Abstract
Although targeting the BCR-ABL tyrosine kinase activity by imatinib mesylate has rapidly become first-line therapy in chronic myeloid leukemia (CML), drug resistance suggests that combination therapy directed to a complementing target may significantly improve treatment results. To identify such potential targets, we used lentivirus-mediated RNA interference (RNAi) as a tool for functional genomics in cell lines as well as primary normal and CML CD34+ cells. In a conditional cell culture model, we demonstrate that RNAi-mediated reduction of SHP2, STAT5, and Gab2 protein expression inhibits BCR-ABL-dependent but not cytokine-dependent proliferation in a dose-dependent manner. Similarly, colony formation of purified primary CML but not of normal CD34+ colony-forming cells is specifically reduced by inhibition of SHP2, STAT5, and Gab2 expression, respectively. In addition, coexpression of both anti-BCR-ABL and anti-SHP2 shRNAs from a single lentiviral vector induces stronger inhibition of colony formation as compared to either shRNA alone. The data indicate that BCR-ABL expression may affect the function of normal signaling molecules. Targeting these molecules may harbor significant therapeutic potential for the treatment of patients with CML.
Introduction
The treatment of BCR-ABL+ chronic myeloid leukemia (CML) with the selective tyrosine kinase inhibitor imatinib mesylate has recently evolved as a new paradigm for molecular-defined anticancer therapy.1 Although superior to conventional drugs such as α-interferon and cytarabine (ara-C) in chronic-phase CML,2 drug resistance, especially in advanced disease, due to mutations in the BCR-ABL kinase domain led to the development of second-generation ATP- or substrate-competitive inhibitors that seem to overcome imatinib resistance for many but not all mutations.3-6 Independent of this rapid progress, imatinib mesylate, and perhaps second-generation inhibitors as well, are not believed to cure CML, indicating some limitations of this exciting therapeutic approach.7
On the other hand, BCR-ABL gene expression has been targeted by RNA interference (RNAi) in cell culture models and in primary CML cells.8-12 Anti-BCR-ABL RNAi inhibits proliferation of BCR-ABL+ cell lines and reduces BCR-ABL mRNA levels in primary CML cells. Interestingly, anti-BCR-ABL RNAi can cooperate with imatinib in inducing cell death in BCR-ABL+ cell lines,10,12 and stable anti-BCR-ABL RNAi can inhibit colony formation of primary CML progenitors in semisolid cultures.12 However, long-term RNAi in primary hematopoietic cells has only been achieved by viral gene transfer strategies, and currently there is no means suitable for clinical application to induce RNAi in hematopoietic cells.
To search for alternative therapeutic targets in CML for potential combination therapy with selective kinase inhibitors, we analyzed the function of SHP2, STAT5, and Gab2 on BCR-ABL-mediated cell proliferation. We considered the constitutive tyrosine phosphorylation of all 3 targets in BCR-ABL+ cells as a biochemical surrogate marker for their potential contribution to BCR-ABL signaling. SHP2-encoded at the PTPN11 locus is an SH2-domain containing hematopoietic protein tyrosine phosphatase, which is involved in full activation of the ERK/MAP kinase pathway by cytokine and receptor tyrosine kinase (RTK) signal transduction.13 In addition, SHP2 can regulate Src family kinases14 and PI-3K activity on RTK and Mpl-receptor signaling.15,16 PTPN11 mutations have been described in Noonan syndrome,17 juvenile myelomonocytic leukemia (JMML),18 and adult acute myeloid leukemia (AML),19 and overexpression of wild-type SHP2 has recently been described in adult leukemia including CML.20 STAT5 belongs to a family of transcription factors that undergo tyrosine phosphorylation on stimulation by several hematopoietic cytokine receptors. STATs subsequently homodimerize, translocate to the nucleus, and activate target gene transcription.21,22 STAT5 has been linked to BCR-ABL-dependent proliferation,23,24 to antiapoptotic signals,25 and to cell cycle regulation via cyclin D2.26 Gab2 is an adaptor protein that associates with several SH2-domain containing proteins, including SHP2, in a phosphotyrosine-dependent manner.27 Importantly, Gab2 has been shown to be essential for transformation by BCR-ABL in murine myeloid cells.28
To analyze the function of SHP2, STAT5, and Gab2 in primary hematopoietic cells we established lentivirus-mediated anti-SHP2, anti-STAT5, and anti-Gab2 RNAi as a tool for reverse genetics in cell lines and CD34+ cells. Using murine TonB cells as a model for inducible BCR-ABL expression that allows the analysis of target gene function in BCR-ABL as compared to cytokine-driven cell proliferation, we demonstrate differential, reversible, and, as shown for Gab2, dose-dependent inhibition of BCR-ABL but not IL-3-dependent cell proliferation. Interestingly, RNAi against BCR-ABL and STAT5, but not against SHP2 and Gab2, reduces cell survival in this model. In primary cells, we demonstrate that colony formation of lentivirally transduced CML CD34+ progenitors is more sensitive to inhibition of SHP2, STAT5, and Gab2 expression than that of normal CD34+ progenitors. This differential sensitivity to RNAi is very similar to that observed in the cell culture model. Finally, the combination of anti-BCR-ABL and anti-SHP2 RNAi cooperates in inhibiting colony formation of CML colony-forming units (CFUs). These data demonstrate for the first time that stable RNAi triggered on lentiviral gene transfer can be used to identify potential targets different from the BCR-ABL tyrosine kinase activity for therapeutic intervention in primary hematopoietic cells. They further suggest that combined molecular targeting of leukemia specific and nonspecific signaling molecules may harbor significant potential for anti-BCR-ABL therapy.
Materials and methods
shRNA synthesis and construction of H1-shRNA expression cassettes
Seven DNA oligonucleotides corresponding to positions 237 to 255, 291 to 309, 594 to 612, 614 to 632, 651 to 669, 714 to 732, and 1569 to 1587, of the sequence of the human PTPN11 gene (GenBank accession no. NM_002834), 6 DNA oligonucleotides corresponding to positions 357 to 376, 371 to 389, 487 to 505, 534 to 552, 620 to 638, and 1005 to 1023 of the sequence of the human Gab2 gene (GenBank accession no. AB018413), as well as 7 DNA oligonucleotides corresponding to positions 837 to 855, 938 to 956, 1151 to 1169, 1243 to 1261, 1367 to 1385, 1460 to 1478, and 1652 to 1670 of the sequence of the human STAT5 gene (GenBank accession no. NM_003152), were subjected to BLAST-homology search and thereafter chemically synthesized including overhang sequences from a 5′ BglII and a 3′ SalI restriction site for cloning purposes (BioSpring, Frankfurt, Germany). The numbering of the first nucleotide of the shRNAs (italics) refers to the ATG start codon. The oligonucleotide sequences of the effective shRNAs used in this study were as follows: FP237SHP2: 5′-GATCCCGTATTACATGGAACATCACTTCAAGAGAGTGATGTTCCATGTAATACTTTTTTGGAAG-3′; RP237SHP2: 5′-TCGACTTCCAAAAAAAGTATTACATGGAACATCACTCTCTTGAAGTGATGTTCCATGTAATACGGG-3′; FP594SHP2: 5′-GATCCCGAAGAATCCTATGGTGGAATTCAAGAGATTCCACCATAGGATTCTTCTTTTTTGGAAG-3′; RP594SHP2: 5′-TCGACTTCCAAAAAAAGAAGAATCCTATGGTGGAATCTCTTGAATTCCACCATAGGATTCTTCGGG-3′; FP714SHP2: 5′-GATCCCGACCACAGATAAAGTCAAATTCAAGAGATTTGACTTTATCTGTGGTCTTTTTTGGAAG-3′; RP714SHP2: 5′-TCGACTTCCAAAAAAAGACCACAGATAAAGTCAAATCTCTTGAATTTGACTTTATCTGTGGTCGGG-3′; FPgab620: 5′-GATCCCGGAGTGCCAGCTTCTCTCATTCAAGAGATGAGAGAAGCTGGCACTCC TTTTTTGGAAG-3′; RPgab620: 5′-TCGACTTCCAAAAAAAGGAGTGCCAGCTTCTCTCATCTCTTGAATGAGAGAAGCTGGCACTCCGGG-3′; FP STAT5-1151: 5′-GATCCCGTACTTCATCATCCAGTACTTCAAGAGAGTACTGGATGATGAAGTACTTTTTTGGAAG-3′; RPSTAT5-1151: 5′-TCGACTTCCAAAAAAGTACTTCATCATCCAGTACTCTCTTGAAGTACTGGATGATGAAGTACGGG-3′.
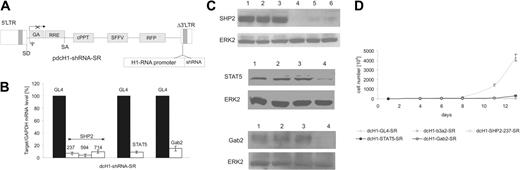
Gene silencing mediated by lentivirus-encoded shRNAs in K562 cells. (A) The shRNA is transcribed from a human H1-RNA promoter inserted into the U3 region of the lentiviral Δ3′-LTR. The vector encodes RFP as a marker gene driven by the SFFV-LTR promoter and harbors a cPPT/CTS sequence. 5′-LTR indicates HIV-1 5′-LTR; Δ3′-LTR, HIV-1 self-inactivating (SIN) 3′-LTR; GA, deleted gag sequence; RRE, Rev responsive element; SD, splice donor site; SA, splice acceptor site; Ψ, packaging signal, SFFV-LTR of spleen focus-forming virus. (B) SHP2, STAT5, and Gab2 mRNA levels were measured by real-time RT-PCR 4 days after lentiviral transduction and normalized in comparison to GAPDH expression. The shRNAs are indicated on top of each bar. mRNA expression of SHP2 (left), STAT5 (middle), and Gab2 (right) in control dcH1-GL4-SR-transduced cells was set to 100% (average of 3 independent experiments). (C) For immunoblotting, cells were transduced with control dcH1-GL4-SR (lane 2), dcH1-b3a2_1-SR (lane 3), anti-SHP2 shRNAs 237, 594, and 714 (top: lanes 4-6), anti-STAT5 shRNA (middle, lane 4), and anti-Gab2 shRNA (bottom, lane 4) and lysed 6 days after transduction, respectively. Lane 1 shows untransduced K562 cells. The immunoblots were probed with anti-SHP2, anti-STAT5, and anti-Gab2 antibodies, respectively, and reprobed with anti-ERK2 antibody as loading control. (D) Number of K562 cells negative for trypan blue after lentiviral transduction with different shRNAs is shown (averages from 3 experiments). One milliliter with 104/mL cells was plated, cultures were split and fed twice a week, and calculated cell numbers are indicated. Triangles depict transduction with dcH1-GL4-SR (control), and circles show transduction with viruses dcH1-b3a2_1-SR, dcH1-237SHP2-SR, dcH1-Gab2-SR, and dcH1-STAT5-SR, respectively.
Gene silencing mediated by lentivirus-encoded shRNAs in K562 cells. (A) The shRNA is transcribed from a human H1-RNA promoter inserted into the U3 region of the lentiviral Δ3′-LTR. The vector encodes RFP as a marker gene driven by the SFFV-LTR promoter and harbors a cPPT/CTS sequence. 5′-LTR indicates HIV-1 5′-LTR; Δ3′-LTR, HIV-1 self-inactivating (SIN) 3′-LTR; GA, deleted gag sequence; RRE, Rev responsive element; SD, splice donor site; SA, splice acceptor site; Ψ, packaging signal, SFFV-LTR of spleen focus-forming virus. (B) SHP2, STAT5, and Gab2 mRNA levels were measured by real-time RT-PCR 4 days after lentiviral transduction and normalized in comparison to GAPDH expression. The shRNAs are indicated on top of each bar. mRNA expression of SHP2 (left), STAT5 (middle), and Gab2 (right) in control dcH1-GL4-SR-transduced cells was set to 100% (average of 3 independent experiments). (C) For immunoblotting, cells were transduced with control dcH1-GL4-SR (lane 2), dcH1-b3a2_1-SR (lane 3), anti-SHP2 shRNAs 237, 594, and 714 (top: lanes 4-6), anti-STAT5 shRNA (middle, lane 4), and anti-Gab2 shRNA (bottom, lane 4) and lysed 6 days after transduction, respectively. Lane 1 shows untransduced K562 cells. The immunoblots were probed with anti-SHP2, anti-STAT5, and anti-Gab2 antibodies, respectively, and reprobed with anti-ERK2 antibody as loading control. (D) Number of K562 cells negative for trypan blue after lentiviral transduction with different shRNAs is shown (averages from 3 experiments). One milliliter with 104/mL cells was plated, cultures were split and fed twice a week, and calculated cell numbers are indicated. Triangles depict transduction with dcH1-GL4-SR (control), and circles show transduction with viruses dcH1-b3a2_1-SR, dcH1-237SHP2-SR, dcH1-Gab2-SR, and dcH1-STAT5-SR, respectively.
The noncomplementary 9-nt-loop sequences are underlined, and each sense oligonucleotide harbors a stretch of T as a pol III transcription termination signal. Corresponding oligonucleotides were annealed and inserted 3′ of the H1-RNA promoter into the BglII/SalI-digested pBlueScript-derived pH1-plasmid to generate pH1-SHP-237, pH1-SHP-594, pH1-SHP-714, pH1-gab620, and pH1-STAT5-1151 as described.29 These shRNAs are complementary to both the human and murine target mRNAs. Plasmids pH1-GL4 (control), pH1-121GFP, pH1-β-GMR, and pH1-b3a2_1 expressing an shRNA covering the fusion sequence of the b3a2-variant of the BCR-ABL gene have been described earlier.9,12,29,30
Construction of lentiviral vectors
pdc-SR (pdc: plasmids resulting in double-copy proviruses) were used to generate lentiviral transgenic plasmids containing H1-siRNA expression cassettes located in the U3 region of the Δ3′-long terminal repeat (LTR).29 To generate the lentiviral pdcH1-siRNA-SR plasmid, the pH1-SHP237, pH1-SHP594, pH1-SHP714, pH1-gab620, and pH1-STAT5-1151 plasmids were digested with SmaI and HincII and the resulting DNA fragments (360 nt) were blunt-end ligated into the SnaBI site of the pdc-SR to generate pdcH1-SHP237-SR, pdcH1-SHP594-SR, pdcH1-SHP714-SR, pdcH1-gab620-SR, and pdcH1-STAT5-1151-SR, respectively (Figure 1A). All lentiviral constructs encode RFPEXPRESS as reporter gene.
To generate an H1-shRNA tandem expression cassette, pH1-b3a2_1 DNA was digested with EcoRI followed by filling in the staggered termini with DNA polymerase I (Klenow), treatment with bacterial alkaline phosphatase, and gel purification. Next, the pH1-SHP237 expression cassette was digested with BamHI and XhoI followed by a DNA polymerase I fill-in reaction. The resulting DNA fragment (∼360 nucleotides) was blunt-end ligated into the linearized pH1-b3a2_1 plasmid to generate pH1-b3a2-SHP237. To generate the lentiviral pdcH1-b3a2-SHP237-SR plasmid, the pH1-b3a2-SHP237 plasmid was digested with BamHI and XhoI followed by a DNA polymerase I fill-in reaction and the resulting DNA fragment (∼700 nucleotides) was blunt-end ligated into the SnaBI site of pdc-SR (Figure 6A). The isolated clone was verified by DNA sequencing.
Cell culture
293T and BHK-21 cells were grown at 37°C in Dulbecco modified Eagle medium (DMEM), 10% FCS, and 2 mM l-glutamine. K562 cells were maintained in RPMI 1640 with 10% FCS. Murine TonB (TonB) cells derived from the BaF3 cell line were cultured as described.31
CD34+ cells from patients with CML in first chronic phase were isolated at the time of initial diagnosis as described.32 Purification resulted in 98% or more purity. G-CSF-primed CD34+ cells were harvested by leukapheresis from 6 healthy volunteers, purified to at least 98% CD34+ content by magnetic cell sorting (Clini MACS, Miltenyi Biotech, Bergisch Gladbach, Germany). Informed consent from the patients and volunteers was obtained in accordance with the Declaration of Helsinki.
Preparation of recombinant lentiviral supernatants and lentiviral transduction
VSV.G-pseudotyped lentiviral particles were generated by calcium phosphate cotransfection of 293T cells, and viral supernatants were concentrated as previously described.32 dcH1-shRNA-SR lentiviral preparations were titered in triplicate by serial dilutions of the concentrated vector stocks on 1 × 105 K562 cells in 24-well plates. The number of RFP+ cells was analyzed 72 hours after transduction by fluorescence-activated cell sorting (FACS) analysis (FACS Calibur, Becton Dickinson, Heidelberg, Germany), and the titers were averaged and typically ranged between 5 and 10 × 107 IU/mL.
Lentiviral supernatants were used to transduce (1) K562, (2) TonB, (3) normal CD34+, and (4) CD34+ CML cells as recently described.32 K562 and normal and CML CD34+ cells were transduced with a multiplicity of infection (MOI) of approximately 4 and 2, respectively. TonB cells were transduced with an MOI of 4 in the presence of muIL-3 (10 ng/mL). Doxycycline (1 μg/mL) was added 2 days after transduction to half of the cells. On day 4 cultures were washed 3 times and resuspended either in the presence of murine IL-3 or with doxycycline in the absence of IL-3. Addition of doxycycline either 2 or 4 days after lentiviral transduction gave identical results.
Colony assays of transduced CD34+ CML and normal CD34+ cells were performed as described earlier.12,30 Then, 5 × 104 to 1 × 105 CD34+ cells were lentivirally transduced, and 1 × 103 cells were plated per methylcellulose culture. Cultures were stimulated with human GM-CSF and IL-3 at either 20 and 10 ng/mL (high stimulation) or 0.2 and 0.1 ng/mL (low stimulation), respectively. In parallel experiments, methylcellulose colony assays of transduced normal and CML CD34+ cells were stimulated with human SCF, G-CSF, and thrombopoietin (Tpo) at either 20 and 10 ng/mL and 10 U/mL (high stimulation) or 0.2 and 0.1 ng/mL and 0.1 U/mL (low stimulation), respectively. Colonies derived from transduced CFUs were identified by fluorescence microscopy.
Real-time RT-PCR
Cytoplasmic RNA was isolated using RNeasy mini-spin columns (Qiagen, Hilden, Germany) from 1 × 106 K562 cells. RNA was reverse transcribed into cDNA in a total volume of 20 μL using the MMLV-reverse transcriptase (Invitrogen, Groningen, Netherlands) and random hexamer primers under standard conditions. Real-time TaqMan reverse transcriptionpolymerase chain reaction (RT-PCR) of BCR-ABL and GAPDH was performed as described previously.9,33 Quantitative RT-PCR of SHP2, Gab2, and STAT5 expression in K562 was analyzed using the following primers: SHP2-FP, 5′-CCCACATCAAGATTCAGAACACT-3′; SHP2-RP, 5′-GCCCGTGATGTTCCATGTAA-3′; Gab2-FP, 5′-GCCGGCACAATACAGAATTCA-3′; Gab2-RP, 5′-GTGAGGCTGCCCTTGGTGT-3′; STAT5-FP, 5′-TGCCATTGACTTGGACAATCC-3′; and STAT5-RP, 5′-AAACCCATCTTCCCCCACC-3′.
The FAM-labeled probes were: SHP2, 5′-TGCCACTTTGGCTGAGTTGGTCC-3′; Gab2, 5′-ACCTCCCCCGCAGCCTGGC-3′; and STAT5, 5′-AGCTGCAGAAGAAGGCGGAGCA-3′.
Proliferation assay and annexin V staining
Cell proliferation was analyzed by trypan blue exclusion assay. Briefly, 1 to 5 × 104 cells/mL were grown in 24-well plates and the number of viable cells was determined after 24 hours and up to 25 days by trypan blue exclusion. In addition, lentivirally transduced TonB cells were analyzed for annexin V expression at day 6 after removal of murine IL-3 and addition of doxycycline to half of cells using the annexin-V-FLUOS staining kit (Roche, Penzberg, Germany) as recommended by the manufacturer.
Immunoblotting
Cellular lysates from K562 and TonB cells were prepared with lysis buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1% NP-40; 1% Triton X-100; 50 mM NaF; 5 mM EDTA; 1 mM PMSF; 1 mM Na2VO4), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to Hybond enhanced chemiluminescence (ECL) nitrocellulose membrane (Amersham Biosciences, Uppsala, Sweden), and immunoblotted with polyclonal rabbit anti-SH-PTP2 antibody (C-18), polyclonal anti-STAT5a (L-20), polyclonal rabbit anti-Gab1 (H-198), and polyclonal anti-ERK2 antibody (C-14) (all antibodies from Santa Cruz Biotechnology, Heidelberg, Germany), polyclonal rabbit anti-Gab2 (Upstate Biotechnology, Dundee, United Kingdom), and polyclonal rabbit anti-STAT3 (Cell Signaling Technology, Beverly, MA), according to the manufacturer's protocol. Chemiluminescence was used for visualization using the ECL Western blotting detection reagents (Amersham Biosciences) according to the manufacturer. Densitometry was performed on scanned images using Adobe Photoshop 7.0 software.
Results
Generation and functional evaluation of anti-SHP2, STAT5, and Gab2 shRNAs
The shRNAs targeting SHP2, STAT5, and Gab2 were rationally designed according to established rules and functionally evaluated using a transient cotransfection assay described earlier.29 This assay is based on FACS analysis of EGFP fluorescence with EGFP encoded along with the target gene on a bi-cistronic transcript. Three, one, and one of several shRNAs tested for SHP2, STAT5, and Gab2 reduced EGFP fluorescence to a similar extent as the anti-GFP shRNA used as a positive control, respectively (data not shown). The shRNAs 237, 594, and 714 for SHP2, 1151 for STAT5, and 620 for Gab2 were used in all further experiments.
To analyze the effects of shRNAs on endogenous target gene expression, lentiviral vectors encoding the respective shRNAs were generated (Figure 1A), and lentiviral supernatants were used to transduce BCR-ABL+ K562 cells at more than 98% efficiency (data not shown). All shRNAs strongly reduced target mRNA and protein expression as determined by real-time RT-PCR and immunoblotting, respectively (Figure 1B-C). The anti-SHP2 and STAT5 shRNAs reduced endogenous mRNA levels by about 90% ± 5%, whereas anti-Gab2 shRNA was slightly less efficient. In addition, protein expression was significantly reduced on expression of each shRNA (Figure 1C). Furthermore, RNAi against all 3 target genes as well as against BCR-ABL inhibited cell proliferation of K562 cells as compared to control shRNA (Figure 1D).
Enhanced sensitivity to gene silencing of SHP2, STAT5, and Gab2 in BCR-ABL-dependent cell proliferation
To specifically analyze the contribution of SHP2, STAT5, and Gab2 to BCR-ABL-mediated proliferation, TonB cells were transduced with high-titer lentiviral supernatants to induce stable RNAi against each of the 3 targets. On transduction to more than 95% (Figure 2A), TonB cells were cultured with murine IL-3 for an additional 4 days to allow for appropriate shRNA expression, and the reduction of target protein expression was determined by immunoblotting (Figure 2B). RNAi against STAT5 and Gab2 reduce STAT5 and Gab2, but not STAT3 and Gab1 protein expression, respectively. After addition of doxycycline to induce BCR-ABL expression to half of the cells and removal of IL-3 from the BCR-ABL+ cultures, the number of viable cells was determined. Anti-BCR-ABL, anti-SHP2, and anti-STAT5 shRNAs efficiently inhibit BCR-ABL-mediated (Figure 2D) but not IL-3-mediated (Figure 2C) cell proliferation with no difference to control shRNAs in this condition. In contrast, the anti-Gab2 shRNA was less effective, causing approximately 75% reduction in cell proliferation. Interestingly, readdition of murine IL-3 to BCR-ABL cultures with reduced target gene expression induces cell recovery as shown in Figure 2E.
To analyze the effects of anti-Gab2 shRNA, which was found to be less effective than anti-SHP2 and anti-STAT5 shRNAs, in more detail, we isolated several independent TonB-Gab2 shRNA clones by limiting dilution and analyzed 3 isolates according to their RFP fluorescence (Figure 3A). Immunoblotting reveals some correlation between RFP fluorescence and inhibition of Gab2 protein expression as determined by densitometry with 67%, 58%, and 30% reduction of Gab2 protein expression in clones with a mean fluorescent intensity (MFI) of about 1500, 500, and 100, respectively (Figure 3B). As shown in Figure 3C, reduction of Gab2 protein expression correlates to inhibition of BCR-ABL-mediated, but not IL-3-mediated proliferation in a dose-dependent manner.
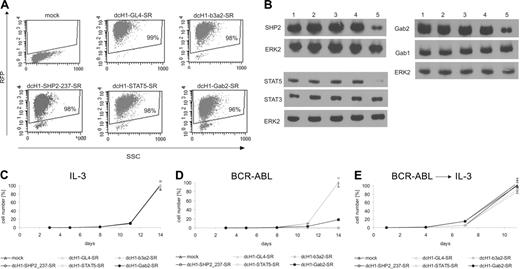
Lentivirus-mediated gene silencing in transduced TonB cells. (A) Dot blots of RFP fluorescence and side scatter (SSC) from TonB cells transduced with dcH1-GL4-SR, dcH1-b3a2-SR, dcH1-237SHP2-SR, dcH1-STAT5-SR, and dcH1-Gab2-SR 4 days after lentiviral transduction in the presence of IL-3. (B) For immunoblotting, TonB cells transduced with control dcH1-GL4-SR (lane 2), dcH1-121gfp-SR (lane3), dcH1-b3a2_1-SR (lane 4), or with anti-STAT5 shRNA, anti-Gab2, or anti-SHP2 shRNA (lane 5 in each blot), respectively, were lysed 4 days after transduction. Lane 1 shows untransduced TonB cells. The immunoblots were probed with anti-SHP2, anti-Gab2, anti-STAT5, anti-STAT3, and anti-Gab1 antibodies as indicated, and reprobed with anti-ERK2 antibody as loading control. (C) Number of trypan blue-negative TonB cells was determined starting 4 days after lentiviral transduction with different shRNAs (set day 0 on x-axis, average from 3 independent experiments) in the presence of IL-3. Triangles depict transduction with dcH1-GL4-SR or mock-transduced cells (control), and the almost identical cell number at day 14 was set 100%. Circles show cell numbers of TonB cells transduced with viruses encoding dcH1-b3a2_1-SR, dcH1-237SHP2-SR, dcH1-Gab2-SR, and dcH1-STAT5-SR as indicated. In a different set of experiments anti-GFP shRNAs were included as an additional control with almost identical results as shown. (D) Number of trypan blue-negative TonB cells was determined in the presence of doxycycline to induce BCR-ABL expression and in the absence of IL-3 (average from 3 independent experiments). Labeling as in panel C. (E) Number of trypan blue-negative TonB cells from cultures shown in panel D after readdition of IL-3 after 4 days (set day 0 on x-axis, averages from 3 experiments). Labeling as in panel C.
Lentivirus-mediated gene silencing in transduced TonB cells. (A) Dot blots of RFP fluorescence and side scatter (SSC) from TonB cells transduced with dcH1-GL4-SR, dcH1-b3a2-SR, dcH1-237SHP2-SR, dcH1-STAT5-SR, and dcH1-Gab2-SR 4 days after lentiviral transduction in the presence of IL-3. (B) For immunoblotting, TonB cells transduced with control dcH1-GL4-SR (lane 2), dcH1-121gfp-SR (lane3), dcH1-b3a2_1-SR (lane 4), or with anti-STAT5 shRNA, anti-Gab2, or anti-SHP2 shRNA (lane 5 in each blot), respectively, were lysed 4 days after transduction. Lane 1 shows untransduced TonB cells. The immunoblots were probed with anti-SHP2, anti-Gab2, anti-STAT5, anti-STAT3, and anti-Gab1 antibodies as indicated, and reprobed with anti-ERK2 antibody as loading control. (C) Number of trypan blue-negative TonB cells was determined starting 4 days after lentiviral transduction with different shRNAs (set day 0 on x-axis, average from 3 independent experiments) in the presence of IL-3. Triangles depict transduction with dcH1-GL4-SR or mock-transduced cells (control), and the almost identical cell number at day 14 was set 100%. Circles show cell numbers of TonB cells transduced with viruses encoding dcH1-b3a2_1-SR, dcH1-237SHP2-SR, dcH1-Gab2-SR, and dcH1-STAT5-SR as indicated. In a different set of experiments anti-GFP shRNAs were included as an additional control with almost identical results as shown. (D) Number of trypan blue-negative TonB cells was determined in the presence of doxycycline to induce BCR-ABL expression and in the absence of IL-3 (average from 3 independent experiments). Labeling as in panel C. (E) Number of trypan blue-negative TonB cells from cultures shown in panel D after readdition of IL-3 after 4 days (set day 0 on x-axis, averages from 3 experiments). Labeling as in panel C.
Reduced viability of TonB cells with decreased STAT5 expression
To analyze whether the decrease in viable cell number of BCR-ABL-expressing TonB cells with reduced target gene expression is due to reduced cell survival, the proportion of apoptotic cells was determined by annexin V staining at day 6 of suspension culture (Figure 4). We did not perform parallel propidium iodide staining because lentivirally transduced TonB cells highly express RFP as indicated in Figure 4. Whereas about 5% of annexin V+ cells were found in IL-3-supplemented cultures independent of reduction of the respective target gene, RNAi against both BCR-ABL and STAT5, but not against SHP2 and Gab2, resulted in a higher proportion of annexin V+ cells (42% and 43% in BCR-ABL as compared to 5%-8%, respectively, in IL3 condition).
Gab2-dependent inhibition of BCR-ABL-mediated cell proliferation. (A) The histogram of RFP fluorescence from 3 independent TonB-Gab2 shRNA clones is shown with high (black line), medium (light gray line), and low (dark gray line) RFP fluorescence. (B) Cellular lysates from the 3 TonB-Gab2 shRNA clones were subjected to immunoblotting with anti-Gab2 and anti-ERK2 antibodies as loading control as indicated. (C) Number of trypan blue-negative TonB-Gab2 shRNA cells in the presence of IL-3 (left) or doxycycline in the absence of IL-3 (right). Triangles depict cell numbers of a control TonB-GL4 shRNA clone with high RFP expression, which was set to 100% on day 14.
Gab2-dependent inhibition of BCR-ABL-mediated cell proliferation. (A) The histogram of RFP fluorescence from 3 independent TonB-Gab2 shRNA clones is shown with high (black line), medium (light gray line), and low (dark gray line) RFP fluorescence. (B) Cellular lysates from the 3 TonB-Gab2 shRNA clones were subjected to immunoblotting with anti-Gab2 and anti-ERK2 antibodies as loading control as indicated. (C) Number of trypan blue-negative TonB-Gab2 shRNA cells in the presence of IL-3 (left) or doxycycline in the absence of IL-3 (right). Triangles depict cell numbers of a control TonB-GL4 shRNA clone with high RFP expression, which was set to 100% on day 14.
Specific inhibition of colony formation of transduced CML progenitors by anti-SHP2, anti-STAT5, and anti-Gab2 shRNAs
To analyze the impact of SHP2, STAT5, and Gab2 on colony formation of normal and CML CD34+ progenitors, purified CD34+ cells were transduced with lentiviruses encoding control or specific shRNAs. To eliminate potential effects of different transduction rates in primary cells on the number of RFP+ colonies, we plated CD34+ cells for each transfection in the presence of optimal or suboptimal concentrations of IL-3 plus GM-CSF. In initial studies we found that colony formation of CD34+ CML cells in early chronic phase is inhibited by both reduction in cytokine stimulation and increase of imatinib concentration although there are some differences between individual CML samples (data not shown). These data indicate that colony formation of CML progenitor cells depends on signaling from both the oncoprotein and cytokine receptors. Therefore, the ratio of transduced, that is, RFP+ colonies under different cytokine stimulation indicates the functional relevance of the respective RNAi-target for CFU colony formation of normal and CML cells, respectively (Table 1).
Effects of lentivirally encoded shRNAs on colony formation of CML CD34+ cells and normal CD34+ cells
. | RFP+ colonies/total colony number, % . | . | . | |
|---|---|---|---|---|
| Sample, dcH1-shRNA-SR . | High cytokine concentration . | Low cytokine concentration . | Comparison of RFP positivity between low and high cytokine condition, % . | |
| CML CD34+ cells | ||||
| A (b3a2) | ||||
| GL4 | 33 | 33 | 100 | |
| b3a2 | 21 | 13 | 62 | |
| 237SHP2 | 30 | 12 | 40 | |
| B (b3a2) | ||||
| GL4 | 23 | 21 | 92 | |
| b3a2 | 17 | 9 | 53 | |
| 237SHP2 | 34 | 17 | 50 | |
| C (b3a2) | ||||
| GL4 | 25 | 21 | 84 | |
| b3a2 | 21 | 10 | 48 | |
| 594SHP2 | 56 | 20 | 36 | |
| 714SHP2 | 11 | 5 | 45 | |
| D (b3a2) | ||||
| GL4 | 44 | 38 | 86 | |
| b3a2 | 33 | 16 | 48 | |
| 714SHP2 | 22 | 11 | 50 | |
| E (b2a2) | ||||
| GL4 | 24 | 20 | 84 | |
| GFP | 22 | 19 | 86 | |
| 594SHP2 | 15 | 5 | 33 | |
| STAT5 | 16 | 6 | 37 | |
| E.2 (b2a2) | ||||
| GL4 | 56 | 51 | 91 | |
| 237SHP2 | 70 | 40 | 57 | |
| Gab2 | 49 | 27 | 55 | |
| STAT5 | 31 | 16 | 52 | |
| F (b2a2) | ||||
| GL4 | 11 | 10 | 91 | |
| GFP | 11 | 10 | 91 | |
| 237SHP2 | 5 | 2 | 40 | |
| 594SHP2 | 3 | 0 | 0 | |
| STAT5 | 7 | 0 | 0 | |
| G (b2a2) | ||||
| GL4 | 38 | 36 | 95 | |
| b3a2 | 46 | 38 | 83 | |
| 237SHP2 | 14 | 2 | 14 | |
| STAT5 | 13 | 4 | 31 | |
| H (b3a2) | ||||
| GL4 | 57 | 50 | 88 | |
| b3a2 | 50 | 23 | 46 | |
| 237SHP2 | 43 | 23 | 53 | |
| 594SHP2 | 58 | 27 | 46 | |
| Gab2 | 25 | 18 | 72 | |
| STAT5 | 64 | 10 | 16 | |
| I (b3a2) | ||||
| GL4 | 32 | 33 | 103 | |
| b3a2 | 6 | 0 | 0 | |
| 237SHP2 | 15 | 0 | 0 | |
| J (b2a2) | ||||
| GL4 | 50 | 54 | 108 | |
| 237SHP2 | 23 | 15 | 65 | |
| Gab2 | 24 | 12 | 50 | |
| STAT5 | 19 | 9 | 47 | |
| K (b2a2) | ||||
| GL4 | 12 | 11 | 92 | |
| 237SHP2 | 63 | 32 | 51 | |
| Gab2 | 24 | 12 | 50 | |
| STAT5 | 33 | 14 | 42 | |
| L (b3a2) | ||||
| GL4 | 34 | 33 | 96 | |
| b3a2 | 11 | 3 | 27 | |
| 237SHP2 | 11 | 6 | 54 | |
| Normal CD34+ cells | ||||
| A | ||||
| GL4 | 47 | 40 | 85 | |
| b3a2 | 34 | 30 | 88 | |
| 237SHP2 | 28 | 26 | 93 | |
| 714SHP2 | 27 | 22 | 82 | |
| B | ||||
| GL4 | 29 | 25 | 86 | |
| 237SHP2 | 9 | 10 | 111 | |
| 594SHP2 | 12 | 13 | 108 | |
| 714SHP2 | 16 | 15 | 94 | |
| Gab2 | 25 | 23 | 92 | |
| C | ||||
| GL4 | 53 | 52 | 98 | |
| b3a2 | 30 | 35 | 117 | |
| 237SHP2 | 27 | 25 | 93 | |
| D | ||||
| GL4 | 34 | 35 | 103 | |
| 237SHP2 | 21 | 21 | 100 | |
| 594SHP2 | 21 | 23 | 109 | |
| Gab2 | 20 | 17 | 85 | |
| STAT5 | 26 | 19 | 73 | |
| E | ||||
| GL4 | 32 | 35 | 109 | |
| 594SHP2 | 28 | 32 | 114 | |
| STAT5 | 42 | 32 | 76 | |
| F | ||||
| GL4 | 28 | 27 | 96 | |
| Gab2 | 25 | 25 | 100 | |
| STAT5 | 16 | 14 | 88 | |
. | RFP+ colonies/total colony number, % . | . | . | |
|---|---|---|---|---|
| Sample, dcH1-shRNA-SR . | High cytokine concentration . | Low cytokine concentration . | Comparison of RFP positivity between low and high cytokine condition, % . | |
| CML CD34+ cells | ||||
| A (b3a2) | ||||
| GL4 | 33 | 33 | 100 | |
| b3a2 | 21 | 13 | 62 | |
| 237SHP2 | 30 | 12 | 40 | |
| B (b3a2) | ||||
| GL4 | 23 | 21 | 92 | |
| b3a2 | 17 | 9 | 53 | |
| 237SHP2 | 34 | 17 | 50 | |
| C (b3a2) | ||||
| GL4 | 25 | 21 | 84 | |
| b3a2 | 21 | 10 | 48 | |
| 594SHP2 | 56 | 20 | 36 | |
| 714SHP2 | 11 | 5 | 45 | |
| D (b3a2) | ||||
| GL4 | 44 | 38 | 86 | |
| b3a2 | 33 | 16 | 48 | |
| 714SHP2 | 22 | 11 | 50 | |
| E (b2a2) | ||||
| GL4 | 24 | 20 | 84 | |
| GFP | 22 | 19 | 86 | |
| 594SHP2 | 15 | 5 | 33 | |
| STAT5 | 16 | 6 | 37 | |
| E.2 (b2a2) | ||||
| GL4 | 56 | 51 | 91 | |
| 237SHP2 | 70 | 40 | 57 | |
| Gab2 | 49 | 27 | 55 | |
| STAT5 | 31 | 16 | 52 | |
| F (b2a2) | ||||
| GL4 | 11 | 10 | 91 | |
| GFP | 11 | 10 | 91 | |
| 237SHP2 | 5 | 2 | 40 | |
| 594SHP2 | 3 | 0 | 0 | |
| STAT5 | 7 | 0 | 0 | |
| G (b2a2) | ||||
| GL4 | 38 | 36 | 95 | |
| b3a2 | 46 | 38 | 83 | |
| 237SHP2 | 14 | 2 | 14 | |
| STAT5 | 13 | 4 | 31 | |
| H (b3a2) | ||||
| GL4 | 57 | 50 | 88 | |
| b3a2 | 50 | 23 | 46 | |
| 237SHP2 | 43 | 23 | 53 | |
| 594SHP2 | 58 | 27 | 46 | |
| Gab2 | 25 | 18 | 72 | |
| STAT5 | 64 | 10 | 16 | |
| I (b3a2) | ||||
| GL4 | 32 | 33 | 103 | |
| b3a2 | 6 | 0 | 0 | |
| 237SHP2 | 15 | 0 | 0 | |
| J (b2a2) | ||||
| GL4 | 50 | 54 | 108 | |
| 237SHP2 | 23 | 15 | 65 | |
| Gab2 | 24 | 12 | 50 | |
| STAT5 | 19 | 9 | 47 | |
| K (b2a2) | ||||
| GL4 | 12 | 11 | 92 | |
| 237SHP2 | 63 | 32 | 51 | |
| Gab2 | 24 | 12 | 50 | |
| STAT5 | 33 | 14 | 42 | |
| L (b3a2) | ||||
| GL4 | 34 | 33 | 96 | |
| b3a2 | 11 | 3 | 27 | |
| 237SHP2 | 11 | 6 | 54 | |
| Normal CD34+ cells | ||||
| A | ||||
| GL4 | 47 | 40 | 85 | |
| b3a2 | 34 | 30 | 88 | |
| 237SHP2 | 28 | 26 | 93 | |
| 714SHP2 | 27 | 22 | 82 | |
| B | ||||
| GL4 | 29 | 25 | 86 | |
| 237SHP2 | 9 | 10 | 111 | |
| 594SHP2 | 12 | 13 | 108 | |
| 714SHP2 | 16 | 15 | 94 | |
| Gab2 | 25 | 23 | 92 | |
| C | ||||
| GL4 | 53 | 52 | 98 | |
| b3a2 | 30 | 35 | 117 | |
| 237SHP2 | 27 | 25 | 93 | |
| D | ||||
| GL4 | 34 | 35 | 103 | |
| 237SHP2 | 21 | 21 | 100 | |
| 594SHP2 | 21 | 23 | 109 | |
| Gab2 | 20 | 17 | 85 | |
| STAT5 | 26 | 19 | 73 | |
| E | ||||
| GL4 | 32 | 35 | 109 | |
| 594SHP2 | 28 | 32 | 114 | |
| STAT5 | 42 | 32 | 76 | |
| F | ||||
| GL4 | 28 | 27 | 96 | |
| Gab2 | 25 | 25 | 100 | |
| STAT5 | 16 | 14 | 88 | |
Cultures were stimulated with high (20 ng/mL GM-CSF, 10 ng/mL IL-3) or low cytokine concentration (0.2 ng/mL GM-CSF, 0.1 ng/mL IL-3). The right column indicates the relative amount of the RFP+ colony ratio under low stimulation (ratio at high concentration set as 100% for each lane).
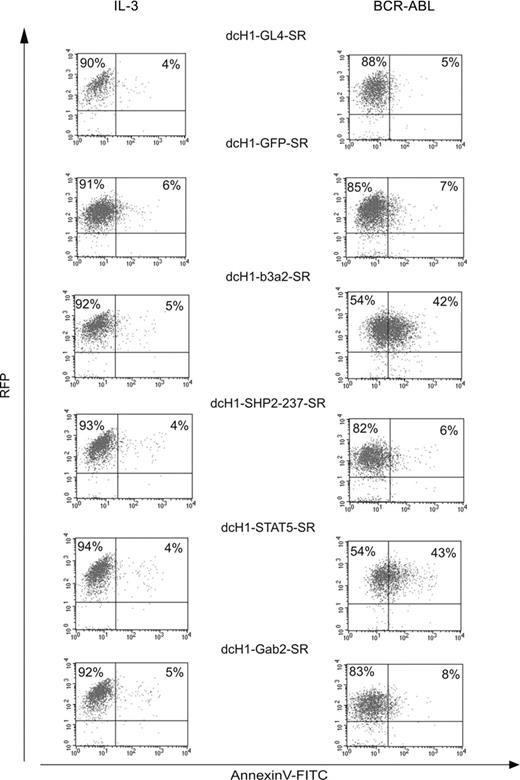
Effects of different cell culture conditions on TonB cells with reduced target gene expression. Lentivirally transduced RFP+ TonB cells were stained for annexin V expression either in the presence of IL-3 (left) or in the presence of doxycycline to induce BCR-ABL and in the absence of IL-3 (right) after 6 days. Dot blots show RFP fluorescence on the y-axis and annexin V-FITC staining on the x-axis.
Effects of different cell culture conditions on TonB cells with reduced target gene expression. Lentivirally transduced RFP+ TonB cells were stained for annexin V expression either in the presence of IL-3 (left) or in the presence of doxycycline to induce BCR-ABL and in the absence of IL-3 (right) after 6 days. Dot blots show RFP fluorescence on the y-axis and annexin V-FITC staining on the x-axis.
As expected, both control shRNAs had no effect on the ratio of transduced RFP+ colonies from CML or normal CFUs under reduced cytokine stimulation (Table 1). In contrast, anti-BCR-ABL shRNA reduced colony formation of transduced CMLs but not normal CFUs under low cytokine stimulation. All 3 anti-SHP2 as well as the anti-STAT5 and anti-Gab2 shRNAs tested reduced the ratio of transduced CML-derived colonies to about 35%, 25%, and 57%, respectively. In contrast, all shRNAs tested had no or only weak (STAT5) effects on normal transduced progenitors (Table 1). The results on differential sensitivity of normal and CML CFUs to silencing of SHP2, STAT5, and Gab2 are summarized in Figure 5A. Finally, normal and CML CD34+ cells were grown in cultures containing high and low concentrations of SCF/G-CSF/Tpo, and anti-β-GMR shRNA against the common β-chain of the receptors for IL-3, GM-CSF, and IL-5 was used as an additional control in this set of experiments (Figure 5B). Again, anti-SHP2, anti-STAT5, and anti-Gab2 shRNAs specifically inhibit colony formation by CML but not normal CFUs. Furthermore, anti-β-GMR RNAi had no impact on colony formation of CD34+ cells when cultures were stimulated with cytokines that induce signaling independent of β-GMR.30 In initial studies individual colonies from 2 CML samples (labeled as A and B) were picked and analyzed for the presence of the BCR-ABL translocation by fluorescence in situ hybridization (FISH). All colonies analyzed were found BCR-ABL+.
Combinatorial RNAi targeting BCR-ABL and SHP2
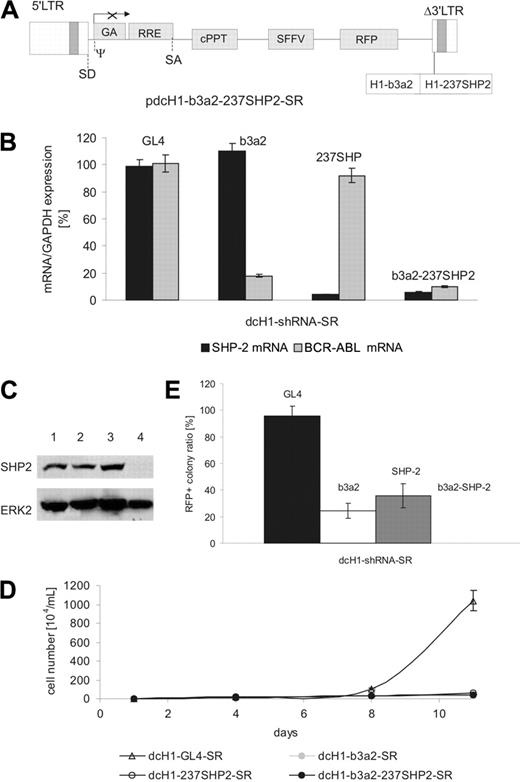
We next studied the effects of simultaneously silencing 2 target genes as a model for combination treatment of CML cells. We constructed a lentiviral plasmid with 2 H1-shRNA expression cassettes in the Δ3′-LTR as schematically shown in Figure 6A. Lentiviral supernatants were generated with titers similar to all lentiviral preparations described. On transduction of K562 cells with lentiviruses expressing either anti-BCR-ABL or anti-SHP2 shRNA alone or both shRNAs from a single vector, the double anti-BCR-ABL plus anti-SHP2 shRNA vector reduced mRNA and protein levels of both target genes with similar efficacy as compared to the respective single shRNA lentiviruses (Figure 6B-C and data not shown). In addition, cell proliferation of transduced K562 cells was inhibited by all 3 viral preparations (Figure 6D).
To analyze combined effects of anti-BCR-ABL and anti-SHP2 shRNAs on CML progenitors, purified CD34+ CML cells were transduced with anti-BCR-ABL and anti-SHP2 shRNA or with lentiviral vectors encoding both shRNAs alone. As shown in Figure 6E, both anti-BCR-ABL and anti-SHP2 shRNA separately reduced the ratio of RFP+ colonies with comparable efficacy as observed before. However, the coexpression of anti-BCR-ABL and anti-SHP2 shRNAs completely blocked colony formation of transduced CML progenitors at suboptimal cytokine concentrations in all 3 samples tested.
Discussion
In CML targeting BCR-ABL tyrosine kinase activity is now well established as an efficient and leukemia-specific therapeutic strategy. Accordingly, second-generation tyrosine kinase inhibitors that provide activity against multiple imatinib-resistant BCR-ABL point mutants have been developed and their function and target specificity are currently under investigation.3-6
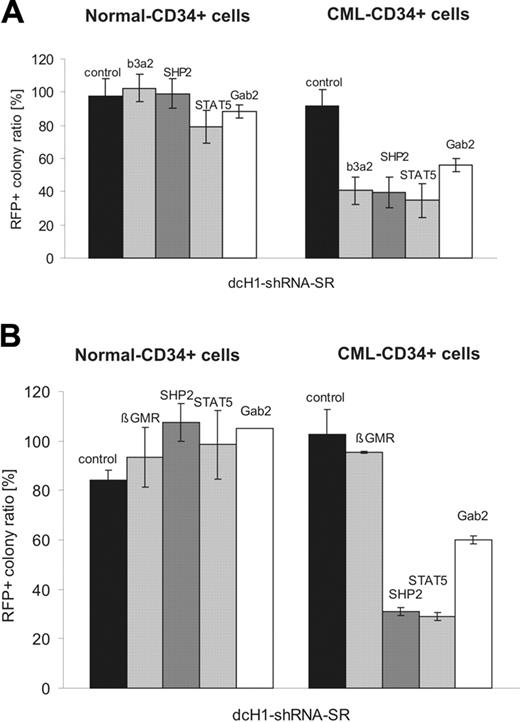
Effects of lentivirus-mediated RNAi on colony-formation of normal and CML CD34+ cells. (A) The diagram shows the relative ratio of RFP+ colonies under low as compared to high cytokine stimulation (IL-3 + GM-CSF) for normal (left) and CML (right) CD34+ progenitor cells. (B) The diagram shows the relative ratio of RFP+ colonies under low as compared to high cytokine stimulation (SCF + G-CSF + TPO) for normal (left) and CML (right) CD34+ progenitor cells as described. The specific RNAi target is indicated on top of each bar. Controls include GL4 and GFP shRNAs.
Effects of lentivirus-mediated RNAi on colony-formation of normal and CML CD34+ cells. (A) The diagram shows the relative ratio of RFP+ colonies under low as compared to high cytokine stimulation (IL-3 + GM-CSF) for normal (left) and CML (right) CD34+ progenitor cells. (B) The diagram shows the relative ratio of RFP+ colonies under low as compared to high cytokine stimulation (SCF + G-CSF + TPO) for normal (left) and CML (right) CD34+ progenitor cells as described. The specific RNAi target is indicated on top of each bar. Controls include GL4 and GFP shRNAs.
An alternative approach to improve drug therapy for CML may focus on the identification of target molecules other than BCR-ABL itself that are required for proliferation of CML cells. Several individual signaling cascades are activated by BCR-ABL, such as signals linked to Ras, PI3-K/Akt, c-Myc, STAT5, and reactive oxygen species, and their functional relevance has been described using forward and reverse genetics in cell culture and murine stem cell transplantation models (for reviews, see Sattler and Griffin34 and Van Etten35 ). However, functional analysis of individual signaling molecules in primary normal as well as leukemic cells has not yet been accomplished, mostly because suitable genetic tools were not available.
We here describe the use of lentivirus-mediated RNAi in primary cells to identify potential therapeutic targets in CML. We selected SHP2, STAT5, and Gab2 based on their constitutive tyrosine phosphorylation in BCR-ABL+ cell lines, suggesting some functional role for aberrant signaling. All 3 belong to different functional classes of proteins (tyrosine phosphatase, transcription factor, adaptor protein) and are linked to 2 independent signaling pathways in BCR-ABL+ cells. We generated 3 different anti-SHP2 shRNAs that all gave identical results indicating the specificity of anti-SHP2 RNAi. In contrast, we could only isolate one shRNA each for STAT5 and Gab2, respectively, and we cannot formally exclude any potential off-target effects for these genes. However, we found similar effects both in murine and human cells, and the expression of the related STAT3 and Gab1 genes was not affected by the respective shRNAs.
Our TonB cell model with inducible BCR-ABL expression clearly demonstrates that different cell culture conditions, that is, IL-3- versus BCR-ABL-driven proliferation, strongly affect cellular responses to RNAi against all 3 targets in genetically identical cells. In the context of BCR-ABL expression, cells display a marked but reversible inhibition of proliferation (Figure 2) that may depend on the actual level of protein expression as shown for Gab2 in 3 independent TonB-Gab2 shRNA clones (Figure 3). Interestingly, reduction of BCR-ABL and STAT5 expression reduces cell viability in the absence of IL-3, whereas RNAi against SHP2 and Gab2 had no effect on cell survival at the time point tested in this study. These data are in line with earlier reports describing apoptotic effects of a dominant-negative STAT5 mutant in BaF3 cells,36 but mainly reduction of BCR-ABL-mediated proliferation in Gab2-/- myeloid cells.28 Finally, overexpression and functional relevance of nonmutated SHP2 in adult leukemia including CML has been described in a recent study by Xu et al.20 In view of these data it is important to note that the mechanisms causing enhanced sensitivity to RNAi in BCR-ABL+ cells are most likely different for each target and need to be characterized individually.
Combinatorial RNAi targeting BCR-ABL and SHP2. (A) Schematic representation of the lentiviral transgene plasmid encoding anti-SHP2 and anti-Bcr-Abl shRNAs. (B) Target mRNA levels were measured by real-time RT-PCR 4 days after lentiviral transduction of K562 cells and normalized in comparison to GAPDH expression. The shRNA encoded by the respective lentivirus is indicated on the top of each bar. (C) Immunoblot of K562 cells transduced with anti-BCR-ABL and an anti-SHP2 plus anti-BCR-ABL shRNA-encoding lentivirus. Cells transduced with control dcH1-GL4-SR (lane 2), dcH1-b3a2_1-SR (lane 3), and b3a2_237SHP2 (lane 4) were lysed 6 days after transduction. Lane 1 shows untransduced K562 cells. The top panel shows an immunoblot with anti-SHP2-specific antibody, and the bottom panel the same membrane reprobed with anti-ERK2 antibodies as loading control. (D) Effects of anti-b3a2, anti-SHP2, and anti-b3a2 plus SHP2 shRNAs on proliferation of K562 cells. Numbers of trypan blue-negative cells are shown after transduction with lentiviruses encoding single (dcH1-GL4-SR, dcH1-b3a2-SR, dcH1-237SHP2-SR) or combined (b3a2-237SHP2) shRNAs as indicated (results from 2 experiments). One milliliter with 104/mL cells was plated, cultures were split and fed twice a week, and calculated cell numbers are indicated. Triangles depict transduction with control virus dcH1-GL4-SR, and circles show transduction with viruses encoding dcH1-b3a2_1-SR, dcH1-237SHP2-SR, and dcH1-b3a2-237SHP2, respectively. (E) Effects of lentivirus-mediated shRNAs on colony formation of CML CD34+ cells. The diagram shows the relative ratio of RFP+ colonies under low as compared to high cytokine stimulation (GM-CSF + IL-3) as in Figure 5.The specific RNAi target is indicated on top of each bar. No RFP+ colonies were found under low cytokine concentration.
Combinatorial RNAi targeting BCR-ABL and SHP2. (A) Schematic representation of the lentiviral transgene plasmid encoding anti-SHP2 and anti-Bcr-Abl shRNAs. (B) Target mRNA levels were measured by real-time RT-PCR 4 days after lentiviral transduction of K562 cells and normalized in comparison to GAPDH expression. The shRNA encoded by the respective lentivirus is indicated on the top of each bar. (C) Immunoblot of K562 cells transduced with anti-BCR-ABL and an anti-SHP2 plus anti-BCR-ABL shRNA-encoding lentivirus. Cells transduced with control dcH1-GL4-SR (lane 2), dcH1-b3a2_1-SR (lane 3), and b3a2_237SHP2 (lane 4) were lysed 6 days after transduction. Lane 1 shows untransduced K562 cells. The top panel shows an immunoblot with anti-SHP2-specific antibody, and the bottom panel the same membrane reprobed with anti-ERK2 antibodies as loading control. (D) Effects of anti-b3a2, anti-SHP2, and anti-b3a2 plus SHP2 shRNAs on proliferation of K562 cells. Numbers of trypan blue-negative cells are shown after transduction with lentiviruses encoding single (dcH1-GL4-SR, dcH1-b3a2-SR, dcH1-237SHP2-SR) or combined (b3a2-237SHP2) shRNAs as indicated (results from 2 experiments). One milliliter with 104/mL cells was plated, cultures were split and fed twice a week, and calculated cell numbers are indicated. Triangles depict transduction with control virus dcH1-GL4-SR, and circles show transduction with viruses encoding dcH1-b3a2_1-SR, dcH1-237SHP2-SR, and dcH1-b3a2-237SHP2, respectively. (E) Effects of lentivirus-mediated shRNAs on colony formation of CML CD34+ cells. The diagram shows the relative ratio of RFP+ colonies under low as compared to high cytokine stimulation (GM-CSF + IL-3) as in Figure 5.The specific RNAi target is indicated on top of each bar. No RFP+ colonies were found under low cytokine concentration.
Very similar to the results seen in the TonB model, primary CML-derived CD34+ colony-forming cells display a higher sensitivity to silencing of SHP2, STAT5, and Gab2 as compared to normal CFUs (Table 1; Figure 5). In our study, we excluded potential effects due to different transduction efficacies by comparing colony formation of transduced and genetically identical CFUs under high and low cytokine stimulation for each sample. Using 2 different cytokine combinations, namely, GM-CSF plus IL-3 and G-CSF plus Tpo plus SCF, we found that primary purified CD34+ CML progenitors are more sensitive to inhibition of gene expression as compared to normal CD34+ CFUs under both conditions tested (Figure 5). Interestingly, data from several BCR-ABL-positive and -negative leukemic cell lines did not reflect the effects of gene silencing observed in normal CD34+ progenitor cells because such cell lines were found to display similar sensitivity to silencing of SHP2, Gab2, and STAT5, respectively (data not shown). Furthermore, anti-SHP2-, anti-Gab2, and anti-STAT5 RNAi inhibits BCR-ABL-driven cell proliferation to a similar extent as leukemia-specific anti-BCR-ABL RNAi.
As described earlier for K562 cells expressing anti-BCR-ABL shRNAs, clonal evolution of lentivirally transduced and shRNA-expressing cells may occur during prolonged periods of cell culture with selection of cells with reduced amounts of gene silencing.12 Interestingly, using our newly developed lentiviral vector capable of expressing 2 different shRNAs, namely, anti-BCR-ABL and anti-SHP2, from a single provirus, we could prevent clonal selection and outgrowth of lentivirally transduced K562 cells (data not shown). In addition, RNAi targeting both BCR-ABL and SHP2 completely blocked colony formation of transduced CML progenitors under low cytokine stimulation, suggesting cooperative effects between silencing of BCR-ABL and SHP2 expression. These data indicate that combined inhibition of target gene expression may indeed be a useful strategy to delay or completely suppress clonal evolution of BCR-ABL+ cells.
In our study, we analyzed colony formation of CD34+ normal and chronic-phase CML progenitor cells in vitro. However, lentiviral transfer of shRNA expression cassettes may provide a strategy to study gene function in more immature cells as well. This could be of some importance because imatinib mesylate seems unable to kill very immature CML progenitor or stem cells.7,37 Identification of CML-specific potential therapeutic targets in such cells may substantially improve drug therapy in CML in the future.
In summary, our data identify SHP2, STAT5, and Gab2 as bona fide candidates for molecularly defined therapeutic approaches in CML. Based on these results, specific small molecules may potentially be developed in the future. However, because signaling molecules usually integrate several upstream signals and may deliver more than one downstream effect by interacting with several signaling components, the identification and specific targeting of individual protein-protein interactions is likely to be required for effective molecularly defined treatment strategies. Alternatively, RNAi itself may be used as a direct therapeutic approach as soon as strategies to trigger stable RNAi in primary cells become available for clinical application. Such options may use either repeated application of RNAi triggers or single-time gene transfer strategies with improved viral vectors in the future. Independent of the potential tool, our data demonstrate that the combined targeting of a leukemia-specific molecule (eg, by imatinib mesylate or anti-BCR-ABL shRNA) and nonspecific molecules (anti-SHP2, anti-STAT5, and anti-Gab2 shRNA) may be highly effective and specific for therapeutic intervention in CML and perhaps other BCR-ABL+ leukemias.
Prepublished online as Blood First Edition Paper, November 8, 2005; DOI 10.1182/blood-2005-08-3087.
Supported in part by grants of the Deutsche Forschungsgemeinschaft (SFB 566), H. W. & J. Hector-Stiftung, and Wilhelm Sanders-Stiftung.
M.S. and A.C. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank T. Hirano (University of Osaka) and B. Neel (Harvard Medical School) for providing us with the human Gab2 and the human SHP2 cDNA, respectively; George Daley (MIT, Cambridge, MA) for providing us with the TonB cell line; and M. Morgan for critical reading of the manuscript.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal