Abstract
Tightly regulated expression of the transcription factor PU.1 is crucial for normal hematopoiesis. PU.1 knockdown mice develop acute myeloid leukemia (AML), and PU.1 mutations have been observed in some populations of patients with AML. Here we found that conditional expression of promyelocytic leukemia-retinoic acid receptor α (PML-RARA), the protein encoded by the t(15;17) translocation found in acute promyelocytic leukemia (APL), suppressed PU.1 expression, while treatment of APL cell lines and primary cells with all-trans retinoic acid (ATRA) restored PU.1 expression and induced neutrophil differentiation. ATRA-induced activation was mediated by a region in the PU.1 promoter to which CEBPB and OCT-1 binding were induced. Finally, conditional expression of PU.1 in human APL cells was sufficient to trigger neutrophil differentiation, whereas reduction of PU.1 by small interfering RNA (siRNA) blocked ATRA-induced neutrophil differentiation. This is the first report to show that PU.1 is suppressed in acute promyelocytic leukemia, and that ATRA restores PU.1 expression in cells harboring t(15;17).
Introduction
The transcription factor PU.1 is expressed at highest levels in granulocytic cells,1-5 and plays a crucial role during myeloid differentiation6,7 ; furthermore, Pu.1-/- mice lack mature myeloid cells.8-10 We recently reported that mutations of the PU.1 gene are found in some patients with acute myeloid leukemia (AML)11 and that decreased Pu.1 expression induces leukemia in mice.12 Others have demonstrated that loss and/or mutation of the gene encoding PU.1 contributes to radiation-induced murine AML.13,14
Acute promyelocytic leukemia (APL) harbors in 98% the translocation t(15;17), fusing a part of the promyelocytic leukemia (PML) gene to the retinoic acid receptor α (RARA) gene to encode PML-RARA.15 All-trans retinoic acid (ATRA) treatment together with chemotherapy is nowadays considered to represent a standard therapy for patients with APL. The response to ATRA induces differentiation of t(15;17) blasts, presumably through degradation of the PML-RARA fusion protein.16 PML-RARA interacts with transcriptional corepressors in an ATRA-sensitive manner, blocking the activation of RARA target genes.17,18 However, this hypothesis so far has hardly connected PML-RARA with transcription factors which are known to be critical for granulocytic differentiation.19 Recently we have identified CEBPB as a PML-RARA-responsive gene mediating granulopoiesis in retinoic acid-induced APL cell differentiation.20
Here, we found that expression of PU.1 is suppressed in human PML-RARA leukemic cells, and that treatment of these cells with ATRA restores PU.1 expression and induces neutrophil differentiation. In primary t(15;17) patient cells, PU.1 was markedly upregulated after treatment with ATRA. Restoring PU.1 in PML-RARA leukemic cells overcame the differentiation block and was necessary and sufficient to trigger neutrophil differentiation. These studies demonstrate that restoration of PU.1 expression is critical to induce differentiation of PML-RARA cells.
Patients, materials and methods
Northern blot analysis
RNA was isolated from cell lines by guanidium isothiocyanate extraction followed by cesium chloride (Ambion, Austin, TX) gradient purification. RNA samples (10 μg) were resolved by agarose formaldehyde gel electrophoresis and transferred to Biotrans nylon membrane (ICN, Irvine, CA). The blots were hybridized with α-32PdCTP-labeled human-specific PU.1 and granulocyte colony-stimulating factor receptor (G-CSFR) probes. The probe for human PU.1 mRNA was a 5′ probe, spanning a 438-bp EcoRI/PstI-fragment of the cDNA,3 and a fragment encompassing bp 289 to 1024 of the human G-CSFR cDNA served as probe for G-CSFR mRNAs.21 Hybridizations were performed at 65°C in 0.5 M sodium phosphate buffer (NaPO4; pH 7.2), 7% sodium dodecyl sulfate (SDS), and 1% bovine serum albumin (BSA) for 15 hours. Membranes were washed twice in 2 × SSC and 0.1% SDS at room temperature for 5 minutes and 3 times in 0.2 × SSC and 0.2% SDS at 65°C for 15 minutes. Autoradiography was performed at -80°C with Kodak MR film (Eastman Kodak, Rochester, NY) and quantitated using Imagequant Phosphorimager software (Molecular Dynamics, Piscataway, NJ). To ensure uniform loading levels and integrity of RNA samples, blots were rehybridized to a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA with a 1.3-kb PstI fragment of GAPDH cDNA.
Quantitative real-time PCR
RNA was extracted from Ficoll-separated mononucleated bone marrow cells. cDNA was synthesized using oligo(dT) 12-18 primers and Superscript II reverse transcriptase (RT) (catalog no. 18064-022; GIBCO BRL, Carlsbad, CA). Primer pairs were designed using the PRIMER Express 1.0 program (PE Applied Biosystems, Foster City, CA). PU.1: forward primer 5′-TGTTACAGGCGTGCAAAATGGAAGG-3′, reverse primer 5′-CTCGTGCGTTTGGCGTTGGTATAGA-3′, and fluorescent probe 5′-FAMCCTCGTCCCCCCTCCATCAGAAGACCTGG-TAMRA-3′; and GAPDH: forward primer 5′-GAAGGTGAAGGTCGGAGT, reverse primer 5′-GAAGATGGTGATGGGATTTC-3′, and fluorescent probe 5′-FAMCAAGCTTCCCGTTCTCAGCCTAMRA-3′. The amplification cycles for PU.1 and GAPDH were 95°C for 10 minutes, followed by 40 cycles with 95°C for 15 seconds and 60°C for 1 minute. The primer and probes for OCT-1, CEBPA, CEBPB, and CEBPE were from Applied Biosystems. The Assay ID for OCT-1 is Hs00 231250_m1; for CEBPA, Hs00269972_s1; for CEBPB, Hs00270923_s1; and for CEBPE, Hs00357657_m1, respectively. The amplification cycles for CEBPA, CEBPB, CEBPE, and OCT-1 were 95°C for 10 minutes, followed by 40 cycles with 97°C for 30 seconds and 60°C for 1 minute. All expression levels (gene of interest and normalization control) in quantitative RT-polymerase chain reaction (PCR) were obtained by the use of standard curves. After normalization, the relative expression levels were calculated.
Immunoblotting
Cells were lysed in RIPA buffer (150 mM NaCL, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris [pH 7.5], and 0.5 mM PMSF). Protein extracts were fractionated on SDS-12% polyacrylamide gels and transferred to nitrocellulose membranes by electroblotting. PU.1, CEBPA, CEBPB, CEBPE, OCT-1, macrophage colony-stimulating factor (M-CSF) receptor, and PML-RARA proteins were detected with rabbit anti-rat polyclonal PU.1 serum (1:500) (catalog no. sc-352; Santa Cruz Biotechnology, Santa Cruz, CA), a rabbit anti-human polyclonal CEBPA serum (1:1000) (catalog no. sc-61; Santa Cruz Biotechnology), a rabbit antihuman polyclonal CEBPB serum (1:1000) (catalog no. sc-150; Santa Cruz Biotechnology), a rabbit anti-human polyclonal CEBPE serum (1:500) (catalog no. sc-158; Santa Cruz Biotechnology), a rabbit anti-human polyclonal OCT-1 serum (1:20) (catalog no. sc-8024; Santa Cruz Biotechnology), a rabbit anti-human polyclonal M-CSF receptor antibody (1:1000) (catalog no. sc-692; Santa Cruz Biotechnology), and a rabbit anti-human polyclonal RARA antibody (1:1000) (catalog no. sc-551x; Santa Cruz Biotechnology), respectively. A monoclonal anti-mouse β-tubulin antibody served as a loading control (catalog no. 1111876; Boehringer Mannheim, Mannheim, Germany), detected with an anti-mouse immunoglobulin G-horseradish peroxidase (IgG-HRP)-conjugated secondary antibody (catalog no. sc-2005; Santa Cruz Biotechnology).
Electrophoretic mobility shift assay
Complementary oligonucleotides were annealed and labeled using γ-p32 ATP and T4 polynucleotide kinase (NEB, Beverly, MA). The M-CSF receptor promoter oligonucleotide (bp -53 to -36) had the sequence 5′-TAA AAG GGG AAG AAG AGG-3′.22 The sequence of the oligonucleotide of the human PU.1 promoter was 5′-CCT GTA TGT AGC GCA AGA GAT TTA TGC AAA CGG GCT GGG GCG-3′; the sequence of the mutated OCT site was 5′-AAG AGA TTT AGA GCT CCG GGC TGG G-3′.
Nuclear extracts (10 μg) were incubated with 20 000 counts per minute (cpm) of double-stranded oligonucleotide in a 20-μL reaction mixture containing 10 mM HEPES (pH 7.9), 50 mM KCl, 0.5 mM EDTA, 1 mM dithiothreitol (DTT), 2.5 mM MgCl2, 10% glycerol, BSA (NEB), 3 μg poly(dI-dC) (Amersham Pharmacia, Piscataway, NJ) at room temperature for 20 minutes. Cells (2-5 × 107) were harvested, pelleted, and resuspended in 1.5 mL cold phosphate-buffered saline (PBS), and the cell suspension was centrifuged. The following procedures were then carried out at 4°C. Cells were resuspended in 1 volume cold buffer A (10 mM HEPES-KOH [pH 7.90], 1.5 mM MgCl2, 10 mM KCL, 0.5 mM DTT, and 0.5 mM PMSF) and were allowed to swell on ice for 15 minutes. The cells were lysed with 10 rapid strokes through a 23- to 26-gauge needle, centrifuged for 20 seconds, and the supernatant fraction was discarded. The pellet was resuspended in 2 of 3 pellet volume cold buffer C (20 mM HEPES-KOH [pH 7.90], 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, and 0.5 mM PMSF) and incubated (rotating end-on-end) at 4°C for 30 minutes. Cellular debris was removed by centrifugation for 5 minutes at 4°C and the supernatant fraction (containing DNA binding proteins) was stored at -80°C. The yield was 50 to 75 μg protein per 106 cells. Protein concentrations were calculated densitophotometrically (Bio-Rad protein assay reagent, catalog no. 500-0006; Bio-Rad, Hercules, CA). For PU.1, CEBPA, CEBPB, and CEBPE supershift experiments, PU.1 polyclonal rabbit serum (catalog no. sc-352x; Santa Cruz Biotechnology), rabbit anti-human polyclonal CEBPA serum (catalog no. sc-61x; Santa Cruz Biotechnology), rabbit anti-human polyclonal CEBPB serum (catalog no. sc-150x; Santa Cruz Biotechnology), and a rabbit anti-human polyclonal CEBPE serum (catalog no. sc-158x; Santa Cruz Biotechnology), respectively, were added.
Transient transfections
For suspension cells, 1 × 107 HT93 cells were transfected by electroporation at 300 V using 10 μg reporter plasmid. For adherent cells, 6 × 104 cells of the human lung cancer line H1299 were seeded in 24-well dishes and transfected after 24 hours using lipofectamine (GIBCO) with reporter plasmid (250 ng), expression vector (40-250 ng), and cytomegalovirus-β-galactosidase (CMV-β-Gal) construct (40 ng). Luciferase assays were carried out 24 hours after transfection. Luciferase activities were normalized for transfection efficiency with the cotransfected CMV-β-Gal construct, using the chemoluminescent reporter gene assay for the detection of beta-galactosidase (Tropix; PE Applied Biosystems, Foster City, CA).
Constructs
The CEBP site at -68 to -64 in the -86-bp PU.1 promoter luciferase construct was mutated from GCAAG to CCTGG, with the mutated base pairs underlined (CEBP mut). The OCT site at -57 to -50 was mutated from ATGCAAA to AGAGCGG (OCT mut). The luciferase construct CEBP+OCT mut had both sites mutated accordingly.
Patient samples
cDNA was available from 141 patients with AML from the Institute of Medical Oncology, University Hospital, Bern, Switzerland, and from the Kumamoto University Hospital, Japan. Informed consent was obtained from all patients according to procedures approved by the respective institutional review boards (decisions of the cantonal ethics committee of Bern, Switzerland, no. 180/94 of 12/26/1994, no. 162/2000 of 8/15/2001, and no. 22/2001 of 7/25/2001; and approval of the ethics committee at the Kumamoto University School of Medicine, Kumamoto, Japan, no. 41/2002). The patient analyzed in Figure 5C with newly diagnosed AML-M3 was treated with 45 mg/m2/d of ATRA alone. No concomitant chemotherapy was given. Peripheral blood was harvested daily and after Ficoll separation RNA was extracted by Qiagen RNeasy Mini Kit (catalog no. 74104; Qiagen, Valencia, CA). Leukemic blasts (percentage of all blood leukocytes) steadily decreased from 51% at day 0 to 25% at day 7.
Chromatin immunoprecipitation
HT93A cells were cross-linked with 0.5% formaldehyde before incubation in 0.125 M glycine. Cells were washed twice with PBS and lysed in cell lysis buffer (10 mM Tris [pH 8], 10 mM NaCl, 0.2% NP-40, 1 mM PMSF, 1 μg of leupeptin and aprotinin/mL). The nuclei were lysed in 1.5 volumes of nuclei lysis buffer (50 mM Tris [pH 8], 10 mM EDTA, 1% SDS, 1 mM PMSF, 1 μg aprotinin/mL, 1 μg leupeptin/mL). The chromatin was sonicated 8 times for 20 seconds, and incubated with 25 μg normal rabbit IgG (Santa Cruz Biotechnology). Protein A agarose (100 μL; Santa Cruz Biotechnology) was added, and the supernatant was treated with 10 μg normal rabbit IgG, 6 μg CEBPA antibody, 6 μg CEBPB antibody, 6 μg CEBPE antibody, or 6 μg OCT-1 antibody (all from Santa Cruz Biotechnology). The lysate-antibody mixture was then incubated with 100 μL protein G beads. Pellets were washed twice with immunoprecipitation (IP) wash I (20 mM Tris [pH 8], 2 mM EDTA, 0.1% SDS, 1% Triton X-100, 1 mM PMSF, 1 mg aprotinin/mL, and 1 mg leupeptin/mL), once with IP wash II (10 mM Tris [pH 8], 0.25 M LiCl, 1 mM EDTA, 1% NP-40, 1% deoxycholate, 1 mM PMSF, 1 mg aprotinin/mL, and 1 mg leupeptin/mL), and twice with Tris-EDTA. The DNA-protein antibody complexes were eluted, and 1 μg RNaseA and 0.3 M NaCl were added before treatment with proteinase K (0.24 mg/mL). DNA was resuspended in 50 μL water, and 2 μL was used for quantitative RT-PCR (qRT-PCR) analysis.
Cell lines with conditional PU.1 expression
Phoenix cells, a human packaging cell line, were transiently transfected using lipofectamine with either the pBabePuro vector or with the PU.1-ER plasmid subcloned into the pBabePuro vector. 104 HT93A cells were incubated in 4 mL supernatant and 5 μg/mL polybrene for 4 hours. A second infection cycle was performed after 24 hours. HT93A cells were grown in 96-well plates, and selection was started 48 hours after the first infection cycle in 0.5 μg/mL puromycin.
Fluorescence-activated cell sorting and cell-cycle analysis
Cells (1 × 106) were incubated in PBS with 2% (wt/vol) BSA on ice for 15 minutes. After washing in PBS, cells were incubated with 2 μL recombinant PE-conjugated mouse anti-human monoclonal CD11b antibody (cat no. 347557; BD Pharmingen, Palo Alto, CA) or mouse anti-human monoclonal G-CSFR antibody (catalog no. 554538; BD Pharmingen) in 50 μL PBS for 60 minutes on ice, washed in PBS, and resuspended in 500 μL PBS with 10% formaldehyde. For cell-cycle analysis, 1 × 106 cells were fixed in 5 mL 70% ethanol/PBS on ice for 1 hour. Cells were washed in PBS, and resuspended for 15 minutes at 37° in 1 mL propidium iodide (PI)-staining solution containing 200 μg PI (Sigma, St Louis, MO). Samples were analyzed on a FACScan flow cytometer (Becton Dickinson, San Jose, CA) using Cellquest software (Becton Dickinson).
RNA interference (siRNA) experiments
Two empirically determined complementary oligonucleotides were designed to establish stably transfected HT93A cell lines expressing PU.1 small interfering RNA (siRNA). Nonhomology with other Ets transcription factors was verified by BLAST (basic local alignment search tool) searches. The 2 sequences were 5′-CGAGCTTCGCCGAGAACAACTTAAGCTTAAGTTGTTCTCGGCGAAGCTCCCCTTTTTG-3′; and 5′-AATTCAAAAAGGGGAGCTTCGCCGAGAACAACTTAAGCTTAAGTTGTTCTCGGCGAAGCTCGGGCC-3′, respectively. The 2 oligonucleotides were annealed and inserted in a modified MSCV-U6 siRNA retroviral expression vector digested with ApaI and EcoRI.23 HT93A cells were separately electroporated with the MSCV-U6-PU.1 vectors and the MSCV-U6 empty vector (siV). Pools of cells were selected with puromycin (1 μg/mL) and expanded for 1 month.
For experiments targeting CEBPA, CEBPB, CEBPE, or OCT-1, 5 × 106 HT93A cells were incubated in 100 μL Amaxa solution V (Nucleofector Kit V; Amaxa, Cologne, Germany) and mixed with 1 μg siRNA. Cells were transfected by electroporation applying Nucleofector technology (software V2.1; Amaxa), and then cultured for 48 hours in the absence or presence of ATRA. The siRNA sequence targeting CEBPA was 5′-GCAAAUCGUGCCUUGUCAUTT-3′, and 5′-AUGACAAGGCACGAUUUGCTC-3′ (siRNA ID no. 146390); for CEBPB it was 5′-CCCACGUGUAACUGUCAGCTT-3′, and 5′-GCUGACAGUUACACGUGGGTT-3′ (siRNA ID no. 114496); for CEBPE it was 5′-GGACCCAGCAUAAUGAUUATT-3′, and 5′-UAAUCAUUAUGCUGGGUCCTG-3′ (siRNA ID no. 10701); and for OCT-1 it was 5′-GCGCAGUUUAUCAUCUCACTT-3′, and 5′-GUGAGAUGAUAAACUGCGCTG-3′ (siRNA ID no. 114254; all from Ambion, Austin, TX). The silencer negative control siRNA no. 2 (Ambion) was used to control for nonspecific effects in siRNA experiments.
Results
PU.1 is suppressed in human myeloid cells conditionally expressing PML-RARA
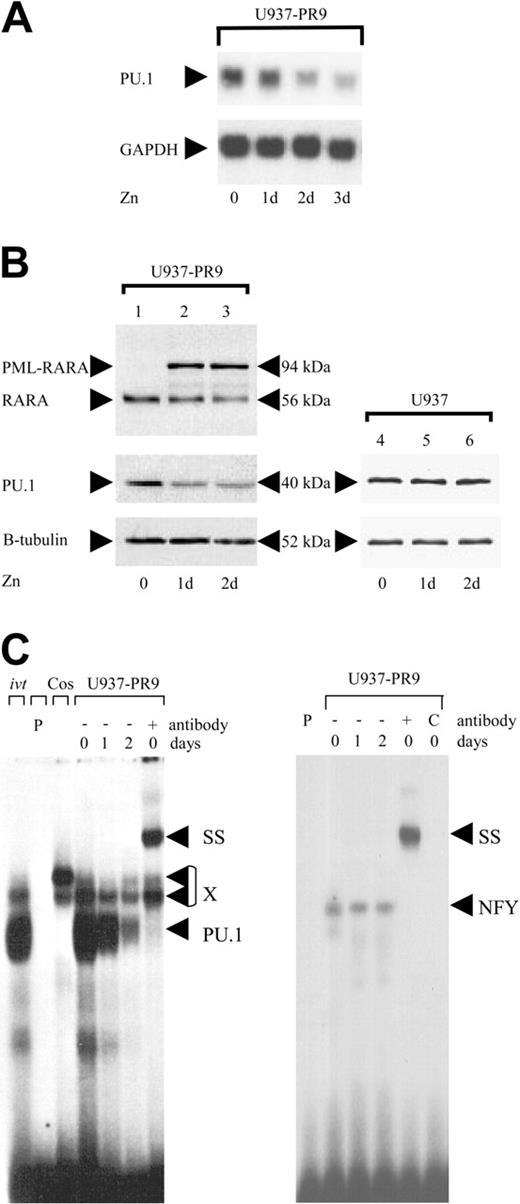
Given the importance of PU.1 in myeloid development, we hypothesized that PML-RARA might suppress PU.1. We analyzed PU.1 expression in U937-PML-RARA cells (U937-PR9), which conditionally express the PML-RARA protein under the control of the zinc-inducible metallothionein promoter.24 Induction of PML-RARA protein with zinc suppressed PU.1 mRNA (Figure 1A) and protein (Figure 1B), and caused a rapid decrease of DNAbinding activity to the PU.1 site in the M-CSF receptor promoter, an important PU.1 target gene22 (Figure 1C). Integrity of the samples was verified by measuring binding activity of the same extracts to an NFY site. Treatment of the parental U937 cells with zinc had no effect on PU.1 expression (Figure 1B). Therefore, expression of the PML-RARA protein can suppress PU.1 expression in human myeloid cells.
PU.1 is suppressed and ATRA restores PU.1 expression in human leukemic PML-RARA cells
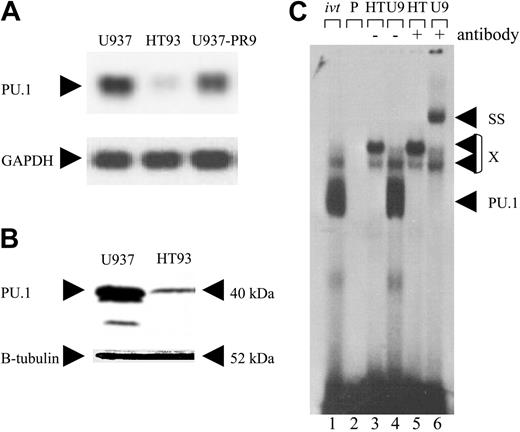
HT93A cells are derived from PML-RARA-positive blasts of a patient with APL.25 We found that PU.1 RNA and protein were markedly decreased in HT93A cells compared with U937 cells, a myeloid leukemic cell line without the t(15;17) (Figure 2A-B). In contrast to U937 cells, nuclear extracts from HT93A cells showed no binding activity to the PU.1 site in the M-CSF receptor promoter (Figure 2C).
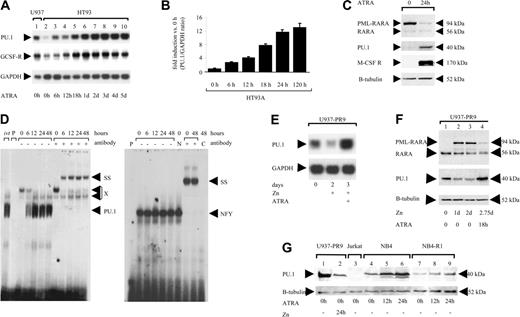
ATRA treatment can overcome the differentiation block in human t(15;17) blasts.15 Interestingly, we found that ATRA treatment increased PU.1 mRNA expression in HT93A cells 8-fold after 18 hours by Northern blot analysis (Figure 3A), and 12-fold after 24 hours by real-time PCR (Figure 3B). ATRA treatment of HT93A cells resulted in loss of the PML-RARA fusion protein after 24 hours (Figure 3C). PU.1 protein was induced concomitant with PML-RARA clearance, as were PU.1 target genes such as the M-CSF receptor (Figure 3C). Electrophoretic mobility shift assay (EMSA) analysis demonstrated a rapid increase in DNA binding activity to the PU.1 site in the M-CSF receptor promoter, as early as 6 hours after ATRA treatment (Figure 3D); therefore, restoring PU.1 expression and activity is an early event after ATRA treatment.
To exclude the possibility that this up-regulation was specific to the HT93A cell line only, we analyzed 2 other leukemic cell lines expressing PML-RARA. In U937 cells, conditional expression of PML-RARA suppressed PU.1 mRNA after 2 days, and treatment with ATRA for an additional 24 hours (in parallel with continued zinc treatment) resulted in a 3-fold increase in PU.1 mRNA and protein levels compared with uninduced U937-PR9 cells (Figure 3E-F). We confirmed that in these cells, treatment with ATRA resulted in clearance of the PML-RARA fusion protein (Figure 3F). Finally, NB4 cells, another cell line derived from a patient with t(15;17),26 also expressed low levels of PU.1, and, as was the case for HT93A and U937-PR9, ATRA treatment of NB4 again induced PU.1 protein (Figure 3G). NB4-R1 cells have acquired resistance to differentiation therapy with ATRA. As in NB4, we observed low levels of PU.1 protein in unstimulated NB4-R1 cells (Figure 3G). However, no increase in PU.1 expression (after normalization to the β-tubulin loading control) was detectable following ATRA treatment. ATRA sensitivity thus appears to be necessary for restoration of PU.1 in cells expressing PML-RARA. In conclusion, we have demonstrated in 2 different PML-RARA leukemic cell lines and a conditional myeloid line that PML-RARA suppresses PU.1 RNA, protein, and DNA binding, and that this suppression is abrogated along with degradation of PML-RARA following treatment with ATRA.
Conditional expression of PML-RARA in myeloid cells suppresses PU.1. (A) Top: U937 cells stably transfected with the PML-RARA cDNA (U937-PR9) under the control of the zinc-inducible metallothionein promoter were analyzed by Northern blot for PU.1 mRNA expression before and after treatment with 100 μM zinc. Bottom: The same blot was hybridized with a GAPDH probe. Zn indicates days of zinc treatment. (B) Western blots using a RARA antibody (top), a PU.1 antibody (middle), and a β-tubulin antibody (bottom). The PML-RARA protein was induced by treating U937-PR9 cells with 100 μM zinc. RARA indicates the endogenous RARA protein as present in U937 cells. Right panel: U937 cells lacking the zinc-inducible construct were treated with zinc to exclude nonspecific effects of zinc. (C) EMSA of binding activity to the PU.1 site in the M-CSF receptor promoter (left panel). U937-PR9 cells were induced with zinc. X represents nonspecific binding commonly observed with this oligonucleotide; ivt; in vitro-translated PU.1 protein is run as a control; P, free probe alone; Cos, nuclear extracts from Cos cells serve as a negative control; SS, supershift with specific antibody; and PU.1, shift of PU.1 protein binding to this particular site. The right panel represents binding of the same extracts to an NFY site as a control for loading and integrity of the extracts. C indicates competition with cold oligo.
Conditional expression of PML-RARA in myeloid cells suppresses PU.1. (A) Top: U937 cells stably transfected with the PML-RARA cDNA (U937-PR9) under the control of the zinc-inducible metallothionein promoter were analyzed by Northern blot for PU.1 mRNA expression before and after treatment with 100 μM zinc. Bottom: The same blot was hybridized with a GAPDH probe. Zn indicates days of zinc treatment. (B) Western blots using a RARA antibody (top), a PU.1 antibody (middle), and a β-tubulin antibody (bottom). The PML-RARA protein was induced by treating U937-PR9 cells with 100 μM zinc. RARA indicates the endogenous RARA protein as present in U937 cells. Right panel: U937 cells lacking the zinc-inducible construct were treated with zinc to exclude nonspecific effects of zinc. (C) EMSA of binding activity to the PU.1 site in the M-CSF receptor promoter (left panel). U937-PR9 cells were induced with zinc. X represents nonspecific binding commonly observed with this oligonucleotide; ivt; in vitro-translated PU.1 protein is run as a control; P, free probe alone; Cos, nuclear extracts from Cos cells serve as a negative control; SS, supershift with specific antibody; and PU.1, shift of PU.1 protein binding to this particular site. The right panel represents binding of the same extracts to an NFY site as a control for loading and integrity of the extracts. C indicates competition with cold oligo.
A DNA sequence in the PU.1 proximal promoter mediates ATRA-induced PU.1 activation
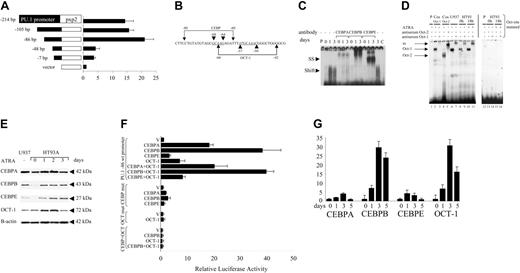
In order to understand the mechanism by which ATRA induced PU.1 expression in cells expressing PML-RARA, we asked whether ATRA induction of PU.1 was mediated by elements in the PU.1 promoter. HT93A cells were transfected with PU.1 promoter constructs and then treated for 18 hours with ATRA. We found that a sequence extending up to 86 bp of the PU.1 promoter upstream of the transcription start site was sufficient to mediate a 21-fold increase of PU.1 promoter-directed luciferase activity in response to treatment with ATRA. In contrast, a construct including 48 bp lost more than 80% of this activation (Figure 4A). We therefore conclude that the region between -86 and -48 of the PU.1 promoter contains 1 or several elements mediating PU.1 activation following ATRA treatment.
Leukemic cells from a patient with APL (HT93 cells) have suppressed PU.1 expression. (A) Northern blot analysis using a PU.1 probe showing myeloid U937 cells lacking the PML-RARA translocation (first lane), t(15;17) HT93A cells (second lane), and uninduced U937-PR9 cells (third lane). The somewhat lower PU.1 expression in U937-PR9 versus U937 cells may be due to leakiness of the system and thus low level expression of PML-RARA in the absence of zinc. The blot was reprobed with a GAPDH probe. (B) Prolonged exposure of a Western analysis using a PU.1 antiserum (top). U937 cells served as a positive control. The blot was reprobed with a β-tubulin antibody (bottom). (C) EMSA studying the binding activity to the PU.1 site in the M-CSF receptor promoter. U937 cells (U9; lanes 4 and 6) were compared with HT93A cells (HT; lanes 3 and 5). ivt indicates in vitro-translated PU.1 protein is run as a control; P, free probe alone; SS, supershift with specific antibody; PU.1, shift of PU.1 protein binding to this particular site; and X, nonspecific binding.
Leukemic cells from a patient with APL (HT93 cells) have suppressed PU.1 expression. (A) Northern blot analysis using a PU.1 probe showing myeloid U937 cells lacking the PML-RARA translocation (first lane), t(15;17) HT93A cells (second lane), and uninduced U937-PR9 cells (third lane). The somewhat lower PU.1 expression in U937-PR9 versus U937 cells may be due to leakiness of the system and thus low level expression of PML-RARA in the absence of zinc. The blot was reprobed with a GAPDH probe. (B) Prolonged exposure of a Western analysis using a PU.1 antiserum (top). U937 cells served as a positive control. The blot was reprobed with a β-tubulin antibody (bottom). (C) EMSA studying the binding activity to the PU.1 site in the M-CSF receptor promoter. U937 cells (U9; lanes 4 and 6) were compared with HT93A cells (HT; lanes 3 and 5). ivt indicates in vitro-translated PU.1 protein is run as a control; P, free probe alone; SS, supershift with specific antibody; PU.1, shift of PU.1 protein binding to this particular site; and X, nonspecific binding.
CEBP family members and OCT-1 activate the PU.1 promoter following ATRA treatment
Detailed analysis of the sequence from -48 to -86 bp of the PU.1 promoter revealed CEBP and OCT consensus binding sites27,28 (Figure 4B). Oligonucleotide probes were designed with either the CEBP or the OCT site. EMSA using the CEBP site probe indicated that ATRA treatment of HT93A cells induced a rapid increase in CEBP binding activity (Figure 4C). Specific antibodies against CEBP family members suggested that there was a small increase in CEBPA binding activity after 24 hours of ATRA treatment, but a much more dramatic increase in binding of CEBPB. Binding by CEBPE was maximal at 3 days (Figure 4C). In addition to the binding site for CEBP family members centered about -68 to -64 bp, the sequence between -48 to -86 bp of the PU.1 promoter also contains an OCT binding site28 centered about -57 to -50 bp (Figure 4B). EMSA using a probe containing this OCT site revealed a strong complex in U937 and HT93A cells, and this binding activity was induced 4-fold after 18 hours of ATRA treatment of HT93A cells (Figure 4D). Comparison with nuclear extracts from COS cells transfected with either OCT-1 or OCT-2 identified this complex to be OCT-1, and mutation of the OCT site abolished binding activity completely (Figure 4D).
ATRA restores PU.1 expression in APL cells. (A) Northern blot analysis of HT93A cells treated with 1 μM ATRA and harvested at the time points indicated. The blot was probed for PU.1 (top), G-CSF receptor (middle), and GAPDH expression (bottom). (B) HT93A cells were treated with 1 μM ATRA. PU.1 mRNA expression was determined by quantitative real-time PCR. Results are given as fold induction of the PU.1/GAPDH ratio compared with the value of HT93A cells before treatment (0 hours). Mean values and SD (error bars) are depicted. (C) Western analysis using a RARA antiserum (top), a PU.1 antibody (second), an M-CSF receptor antibody (third), and a β-tubulin antiserum (bottom). HT93A cells were harvested before and after ATRA treatment for 24 hours. (D) EMSA of DNA-binding activity to the PU.1 site in the M-CSF receptor promoter. HT93A cells were stimulated with 1 μM ATRA and harvested at the time points indicated. ivt indicates in vitro-translated PU.1 protein; P, free probe alone; SS, supershift with specific antibody; PU.1, shift of PU.1 protein binding to this particular site; and X, nonspecific binding. The panel on the right represents binding of the same extracts to an NFY site as a control for loading and integrity of the extracts. N indicates treatment of the extracts with normal rabbit serum; and C, competition with cold oligo. (E) ATRA treatment overcomes the suppressive effect of PML-RARA on PU.1 expression. Northern blot showing PU.1 mRNA expression of U937-PML/RARA (U937-PR9) cells (top). Lane 1 depicts U937-PML-RARA cells in the absence of zinc or ATRA. Lane 2 shows U937-PR9 cells after 2 days of treatment with zinc (100 μM); lane 3 demonstrates U937-PR9 cells after 2 days of treatment with zinc (100 μM) and also treated with both ATRA (1 μM) and zinc for the last 24 hours. The same blot was probed for GAPDH (bottom). (F) Western blot analysis of whole-cell lysates of U937-PR9 cells. Lane 1 represents unstimulated cells. Cells were induced with zinc for 1 and 2 days (lanes 2 and 3). In lane 4 cells were treated for 48 hours with zinc, and then for 18 hours with both zinc and ATRA. Top: Staining with a RARA antibody detecting the fusion protein PML-RARA and the endogenous RARA. Middle: Probing the same blot with an antibody-detecting PU.1 protein. Bottom: The blot was restained for β-tubulin. (G) ATRA sensitivity is a prerequisite for PU.1 restoration in APL cells. Whole-cell lysates were analyzed by Western blot using a PU.1 antibody. Unstimulated U937-PR9 cells (lane 1) served as a positive control and Jurkat cells as a negative control (lane 3). Lane 2 shows U937-PR9 cells after 24 hours of PML-RARA induction. NB4 cells (lanes 4 to 6) and the ATRA-resistant APL cell line NB4-R1 (lanes 7 to 9) were induced for 12 or 24 hours with 1 μM ATRA. The blot was restained for β-tubulin (bottom).
ATRA restores PU.1 expression in APL cells. (A) Northern blot analysis of HT93A cells treated with 1 μM ATRA and harvested at the time points indicated. The blot was probed for PU.1 (top), G-CSF receptor (middle), and GAPDH expression (bottom). (B) HT93A cells were treated with 1 μM ATRA. PU.1 mRNA expression was determined by quantitative real-time PCR. Results are given as fold induction of the PU.1/GAPDH ratio compared with the value of HT93A cells before treatment (0 hours). Mean values and SD (error bars) are depicted. (C) Western analysis using a RARA antiserum (top), a PU.1 antibody (second), an M-CSF receptor antibody (third), and a β-tubulin antiserum (bottom). HT93A cells were harvested before and after ATRA treatment for 24 hours. (D) EMSA of DNA-binding activity to the PU.1 site in the M-CSF receptor promoter. HT93A cells were stimulated with 1 μM ATRA and harvested at the time points indicated. ivt indicates in vitro-translated PU.1 protein; P, free probe alone; SS, supershift with specific antibody; PU.1, shift of PU.1 protein binding to this particular site; and X, nonspecific binding. The panel on the right represents binding of the same extracts to an NFY site as a control for loading and integrity of the extracts. N indicates treatment of the extracts with normal rabbit serum; and C, competition with cold oligo. (E) ATRA treatment overcomes the suppressive effect of PML-RARA on PU.1 expression. Northern blot showing PU.1 mRNA expression of U937-PML/RARA (U937-PR9) cells (top). Lane 1 depicts U937-PML-RARA cells in the absence of zinc or ATRA. Lane 2 shows U937-PR9 cells after 2 days of treatment with zinc (100 μM); lane 3 demonstrates U937-PR9 cells after 2 days of treatment with zinc (100 μM) and also treated with both ATRA (1 μM) and zinc for the last 24 hours. The same blot was probed for GAPDH (bottom). (F) Western blot analysis of whole-cell lysates of U937-PR9 cells. Lane 1 represents unstimulated cells. Cells were induced with zinc for 1 and 2 days (lanes 2 and 3). In lane 4 cells were treated for 48 hours with zinc, and then for 18 hours with both zinc and ATRA. Top: Staining with a RARA antibody detecting the fusion protein PML-RARA and the endogenous RARA. Middle: Probing the same blot with an antibody-detecting PU.1 protein. Bottom: The blot was restained for β-tubulin. (G) ATRA sensitivity is a prerequisite for PU.1 restoration in APL cells. Whole-cell lysates were analyzed by Western blot using a PU.1 antibody. Unstimulated U937-PR9 cells (lane 1) served as a positive control and Jurkat cells as a negative control (lane 3). Lane 2 shows U937-PR9 cells after 24 hours of PML-RARA induction. NB4 cells (lanes 4 to 6) and the ATRA-resistant APL cell line NB4-R1 (lanes 7 to 9) were induced for 12 or 24 hours with 1 μM ATRA. The blot was restained for β-tubulin (bottom).
These increases in DNA binding activity of CEBP and OCT-1 were paralleled by similar changes in levels of protein. ATRA treatment of HT93A cells induced CEBPB and CEBPE proteins after 24 hours. Interestingly, the level of CEBPA protein was slightly decreased after 3 days, at a time when CEBPE protein peaked (Figure 4E). Similarly, Western blot analysis verified a 2.4-fold increase of OCT-1 protein in HT93A cells after 2 days of ATRA treatment (Figure 4E). Therefore, the temporal pattern of DNA binding activity of several CEBP family members is similar to the levels of protein observed in these cells following ATRA induced neutrophil differentiation, as is the case for OCT-1 as well.
We next analyzed the potential of CEBP and OCT-1 to activate the -86-bp PU.1 promoter in transient transfections in adherent H1299 cells. Among CEBP family members, CEBPB preferentially activated this promoter construct up to 38-fold; in contrast, CEBPB failed to activate the -48-bp PU.1 promoter construct lacking the CEBP site (data not shown). OCT-1 activated the promoter to a lesser extent, and cotransfection of CEBP and OCT-1 failed to demonstrate any evidence for synergistic activation of the promoter, suggesting that CEBP factors and OCT-1 act independently on the PU.1 promoter (Figure 4F). Mutation of the CEBP site or the OCT site abolished CEBP- or OCT-1-mediated activation of the -86-bp PU.1 promoter construct, respectively (Figure 4F). Mutation of both sites abolished ATRA activation completely.
In order to ultimately assess the composition of the DNA-protein complex in ATRA-treated APL cells, chromatin immunoprecipitation (ChIP) experiments were performed with quantitative RT-PCR for CEBPA, CEBPB, CEBPE, and OCT-1 (Figure 4G). We found that binding activities of CEBPA (maximum 4.1-fold at day 3) and CEBPE (maximum 4.0-fold at day 3) to the PU.1 promoter are induced to a small extent in HT93A cells after ATRA treatment. In contrast, binding activity of CEBPB was strongly induced (7.0-fold at day 1, 29.4-fold at day 3, and 23.8-fold at day 5). Thus, CEBPB binding appears to be induced predominantly in APL cells after ATRA treatment. In addition, OCT-1 binding was strongly induced in HT93A cells after ATRA treatment (6.7-fold at day 1, 30.5-fold at day 3, and 16.1-fold at day 5). These data suggest that CEBPB and OCT-1 mediate activation of the PU.1 promoter following ATRA treatment in APL cells.
PU.1 RNA is inversely correlated with PML-RARA transcripts in primary APL cells, and strongly induced following ATRA treatment. Because all of the previous studies were conducted in cell lines, we next attempted to correlate these findings with levels of expression in primary AML cells from patients. We analyzed RNA levels of CEBPA, CEBPB, and CEBPC (Figure S1; see the Supplemental Figures link at the top of the online article, at the Blood website), as well as PML-RARA, OCT-1, and PU.1 using real-time qPCR derived from blasts at diagnosis from 141 patients with AML, as well as from total normal bone marrow, CD34+-selected hematopoietic stem cells (mean purity of 98%), granulocytes, and monocytes from 3 healthy volunteers. Mature neutrophils expressed 16-fold and monocytes 8.5-fold more PU.1 than undifferentiated CD34+ cells. In patients with AML, the t(15;17) group (n = 18) had approximately half the PU.1 RNA compared with patients with AML-M2 and AML-M4, but these differences were not statistically significant, perhaps due to the heterogeneity of the patient samples. In addition, contamination of blast samples with even small percentages of nonleukemic cells is particularly confounding, given that any contaminating mature myeloid cells will express 8- to 16-fold more PU.1 than AML blasts.
CEBP factors and OCT-1 induce activity of the proximal PU.1 promoter in HT93A APL cells following ATRA stimulation. (A) A series of PU.1 promoter constructs were ligated to the luciferase gene in the pxp2 vector. HT93A cells were transfected with equal amounts of each of the deletion constructs and induced for 18 hours with 1 μM ATRA. The results are given as fold activation compared with HT93A cells transfected with the empty pxp2 vector only. (B) The sequence between -48 bp and -86 bp of the human PU.1 promoter contains a CEBP and an octamer consensus binding site (with the sites underlined and italicized). The brackets demonstrate the sequences used in the EMSA assays. (C) EMSA studying binding activity to the CEBP consensus site present in the PU.1 promoter with the CEBP probe (-80 to -60 bp). HT93A cells were stimulated with 1 μM ATRA and harvested at the time points indicated. SS indicates supershift using specific antibodies against CEBPA, CEBPB, and CEBPE. Specificity of the antibodies was verified by supershifting extracts from K562 cells transfected with human CEBPA, CEBPB, or CEBPE cDNAs; no cross-reactivity was observed (data not shown). C indicates competition with cold oligo. (D) EMSA of binding to the OCT site in the PU.1 promoter before and after 18 hours of stimulation with 1 μM ATRA (left panel). The probe is depicted in panel B as “OCT-1” extending from -66 to -42. Cos OCT-1 indicates Cos cells transfected with an OCT-1 expression plasmid as a positive control (lanes 2 and 3). Cos OCT-2 indicates Cos cells transfected with an OCT-2 expression plasmid as a positive control (lanes 4 and 5). Supershifts are depicted for OCT-1 (lanes 3, 7, 9, 11) and OCT-2 (lane 5). U937 indicates binding activity in lane 6, and supershift with OCT-1 in lane 7; HT93A, increase of binding activity before and after induction with ATRA (lanes 8 and 10), with OCT-1 supershifts in lanes 9 and 11; and P, free probe alone. Right panel shows EMSA with a probe with a mutation in the OCT site; no binding activity was detectable with this probe. (E) Western blot analysis of HT93A cells treated with 1 μM ATRA using a CEBPA antibody (top), a CEBPB antibody (second), a CEBPE antibody (third), an OCT-1 antibody (fourth), and a β-actin antibody (bottom). (F) H1299 cells were transiently transfected with the -86-bp PU.1 promoter (top 8 bars) and pcDNA3 vector (V) or equal amounts of expression plasmids for CEBPA, CEBPB, CEBPE, and/or OCT-1. OCT-1 was always cotransfected with equimolar amounts of BOB-1. Results are given as fold induction of the -86-bp promoter alone (to P = 1-fold). CEBP mut bars represent transfections with the -86-bp construct with the CEBP site mutated, whereas OCT mut bars are experiments with the -86-bp construct with the OCT site mutated. The bottom 4 bars are transfections with the -86-bp construct with both sites mutated (CEBP+OCT). (G) ChIP assay of the PU.1 promoter in HT93A cells treated with 1 μM ATRA. Depicted are the results of the quantitative RT-PCR for CEBPA, CEBPB, CEBPE, and OCT-1 binding to the PU.1 promoter at days 0, 1, 3, and 5 after ATRA treatment. (A, F-G) Mean values and SD (error bars) are depicted.
CEBP factors and OCT-1 induce activity of the proximal PU.1 promoter in HT93A APL cells following ATRA stimulation. (A) A series of PU.1 promoter constructs were ligated to the luciferase gene in the pxp2 vector. HT93A cells were transfected with equal amounts of each of the deletion constructs and induced for 18 hours with 1 μM ATRA. The results are given as fold activation compared with HT93A cells transfected with the empty pxp2 vector only. (B) The sequence between -48 bp and -86 bp of the human PU.1 promoter contains a CEBP and an octamer consensus binding site (with the sites underlined and italicized). The brackets demonstrate the sequences used in the EMSA assays. (C) EMSA studying binding activity to the CEBP consensus site present in the PU.1 promoter with the CEBP probe (-80 to -60 bp). HT93A cells were stimulated with 1 μM ATRA and harvested at the time points indicated. SS indicates supershift using specific antibodies against CEBPA, CEBPB, and CEBPE. Specificity of the antibodies was verified by supershifting extracts from K562 cells transfected with human CEBPA, CEBPB, or CEBPE cDNAs; no cross-reactivity was observed (data not shown). C indicates competition with cold oligo. (D) EMSA of binding to the OCT site in the PU.1 promoter before and after 18 hours of stimulation with 1 μM ATRA (left panel). The probe is depicted in panel B as “OCT-1” extending from -66 to -42. Cos OCT-1 indicates Cos cells transfected with an OCT-1 expression plasmid as a positive control (lanes 2 and 3). Cos OCT-2 indicates Cos cells transfected with an OCT-2 expression plasmid as a positive control (lanes 4 and 5). Supershifts are depicted for OCT-1 (lanes 3, 7, 9, 11) and OCT-2 (lane 5). U937 indicates binding activity in lane 6, and supershift with OCT-1 in lane 7; HT93A, increase of binding activity before and after induction with ATRA (lanes 8 and 10), with OCT-1 supershifts in lanes 9 and 11; and P, free probe alone. Right panel shows EMSA with a probe with a mutation in the OCT site; no binding activity was detectable with this probe. (E) Western blot analysis of HT93A cells treated with 1 μM ATRA using a CEBPA antibody (top), a CEBPB antibody (second), a CEBPE antibody (third), an OCT-1 antibody (fourth), and a β-actin antibody (bottom). (F) H1299 cells were transiently transfected with the -86-bp PU.1 promoter (top 8 bars) and pcDNA3 vector (V) or equal amounts of expression plasmids for CEBPA, CEBPB, CEBPE, and/or OCT-1. OCT-1 was always cotransfected with equimolar amounts of BOB-1. Results are given as fold induction of the -86-bp promoter alone (to P = 1-fold). CEBP mut bars represent transfections with the -86-bp construct with the CEBP site mutated, whereas OCT mut bars are experiments with the -86-bp construct with the OCT site mutated. The bottom 4 bars are transfections with the -86-bp construct with both sites mutated (CEBP+OCT). (G) ChIP assay of the PU.1 promoter in HT93A cells treated with 1 μM ATRA. Depicted are the results of the quantitative RT-PCR for CEBPA, CEBPB, CEBPE, and OCT-1 binding to the PU.1 promoter at days 0, 1, 3, and 5 after ATRA treatment. (A, F-G) Mean values and SD (error bars) are depicted.
Therefore, in order to better assess whether PML-RARA suppresses PU.1 in patients with primary APL, we compared RNA levels of PML-RARA, PU.1, and OCT-1 in the primary cells of t(15;17) patients only (Figure 5A). We observed an inverse correlation between the amount of PML-RARA and PU.1 transcripts that was highly statistically significant (r = -.912), and a correlation between PU.1 and OCT-1 mRNA levels (r = .907). Furthermore, analysis of OCT-1 RNA expression in all 141 AML patient samples (Table S2) demonstrated that it was selectively decreased in the subgroup of 18 t(15;17) AML-M3 patients (Figure S2). These results suggest that both PU.1 and OCT-1 are suppressed in patients with APL.
In order to test whether PU.1 RNA was induced in primary samples following ATRA treatment, we measured RNA expression of PU.1 by quantitative RT-PCR in primary cells from a patient with newly diagnosed APL (Figure 5B). ATRA without chemotherapy was given in a daily dosage of 45 mg/m2. We observed that the kinetics of change in PU.1 RNA expression following ATRA treatment occurred in vivo later than in vitro. We observed a steady increase of PU.1 mRNA to 8-fold after 24 hours, reaching a plateau of about 65-fold after 3 days of ATRA treatment (Figure 5B). Since we had observed that CEBP family members and OCT-1 mediated up-regulation of the PU.1 promoter following ATRA treatment of PML-RARA cells (Figure 4), we concomitantly measured RNA of CEBP family members and OCT-1 in this patient. Among CEBP family members, we observed an up to 28-fold increase in CEBPB and 12-fold increase in CEBPA mRNA after 5 days. CEBPE mRNA increased to a plateau of 25-fold after 5 and 7 days. Both CEBPB and OCT-1, and to a lesser extent, CEBPA, were increased by day 2, preceding the increase in PU.1 RNA at day 3. In summary, treatment of an APL patient with ATRA in vivo led to first an induction of CEBPA, CEBPB, and OCT-1, factors that mediate ATRA activation of the PU.1 promoter, prior to the increase in PU.1 RNA.
PU.1 expression in primary APL cells: inverse correlation with PML-RARA at diagnosis, and induction of expression following treatment with ATRA in vivo. (A) Correlation of PU.1 and OCT-1 mRNA expression and inverse correlation between PML-RARA vs. PU.1 and OCT-1 transcripts in bone marrow cells at diagnosis of 9 patients with newly diagnosed t(15;17) AML-M3 as assessed by quantitative real-time PCR. The median value was determined as 100% for PML-RARA, OCT-1, and PU.1, respectively. The percentage of blasts was more than 90% in all patients. The correlation coefficient PML-RARA versus OCT-1 was r = -0.887, for PML-RARA versus PU.1 it was r = -0.912, and for OCT-1 versus PU.1 it was r = 0.907. (Different symbols indicate different patients.) (B) RNA expression was determined by quantitative RT-PCR of CEBPA, PU.1, CEBPB, OCT-1, and CEBPE in primary cells from a patient with newly diagnosed APL treated with orally given ATRA (45 mg/m2). No additional chemotherapy was given. The cells analyzed on a daily basis were obtained from peripheral blood (Table S1). Error bars depict the standard deviation as the result of triplicate mRNA determinations.
PU.1 expression in primary APL cells: inverse correlation with PML-RARA at diagnosis, and induction of expression following treatment with ATRA in vivo. (A) Correlation of PU.1 and OCT-1 mRNA expression and inverse correlation between PML-RARA vs. PU.1 and OCT-1 transcripts in bone marrow cells at diagnosis of 9 patients with newly diagnosed t(15;17) AML-M3 as assessed by quantitative real-time PCR. The median value was determined as 100% for PML-RARA, OCT-1, and PU.1, respectively. The percentage of blasts was more than 90% in all patients. The correlation coefficient PML-RARA versus OCT-1 was r = -0.887, for PML-RARA versus PU.1 it was r = -0.912, and for OCT-1 versus PU.1 it was r = 0.907. (Different symbols indicate different patients.) (B) RNA expression was determined by quantitative RT-PCR of CEBPA, PU.1, CEBPB, OCT-1, and CEBPE in primary cells from a patient with newly diagnosed APL treated with orally given ATRA (45 mg/m2). No additional chemotherapy was given. The cells analyzed on a daily basis were obtained from peripheral blood (Table S1). Error bars depict the standard deviation as the result of triplicate mRNA determinations.
Restoring PU.1 expression in human PML-RARA cells induces granulocytic differentiation
Based on our findings that PML-RARA suppresses and that ATRA restores expression of PU.1, we hypothesized that restoring PU.1 expression in PML-RARA leukemic cells would induce granulocytic differentiation. HT93A APL cells were transduced with a retrovirus expressing the human PU.1 protein fused to the estrogen receptor (PU.1-ER), and the presence of the PU.1-ER protein was verified using PU.1 antiserum (Figure 6A). In the absence of treatment with β-estradiol, the PU.1-ER remains in the cytoplasm and is not transcriptionally active. Conditional induction of nuclear PU.1 protein in these cells (HT-PER) with β-estradiol induced a 6-fold increase of M-CSF receptor protein after 4 days. In order to assess whether nuclear translocation of PU.1-ER led to granulocytic differentiation, we measured surface expression of CD11b and the granulocyte CSF (G-CSF) receptor, both of which are up-regulated during neutrophil differentiation. Both proteins were induced after 4 days of β-estradiol treatment as determined by flow cytometry (Figure 6B). In contrast, no changes were seen in the HT93A vector-transduced control line (HT-V). Cell-cycle analysis using propidium iodide staining demonstrated a decrease of cells in S phase from 25.5% to 5.5% and an increase of cells in G0-G1 stage from 64.8% to 82.6% after 2 days of β-estradiol treatment (Figures 6C and S3). The effect of ATRA treatment and PU.1 induction in HT93A cells on the cell cycle were comparable (Figure 6C). Interestingly, the effect of PU.1 induction by estradiol treatment in HT-PER cells decreased growth in a way very similar to the effect of ATRA treatment in these cells. Finally, Wright-Giemsa staining of HT-PER cells after 6 days of β-estradiol treatment demonstrated morphologic differentiation from promyelocytes at day 0 to polymorphonuclear cells with neutrophil appearance at day 6 (Figure 6D). Similar results were observed in 3 independent HT-PER clones. The morphologic changes over time were comparable with the effects observed after ATRA treatment of HT-PER cells. In contrast, no changes were observed in HT-V cells. We therefore conclude that restoring functional PU.1 protein in PML-RARA leukemic cells is sufficient to induce terminal granulocytic differentiation.
Restoring PU.1 expression in PML-RARA leukemic cells induces granulocytic differentiation. (A) Western blot for PU.1 (top) detecting the PU.1-ER fusion protein in HT93A cells transduced with a PU.1-ER-expressing pBabePuro retrovirus (HT-PER) or with the vector alone (HT-V). Cells were treated with 1 μM β-estradiol as indicated. C indicates that Cos cells serve as a negative control and U937 cells as a positive control for endogenous PU.1 expression. The blot was also stained with an antibody against the M-CSF receptor (middle) and β-tubulin (bottom). (B) HT-PER cells and the control line HT-V were induced for 6 days with 1 μM β-estradiol. Fluorescence-activated cell sorting (FACS) analysis was performed at day 0 (left panels) and day 6 (right panels) using a PE-conjugated CD11b antibody (top panels) or a G-CSF receptor antibody (bottom panels). (C) HT-PER and HT-V cells were treated with 1 μM β-estradiol (Est) or 1 μM ATRA. Cell-cycle analysis was performed using propidium iodide staining at day 0 and 2. ▪ indicates the percentage of cells in G0-G1 stage, whereas □ indicates cells in G2-M stage and  indicates cells in S phase. Median values are shown and error bars depict standard deviation from 3 independent experiments. (D) Wright-Giemsa staining of HT93A cells expressing the PU.1-ER fusion (HT-PER cells). The top 3 panels show unstimulated HT-PER cells (left panel), after 6 days of ATRA treatment indicating that the cells still differentiate toward neutrophils after having established the stable estrogen inducible system (middle panel), and after 6 days of treatment with 1 μM β-estradiol indicating differentiation toward neutrophils after 6 days (right panel). The bottom 3 panels depict the HT-V control line, which fails to show morphologic changes after β-estradiol treatment (right panel), but differentiates toward neutrophils after 6 days of ATRA treatment (middle panel). Images were visualized using a Zeiss Axioplan 2 microscope (Carl Zeiss, Oberkochen, Germany) and a 100 ×/2 numeric aperture objective. Images were acquired using a Zeiss AxioCam camera.
indicates cells in S phase. Median values are shown and error bars depict standard deviation from 3 independent experiments. (D) Wright-Giemsa staining of HT93A cells expressing the PU.1-ER fusion (HT-PER cells). The top 3 panels show unstimulated HT-PER cells (left panel), after 6 days of ATRA treatment indicating that the cells still differentiate toward neutrophils after having established the stable estrogen inducible system (middle panel), and after 6 days of treatment with 1 μM β-estradiol indicating differentiation toward neutrophils after 6 days (right panel). The bottom 3 panels depict the HT-V control line, which fails to show morphologic changes after β-estradiol treatment (right panel), but differentiates toward neutrophils after 6 days of ATRA treatment (middle panel). Images were visualized using a Zeiss Axioplan 2 microscope (Carl Zeiss, Oberkochen, Germany) and a 100 ×/2 numeric aperture objective. Images were acquired using a Zeiss AxioCam camera.
Restoring PU.1 expression in PML-RARA leukemic cells induces granulocytic differentiation. (A) Western blot for PU.1 (top) detecting the PU.1-ER fusion protein in HT93A cells transduced with a PU.1-ER-expressing pBabePuro retrovirus (HT-PER) or with the vector alone (HT-V). Cells were treated with 1 μM β-estradiol as indicated. C indicates that Cos cells serve as a negative control and U937 cells as a positive control for endogenous PU.1 expression. The blot was also stained with an antibody against the M-CSF receptor (middle) and β-tubulin (bottom). (B) HT-PER cells and the control line HT-V were induced for 6 days with 1 μM β-estradiol. Fluorescence-activated cell sorting (FACS) analysis was performed at day 0 (left panels) and day 6 (right panels) using a PE-conjugated CD11b antibody (top panels) or a G-CSF receptor antibody (bottom panels). (C) HT-PER and HT-V cells were treated with 1 μM β-estradiol (Est) or 1 μM ATRA. Cell-cycle analysis was performed using propidium iodide staining at day 0 and 2. ▪ indicates the percentage of cells in G0-G1 stage, whereas □ indicates cells in G2-M stage and  indicates cells in S phase. Median values are shown and error bars depict standard deviation from 3 independent experiments. (D) Wright-Giemsa staining of HT93A cells expressing the PU.1-ER fusion (HT-PER cells). The top 3 panels show unstimulated HT-PER cells (left panel), after 6 days of ATRA treatment indicating that the cells still differentiate toward neutrophils after having established the stable estrogen inducible system (middle panel), and after 6 days of treatment with 1 μM β-estradiol indicating differentiation toward neutrophils after 6 days (right panel). The bottom 3 panels depict the HT-V control line, which fails to show morphologic changes after β-estradiol treatment (right panel), but differentiates toward neutrophils after 6 days of ATRA treatment (middle panel). Images were visualized using a Zeiss Axioplan 2 microscope (Carl Zeiss, Oberkochen, Germany) and a 100 ×/2 numeric aperture objective. Images were acquired using a Zeiss AxioCam camera.
indicates cells in S phase. Median values are shown and error bars depict standard deviation from 3 independent experiments. (D) Wright-Giemsa staining of HT93A cells expressing the PU.1-ER fusion (HT-PER cells). The top 3 panels show unstimulated HT-PER cells (left panel), after 6 days of ATRA treatment indicating that the cells still differentiate toward neutrophils after having established the stable estrogen inducible system (middle panel), and after 6 days of treatment with 1 μM β-estradiol indicating differentiation toward neutrophils after 6 days (right panel). The bottom 3 panels depict the HT-V control line, which fails to show morphologic changes after β-estradiol treatment (right panel), but differentiates toward neutrophils after 6 days of ATRA treatment (middle panel). Images were visualized using a Zeiss Axioplan 2 microscope (Carl Zeiss, Oberkochen, Germany) and a 100 ×/2 numeric aperture objective. Images were acquired using a Zeiss AxioCam camera.
Blocking of PU.1 expression by siRNA in PML-RARA leukemic cells prevents ATRA-induced granulocytic differentiation
Our findings demonstrated that expression of PU.1 can induce granulocytic differentiation of PML-RARA cells, but other transcription factors, notably the CEBP family, have also been shown to produce this effect.29 Therefore, in order to demonstrate that expression of PU.1 is required for ATRA-induced differentiation of PML-RARA cells, we transduced HT93A cells with a retrovirus expressing a PU.1 siRNA sequence (siHT), and 2 pools of cells expanded under antibiotic selection were used for this analysis. In contrast to the siHT-V (vector control) cells, we observed only a slight increase in PU.1 protein in pools expressing PU.1 siRNA after 2 days of ATRA (Figure 7A). Furthermore, HT93A pools expressing the PU.1 siRNA were blocked in their ability to undergo granulocytic differentiation after 5 days of ATRA as assessed both by surface expression of the granulocytic markers CD11b and G-CSF receptor (Figure 7B), as well as by morphology (Figure 7C), whereas neutrophil differentiation was observed after 5 days in siHT-V cells. We thus conclude that activation of PU.1 expression is required for ATRA-mediated neutrophil differentiation of PML-RARA leukemic cells.
In order to determine whether OCT-1, CEBPA, CEBPB, and CEBPE are all required for PU.1 expression in ATRA-treated APL cells, siRNA experiments were performed. HT93A cells were transiently transfected with siRNAs against OCT-1 and CEBPs, and PU.1 mRNA expression was determined after 48 hours of ATRA treatment (and transfection). We found that PU.1 expression was reduced 13-fold by OCT-1 siRNA, and 3.5-fold by CEBPB siRNA after ATRA treatment as compared with a control siRNA construct (Figure 7D). Blocks of both OCT-1 and CEBPB by specific siRNAs abolished PU.1 mRNA expression to levels as observed in unstimulated HT93A cells. In contrast, siRNAs against CEBPA or CEBPE led to only a minor reduction of PU.1 mRNA expression (2-fold and 1.8-fold, respectively). We conclude that ATRA-induced activation of PU.1 in APL cells is mediated by CEBPB and OCT-1.
Reduction of PU.1 expression by siRNA blocks ATRA-induced differentiation of HT93A cells. (A) HT93A cells were stably transfected with an MSCV retrovirus expressing PU.1 siRNA to reduce PU.1 expression (siPU.1-1 and siPU.1-2). Two pools of cells were expanded and analyzed after 1 month. Western blot for PU.1 demonstrates that treatment with 1 μM ATRA induced only small amounts of PU.1 protein in siPU.1 cells after 2 days compared with vector-transfected siV cells. (B) siPU.1 cells (pools siPU.1-1 and siPU.1-2) were induced with 1 μMATRAfor 5 days. FACS analysis was performed at day 0 (d0) and day 5 (d5) using a G-CSF receptor antibody. Parental HT93A cells transfected with the siRNA vector (siV) served as a positive control (right panel). (C) Wright-Giemsa staining of siPU.1 cells expressing PU.1 siRNA is shown before and after 5 days of ATRA treatment, indicating that siPU.1 cells morphologically fail to respond to ATRA treatment. Right panel: ATRA treatment induces neutrophil differentiation in the siV vector-transfected HT93A cells. (D) HT93A cells were transiently transfected with siRNAs against OCT-1, CEBPA, CEBPB, or CEBPE. PU.1 mRNA expression was determined by RT-PCR after 48 hours of ATRA treatment (and transfection). V indicates the silencer negative control siRNA no. 2 (Ambion) was used to control for nonspecific effects in siRNAexperiments.
Reduction of PU.1 expression by siRNA blocks ATRA-induced differentiation of HT93A cells. (A) HT93A cells were stably transfected with an MSCV retrovirus expressing PU.1 siRNA to reduce PU.1 expression (siPU.1-1 and siPU.1-2). Two pools of cells were expanded and analyzed after 1 month. Western blot for PU.1 demonstrates that treatment with 1 μM ATRA induced only small amounts of PU.1 protein in siPU.1 cells after 2 days compared with vector-transfected siV cells. (B) siPU.1 cells (pools siPU.1-1 and siPU.1-2) were induced with 1 μMATRAfor 5 days. FACS analysis was performed at day 0 (d0) and day 5 (d5) using a G-CSF receptor antibody. Parental HT93A cells transfected with the siRNA vector (siV) served as a positive control (right panel). (C) Wright-Giemsa staining of siPU.1 cells expressing PU.1 siRNA is shown before and after 5 days of ATRA treatment, indicating that siPU.1 cells morphologically fail to respond to ATRA treatment. Right panel: ATRA treatment induces neutrophil differentiation in the siV vector-transfected HT93A cells. (D) HT93A cells were transiently transfected with siRNAs against OCT-1, CEBPA, CEBPB, or CEBPE. PU.1 mRNA expression was determined by RT-PCR after 48 hours of ATRA treatment (and transfection). V indicates the silencer negative control siRNA no. 2 (Ambion) was used to control for nonspecific effects in siRNAexperiments.
Discussion
In this study, we found that the transcription factor PU.1 is suppressed in human leukemic cells harboring the PML-RARA fusion protein, and that treatment with ATRA restored PU.1 expression and induced neutrophil differentiation in t(15;17) leukemic cells both in vitro and in primary patient cells. Furthermore, we demonstrated that conditional expression of PU.1 in PML-RARA leukemic cells is sufficient to trigger granulocytic differentiation and that reduction of PU.1 protein by siRNA blocked ATRA-induced neutrophil differentiation of PML-RARA leukemic cells. These findings propose a model that PU.1 is essential for proceeding from the promyelocytic stage, at which t(15;17) cells are arrested, to mature granulocytes.
PU.1 is absolutely required for proper development of multiple hematopoietic cell lineages, and PU.1-deficient mice lack mature myeloid cells.8,9 In addition to this critical role in normal myeloid development, PU.1 has been strongly implicated in the pathogenesis of experimental leukemias in mice. Increased expression of PU.1 in early erythroid cells leads to erythroleukemia,30 and graded reduction of PU.1 to 20% of wild-type levels induces AML in mice.12 Furthermore, deletion or mutation of PU.1 has been described in radiation-induced AML.13,14
In humans, the role of PU.1 in the pathogenesis is less clear. We described heterozygous mutations of the PU.1 gene in patients primarily of Japanese descent with various subtypes of AML, but not in PML-RARA-positive AML with the t(15;17).11 However, we and others were unable to confirm these findings in patients of North American or Northern European descent, suggesting that ethnic differences might account for these discrepancies.11,14,31 Here, we present further evidence for the role of disruption of PU.1 function in human AML. Suppression of PU.1 expression in APL appears to be a novel mechanism by which PU.1 is affected in human AML.
Previous in vitro studies have shown that lymphoid- and myeloid-specific activity of the PU.1 promoter is determined by the combinatorial action of OCT and ETS transcription factors.27,28,32 We identified the sequence extending 86 bp upstream of the PU.1 transcription start site to be sufficient to mediate ATRA-induced activation of the PU.1 promoter in PML-RARA leukemic cells. This points to PU.1 as an ATRA responsive gene, and that ATRA overcomes the PML-RARA-mediated inhibition of the PU.1 promoter. While ATRA has been shown to induce the expression of a number of granulocytic genes, to date only the CEBP proteins, namely CEBPB and CEBPE, have been shown to be ATRA responsive in PML-RARA cells and been capable of inducing granulocytic differentiation, and both have elements in their promoters which mediate responsiveness to PML-RARA in the presence of ATRA.20,33 Our findings identify PU.1 as another ATRA-responsive gene that can induce granulopoiesis. In this case, it appears that ATRA induction of PU.1 in PML-RARA cells is mediated by up-regulation of the CEBP proteins, as well as OCT-1, which together can activate the PU.1 promoter. Studies of the expression of these transcription factors in patients with APL treated with ATRA are consistent with this model (Figure 5B).
Finally, these results demonstrate that suppression of PU.1 is an important mechanism leading to the maturation block at the promyelocytic stage seen in human APL. In murine models in which the PML-RARA gene is not expressed, decreased expression, loss of heterozygosity, or mutation of PU.1 is associated with development of AML.13,14 These studies suggest that restoring PU.1 expression may represent a possible therapeutic modality leading to differentiation of AML cells.
Prepublished online as Blood First Edition Paper, December 13, 2005; DOI 10.1182/blood-2005-07-3068.
Supported by a grant from the Swiss National Science Foundation (SF 3100A0-100445) to B.U.M., SF 310000-109388 to T.P., and grants from the National Institutes of Health CA66996 and CA41456 to D.G.T.
B.U.M. designed and performed research and wrote the paper; T.P. performed research and wrote the paper; J.F. performed research; V.P. performed research; M.F.F. contributed patient samples; N.A. contributed patient samples; U.B. contributed vital material and wrote the paper; and D.G.T. analyzed data and wrote the paper.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We wish to thank Alan Friedman for the pBabePuro vector, Hui-Min Chen and Pu Zhang for the PU.1 promoter constructs, Pam Silver for the MSCV-U6 siRNA vector, Yuji Yamaguchi for the HT93A cell line, Pier Guiseppe Pelicci for the U937-PR9 cell line, Estelle Duprez for cell line reagents, and Dong-Er Zhang and Atsushi Iwama for valuable suggestions.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal