMSCV) retroviral vectors (Clontech, Palo Alto, CA) yielding pMSCVpuro-flagMeis, pMSCVhygro-HA-Hoxa9, and pMSCVpuro-flagMyb. For creation of Myb* a silent mutation was introduced changing bases 151 to 156 of c-Myb from CTG AAG to CTC AAA by site-directed mutagenesis with the Quick Change mutagenesis system (Stratagene, La Jolla, CA). A dominant-negative version of c-Myb was constructed by a C-terminal fusion of the c-Myb DNA binding domain (amino acids 1-200) to the KRAB repressor domain (amino acids 1-74). For production of anti-Myb small inhibitory RNA (siRNA) an oligonucleotide targeting c-Myb nucleotides 148 to 161 (AAGCTGAAGAAGCTGGTGGAA) was designed and introduced into the “self-inactivating” retroviral vector pSIRENretroQ (Clontech) according to the instructions of the manufacturer. After integration into the genomic DNA this construct provides a continuous source of small hairpin RNA (shRNA) for intracellular production of specific siRNA. pMSCVneo-flagE2AHLF was a laboratory stock. All inserts generated by PCR were controlled by sequencing.
Tissue culture, transduction procedures, and methocel replating assays
Generation of conditionally transformed hematopoietic cells (MLL-ENL-ERtm cells) is described in Zeisig et al.31 These cells were maintained in RPMI1640/GlutamaxI (Invitrogen, Karlsruhe, Germany) supplemented with fetal calf serum (FCS; 10%), interleukin (IL)-3, IL-6, granulocyte-macrophage colony-stimulating factor (GM-CSF) (each 10 ng/mL), stem-cell factor (SCF; 100 ng/mL), and 4-hydroxy-tamoxifene (4-OHT; 100 nM). In the presence of 4-OHT, MLL-ENL-ERtm cells proliferate indefinitely as early myeloid precursors. Without 4-OHT the cells revert and initiate a normal program of differentiation.
The ecotropic packaging cell line Phoenix was obtained from Gary Nolan (Stanford, CA) and cultured as outlined at http://www.stanford.edu/group/nolan/. High-titer retrovirus supernatants were produced by transient transfection of Phoenix-E cells by a standard Ca-phosphate precipitation method. Viral titers usually reached approximately 5 × 106 cfu/mL. Infection was done by spinoculation (2500g, 35°C) followed by selection with the appropriate antibiotics (G418, 1 mg/mL; puromycin, 0.6 μg/mL; and hygromycin, 100 μg/mL).
Serial replating assays were done according to Zeisig et al.31 In short, primary bone marrow cells were isolated from 5-fluorouracil (150 mg/kg)-treated donor mice. After retroviral transduction the cells were cultured and replated every 5 to 7 days in MethoCult (M3234) methylcellulose medium (Stem Cell Technologies, Vancouver, BC, Canada) under appropriate antibiotic selection. Recombinant mouse cytokines (Strathmann Biotech, Hannover, Germany) were present in the following concentrations: IL-3, IL-6, and GM-CSF, 10 ng/ml; SCF, 100 ng/mL. Replating assays were performed at least 3 times per construct. Generation, maintenance and genotyping of Mybkd mice has been described in Emambokus et al.37
Western blot and antibodies
For the detection of tagged HA-Hoxa9, flag-Meis, flag-cMyb, and derivatives nuclear extracts were prepared from transfected packaging cells in a high salt elution buffer (300 mM NaCl, 20 mM HEPES [pH 7.5], 0.5 mM EDTA, 0.1% Triton X-100, 0.5 mM sodium vanadate, 2 mM NaF, 2 mM DTT, 0.2 mM PMSF, 20 μg/mL leupeptin, and 40 μg/mL pepstatin A). After separation on standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, the proteins were blotted onto nitrocellulose in 10 mM CAPS (pH 11), 10% methanol. The detection was done with monoclonal anti-flag and anti-HA antibodies (SIGMA, Taufkirchen, Germany). Fluorophore-labeled antibodies for fluorescence-activated cell sorting (FACS) analysis (isotype control, c-kit, Gr-1) were purchased from BD Biosciences (Palo Alto, CA) and used according to the recommendations of the manufacturer.
RT-PCR, qRT-PCR, and array analysis
Total RNA was isolated by ion-exchange chromatography with kits from Qiagen (Hilden, Germany) according to the recommended protocols. RNAs were quantified by quantitative reverse transcription-PCR (qRT-PCR) according to the ΔΔCt method after reverse transcription of total RNA with random hexamer primers. Samples were normalized to β-actin and amplification products were quantified by SYBR green (Stratagene, La Jolla, CA). Primers spanning introns were used as follows: c-Myb: fwd: gaataaaggagctggagttgctc, rev: gtgcatctaagcccgagctttc; Hoxa9: fwd: cggccttatggcattaaacctg, rev: gagcgagcatgtagccagttg; and Meis1: fwd: cctctgcactcgcatcagtac, rev: gtttggcgaacaccgctatatc.
Target preparation and hybridization of microarrays was conducted as described in the Affymetrix GeneChip Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA). Briefly, total RNA was converted to first-strand cDNA primed by a poly(T) oligomer that incorporated the T7 promoter. Second-strand cDNA synthesis was followed by in vitro transcription for linear amplification of each transcript and incorporation of biotinylated CTP and UTP. The cRNA products were fragmented to 200 nucleotides or less, heated at 99°C for 5 minutes, and hybridized for 16 hours at 45°C to the microarrays. The microarrays were then washed at low (6 × salt sodium phosphate EDTA buffer (SSPE) and high (100 mM MES, 0.1 M NaCl) stringency and stained with streptavidin-phycoerythrin. Fluorescence was amplified by adding biotinylated antistreptavidin and an additional aliquot of streptavidin-phycoerythrin stain. A confocal scanner was used to collect fluorescence signal at 3-μm resolution after excitation at 570 nm. The average signal from 2 sequential scans was calculated for each microarray feature.
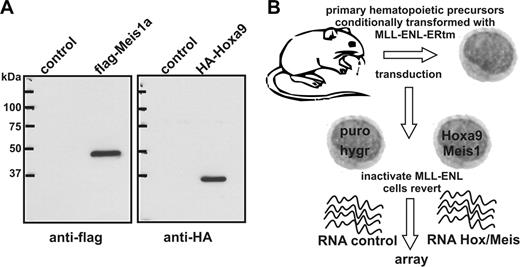
Experimental setup for identification of Hoxa9/Meis1 target genes. (A) Immunoblot of nuclear extracts from packaging cells used to produce pMSCV-HA-Hoxa9 and pMSCV-flag-Meis1 viruses. Control indicates cells transfected with empty vector. (B) Schematic representation of experimental outline. MLL-ENL-ERtm is a fusion of MLL-ENL with the ligand binding domain of a mutated estrogen receptor.31 MLL-ENL-ERtm is only active in the presence of 4-OHT.
Experimental setup for identification of Hoxa9/Meis1 target genes. (A) Immunoblot of nuclear extracts from packaging cells used to produce pMSCV-HA-Hoxa9 and pMSCV-flag-Meis1 viruses. Control indicates cells transfected with empty vector. (B) Schematic representation of experimental outline. MLL-ENL-ERtm is a fusion of MLL-ENL with the ligand binding domain of a mutated estrogen receptor.31 MLL-ENL-ERtm is only active in the presence of 4-OHT.
Affymetrix Microarray Suite 5.0 was used to quantitate expression levels for targeted genes; default values provided by Affymetrix were applied to all analysis parameters. Border pixels were removed, and the average intensity of pixels within the 75th percentile was computed for each probe. The average of the lowest 2% of probe intensities occurring in each of 16 microarray sectors was set as background and subtracted from all features in that sector. Probe pairs were scored positive or negative for detection of the targeted sequence by comparing signals from the perfect match and mismatch probe features. The number of probe pairs meeting the default discrimination threshold (τ= 0.015) was used to assign a call of absent, present, or marginal for each assayed gene, and a P value was calculated to reflect confidence in the detection call. A weighted mean of probe fluorescence (corrected for nonspecific signal by subtracting the mismatch probe value) was calculated using the 1-step Tukey biweight estimate. This signal value, a relative measure of the expression level, was computed for each assayed gene. Global scaling was applied to allow comparison of gene signals across multiple microarrays: after exclusion of the highest and lowest 2%, the average total chip signal was calculated and used to determine what scaling factor was required to adjust the chip average to an arbitrary target of 150. All signal values from 1 microarray were then multiplied by the appropriate scaling factor.
Results
Identification of genes regulated by Hoxa9 and Meis1
Our strategy to discover genes that are controlled by Hoxa9 and Meis1 was based on the use of conditionally transformed MLL-ENL-ERtm cells. In the presence of 4-OHT, active MLL-ENL protein immortalizes hematopoietic precursors through up-regulation of endogenous Hoxa9 and Meis1. To achieve constitutive expression of these genes MLL-ENL-ERtm cells were transduced simultaneously with 2 pMSCV retroviruses encoding flag-Meis1 and HA-Hoxa9 (Figure 1A) followed by antibiotic selection. Infection with empty puro/hygro viruses served as control. Withdrawal of 4-OHT led to inactivation of MLL-ENL, and as a consequence, the cells reverted to a normal phenotype. Without 4-OHT control cells stopped proliferation after approximately 5 days, whereas Hoxa9/Meis1-transduced cells continued to grow (Zeisig et al31 ) The RNA inventory of cells with integrated Hoxa9/Meis1 proviruses was compared on 430A Affymetrix arrays with RNA isolated from the respective control cells (Figure 1B). For hybridization RNA was isolated 72 hours after 4-OHT withdrawal from 3 independently established cell populations transduced either with Hoxa9/Meis1 or empty viruses. In total, 360 genes could be identified that showed expression level differences 2-fold and greater in at least 2 of 3 pairwise comparisons. After elimination of duplicate probe sets and expressed sequence tag (EST) sequences 70 genes were found to be significantly up-regulated by Hoxa9/Meis1 (Table S1, available on the Blood website; see the Supplemental Tables link at the top of the online article) and 92 genes showed a decreased expression in Hoxa9/Meis1-transduced cells versus control cells (Table S2). Genes with the strongest response to Hoxa9/Meis1 (> 2.5-fold up-regulated) are listed in Table 1. Detection of a 7.1-fold increase in Hoxa9 mRNA concentration confirmed successful transduction and data analysis. Ectopic Meis1 could not be identified since the A430 probe set for this gene exclusively locates to the untranslated 3′ end of the mRNA that was not present in the retroviral construct. Strikingly, the known hematopoietic oncogene c-Myb and a known c-Myb target gene (glutathione-S-transferase μ) were highly over-represented in RNA samples of Hoxa9/Meis1 transduced cells (Table 1). In addition, B-Myb and the gene for the kinase Wee1, another c-Myb target, were also up-regulated by Hoxa9/Meis1 (Table S1). Several genes normally specific for cells of neuronal origin (Schip1, Enc1, and Nedd4) appeared also to be targets of Hoxa9/Meis1 in hematopoietic cells. Conversely, genes down-regulated by Hoxa9/Meis1 mostly coded for proteins necessary for function or development of mature myeloid cells. This reflects the fact that control cells started terminal differentiation, whereas Hoxa9/Meis1 cells remained blocked at an early precursor stage (Zeisig et al31 and not shown).
Genes up-regulated in Hoxa9/Meis1-transduced cells versus control
Fold change . | Gene . | Name/comment . | Affymetrix ID . |
|---|---|---|---|
| 7.1 | Hoxa9 | Homeobox A9* | 1421579_at |
| 6.8 | Arhj | ras homolog gene family, member J, small GTPase | 1418892_at |
| 4.0 | LOC235036 | Peter pan homolog; second-step splicing factor 1 | 1423703_at |
| 3.8 | Klf9 | Kruppel-like factor 9, GC box binding transcription factor | 1456341_a_at |
| 3.6 | Tst | Thiosulfate sulfurtransferase, mitochondrial | 1448609_at |
| 3.4 | AW545363 | CD40 ligand-activated specific transcript 3 | 1425373_a_at |
| 3.4 | Myb | Myeloblastosis oncogene | 1422734_a_at |
| 3.3 | Nucb2 | Nucleobindin 2, overexpressed in non-Hodgkin lymphoma | 1418355_at |
| 3.3 | Cd28 | CD28 antigen, T-cell marker | 1437025_at |
| 3.2 | Mylk | Myosin, light polypeptide kinase | 1425505_at |
| 3.1 | Schip 1 | Schwannomin interacting protein 1, neural cell specific | 1423025_a_at |
| 2.8 | Gstm 1 | Glutathione S-transferase μ1, myb target gene | 1448330_at |
| 2.7 | Ide | Insulin degrading enzyme | 1453988_a_at |
| 2.6 | Trip 13 | Thyroid hormone receptor interactor 13 | 1429294_at |
| 2.6 | Pde7a | Phosphodiesterase 7A | 1451839_a_at |
| 2.5 | Enc 1 | Ectodermal-neural cortex 1 | 1454904_at |
| 2.5 | Bcat 1 | Branched chain aminotransferase 1, cytosolic | 1450871_a_at |
| 2.5 | Nedd4 | Neural precursor expressed, developmentally down-regulated | 1450431_a_at |
| 2.5 | Dutp | Deoxyuridine triphosphatase | 1419270_a_at |
Fold change . | Gene . | Name/comment . | Affymetrix ID . |
|---|---|---|---|
| 7.1 | Hoxa9 | Homeobox A9* | 1421579_at |
| 6.8 | Arhj | ras homolog gene family, member J, small GTPase | 1418892_at |
| 4.0 | LOC235036 | Peter pan homolog; second-step splicing factor 1 | 1423703_at |
| 3.8 | Klf9 | Kruppel-like factor 9, GC box binding transcription factor | 1456341_a_at |
| 3.6 | Tst | Thiosulfate sulfurtransferase, mitochondrial | 1448609_at |
| 3.4 | AW545363 | CD40 ligand-activated specific transcript 3 | 1425373_a_at |
| 3.4 | Myb | Myeloblastosis oncogene | 1422734_a_at |
| 3.3 | Nucb2 | Nucleobindin 2, overexpressed in non-Hodgkin lymphoma | 1418355_at |
| 3.3 | Cd28 | CD28 antigen, T-cell marker | 1437025_at |
| 3.2 | Mylk | Myosin, light polypeptide kinase | 1425505_at |
| 3.1 | Schip 1 | Schwannomin interacting protein 1, neural cell specific | 1423025_a_at |
| 2.8 | Gstm 1 | Glutathione S-transferase μ1, myb target gene | 1448330_at |
| 2.7 | Ide | Insulin degrading enzyme | 1453988_a_at |
| 2.6 | Trip 13 | Thyroid hormone receptor interactor 13 | 1429294_at |
| 2.6 | Pde7a | Phosphodiesterase 7A | 1451839_a_at |
| 2.5 | Enc 1 | Ectodermal-neural cortex 1 | 1454904_at |
| 2.5 | Bcat 1 | Branched chain aminotransferase 1, cytosolic | 1450871_a_at |
| 2.5 | Nedd4 | Neural precursor expressed, developmentally down-regulated | 1450431_a_at |
| 2.5 | Dutp | Deoxyuridine triphosphatase | 1419270_a_at |
Listed are genes (excluding ESTs) with a change greater than 2.5-fold. If genes were represented by multiple probe sets on the array only the probe set with the highest change in expression level is shown.
The expression of Meis 1 could not be detected on the array since all Meis-specific probe sets are located in the 3′ UTR that was not present in the expression construct.
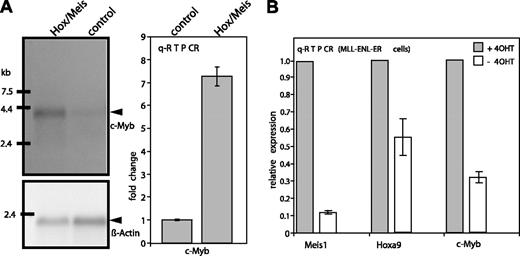
To verify the changes of c-Myb RNA levels in response to Hoxa9/Meis1 Northern blots and qRT-PCR were performed (Figure 2A). Both experiments confirmed the array result. When normalized to actin, qRT-PCR detected approximately 7-fold more c-Myb in Hoxa9/Meis1 versus control cells and this difference was corroborated by a corresponding up-regulation of c-Myb RNA in the blotting experiment. Since Hoxa9 and Meis1 are targets of MLL-ENL, c-Myb should also be, albeit indirectly, controlled by this protein. In order to test this proposition RNA was isolated from MLL-ENL-ERtm cells before and 96 hours after inactivation of MLL-ENL by withdrawal of 4-OHT. Changes in Meis1, Hoxa9, and c-Myb expression were analyzed by qRT-PCR (Figure 2B). In support of a hierarchic regulation cascade the inactivation of MLL-ENL led to a coordinated down-regulation of Meis1, Hoxa9 (8-fold and 2-fold, respectively), and c-Myb (approximately 3-fold).
Verification of array results. (A) Northern blot and qRT-PCR. RNA from Hoxa9/Meis1-transduced MLL-ENL-ERtm cells was isolated 72 hours after inactivation of MLL-ENL and compared with control RNA from cells transduced with empty vectors. Hybridization was done with c-Myb- and β-Actin-specific probes. Specific bands are labeled. qRT-PCR was performed with an intron-spanning primer pair, and expression levels were determined with the ΔΔCt method normalizing to actin. Means and standard deviations of triplicates are given. c-Myb RNA concentration in control cells was arbitrarily set to 1. (B) qRT-PCR testing for changes in Meis1, Hoxa9, and c-Myb expression in MLL-ENL-ERtm cells after inactivation of MLL-ENL. RNA was isolated from cells in the presence of 4-OHT (0-hour value, ▪), and 96 hours after 4-OHT withdrawal (□). The experiment was conducted as before except that 0-hour expression values were set arbitrarily to 1.
Verification of array results. (A) Northern blot and qRT-PCR. RNA from Hoxa9/Meis1-transduced MLL-ENL-ERtm cells was isolated 72 hours after inactivation of MLL-ENL and compared with control RNA from cells transduced with empty vectors. Hybridization was done with c-Myb- and β-Actin-specific probes. Specific bands are labeled. qRT-PCR was performed with an intron-spanning primer pair, and expression levels were determined with the ΔΔCt method normalizing to actin. Means and standard deviations of triplicates are given. c-Myb RNA concentration in control cells was arbitrarily set to 1. (B) qRT-PCR testing for changes in Meis1, Hoxa9, and c-Myb expression in MLL-ENL-ERtm cells after inactivation of MLL-ENL. RNA was isolated from cells in the presence of 4-OHT (0-hour value, ▪), and 96 hours after 4-OHT withdrawal (□). The experiment was conducted as before except that 0-hour expression values were set arbitrarily to 1.
c-Myb is essential for MLL-ENL-mediated transformation
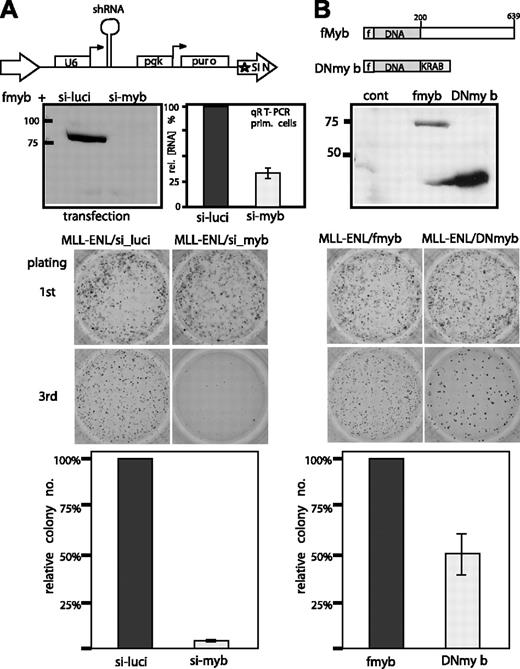
Next, we wanted to determine if c-Myb up-regulation is important for the oncogenic activity of Hoxa9 and Meis1 and therefore indirectly also for the transforming function of MLL fusion proteins. To this end hematopoietic precursor cells were transduced with MLL-ENL and either anti-Myc inhibitory RNA (RNAi) or a construct expressing a dominant-negative version of c-Myb. Transformation was tested by a methylcellulose-replating assay. This test relies on the fact that an oncogenic MLL fusion protein inhibits hematopoietic differentiation. Therefore, transformed immature precursors will continuously multiply and form colonies even after several replating rounds, whereas nontransformed cells will terminate proliferation. For production of RNAi targeted against c-Myb an oligonucleotide capable of forming a hairpin structure and with a sequence corresponding to a suitable target site in c-Myb was cloned into pSIRENretroQ (Figure 3A). This retroviral vector carries a deletion in the 3′ long terminal repeat (LTR) that will be copied to the 5′ LTR after genomic integration (self-inactivating virus [SIN]). An integrated sh cassette will therefore be exclusively transcribed under control of a vector-borne RNA polymerase III promoter (U6 snRNA promoter). After processing by endogenous enzymes a specific siRNA is produced that causes a degradation of the cognate mRNA. After testing several possible target sites for c-Myb a siRNA sequence was identified that enabled efficient knockdown of c-Myb as detected by Western blot in transiently transfected cells (Figure 3A). Transduction of primary hematopoietic precursors with this construct caused a knockdown of endogenous c-Myb RNA to about 33% ± 5% of control levels. A luciferase-specific shRNA served as control. Cotransduction of primary hematopoietic precursors with MLL-ENL and the shRNA vector did not cause any immediate toxicity, as colony numbers achieved by control cells and c-Myb knockdown cells were comparable in the first round of plating. However, after the second replating, c-Myb-deprived cells formed very few colonies (< 5% of controls; n = 3), indicating a loss of reproductive capacity. In a second approach precursor cells were transduced with MLL-ENL and a vector coding for a fusion of the flag-tagged c-Myb DNA-binding domain (amino acids 1-200) with the KRAB repressor motif (Figure 3B). Also, this dominant-negative c-Myb derivative caused a slight but significant reduction of colony numbers to about 50% ± 20% (n = 3) of control assays after the second replating.
Effect of c-Myb inhibition on MLL-ENL mediated transformation. (A) A retroviral construct in a “self-inactivating” (SIN) viral backbone (pSIRENretroQ) expressing a small hairpin RNA (shRNA) specifically directing against c-Myb was tested for efficacy by cotransfection with a flag-c-Myb expression clone. A similar virus with a luciferase-directed shRNA sequence was used as control. shRNAs are processed in the cell to siRNA. Knockdown was verified by Western blotting with anti-flag antibodies (top panel) and by transduction of primary hematopoietic cells with Myb shRNA viruses followed by qRT-PCR of c-Myb RNA (bar graph gives mean and standard deviation of 3 samples). Primary hematopoietic precursors were simultaneously transduced with a pMSCV-MLL-ENL virus and either the luciferase or the c-Myb shRNA construct. The cells were tested for enhanced replicative capacity as a marker for transformation in a methocel serial-replating assay under appropriate antibiotic selection. First-round and third-round colonies are shown in a representative example of 3 independent experiments. A numeric evaluation of relative colony numbers (n = 3) is shown in the bottom panel. (B) In a parallel experiment, c-Myb activity was reduced by coexpression of a dominant-negative c-Myb derivative (DNmyb). For this purpose the DNA binding domain of c-Myb (amino acids 1-200) was fused to a KRAB repressor domain. Correct expression of the flag-tagged construct was verified by an anti-flag immunoblot. Representative colony formation of cells expressing MLL-ENL and DNmyb or as control MLL-ENL and flag-c-Myb is shown as in panel A. Colonies were phtographed with a Canon Coolpix 990 camera (Canon, Tokyo, Japan) in nanomode after staining with iodonitro tetrazolim violet. Image processing was done with standard Windows (Microsoft, Redmond, WA) processing software.
Effect of c-Myb inhibition on MLL-ENL mediated transformation. (A) A retroviral construct in a “self-inactivating” (SIN) viral backbone (pSIRENretroQ) expressing a small hairpin RNA (shRNA) specifically directing against c-Myb was tested for efficacy by cotransfection with a flag-c-Myb expression clone. A similar virus with a luciferase-directed shRNA sequence was used as control. shRNAs are processed in the cell to siRNA. Knockdown was verified by Western blotting with anti-flag antibodies (top panel) and by transduction of primary hematopoietic cells with Myb shRNA viruses followed by qRT-PCR of c-Myb RNA (bar graph gives mean and standard deviation of 3 samples). Primary hematopoietic precursors were simultaneously transduced with a pMSCV-MLL-ENL virus and either the luciferase or the c-Myb shRNA construct. The cells were tested for enhanced replicative capacity as a marker for transformation in a methocel serial-replating assay under appropriate antibiotic selection. First-round and third-round colonies are shown in a representative example of 3 independent experiments. A numeric evaluation of relative colony numbers (n = 3) is shown in the bottom panel. (B) In a parallel experiment, c-Myb activity was reduced by coexpression of a dominant-negative c-Myb derivative (DNmyb). For this purpose the DNA binding domain of c-Myb (amino acids 1-200) was fused to a KRAB repressor domain. Correct expression of the flag-tagged construct was verified by an anti-flag immunoblot. Representative colony formation of cells expressing MLL-ENL and DNmyb or as control MLL-ENL and flag-c-Myb is shown as in panel A. Colonies were phtographed with a Canon Coolpix 990 camera (Canon, Tokyo, Japan) in nanomode after staining with iodonitro tetrazolim violet. Image processing was done with standard Windows (Microsoft, Redmond, WA) processing software.
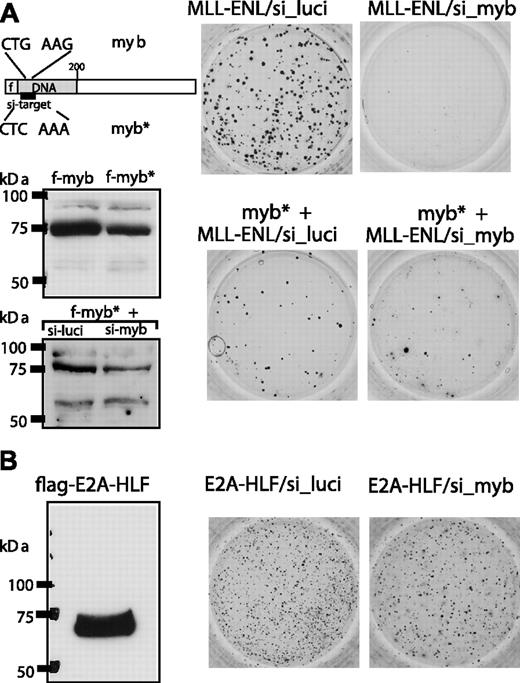
Specificity controls for siRNA mediated c-Myb knockdown. (A) A flag-tagged c-Myb mutant (Myb*) with a silent mutation in the siRNA target sequence was constructed by site-directed mutagenesis. Myb* was inserted into pMSCV-MLL-ENL under control of the plasmid-borne phosphoglycerate kinase promoter and tested for expression by anti-flag immunoblot with wild-type c-Myb as control. When tested under identical conditions, Myb* was significantly more resistant than wild-type Myb to the knockdown effect of anti-Myb shRNA (compare with Figure 3A). The constructs coding for MLL-ENL or MLL-ENL/Myb* were cotransduced with shRNA expression constructs as indicated into hematopoietic precursors and tested for colony-forming ability after serial replating. A representative example of third-round colonies is shown. (B) The effect of shRNA-mediated c-Myb suppression on E2A-HLF-mediated transformation was tested in an experiment analogous to that in panel A.
Specificity controls for siRNA mediated c-Myb knockdown. (A) A flag-tagged c-Myb mutant (Myb*) with a silent mutation in the siRNA target sequence was constructed by site-directed mutagenesis. Myb* was inserted into pMSCV-MLL-ENL under control of the plasmid-borne phosphoglycerate kinase promoter and tested for expression by anti-flag immunoblot with wild-type c-Myb as control. When tested under identical conditions, Myb* was significantly more resistant than wild-type Myb to the knockdown effect of anti-Myb shRNA (compare with Figure 3A). The constructs coding for MLL-ENL or MLL-ENL/Myb* were cotransduced with shRNA expression constructs as indicated into hematopoietic precursors and tested for colony-forming ability after serial replating. A representative example of third-round colonies is shown. (B) The effect of shRNA-mediated c-Myb suppression on E2A-HLF-mediated transformation was tested in an experiment analogous to that in panel A.
To control for potential off-target siRNA effects the transformation assay was repeated with cells that expressed Myb*, a flag-c-Myb version with a silent mutation destroying the siRNA target site (Figure 4). Myb* was inserted into the MLL-ENL expressing retrovirus under control of the vector-encoded phospho-glycerate-kinase promoter to ensure coexpression in transduced cells. Correct expression of Myb* was checked by Western blot. Compared with unaltered Myb, Myb* was considerably more resistant to the knockdown effect of cotransfected anti-Myb shRNA. Although MLL-ENL/Myb* cells consistently generated fewer colonies compared with controls, these cells were resistant against c-Myb knockdown as similar numbers of third-round colonies (± 20%) developed regardless of the presence of c-Myb shRNA. To exclude any further nonspecific effects by c-Myb shRNA, transformation by the unrelated oncoprotein E2A-HLF was tested. E2A-HLF does not work through direct up-regulation of Hoxa9 or Meis1 and should be less affected by c-Myb depletion. Indeed, E2A-HLF transformed cells did not respond to cotransduced c-Myb shRNA in replating assays (Figure 4B).
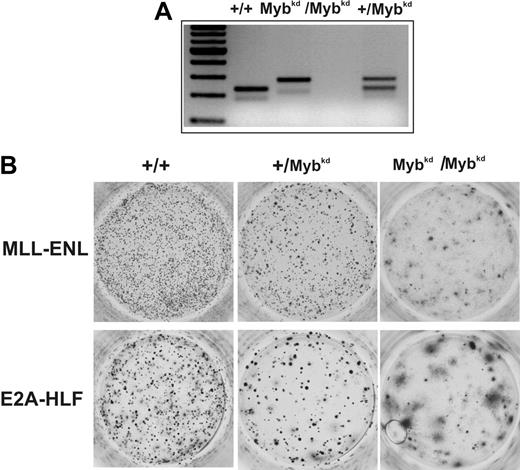
As an alternative to short-term knockdown techniques, we wanted to test the importance of c-Myb for hematopoietic-cell transformation in a c-Myb-deficient genetic background. Myb-null mice die in utero due to severe hematopoietic defects.38 However, it has been shown that mice with a hypomorphic c-Myb allele are viable. Integration of a floxed neo cassette into intron 6 of the c-Myb gene (cMybkd) reduces c-Myb protein to about 5% to 10% of the amount present in wild-type mice.37 We used this type of Myb-neo-tagged mice to investigate the necessity of c-Myb for transformation by MLL-ENL and E2A-HLF (Figure 5). Heterozygous +/c-Mybkd mice were intercrossed and offspring was used to isolate an HSC-enriched population. The cells were genotyped by PCR, transduced with MLL-ENL or E2A-HLF, and assayed in methocel. In contrast to short-term knockdown by shRNA, HSCs from a general Myb-deficient background were more severely inhibited in their proliferative capacity. In this model colony-formation was severely impaired after transduction not only with MLL-ENL but also with E2A-HLF. Third-round colonies were present in reduced numbers and remaining colonies appeared diffuse, indicating enhanced differentiation in the absence of sufficient c-Myb.
Effect of a c-Myb-deficient genetic background on hematopoietic transformation. Mice heterozygous for a hypomorphic allele of c-Myb (Mybkd) were intercrossed and the genotype of the offspring was determined by PCR. Mybkd is a floxed-neo cassette inserted into intron 6 of the c-Myb gene.37 This modification leads to a reduction of c-Myb protein expression to approximately 5% to 10% of the wild-type level. Hematopoietic precursors from 5-FU-treated bone marrow harvested from 8- to 10-week-old mice of the indicated phenotype were transduced with MLL-ENL- or E2A-HLF-expressing retroviruses. Cells were subject to standard serial replating in methocel. A typical example of third-round colonies is shown. In total the experiment was performed 3 times.
Effect of a c-Myb-deficient genetic background on hematopoietic transformation. Mice heterozygous for a hypomorphic allele of c-Myb (Mybkd) were intercrossed and the genotype of the offspring was determined by PCR. Mybkd is a floxed-neo cassette inserted into intron 6 of the c-Myb gene.37 This modification leads to a reduction of c-Myb protein expression to approximately 5% to 10% of the wild-type level. Hematopoietic precursors from 5-FU-treated bone marrow harvested from 8- to 10-week-old mice of the indicated phenotype were transduced with MLL-ENL- or E2A-HLF-expressing retroviruses. Cells were subject to standard serial replating in methocel. A typical example of third-round colonies is shown. In total the experiment was performed 3 times.
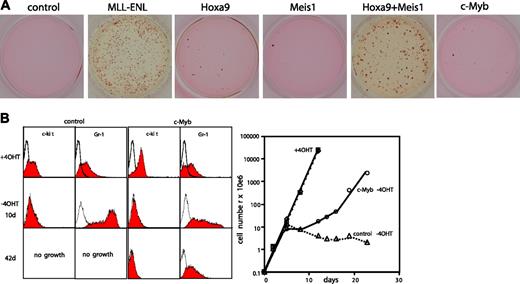
Transformation properties of c-Myb. (A) Serial replating assay with hematopoietic precursors transduced with empty virus or viruses coding for MLL-ENL, Hoxa9, Meis1, and c-Myb singly or in combination as indicated. Typical results of third-round colonies are shown. (B) MLL-ENL-ERtm cells were transduced with c-Myb or empty control virus. Surface markers c-kit and Gr-1 as well as proliferation rates of the resulting cells were analyzed by FACS (left panel) and cell counts (right panel) before and after inactivation of MLL-ENL by withdrawal of 4-OHT.
Transformation properties of c-Myb. (A) Serial replating assay with hematopoietic precursors transduced with empty virus or viruses coding for MLL-ENL, Hoxa9, Meis1, and c-Myb singly or in combination as indicated. Typical results of third-round colonies are shown. (B) MLL-ENL-ERtm cells were transduced with c-Myb or empty control virus. Surface markers c-kit and Gr-1 as well as proliferation rates of the resulting cells were analyzed by FACS (left panel) and cell counts (right panel) before and after inactivation of MLL-ENL by withdrawal of 4-OHT.
c-Myb is not sufficient for transformation
c-Myb has an established role as a hematopoietic oncogene. Therefore, we wanted to test the possibility that the transforming activity of a combined expression of Hoxa9 and Meis1 is due mostly or even exclusively to up-regulation of c-Myb. For this purpose the transforming potential of c-Myb as single agent was tested by serial replating. Primary hematopoietic precursors were transduced with retroviral constructs coding for c-Myb and as controls with empty viruses, MLL-ENL, Hoxa9, and Meis1 individually and in combination (Figure 6A). Cells expressing MLL-ENL and Hoxa9 plus Meis1 had extended proliferative potential as indicated by colony formation in the third round of replating. In contrast, the transduction with Hoxa9, Meis1, or c-Myb alone was not sufficient to induce a differentiation block under these conditions. This was not simply a problem of protein expression, as the respective constructs had been verified in Western blot experiments (Figures 1,5). To further address this question the consequences of c-Myb overexpression for hematopoietic differentiation were analyzed. For this purpose MLL-ENL-ERtm cells were transduced with c-Myb, and 4-OHT was washed out to initiate differentiation. Cellular maturation was followed by FACS analysis and cell counts (Figure 6B-C). Expression of c-Myb increased the levels of the early hematopoietic marker c-kit but had no effect on proliferation in the presence of 4-OHT. After withdrawal of 4-OHT control cells ceased growth after 5 days, lost c-kit expression, and gained high levels of the maturation marker Gr-1. In contrast, c-Myb cells experienced only a transient lag in proliferation. Upon prolonged culture a c-kit-, Gr-1low subpopulation emerged that sustained growth in culture for more than 2 months, indicating a c-Myb-dependent inhibition of differentiation.
Discussion
In this report we identify the genomic homolog of the avian myeloblastosis virus oncogene c-Myb as an important downstream target of Hoxa9 and Meis1. siRNA-mediated knockdown of c-Myb RNA to about one-third of the endogenous levels specifically inhibited Hoxa9/Meis1-induced transformation. A more severe depletion of c-Myb of 5% to 10% of normal amounts by genetic means37 was also able to impair hematopoietic transformation by the unrelated oncogene E2A-HLF. Finally, c-Myb could recapitulate some aspects of the phenotype imposed on hematopoietic cells by overexpression of Hoxa9 and Meis1. In summary, these results suggest that Hoxa9 and Meis1 use c-Myb as the entry point to influence central growth control. Support for an intricate connection of c-Myb and homeobox genes comes from experiments recently performed by Dorsam et al.34 In this study c-Myb was up-regulated in response to the introduction of Hoxa9 into U937 and K562 leukemia cell lines, and expression of Hoxa9 and c-Myb appeared to be correlated in primary human acute myeloid leukemia (AML) samples. According to a global survey of gene expression in normal tissues (http://symatlas.gnf.org/SymAtlas/), c-Myb is most abundant in CD34+ HSCs. In this respect our results are compatible with recent work by Wang et al that detects a stem-cell gene-expression signature in Hoxa9-immortalized cell lines transduced with Meis1.33 In contrast to this report, however, we could not find a significant up-regulation of the HSC receptor tyrosine kinase Flt-3 by Hoxa9/Meis1. This is probably due to a different target-cell type, as our cells were cultivated in cytokines, including granulocyte-macrophage-CSF (GM-CSF). According to Wang et al.33 Meis1 does not induce Flt3 expression in Hoxa9-immortalized progenitors in the presence of GM-CSF. The genetic program governed by homeobox genes therefore appears to be highly adaptable to the needs of the respective cellular environment.
Despite the fact that Hoxa9 and Meis1 are direct downstream targets of MLL fusion proteins c-Myb was not detected in our initial screen for MLL-ENL targets. This fact can be explained by the hierarchy of the gene-regulation events and the experimental setup. Originally, MLL-ENL-controlled genes were identified after inactivation of a conditional MLL-ENL protein. In order for c-Myb to show a response to a loss of MLL-ENL function, the available Hoxa9 and Meis1 proteins have to be degraded sufficiently to allow for c-Myb down-regulation. Reassessment of this question by qRT-PCR at a later time-point after MLL-ENL inactivation confirmed c-Myb also as an indirect MLL-ENL target. In summary, our results suggest a potential oncogenic pathway that transmits growth signals from upstream Hox regulators like the MLL fusions to Hox and Meis proteins and finally to c-Myb. c-Myb alone was not sufficient for a full transformation of hematopoietic cells. Therefore, other genes have to participate in this pathway.
Unexpectedly, several supposedly neuronal-specific genes were also among the identified Hoxa9/Meis1 targets, a hint that similar mechanisms might control early blood and neuronal development. This suggestion is supported by the fact that Meis1 is highly overexpressed in a subset of neuroblastomas with a particularly dismal prognosis.39 Interestingly, c-Myb and B-Myb have also been detected in neuroblastoma.40 The presence of B-Myb was negatively correlated with survival,41 and c-Myb was selected as cellular target for experimental neuroblastoma treatment with antisense deoxyribonucleotides.42 Further studies of these surprising similarities seem to be warranted.
Finally, it will be very interesting to learn how homeobox genes cause up-regulation of c-Myb, and if this mechanism will be suitable for therapeutic intervention during leukemia treatment. Regulated expression of c-Myb is not achieved by classic promoter modulation but mainly through transcriptional attenuation. An elongation-pause site in the first intron causes a premature transcriptional stop that is relieved by as-yet completely unknown mechanisms.43 Therefore, a straightforward activation by binding of Hox/Meis to the basal c-Myb promoter seems unlikely. Future studies will have to clarify this point.
Prepublished online as Blood First Edition Paper, February 28, 2006; DOI 10.1182/blood-2005-12-5014.
Supported by DFG grants SL27/6 and SFB473/D2 to R.K.S., Deutsche Krebshilfe BO10-2123 and EU-RIGHT to A.B. and R.K.S. Specialized Center of Research (SCOR) grant from the Leukemia and Lymphoma Society and National Institutes of Health grants CA-78815 and CA-92251 to J.L.H., and a Wellcome Trust grant to J.F. Equipment funding from Jose-Carreras-Stiftung and Curt-Bohnewand-Fond is gratefully acknowledged.
J.L.H. and C.B.B. contributed equally to this work.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal