Abstract
The reduced folate carrier (RFC) is the dominant route for the uptake of various antifolates including PT523, a potent dihydrofolate reductase inhibitor (Ki = 0.35 pM) and an excellent transport substrate of the RFC (Kt = 0.7 μM). Here, we describe the multiple mechanisms of RFC inactivation in human leukemia PT523-resistant cells originally harboring 3 RFC alleles. Cellular exposure to gradually increasing PT523 concentrations resulted in sublines displaying up to 3500-fold resistance to various hydrophilic antifolates that rely on RFC for their cellular uptake. Antifolate-resistant cells lost RFC gene expression (65%-99% loss) due to impaired promoter binding of various transcription factors that regulate RFC gene expression. Additionally, DNA sequencing revealed that PT523-resistant cells contained a cluster of 4 nearly consecutive mutations residing on a single RFC allele including L143P, A147V, R148G, and Q150Stop. Southern blot analysis established the loss of an RFC allele in PT523-resistant cells. These alterations resulted in markedly decreased RFC protein levels (∼80%-99% loss) and consequently impaired [3H]methotrexate transport (87%-99% loss). This study provides the first evidence that acquisition of PT523 resistance in human leukemia cells harboring 3 RFC alleles is due to multiple coexisting alterations including transcriptional silencing, inactivating mutations, and RFC allele loss.
Introduction
Reduced folate cofactors are essential vitamins that serve as 1-carbon donors in the de novo biosynthesis of purines and thymidylate and are therefore crucial for DNA replication in mammalian cells.1 The first class of cytotoxic antimetabolites introduced nearly 60 years ago was folic acid analogs (ie, antifolates).2 These antifolates, which originally included aminopterin and methotrexate (MTX), achieved remissions in acute lymphoblastic leukemia (ALL).2 Since then, MTX continues to play an important role in the treatment of various hematologic and nonhematologic malignancies including ALL, lymphomas, breast cancer, osteogenic sarcoma, colorectal cancer, and choriocarcinoma.3
Because mammalian cells are devoid of folic acid biosynthesis, they must take up folate cofactors from exogenous sources by specific transport systems, primarily through the reduced folate carrier (RFC).4,5 RFC displays a high affinity to various hydrophilic antifolates and mediates their efficient cellular uptake via an anion-exchange mechanism.6,7 However, alterations in drug transport frequently underlie antifolate resistance.5,8,9 Defective membrane transport is a well-established mechanism of resistance to MTX and various novel antifolates in cultured human cell lines.5,8,9 Among novel antifolates is PT523, which exerts its cytotoxic effect via potent inhibition (Ki = 0.35 pM) of dihydrofolate reductase (DHFR).10 Furthermore, PT523 is efficiently transported into mammalian cells via the RFC with a very high affinity to this transporter (Ki = 0.7 μM, determined by inhibition of [3H]MTX uptake).11 Alterations in membrane transport in antifolate-resistant human cell lines can manifest as decreased folate/antifolate translocation (ie, reduced Vmax), decreased affinities for the antifolate transport substrate (increased Km), or a combination of the two.5,8,9,12,13 Recent studies with multiple antifolate-resistant human tumor cell lines have demonstrated that such alterations in drug transport are frequently due to RFC inactivating mutations resulting in a structurally altered transporter.9,12,13 Loss of RFC gene expression due to promoter methylation has been described at least in one inherently MTX-resistant human breast cancer cell line.14 Recently, we have shown that RFC gene silencing frequently occurs due to loss of function of various transcription factors that physiologically regulate RFC gene expression regardless of the presence of inactivating RFC mutations.15-17 Furthermore, these studies have clearly shown that RFC promoter methylation is not a contributing mechanism of antifolate resistance in leukemia in vitro and in vivo.16
In the present study we characterized the molecular basis of the loss of antifolate transport in human CCRF-CEM leukemia cells that acquired stepwise resistance to the novel antifolate PT523. We find that multiple mechanisms of RFC gene inactivation coexist including nonsense and missense mutations, allele loss as well as transcriptional silencing via loss of function of several transcription factors that normally promote RFC gene expression. Hence, whereas various studies focused on the characterization of a single modality of antifolate resistance, we show here that multiple molecular mechanisms of PT523 resistance simultaneously coexist that clearly contribute to the disruption of transporter expression and function.
Materials and methods
Materials
[3H]MTX (23.4 Ci/mmol [865.8 GBq]) was obtained from Moravek Biochemicals (Brea, CA), purified prior to use by thin layer chromatography and stored at -80°C. MTX was from Teva Pharmaceuticals (Petach Tikva, Israel). The various antifolate drugs were generous gifts from the following sources: PT523, Dr W. T. McCulloch (Sparta Pharmaceuticals, Horsham, PA); GW1843U89, Dr G. Smith (GlaxoWelcome, Research Triangle Park, NC); ZD9331, Dr A. Jackman, (Institute of Cancer Research, Sutton, United Kingdom); pemetrexed (MTA, Alimta, LY231514) and lometrexol (DDATHF; Eli Lilly, Indianapolis, IN); trimetrexate (TMQ), Dr D. Fry (Warner-Lambert Parke-Davis, Ann Arbor, MI).
Tissue culture and antifolate drug selection
Human CCRF-CEM leukemia cells were maintained in RPMI 1640 medium containing 2.3 μM folic acid (Biological Industries, Beth-Haemek, Israel) supplemented with 10% fetal calf serum (Gibco, Karlsruhe, Germany), 2 mM glutamine, 100 U/mL penicillin G, and 100 μg/mL streptomycin sulfate (Biological Industries). Stepwise selections were performed in gradually increasing PT523 concentrations over a period of 4 to 8 months as follows. Parental CCRF-CEM (wild-type [WT]) cells (2 × 105/T25 flask containing 5 mL growth medium) were first exposed to a PT523 concentration (0.43 nM) that is equal to the 50% drug inhibitory concentration (IC50). Following 3 to 6 days of growth at 37°C when the total cell number doubled, the antifolate concentration was gradually increased by 25% to 100% increments and selection was terminated at 2.5 μM. This resulted in the establishment of the sublines PTR0.003, PTR0.05, PTR0.5, and PTR2.5 growing at PT523 concentrations of 0.003, 0.05, 0.5, and 2.5 μM, respectively. CCRF-CEM 7A cells that highly overexpress the RFC were cultured in folic acid-free medium containing 10% dialyzed fetal calf serum and supplemented with 0.25 nM leucovorin.18
Analysis of intracellular folate pools
Suspension cell cultures were grown for at least 3 passages in antifolate-free medium. Thereafter, exponentially growing cells (2.5 × 107) were harvested and washed 3 times in PBS, counted, centrifuged, snap-frozen in liquid nitrogen as cell pellets, and stored at -80°C until analysis. Intracellular reduced folate pools were analyzed according to a method based on enzymatic cycling of reduced folates to N5, N10-methylenetetrahydrofolate (CH2THF) followed by entrapment into a stable ternary complex with Lactobacillus casei TS and [6-3H]-F-dUMP (20 Ci/mmol [740 GBq]; Moravek Biochemicals) as previously described.19,20
Antifolate growth inhibition
Parental cells and their various antifolate-resistant sublines were first grown in normal growth medium lacking antifolates for 5 to 7 cell doublings. Thereafter, cells were seeded in 96-well plates (3 × 104/well) in growth medium (0.15 mL/well) containing increasing concentrations of the different antifolates. After 3 days of incubation at 37°C, cell numbers were determined by viability count using trypan blue exclusion.
Isolation of gDNA and Southern blot analysis
High-molecular-weight gDNA was extracted from the various cell lines using either standard phenol-chloroform extraction or using the Roche (Basel, Switzerland) high pure polymerase chain reaction (PCR) template preparation kit. To determine RFC gene copy numbers, gDNA was digested with several restriction enzymes and fractionated by electrophoresis on 0.8% agarose gels, depurinated, denatured, and neutralized as previously described.13 The DNA was then transferred to a ZetaProbe-GT nylon membrane (Bio-Rad, Hercules, CA) and UV cross-linked (1200 rad). A 538-bp intronic probe corresponding to human RFC genomic positions c241212-213499 (accession no. 7717445; nucleotides 634-1171 in hRFC cDNA, accession number U19720 [NCBI Nucleotide]) was prepared by PCR and T/A cloning. [32P] labeling of this genomic RFC probe was performed using a random hexamer priming kit according to the instructions of the manufacturer (Biological Industries). Hybridization was performed at 65°C for about 15 hours in an EZ-hybridization buffer (Biological Industries) after which the membrane was washed under high-stringency conditions.13 The membrane was then exposed to an x-ray film and the intensity of the RFC band was quantified by scanning densitometry and normalized to an internal repetitive DNA band observed after ethidium bromide staining.
Genomic PCR-SSCP analysis
High-molecular-weight gDNA was extracted from parental CCRF-CEM cells and their various antifolate-resistant sublines using the Roche high pure PCR template preparation kit. Genomic PCR was performed using 9 oligonucleotide primer pairs spanning the entire RFC coding region that contains 5 coding exons (exons 2-6).9,21 gDNA samples (10-100 ng) were amplified with Taq polymerase (Promega, Madison, WI) in a reaction buffer (50 μL) consisting of 10 pmol of each primer, 0.2 mM nonradioactive dNTPs, and 10 μCi (0.37 MBq) [32P]dCTP in 1 × reaction buffer (Promega) supplemented with 1.5 mM MgCl2 and 5% DMSO. The conditions of the PCRs as well as those of the single-strand conformational polymorphism (SSCP) analysis were as previously detailed.9,21
T/A cloning of PCR products
After genomic PCR, the products were gel-purified (Qiagen QIAquick Gel Extraction; Qiagen, Hilden, Germany), incubated overnight at 4°C in 10 mL 5 × reaction buffer (Promega) containing Easy-pGEM-T plasmid and T4 ligase according to the instructions of the manufacturer. The recombinant plasmid was transformed into competent DH5α Escherichia coli. The recombinant plasmid was purified using the Promega Wizard Plus SV Minipreps DNA purification system and was subsequently analyzed by PCR-SSCP or DNA sequencing or both.
DNA sequencing
PCR products obtained with DNA from parental and PT523-resistant cells showing an altered RFC electrophoretic mobility pattern on several independent genomic PCR-SSCP analyses were fractionated on 1.5% agarose gels and purified using a DNA purification kit (Qiagen). DNA sequencing was performed on purified genomic PCR products by the fluorescent dideoxy chain termination method using an ABI377 sequencer. At least 3 independent PCRs and DNA sequence determinations were performed (using different DNA preparations) to determine a definitive mutation.
Isolation of RNA and Northern blot analysis
Exponentially growing cells (1-3 × 107 cells) were harvested by centrifugation, washed with PBS, and total RNA was extracted using the EZ-RNA kit according to the instructions of the manufacturer (Biological Industries). Aliquots of total RNA (30 μg) from parental CCRF-CEM cells and their various sublines were denatured at 68°C for 10 minutes in a MOPS-formamide buffer13 and fractionated by electrophoresis on 1.2% agarose gels containing formaldehyde for 6 hours at 3 V/cm. Resolved RNA was then blotted onto Zeta-Probe-GT nylon membrane (Bio-Rad) and immobilized onto the nylon membrane by UV cross-linking at 1200 rad. The membrane was first incubated at room temperature for 15 minutes in a solution of 5% acetic acid, stained for 5 minutes with 0.04% methylene blue in 0.5 M sodium acetate pH 5.2, and finally rinsed in distilled water for 10 minutes. The blot was then hybridized with a gel-purified, [32P]oligolableled (using a random hexamer priming kit from Biological Industries) full-length human RFC cDNA probe. Hybridization was performed overnight at 68°C in an EZ-hybridization solution (Biological Industries) after which the membrane was washed under high stringency conditions.13 The membrane was then exposed to an x-ray film and the intensity of the RFC band was quantified by scanning densitometry and normalization to the methylene blue stained 18S and 28S rRNA bands.
Antifolate growth inhibition of parental human leukemia CCRF-CEM cells and their various PT523-resistant sublines. Exponentially growing parental CCRF-CEM (WT, ○) cells and their various PT523-resistant sublines PTR0.03 (•), PTR0.05 (▪), and PTR0.5 (▴) were exposed to increasing concentrations of GW1843 (A) or pemetrexed (MTA) (B) as detailed in “Materials and methods.” Following 3 days of incubation at 37°C, viable cell counting was performed using trypan blue exclusion. Values depicted represent means ± SD of 3 to 5 independent experiments.
Antifolate growth inhibition of parental human leukemia CCRF-CEM cells and their various PT523-resistant sublines. Exponentially growing parental CCRF-CEM (WT, ○) cells and their various PT523-resistant sublines PTR0.03 (•), PTR0.05 (▪), and PTR0.5 (▴) were exposed to increasing concentrations of GW1843 (A) or pemetrexed (MTA) (B) as detailed in “Materials and methods.” Following 3 days of incubation at 37°C, viable cell counting was performed using trypan blue exclusion. Values depicted represent means ± SD of 3 to 5 independent experiments.
DNA sequencing
Genomic PCR products obtained with DNA from antifolate-resistant sublines showing an altered electrophoretic mobility pattern of the RFC gene in 3 independent experiments were fractionated on 1% agarose gels and purified using the gel purification kit of Qiagen. These genomic PCRs were performed with nonradioactive dNTPs. DNA sequencing was performed on purified RFC fragments by the fluorescent dideoxy chain termination method using anABI 377 sequencer (DNA sequencing unit, the Weizmann Institute, Rehovot, Israel). At least 3 independent genomic PCRs and sequencing performed on different days were done to determine a definitive mutation.
[3H]MTX transport
To determine the ability of the various antifolate-resistant sublines to take up antifolates we measured the initial influx rates of [3H]MTX relative to parental CCRF-CEM cells. Cells (2 × 107) from the mid-log phase of growth were washed 3 times in transport buffer consisting of HEPES-buffered saline solution (HBSS)9,18 and incubated at 37°C for 3 minutes in the same buffer (1-mL suspensions) containing 2 μM [3H]MTX. Transport controls contained a 500-fold excess (1 mM) of unlabeled MTX. The transport of [3H]MTX was terminated by the addition of 10 mL ice-cold HBSS. Then, the cell suspension was centrifuged at 500g for 5 minutes at 4°C and the cell pellet was washed twice with 10 mL ice-cold transport buffer. The final cell pellet was suspended in water and processed for scintillation counting.
Western blot analysis
To determine the levels of RFC expression in the various antifolate-resistant cell lines, we first isolated microsomes as previously described.22,23 Microsomal proteins were resolved by electrophoresis on 10% polyacrylamide gels containing SDS and electroblotted onto nitrocellulose nylon membrane (Schleicher and Schuell, Keene, NH). The blots were then blocked for 1 hour at room temperature in TBS buffer (150 mM NaCl, 0.5% Tween 20, and 10 mM Tris-Cl at pH 8.0) containing 1% skim milk. The blots were reacted with a polyclonal antiserum (1:700) prepared in mice against a human RFC peptide.9 Blots were then rinsed in the same buffer for 10 minutes at room temperature and reacted with horseradish peroxidase-conjugated goat anti-mouse IgG (1:8000 dilution, Jackson ImmunoResearch Labs, West Grove, PA) for 1 hour at room temperature. Following three 10-minute washes in TBS at room temperature, enhanced chemiluminescence detection was performed according to the manufacturer's instructions (Biological Industries). Protein content was determined using the Bio-Rad protein assay of Bradford.24
Results
Cross-resistance to various antifolates in PT523-resistant variants
The gradual acquisition of stepwise resistance to PT523 was consistently accompanied by increasing levels of cross-resistance to various hydrophilic antifolates that use RFC for their cellular entry; these include GW1843 (Figure 1A), pemetrexed (MTA; Figure 1B) as well as MTX, lometrexol (DDATHF), and ZD9331 (Table 1). Hence, the gradual increase in PT523 concentrations resulted in up to about 3500-fold resistance to this novel antifolate and up to 900-fold cross-resistance to hydrophilic antifolates, which primarily target single folate-dependent enzymes including DHFR (PT523 and MTX), thymidylate synthase (TS; GW1843 and ZD9331), and glycinamide ribonucleotide formyltransferase (GARTF; lometrexol) or multiple enzymes including TS, GARTF, and DHFR (pemetrexed; Figure 1; Table 1). In contrast, the gradual acquisition of PT523 resistance was inversely associated with a gradual increase in the collateral sensitivity to trimetrexate (TMQ), a lipophilic antifolate that like its homologue piritrexim enters cells by diffusion and is therefore independent of the RFC for its entry25,26 ; PT523-resistant variants displayed up to 9-fold hypersensitivity to TMQ (Table 1).
Antifolate growth inhibition in parental CCRF-CEM cells and their PT523-resistant sublines
Antifolate . | WT . | PTR0.003 . | PTR0.05 . | PTR0.5 . |
|---|---|---|---|---|
| PT523 | 0.43 ± 0.19 | 3.7 ± 2 (8.7) | 91.7 ± 15.1 (216) | 1471 ± 251 (3461) |
| MTX | 0.86 ± 0.5 | 9.7 ± 5.8 (11.3) | 28.1 ± 16.1 (33) | 512 ± 153 (593) |
| GW1843 | 0.5 ± 0.1 | 2.1 ± 0.4 (4.1) | 26.5 ± 5.3 (53) | 440 ± 167 (881) |
| Pemetrexed (MTA) | 8.1 ± 3.1 | 24.1 ± 0.6 (3) | 208 ± 34.2 (25) | 1063 ± 264 (131) |
| Lometrexol (DDATHF) | 13.5 ± 6.9 | 31.6 ± 8.9 (2.3) | 595 ± 126 (44) | 6945 ± 1362 (514) |
| ZD9331 | 17.6 ± 4.6 | 62 ± 3.9 (3.5) | 874 ± 101 (50) | 4557 ± 984 (259) |
| TMQ | 15.4 ± 6.1 | 3.2 ± 1.4 (0.21) | 2.7 ± 0.5 (0.17) | 1.7 ± 0.4 (0.11) |
Antifolate . | WT . | PTR0.003 . | PTR0.05 . | PTR0.5 . |
|---|---|---|---|---|
| PT523 | 0.43 ± 0.19 | 3.7 ± 2 (8.7) | 91.7 ± 15.1 (216) | 1471 ± 251 (3461) |
| MTX | 0.86 ± 0.5 | 9.7 ± 5.8 (11.3) | 28.1 ± 16.1 (33) | 512 ± 153 (593) |
| GW1843 | 0.5 ± 0.1 | 2.1 ± 0.4 (4.1) | 26.5 ± 5.3 (53) | 440 ± 167 (881) |
| Pemetrexed (MTA) | 8.1 ± 3.1 | 24.1 ± 0.6 (3) | 208 ± 34.2 (25) | 1063 ± 264 (131) |
| Lometrexol (DDATHF) | 13.5 ± 6.9 | 31.6 ± 8.9 (2.3) | 595 ± 126 (44) | 6945 ± 1362 (514) |
| ZD9331 | 17.6 ± 4.6 | 62 ± 3.9 (3.5) | 874 ± 101 (50) | 4557 ± 984 (259) |
| TMQ | 15.4 ± 6.1 | 3.2 ± 1.4 (0.21) | 2.7 ± 0.5 (0.17) | 1.7 ± 0.4 (0.11) |
Values represent IC50, the drug concentration that inhibits cell growth by 50% following a 72-hour antifolate drug exposure. Results depicted are the mean (nM) ± SEM of 3 experiments.
Values in parentheses represent fold of antifolate resistance relative to parental cells.
Initial rates of [3H]MTX uptake in CCRF-CEM cells and their various PT523-resistant sublines. Exponentially growing cells were washed, resuspended in a HEPES-buffered saline solution at pH 7.4 containing 2 μM [3H]MTX, and incubated at 37°C for 3 minutes. Cells were then washed 3 times with ice-cold buffer, centrifuged, lysed, and processed for scintillation counting as described in “Materials and methods.” Influx rates are given as pmol [3H]MTX/min/107 cells and are presented as the means ± SD of 3 to 6 independent experiments performed on different days. The influx rate of [3H]MTX in parental (WT) cells was 3.9 ± 0.64 pmol [3H]MTX/min/107 cells. The influx rate of WT cells was referred to as 100% and the residual percentage of [3H]MTX transport in the various sublines relative to parental cells is also depicted.
Initial rates of [3H]MTX uptake in CCRF-CEM cells and their various PT523-resistant sublines. Exponentially growing cells were washed, resuspended in a HEPES-buffered saline solution at pH 7.4 containing 2 μM [3H]MTX, and incubated at 37°C for 3 minutes. Cells were then washed 3 times with ice-cold buffer, centrifuged, lysed, and processed for scintillation counting as described in “Materials and methods.” Influx rates are given as pmol [3H]MTX/min/107 cells and are presented as the means ± SD of 3 to 6 independent experiments performed on different days. The influx rate of [3H]MTX in parental (WT) cells was 3.9 ± 0.64 pmol [3H]MTX/min/107 cells. The influx rate of WT cells was referred to as 100% and the residual percentage of [3H]MTX transport in the various sublines relative to parental cells is also depicted.
Markedly impaired [3H]MTX transport and diminished intracellular folate pools in the PT523-resistant variants
The pattern of high-level cross-resistance to various hydrophilic antifolates that use RFC for their cellular entry but collateral hypersensitivity to the lipophilic antifolate TMQ was consistent with an impaired antifolate uptake in the PT523-resistant variants. Hence the initial rates of [3H]MTX transport were determined; the acquisition of only 9-fold PT523 resistance in the PTR0.003 variant (Table 1) was associated with as high as about 87% loss of [3H]MTX influx, relative to parental cells (Figure 2).
Further stepwise selection in increasing PT523 concentrations resulted in a further gradual decrease in [3H]MTX transport thereby attaining an approximate 99% loss of [3H]MTX influx in PTR0.5 and PTR2.5 cells (Figure 2). This profound impairment of MTX uptake consequently resulted in up to 92% depletion in the total cellular folate pools in PTR0.5 cells (Table 2); the shrunken cellular reduced folate pool was consistent with the prominent hypersensitivity of PT523-resistant variants to the lipid-soluble antifolate, TMQ (Table 1).
Intracellular reduced folate pools in parental CCRF-CEM cells and their PT523-resistant sublines
Cell line . | CH2THF + THF . | 5-CH3THF . | DHF . | 10-CHO-THF . | Total . |
|---|---|---|---|---|---|
| WT | 24 ± 1 | 9.9 ± 3.1 | 0.3 ± 0.1 | 37 ± 2 | 70 ± 3 |
| PTR0.003 | 19.9 ± 2.7 | 3.6 ± 0.4 | 2.1 ± 1.0 | 18.1 ± 5.5 | 42.4 ± 6.5 |
| PTR0.05 | 13.9 ± 1.3 | 2.4 ± 0.4 | 11.3 ± 1.8 | 7.3 ± 2.4 | 34.9 ± 3.2 |
| PTR0.5 | 1.6 ± 0.3 | 0.4 ± 0.2 | 3.0 ± 0.9 | 0.8 ± 0.3 | 5.8 ± 0.5 |
Cell line . | CH2THF + THF . | 5-CH3THF . | DHF . | 10-CHO-THF . | Total . |
|---|---|---|---|---|---|
| WT | 24 ± 1 | 9.9 ± 3.1 | 0.3 ± 0.1 | 37 ± 2 | 70 ± 3 |
| PTR0.003 | 19.9 ± 2.7 | 3.6 ± 0.4 | 2.1 ± 1.0 | 18.1 ± 5.5 | 42.4 ± 6.5 |
| PTR0.05 | 13.9 ± 1.3 | 2.4 ± 0.4 | 11.3 ± 1.8 | 7.3 ± 2.4 | 34.9 ± 3.2 |
| PTR0.5 | 1.6 ± 0.3 | 0.4 ± 0.2 | 3.0 ± 0.9 | 0.8 ± 0.3 | 5.8 ± 0.5 |
Values represent the mean (pmol/mg protein) ± SEM from 6 determinations performed on 3 separate samples.
Identification of multiple RFC mutations in PT523-resistant cells
The antifolate-resistance phenotype in PTR0.5 cells was stable for at least 6 months in PT523-free medium, thereby suggesting the presence of stable genomic alterations such as inactivating RFC mutations. Thus, genomic PCR-SSCP analysis was performed to screen for potential RFC mutations.9 Whereas the sublines PTR0.003 and PTR0.05 had a parental SSCP pattern, PTR0.5 and PTR2.5 displayed an altered pattern in RFC exon 3 (Figure 3 arrowhead).
Genomic PCR-SSCP analysis of RFC exon 3 in parental cells and their PT523-resistant sublines. High-molecular-weight gDNA (∼50 ng) isolated from parental CCRF-CEM cells and their various antifolate-resistant sublines was PCR amplified in the presence of [32P]dCTP using oligonucleotide primers targeting exon 3. The PCR products were resolved by electrophoresis on nondenaturing, 8% polyacrylamide gels, as described in “Materials and methods.” The arrowhead shown on the right side depicts the band with the altered electrophoretic mobility.
Genomic PCR-SSCP analysis of RFC exon 3 in parental cells and their PT523-resistant sublines. High-molecular-weight gDNA (∼50 ng) isolated from parental CCRF-CEM cells and their various antifolate-resistant sublines was PCR amplified in the presence of [32P]dCTP using oligonucleotide primers targeting exon 3. The PCR products were resolved by electrophoresis on nondenaturing, 8% polyacrylamide gels, as described in “Materials and methods.” The arrowhead shown on the right side depicts the band with the altered electrophoretic mobility.
Cloning of the genomic PCR products and sequencing of multiple clones revealed that one RFC allele contained 4 single nucleotide substitutions in both PTR0.5 and PTR2.5 cells; it should be emphasized that whereas CCRF-CEM cells contained 3 RFC alleles, its PTR0.5 and PTR2.5 sublines harbored one mutant allele with 4 mutations and 2 normal RFC alleles (Table 3). Specifically, these variants harbored a cluster of 4 nearly consecutive mutations including L143P, A147V, R148G, and Q150Stop (Table 3). These mutations were predicted to reside in the fourth transmembrane domain (TMD) and in intracellular loop 4. Hence whereas PTR0.5 and PTR2.5 cells contained a cluster of 4 mutations on a single RFC allele including a premature translation termination, this mutant allele was undetectable by both genomic PCR-SSCP analysis and DNA sequencing in the preceding variants PTR0.05 and PTR0.003 (Figure 3).
Summary of mutations in the various PT523-resistant sublines
Nucleotide alteration* . | Amino acid alteration . | Transporter location . |
|---|---|---|
| Thymidine 523 to cytosine (T523C) | Leu143Pro | TMD4 |
| C535T | Ala147Val | L4 |
| C537G | Arg148Gly | L4 |
| C543T | Gln150Stop | L4 |
Nucleotide alteration* . | Amino acid alteration . | Transporter location . |
|---|---|---|
| Thymidine 523 to cytosine (T523C) | Leu143Pro | TMD4 |
| C535T | Ala147Val | L4 |
| C537G | Arg148Gly | L4 |
| C543T | Gln150Stop | L4 |
Nucleotide positions are numbered according to accession number U19720
Loss of RFC gene and protein expression in PT523-resistant variants
Because a profound loss (87%-99%) of [3H]MTX transport occurred either in the absence (PTR0.003 and PTR0.05) or presence of heterozygous RFC mutations (PTR0.5 and PTR2.5), RFC mRNA and protein levels were determined by Northern (Figure 4) and Western blot analyses, respectively (Figure 5).
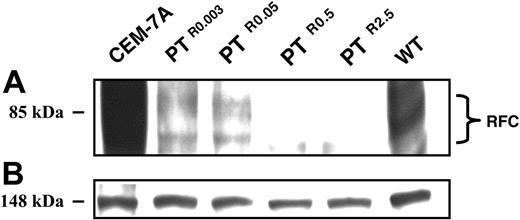
Relative to parental CCRF-CEM cells, PTR0.003, PTR0.05, PTR0.5, and PTR2.5 displayed a gradual decrease in RFC mRNA levels with residual transcript levels of about 35%, 20%, 5%, and 1%, respectively (Figure 4A); CEM-7A cells with 50-fold elevation in RFC mRNA levels due to RFC gene amplification23 were used as a positive control (Figure 4A lane a). The identical levels of 28S rRNA confirmed that equal amounts of RNA were being analyzed in the various PT523-resistant sublines (Figure 4B). Consequently, a prominent decrease in RFC protein expression was already apparent in PTR0.003 and PTR0.05 cells, whereas PTR0.5 and PTR2.5 cells showed a nearly complete loss of RFC protein expression (Figure 5A); the equal levels of an unrelated 148-kDa protein observed on Panceau S staining of the Western blot confirmed that equal amounts of membrane proteins were being analyzed in the various antifolate-resistant sublines (Figure 5B).
RFC allele loss in PT523-resistant variants
Because at low levels of PT523 resistance (eg PTR0.05) cells did not contain a mutant allele but surprisingly displayed an 80% decrease in RFC mRNA levels (Figure 4A) and a 91% loss of MTX transport (Figure 2), we explored the possibility that a WT RFC allele was deleted. Using a [32P]-labeled genomic RFC probe, Southern blot analysis revealed that all the PT523-resistant variants had an approximate 50% decrease in the intensity of the RFC band, thereby establishing an RFC allele loss (Figure 6A). Furthermore, following digestion of gDNA with 4-cutter restriction enzymes (MfeI and MspI), Southern blot analysis revealed that WT CCRF-CEM cells had a 3-band hybridization pattern (Figure 6C). In contrast, PT523-resistant variants showed only a single-band pattern (Figure 6C arrowhead), thereby suggesting that RFC allele loss was associated with genomic rearrangements. Ethidium bromide staining of an about 1.5-kb band representing a repetitive DNA sequence was used to normalize for loading differences (Figure 6B).
Discussion
The acquisition of resistance to the novel antifolate PT523 via loss of antifolate transport results from several molecular mechanisms including mutational RFC inactivation, allele loss, and transcriptional silencing as detailed.
Northern blot analysis of RFC mRNA levels in parental CCRF-CEM cells and their PT523-resistant sublines. Total RNA (30 μg, except for RFC overexpressing CEM-7A cells, which contained only 6 μg) from WT cells and the various PT523-resistant sublines was fractionated by electrophoresis on 1.2% agarose gels containing formaldehyde, blotted onto Zeta-Probe-GT nylon membrane (Bio-Rad) and probed with a [32P]oligolabeled full-length RFC cDNA13 (A). The blots were then washed under high stringency conditions and exposed to x-ray films. The intensity of the 3-kb RFC transcript was determined by scanning densitometry and normalized to the methylene blue staining of the 18S and 28S rRNA bands (B). The ratio of the intensity of the RFC transcript in each cell line versus WT cells is depicted in the bottom of panel A.
Northern blot analysis of RFC mRNA levels in parental CCRF-CEM cells and their PT523-resistant sublines. Total RNA (30 μg, except for RFC overexpressing CEM-7A cells, which contained only 6 μg) from WT cells and the various PT523-resistant sublines was fractionated by electrophoresis on 1.2% agarose gels containing formaldehyde, blotted onto Zeta-Probe-GT nylon membrane (Bio-Rad) and probed with a [32P]oligolabeled full-length RFC cDNA13 (A). The blots were then washed under high stringency conditions and exposed to x-ray films. The intensity of the 3-kb RFC transcript was determined by scanning densitometry and normalized to the methylene blue staining of the 18S and 28S rRNA bands (B). The ratio of the intensity of the RFC transcript in each cell line versus WT cells is depicted in the bottom of panel A.
Western blot analysis of RFC expression in CCRF-CEM cells and their PT523-resistant sublines. Microsomes were isolated from exponentially growing cells and proteins were extracted using a Triton X-100-containing buffer as detailed in “Materials and methods.” Aliquots of microsomal proteins (75 μg) from RFC overxepressing CEM-7A cells, WT cells, and their PT523-resistant sublines were resolved by electrophoresis on 10% polyacrylamide gels containing SDS and electroblotted onto a Nytran nylon membrane (Schleicher and Schuell). Then, the blots were reacted with a peptide antiserum against a C-terminal peptide of human RFC9 and detected by enhanced chemiluminescence (A). An internal 148-kDa protein detected during the Panceau S staining was used to normalize for any differences in protein loading (B).
Western blot analysis of RFC expression in CCRF-CEM cells and their PT523-resistant sublines. Microsomes were isolated from exponentially growing cells and proteins were extracted using a Triton X-100-containing buffer as detailed in “Materials and methods.” Aliquots of microsomal proteins (75 μg) from RFC overxepressing CEM-7A cells, WT cells, and their PT523-resistant sublines were resolved by electrophoresis on 10% polyacrylamide gels containing SDS and electroblotted onto a Nytran nylon membrane (Schleicher and Schuell). Then, the blots were reacted with a peptide antiserum against a C-terminal peptide of human RFC9 and detected by enhanced chemiluminescence (A). An internal 148-kDa protein detected during the Panceau S staining was used to normalize for any differences in protein loading (B).
Introduction of a premature translation termination codon into the RFC gene
PTR0.5 and PTR2.5 cells contained an allele with 3 single amino acid substitutions and in addition, a premature translation termination. Recent studies have indicated that an mRNA species containing a stop codon is rapidly degraded via a quality control mechanism known as nonsense-mediated mRNA decay (NMD).27,28 The objective of the cells in using this quality control mechanism is to destroy mRNA species that contain premature termination codons such that only full-length, functional proteins are produced. Thus, it is most likely that the nonsense RFC mutation present in these PT523-resistant cells is recognized by the NMD machinery that presumably degrades this mutant RFC transcript.
Loss of a WT RFC allele
Southern blot analysis revealed that all the PT523-resistant sublines displayed a genomic deletion of a WT RFC allele. This RFC allele loss is consistent with 2 previous studies. The first involves MTX-resistant human erythroleukemia cells in which a nearly complete loss of RFC transcripts was observed due to a genomic deletion of RFC alleles.29 Interestingly, genomic deletion of RFC alleles in these erythroleukemia cells was associated with chromosomal aberrations including translocations. Consistently, in the current study we also find that RFC allele loss was associated with genomic rearrangements. The second study focused on cervical carcinoma HeLa cells exposed to MTX selection after treatment with the chemical mutagen ethylmethanesulfonate; these cells showed a complete deletion of the genomic RFC locus.30 In the current study, deletion of a WT RFC allele in PT523-resistant cells clearly contributed to the loss of RFC mRNA in PTR0.5 and PTR2.5 cells. It should be further noted that RFC deletions were already present during the early stages of PT523 selection (eg, PTR0.05) in which RFC transcript levels were decreased by 80%, whereas no mutations were detectable. These findings reinforce the contributing role genomic RFC deletions may play in the loss of RFC gene expression.
Southern blot analysis of RFC gene copy number in parental WT cells and their PT523-resistant variants. High-molecular-weight gDNA (15 μg) was codigested with MfeI and XmaI (a 6-cutter; A) or MfeI and MspI (a 4-cutter; C), fractionated by electrophoresis on 0.8% agarose gels, and transferred to Zeta-Probe-GT nylon membranes. The blots were then hybridized with a [32P]labeled 5′-genomic hRFC probe as detailed in “Materials and methods.” Following hybridization at 65°C, the membranes were washed under high stringency conditions and exposed to x-ray film. The intensity of the RFC band in panels A and C was estimated by scanning densitometry and normalization to an approximate 1.5-kb repetitive DNA band observed after ethidium bromide staining (B). The band obtained in panel A and the middle band observed in panel C (arrowhead) are both approximately 600 bp in length. The arrows shown on the right side in panel C denote the WT bands that were lost in the PT523-resistant cells.
Southern blot analysis of RFC gene copy number in parental WT cells and their PT523-resistant variants. High-molecular-weight gDNA (15 μg) was codigested with MfeI and XmaI (a 6-cutter; A) or MfeI and MspI (a 4-cutter; C), fractionated by electrophoresis on 0.8% agarose gels, and transferred to Zeta-Probe-GT nylon membranes. The blots were then hybridized with a [32P]labeled 5′-genomic hRFC probe as detailed in “Materials and methods.” Following hybridization at 65°C, the membranes were washed under high stringency conditions and exposed to x-ray film. The intensity of the RFC band in panels A and C was estimated by scanning densitometry and normalization to an approximate 1.5-kb repetitive DNA band observed after ethidium bromide staining (B). The band obtained in panel A and the middle band observed in panel C (arrowhead) are both approximately 600 bp in length. The arrows shown on the right side in panel C denote the WT bands that were lost in the PT523-resistant cells.
Down-regulation of RFC gene expression via transcriptional silencing
PT523-resistant cells were characterized by loss of function of various transcription factors that promote transcriptional transactivation of the RFC gene under physiologic conditions.16 Thus, electrophoretic mobility shift analysis with oligonucleotides representing consensus-binding sites for various transcriptions factors revealed markedly decreased binding16,31 ; these cis-acting elements include CRE, E-box, Mzf-1, GC-box, and AP-2. This was apparently due to decreased levels of phosphorylated CREB-1, USF-1, and AP-2α.16 Furthermore, these results are in accord with our recent studies demonstrating the loss of function of multiple transcription factors in multiple cell lines with resistance to various antifolates including PT523.15,16,31 Surprisingly however, whereas GC-box binding was severely abolished in PTR0.5 cells, normal Sp1 levels were maintained. Hence, based on a recent paper from our laboratory,17 it is likely that posttranslational modifications such as inhibitory phosphorylation may take place in PT523-resistant cells that disrupt the transactivation capability of Sp1 thereby resulting in down-regulation of RFC gene expression. It should be emphasized that in the current study, a thorough DNA methylation analysis of the RFC promoter region in the various PT523-resistant sublines failed to identify any promoter methylation using both the bisulfite sequencing assay as well as methylation-sensitive restriction enzymes on Southern blot analysis. This finding is in accord with our previous study that also failed to identify any RFC promoter methylation with multiple antifolate-resistant cell lines in which RFC gene expression was prominently repressed.16 This is further consistent with the lack of promoter methylation in 40 DNA specimens from newly diagnosed and relapsed childhood acute lymphoblastic leukemia patients (Y.K. and Y.G.A., unpublished data, October 2002).
Here we identified an extremely rare cluster of 4, nearly consecutive, single nucleotide substitutions within a stretch of 20 nucleotides in exon 3 of the RFC gene in PT523-resistant cells. This finding is consistent with our recent studies with GW1843-resistant13 and MTX-resistant human leukemia cells,9 in which 3 single nucleotide substitutions were observed within 52 and 119 nucleotides in RFC exons 2 and 3, respectively. Based on a spontaneous mutation rate of about 10-7 in mammalian cells, the probability of emergence of 3 to 4 independent RFC mutations residing on a single allele is an extremely rare event (∼10-21-10-28 per gene/generation). However, these cell lines suffered a severe folate deprivation resulting from increasing antifolate pressure; this was reflected in their diminished folate pools comprising as low as 8.3% of parental cells.9 Recent studies have shown that under conditions of folate deprivation, multiple genetic abnormalities including nucleotide misincorporation may be a frequent aberration.32 Thus, it is most likely that during DNA replication under conditions of folate deprivation and consequent purine-pyrimidine pool imbalance, DNA polymerase α may introduce a relatively short stretch of misincorporated nucleotides. Based on an elongation rate of DNA polymerase α estimated at 2200 nucleotides/min, the synthesis of such short stretches of mutant DNA during the S phase may be a matter of a few seconds at most. Interestingly, apart from the nonsense mutation, the rest of the single amino acid substitutions in the PT523-resistant cells appear to have by themselves, adverse functional consequences on MTX transport. Thus, Leu143Pro, the first among the 4 mutations resides in the end of TMD4.33 In this respect, it was demonstrated that the insertion of a proline residue disrupts transmembrane α-helices, especially when this imino acid is present in the end of the TMD.34 Indeed, the introduction of this potent α-helix breaker in TMD4 of the murine RFC in MTX-resistant L1210 leukemia cells resulted in the complete loss of MTX transport due to the lack of carrier mobility.35 Furthermore, the third mutation in PT523-resistant cells involved an Arg148Gly substitution in the fourth intracellular loop that links TMD4 to TMD5. The loss of this positively charged amino acid residue in this loop could also inflict a transport alteration as shown by Roy et al.36 Thus, even under rare conditions of stop codon read-through, this mutant RFC allele presumably preserved the potency of imparting loss of antifolate transport. Hence the presence of such a mutant RFC allele clearly conferred on individual clones, a selective growth advantage resulting in their expansion and dominance.
Aneuploidy is one of the established hallmarks of cancer. Among the different chromosomal abnormalities are various trisomies. Owing to nonrandom gains of chromosomes, trisomies are present in many cases of leukemia and other malignancies.37 Because the human RFC gene maps to a telomeric region in chromosome 21 (21q22.3),38 the well-established trisomies of chromosome 21 will necessarily result in the gain of extra allele(s) of the RFC gene. Hence acquisition of antifolate resistance in such human leukemia cells harboring 3 or more copies of the RFC gene may be associated with various quantitative and qualitative alterations that disrupt the maintenance of the proper gene copy number, gene expression, and the functionality of the encoded antifolate influx transporter. Consistently, our current study is the first to establish the simultaneous coexistence of both genomic deletions, transcriptional silencing as well as mutational inactivation in human leukemia cells harboring 3 copies of the RFC gene. Hence, these findings warrant further clinical evaluation of the frequency of emergence of these RFC gene inactivation modalities in specimens from patients with ALL harboring trisomy 21 before and after MTX-containing chemotherapy. Importantly however, the finding here that leukemia cells displaying as high as 3500-fold resistance to certain hydrophilic antifolates but at the same time 9-fold hypersensitivity to the lipophilic antifolate TMQ offers a potentially efficient treatment strategy of antifolate-resistant leukemias.
Prepublished online as Blood First Edition Paper, December 20, 2005; DOI 10.1182/blood-2005-10-4048.
Supported by research grants of the Israel Cancer Association and the Star Foundation (Y.G.A.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Prof David G. Priest and Marlene A. Bunni for the folate pool analysis.


![Figure 2. Initial rates of [3H]MTX uptake in CCRF-CEM cells and their various PT523-resistant sublines. Exponentially growing cells were washed, resuspended in a HEPES-buffered saline solution at pH 7.4 containing 2 μM [3H]MTX, and incubated at 37°C for 3 minutes. Cells were then washed 3 times with ice-cold buffer, centrifuged, lysed, and processed for scintillation counting as described in “Materials and methods.” Influx rates are given as pmol [3H]MTX/min/107 cells and are presented as the means ± SD of 3 to 6 independent experiments performed on different days. The influx rate of [3H]MTX in parental (WT) cells was 3.9 ± 0.64 pmol [3H]MTX/min/107 cells. The influx rate of WT cells was referred to as 100% and the residual percentage of [3H]MTX transport in the various sublines relative to parental cells is also depicted.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/8/10.1182_blood-2005-10-4048/4/m_zh80080694130002.jpeg?Expires=1769690494&Signature=nPaffmojIlQQIE6hEuHErQkfFuu5odkXfYRtxm-yKN4Syms2aQhQHy70jTdCJ-6sQjdA~jAN2XUke-KK7i3HcUJ8hguPn7RN6bMkZK2jYfnEUZT1aEVXnkvLm6PkVhlHnVTsddUKVOOHJpxlUZuLaACf~qcTwHUQIB7cHTBUEZptfmOBERdJEbnJSBK3QAWZmJrTfXqyDZTnosZ4Lqk2szSlW0tExhibUdEPx3uRNyd2ljymKI0o53GDWwvRyOr7SmibCf5LYBjxri6F1bRnGg7lHpfR0H4JrfRLooxF5F7XRzANuXDz5K67mvw2Lpid4e5Kf0sCP1p9SBwRSGgCPQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Genomic PCR-SSCP analysis of RFC exon 3 in parental cells and their PT523-resistant sublines. High-molecular-weight gDNA (∼50 ng) isolated from parental CCRF-CEM cells and their various antifolate-resistant sublines was PCR amplified in the presence of [32P]dCTP using oligonucleotide primers targeting exon 3. The PCR products were resolved by electrophoresis on nondenaturing, 8% polyacrylamide gels, as described in “Materials and methods.” The arrowhead shown on the right side depicts the band with the altered electrophoretic mobility.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/8/10.1182_blood-2005-10-4048/4/m_zh80080694130003.jpeg?Expires=1769690494&Signature=U8XYKNn88l0sqeHo8djS1F~0bL4LTrgmDS2pG4FkkuPw3Z2CnQ1LwKJUodQDUkH7MAj4tERqV1ENcCne1RThm8omYJPcpjlIlVMk9HgHx~OQNW41aS6553cyuvM8EPnWj9BcBY1GAt4-LAnSrzWQJZHDVtNnbiAYMmtc-nwn7G6TNnFQO8HCFyEksTgbePYYYC1nQCHDvbPbRuFb9NkbcqPWW4NXpXvpGKjTHUpS3N~Hpkq93j6UdvwDdLt0bbMhjlm27ZA-xvkt5PajJmWCNol5G-M3x4k45xVVoi2zVkzzjy5UUQzLxu3qPzcHH-u1shLIJJ0MDXZJHXe8lbco9w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Northern blot analysis of RFC mRNA levels in parental CCRF-CEM cells and their PT523-resistant sublines. Total RNA (30 μg, except for RFC overexpressing CEM-7A cells, which contained only 6 μg) from WT cells and the various PT523-resistant sublines was fractionated by electrophoresis on 1.2% agarose gels containing formaldehyde, blotted onto Zeta-Probe-GT nylon membrane (Bio-Rad) and probed with a [32P]oligolabeled full-length RFC cDNA13 (A). The blots were then washed under high stringency conditions and exposed to x-ray films. The intensity of the 3-kb RFC transcript was determined by scanning densitometry and normalized to the methylene blue staining of the 18S and 28S rRNA bands (B). The ratio of the intensity of the RFC transcript in each cell line versus WT cells is depicted in the bottom of panel A.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/8/10.1182_blood-2005-10-4048/4/m_zh80080694130004.jpeg?Expires=1769690494&Signature=n98TFG8v9CmfT0mV~Y55YS4iuvXXw2lR1Nqt4AQtOL5LbmJ3~UIwnWpcbvfBJpG1fF7JlkywqH9nYFY7O~IDMxiNcWco8uDOF~B9DnnyxDFrmneQH9-cdqbjew-t395np-F3UW6A7gfHB5YX5VywMYixzxKfH0MBoluV2on1mOQm32Jyh7-QCNg51wLijUzXfKCQQ02VYQFY8V6VZnwj3o5ENJPnm3cpVgJK83P~srcsFi05d-PxEAW1bEtCAkT2AcoWY1FfHdar2s48w121jBpez0UN9JWp5faKHxZjjhEdU4wNq1HLMwm09~LDeJsBICNDz2fAmFTH-JiL1RgvSg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Southern blot analysis of RFC gene copy number in parental WT cells and their PT523-resistant variants. High-molecular-weight gDNA (15 μg) was codigested with MfeI and XmaI (a 6-cutter; A) or MfeI and MspI (a 4-cutter; C), fractionated by electrophoresis on 0.8% agarose gels, and transferred to Zeta-Probe-GT nylon membranes. The blots were then hybridized with a [32P]labeled 5′-genomic hRFC probe as detailed in “Materials and methods.” Following hybridization at 65°C, the membranes were washed under high stringency conditions and exposed to x-ray film. The intensity of the RFC band in panels A and C was estimated by scanning densitometry and normalization to an approximate 1.5-kb repetitive DNA band observed after ethidium bromide staining (B). The band obtained in panel A and the middle band observed in panel C (arrowhead) are both approximately 600 bp in length. The arrows shown on the right side in panel C denote the WT bands that were lost in the PT523-resistant cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/8/10.1182_blood-2005-10-4048/4/m_zh80080694130006.jpeg?Expires=1769690494&Signature=4tjMsT65aPk4kn3E-GKbWH44ef8pFfQaEKSDJv-IhKh9LOLD7k6GqC13pFoeEX2BIOp1c4CIcbi4HV1kLJn-4-mpFOkoE2nBEzfjOUjmVHea6GoduSrARJcHPxR1Qb3ZjKddQC0JvpLmpgvznQ1cNlTy~5uBF795nsxyhpEt-qPvOt3cFf8Q6zjXRSFiBaQ2tQIiXuOy5B6kirqHjABanJB~l2h9WTyTNYPVL1lP3aDjUSL1VhUBTNnDXCAvav9R7tOSr2NIa-eja74IlimnkH-VYShd8Z68xNjGD7~3jp-5ODKJ5bBDbsRRfRUJa3BwIO3Vzi1kCag9GVAPQ92QSg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal