Abstract
Vα14i natural killer T (NKT)–cell function has been implicated in a number of disease conditions. The molecular events that drive Vα14i NKT-cell development remain elusive. We recently showed that T-bet is required for the terminal maturation of these cells. Here we identify some of the genetic targets of T-bet during Vα14i NKT-cell lineage development. Microarray gene-expression analyses on developing Vα14i NKT cells were performed and provide a molecular framework to study these maturation events. In vitro ectopic expression of T-bet in immature Vα14i NKT cells, which do not yet express T-bet, was sufficient to promote Vα14i NKT-cell maturation, driving the expression of multiple genes, including those that participate in migration, survival, and effector functions. By regulating the expression of T-helper 1 (Th1)–associated cytokines, chemokines, chemokine receptors, and molecules involved in cytolysis, T-bet defines the unique lineage attributes of mature Vα14i NKT cells and acts to link these attributes to a developmental process.
Introduction
Natural killer T (NKT) cells are a unique lymphocyte lineage that coexpresses a rearranged T-cell receptor (TCR) in association with the CD3 complex as well as several receptors first identified on bona fide NK cells, such as NK1.1, the interleukin-2 (IL-2)/15Rβ chain (CD122) and various Ly49 molecules.1 Most NKT cells in the mouse express a semi-invariant TCR composed of a Vα14-Jα18 rearrangement that preferentially associates with either Vβ8, Vβ7, or Vβ2,2 and are positively selected by CD1d, a nonclassical major histocompatibility (MHC) class I molecule. We refer to this CD1d reactive subgroup as Vα14 invariant (Vα14i) NKT cells to distinguish them from other populations of NKT cells that have been defined.3,4 The early and potent cytokine secretion by Vα14i NKT cells following TCR stimulation may provide an important link between the innate and adaptive immune systems.3,5 Consistent with this, Vα14i NKT cells appear to be important for responses to tumors, infectious agents, the maintenance of self tolerance, and the prevention of autoimmunity.1,3
Recent progress has been made in identifying Vα14i NKT-cell developmental intermediates.6-9 Vα14i NKT cells develop from CD4+CD8+ double-positive (DP) Vα14-Jα18 TCR+ thymocyte precursors.6,10-12 Following positive selection by CD1d molecules, Vα14i NKT cells undergo a maturation process defined by the sequential up-regulation of diverse markers and the late fate specification of the NKT-cell lineage.6-9 NKT-cell precursors can be identified on the basis of TCR specificity in the thymus of young mice using the CD1d tetramer. These cells are phenotypically indistinguishable from conventional mature lymphocytes, being HSAlow, CD44low, and NK1.1–.7 This phenotype is defined as stage 1.13 Ontogeny and transfer studies defined a developmental sequence from CD44low NK1.1– (stage 1) to CD44high NK1.1– (stage 2). At this latter stage, Vα14i NKT cells are still considered immature but can leave the thymus and colonize peripheral organs.7,8 A final maturation step that occurs either in the thymus or in the peripheral organs is accompanied by the expression of NK1.1, Ly49 receptors and CD122 (stage 3).7,8,14 Interestingly, these 3 developmental intermediates each have different cytokine secretion potential. During progression through these developmental stages, the cells show a T-helper (Th) Th2 to Th1 progression, similar to that reported for developing human NK cells.15
Previous studies have identified several transcription factors and signaling pathways as being essential for the development and functional differentiation of Vα14i NKT cells.13 Despite all these data, the cellular and molecular mechanisms involved in the process of Vα14i NKT-cell commitment and maturation remain poorly understood.
We recently reported that the final fate specification of Vα14i TCR+ precursors to the NKT-cell lineage requires the transcription factor T-bet,16 a factor originally implicated in the Th1-cell lineage commitment of CD4+ T cells.17 However, it is not clear how this single transcription factor might drive Vα14i NKT-cell maturation.
In this study, we have used 2 distinct but complementary approaches to better understand Vα14i NKT-cell development. First, to begin to uncover the molecular mechanisms involved in Vα14i NKT-cell development, we have identified genes that are differentially expressed at different developmental stages. Using DNA microarrays on sorted ex vivo Vα14i NKT cells, we show that Vα14i NKT-cell maturation in the thymus is associated with multiple changes in the expression of genes involved in cell migration (including both chemokines and their receptors), cell survival, cytokine production, and cytotoxicity. Second, to better understand how T-bet directs terminal Vα14i NKT-cell maturation, we have assessed how gene-expression patterns change in Vα14i NKT cells when T-bet activity is manipulated in vitro. We demonstrate that the transcription factor T-bet acts as a master regulator, linking the development of Vα14i NKT cells with the acquisition of their specialized functions. Altogether, our data provide a molecular framework defining the developmental progression of Vα14i NKT-cell precursors to mature Vα14i NKT cells and reveal a critical and multifaceted role for T-bet during this developmental process.
Materials and methods
Mice
C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME). T-bet–/– mice backcrossed to the C57BL/6 background for 8 generations have been described previously.17 Mice were housed at the University of Colorado Health Science Center under specific pathogen-free conditions. Handling of mice and experimental procedures were in accordance with institutional requirements for animal care and use.
Flow cytometry
Cells were stained with CD1d tetramers as described.18 Monoclonal antibodies (mAbs) used in this study for flow cytometry included fluorescein isothiocyanate (FITC)–labeled anti-NK1.1 clone PK1316, CyChrome-labeled anti-TCRβ clone H57-597, allophycocyanin (APC)–labeled anti-CD44 clone IM7, APC-labeled anti-CD38 clone 90, FITC-labeled anti-CD94 clone 18d3, biotin-labeled anti-CD244.2 clone 2B4, biotin-labeled anti-NKG2D clone MI-6, biotin-labeled anti-CD122 clone 5H4, FITC-labeled anti-Ly6C clone HK1.4 (Southernbiotech, Birmingham, AL), PE-Cy7–labeled anti–interferon γ (IFNγ) clone XMG1.2, PE-Cy7–labeled Rat immunoglobulin G1 (IgG1) isotype control, APC-labeled anti–granzyme B clone GB11 (Caltag, Burlingame, CA), APC-labeled mouse IgG1 isotype control (Caltag), and Cychrome-labeled streptavidin. Antibodies were purchased from eBioscience (San Diego, CA) except where indicated.
Microarray data analysis
Total RNA was prepared using the PicoPure RNA isolation Kit (Arcturus, Mountain View, CA) following manufacturer's instructions. RNA was amplified for 2 cycles using the RiboAmp RNA Amplification Kit (Arcturus) as described in the manufacturer's protocol and labeled using the BioArray High Yield RNA transcript labeling Kit (Enzo, Farmingdale, NY). All the samples were amplified and labeled using identical methods such as to permit comparison between samples. RNA processing and hybridization to the Affymetrix gene chip MOE 430A (http://www.affymetrix.com) was performed according to the manufacturer's recommendation by the Gene Expression Core Facility at the University of Colorado. Microarray data were analyzed by using Genespring 6.2 software (ChemAgilent, Santa Clara, CA). Gene-expression values below 0.01 were set to 0.01. Each measurement was divided by the 50th percentile of all measurements in that sample. Each gene value was divided by the median of its measurements in all samples. If the median of the raw values was below 10, each measurement for that gene was divided by 10. Two independent sets of data from 2 independent cell sorts were grouped together as replicates for analysis and visualization. For the 3 developmental stages of Vα14i NKT cells, the analysis was further restricted to genes expressed with a minimum normalized value of 0.1 and flagged as “present” in at least 2 of 6 samples. Filter on “Fold Change” finds genes based on a comparison of 2 samples or conditions. Robust changes in gene expression were identified for transcripts with a fold change of more than 2 for increases and less than 2 for decreases between two conditions.
Real-time PCR
Total mRNA was extracted from sorted cells using TRIzol solution (Invitrogen, Carlsbad, CA). RNA was treated with the DNA-free kit (Ambion, Austin, TX) to remove contaminating DNA from RNA preparations. Reverse transcription was carried out by using the SuperScript III Kit (Invitrogen) following the manufacturer's instructions. The amount of amplicon generated during the polymerase chain reaction (PCR) was monitored with a DNA engine Opticon 2 apparatus (MJ Research, Waltham, MA) by using gene specific primers and probes and the Platinum Quantitative PCR SuperMix UDG (Invitrogen) or alternatively the SyBR green quantitative PCR SuperMix UDG (Invitrogen). The sequences of the primers and probes have been published16,19 or are as follows: CD122 forward 5′-CGTCCATGCCAAGTCGAAC-3′, CD122 reverse 5′-GATGCCTGCCTCACAAGAGTT-3′, CD122 probe 5′ FAM-TGCGACACTGGAACAAAACCTGTGAGC-TAMRA-3′; CD95L forward 5′-TACCACCGCCATCACAACC-3′, CD95L reverse 5′-GATTTGTGTTGTGGTCCTTCTTCTT-3′, CD95L probe 5′ FAM-CACTGCCGCCACTGACCCCTC-TAMRA 3′; CxCR3 forward 5′-GCTGCTGTCCAGTGGGTTTT-3′, CxCR3 reverse 5′-AGTTGGATGTTGAACAAGGCGC-3′, CxCR3 probe 5′ FAM-CCCTGGCCTCTGCAAAGTGGCA-TAMRA 3′; granzyme B forward 5′-AAACGTGCTTCCTTTCGGG-3′, granzyme B reverse 5′-GAAACTATGCCTGCAGCCACT-3′, granzyme B probe 5′ FAM-TTCTGGAGGCCCGCTTGTGTGTAAA-TAMRA 3′; CCR5 forward 5′-ACCTGCTCAACCTGGCCAT-3′, CCR5 reverse 5′-TGCAGCATAGTGAGCCCAGA-3′, CCR5 probe 5′ FAM-TCTGACCTGCTCTTCCTGCTCACACTACC-TAMRA 3′. Perforin forward 5′-GCAGCTGAGAAGACCTATCAGGAC-3′; perforin reverse 5′-TCTGAGCGCCTTTTTGAAGTC-3′; RANTES (regulated on activation, normal T expressed and secreted) forward 5′-ACACCACTCCCTGCTGCTTT-3′; RANTES reverse 5′-AAATACTCCTTGACGTGGGCA-3′; RANTES probe 5′-FAM-CCTACCTCTCCCTCGCGCTGCC-TAMRA 3′.
Retroviral transduction
The retroviral LTR (long terminal repeat) plasmids previously described20-22 were cotransfected into 293T cells together with the retroviral packaging vector pCL-Eco. Freshly isolated thymocytes from T-bet–/– mice were depleted of CD8+ cells using anti-CD8 conjugated beads (Miltenyi Biotec, Auburn, CA) and cultured with recombinant murine (rm) IL-7 (12.5 ng/mL; R&D systems, Minneapolis, MN). The cells were then spin-infected on days 2 and 3 with control or T-bet–expressing retroviruses.21,22 After infection the cells were again cultured in the presence of rmIL-7 for an additional 2 to 3 days before the cells were stained with anti-TCRβ mAb and CD1d tetramers. Green fluorescent protein–positive (GFP+) tetramer+ cells were sorted with a MoFlo flow cytometer (Cytomation, Fort Collins, CO). For the infection of wild-type Vα14i NKT cells, thymocytes from C57BL/6 mice were CD8 depleted and placed in culture with recombinant human (rh) IL-15 (50 ng/mL; R&D Systems) and infected as described with control, dominant-negative (DN) T-bet, or DN Tbx13–expressing retroviruses.
In vitro cytotoxicity assay (VITAL assay)
The VITAL assay has been previously described using different target and/or effector cells.23 Briefly, A20 mCD1 target cells were loaded with 4-deoxyGalCer (150 ng/mL) or not by incubation for 90 minutes followed by labeling with either CFSE or CMTMR (Molecular Probes, Eugene, OR). Labeled cells were then combined in a 1:1 ratio and plated in 96-well U-bottomed plates (5 × 103 of each population per well). Effector cells were retrovirally transduced NKT cells from T-bet–/– mice that were sorted following transduction for both GFP and CD1 tetramer+ staining, stimulated in the presence of anti-CD3/anti-CD28 for 48 hours and expanded in IL-2 (40 U/mL) for 5 to 10 days prior to being harvested for the cytotoxicity assay. They were added to the wells with the target A20mCD1 cells in the indicated effector-target (E/T) ratios, in triplicate. Following a 2.5-hour incubation at 37°C, cells were harvested, resuspended in 5 μg/mL propidium iodide (Sigma-Aldrich, St Louis, MO), and analyzed by flow cytometry. Analysis was gated on propidium iodide–negative cells only. Percentage of specific lysis was calculated as follows: adjusted % survival = 100 × (% survival ÷ mean % survival in the absence of effectors), where % survival = % antigen loaded A20mCD1 (CFSE+) ÷ % unloaded A20mCD1(CMRMR+).
Results
Gene-expression profile of developing Vα14i NKT cells
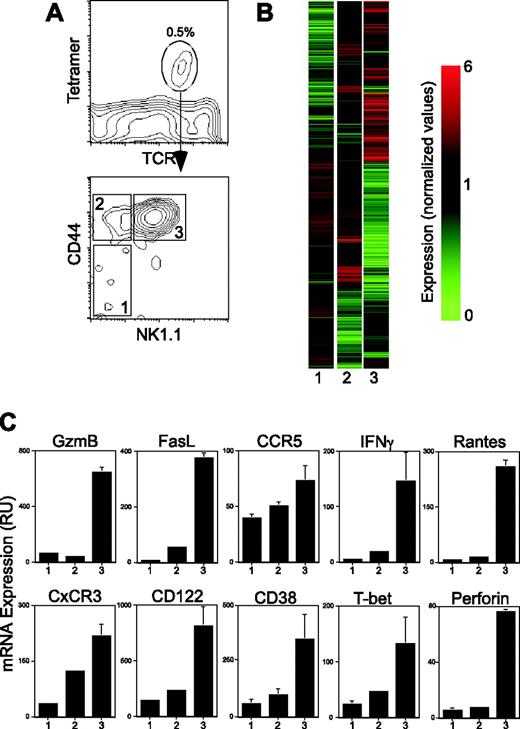
To identify genes that are expressed in mature mouse Vα14i NKT cells (CD44high NK1.1+) as well as to define how these genes are regulated during Vα14i NKT-cell maturation, we sorted CD1d tetramer+ thymocytes derived from C57BL/6 mice into 3 developmental stages based on the expression of the CD44 and NK1.1 markers (Figure 1A). Gene expression was analyzed using DNA microarrays. The change in expression for each gene was computed and is presented as normalized intensity versus the 3 developmental stages (Figure 1B). Genes down-regulated during development appear in green, while genes up-regulated are in red. This analysis includes only genes that were repressed or induced at least 1.8-fold between the expression levels found in developmental stages 2 and 3 relative to the levels of expression in stage 1, in 2 of 2 comparisons.
We found that the expression levels of 597 genes (out of 14 000 genes analyzed) changed significantly in the course of Vα14i NKT-cell development (Figure 1B and supplemental Table S1; see the Supplemental Data Set link at the top of the online article on the Blood website). To simplify the analysis we chose to concentrate on genes whose expression has been associated with Vα14i NKT-cell biology (Table 1). As expected from the cell-sorting strategy, we found up-regulation of the CD44 and NK1.1 mRNA levels in the respective populations. Furthermore, we independently confirmed the array data by measuring the expression levels of several different genes using quantitative real-time PCR analysis (Figure 1C). In agreement with the DNA array data (Table 1), quantitative PCR analysis of granzyme B, FasL, CCR5, IFNγ, CxCR3, CD122, CD38, perforin, RANTES, and T-bet mRNA levels showed increased expression of all these genes with maturation (Figure 1C).
Normalized signal values for selected genes from Affymetrix MOE430A Genechip array
. | . | Developmental stage . | . | . | ||
|---|---|---|---|---|---|---|
| Protein encoded by gene . | Probe ID . | CD44- NK1.1- . | CD44+ NK1.1- . | CD44+ NK1.1+ . | ||
| Cytokines and receptors | ||||||
| IFNγ* | 1425947 | 0.460 | 1.557 | 8.114 | ||
| IL-13† | 1420802 | 2.466 | 0.459 | 0.126 | ||
| IL-10 | 1450330 | 1.103 | 1.135 | 0.559 | ||
| IL-4 | 1449864 | 1.276 | 1.090 | 0.863 | ||
| IL-21† | 1450334 | 1.243 | 1.104 | 0.195 | ||
| CD122* | 1448759 | 0.675 | 1.222 | 3.986 | ||
| Cytotoxicity | ||||||
| Granzyme B* | 1419060 | 0.327 | 1.700 | 11.41 | ||
| Perforin | 1451862 | 0.585 | 1.095 | 1.238 | ||
| FasL* | 1449235 | 0.285 | 1.695 | 9.186 | ||
| Chemokines and receptors | ||||||
| CxCL2 (MIP2α) | 1449984 | 0.354 | 1.250 | 1.440 | ||
| CxCL10 (IP10)* | 1418930 | 0.758 | 1.318 | 2.187 | ||
| XCL1 (lymphotactin)* | 1419412 | 0.764 | 2.327 | 16.53 | ||
| CCL4 (MIP1β) | 1421578 | 0.576 | 1.285 | 1.326 | ||
| CCL5 (RANTES)* | 1418126 | 0.438 | 2.275 | 40.82 | ||
| CCR1 | 1419610 | 0.151 | 2.087 | 0.482 | ||
| CCR5* | 1424727 | 0.847 | 0.999 | 2.833 | ||
| CCR7† | 1423466 | 1.494 | 0.317 | 0.068 | ||
| CCR9† | 1421920 | 2.591 | 0.703 | 0.124 | ||
| CxCR3* | 1449925 | 0.749 | 1.549 | 3.937 | ||
| CxCR6 | 1422812 | 0.551 | 1.442 | 1.399 | ||
| NK receptors | ||||||
| Klb1c (NK1.1)* | 1449570 | 0.388 | 1.601 | 9.780 | ||
| Klrk1 (NKG2D)* | 1450495 | 0.659 | 1.504 | 4.387 | ||
| Klrc2/3 (NKG2C)* | 1425005 | 0.107 | 3.291 | 24.33 | ||
| Klrd1 (CD94)* | 1460245 | 0.262 | 1.919 | 6.639 | ||
| Klra3 (Ly49C)* | 1425436 | 0.254 | 1.778 | 3.674 | ||
| Klra5 (Ly49E)* | 1420789 | 0.408 | 1.935 | 24.39 | ||
| Klra7 (Ly49G2)* | 1426171 | 0.264 | 1.822 | 8.525 | ||
| Klra20 (Ly49T)* | 1451942 | 0.160 | 1.566 | 2.644 | ||
| gp49b | 1420394 | 0.561 | 1.924 | 1.507 | ||
| CD160* | 1420066 | 0.836 | 1.455 | 2.572 | ||
| CD2* | 1418770 | 0.979 | 0.935 | 2.252 | ||
| Slamf4 (2B4)* | 1449991 | 0.316 | 1.525 | 12.83 | ||
| Stat proteins | ||||||
| Stat4* | 1448713 | 0.192 | 1.357 | 12.53 | ||
| Stat5b | 1422103 | 1.354 | 0.452 | 0.615 | ||
| Activation/memory | ||||||
| Ly6C* | 1421571 | 0.742 | 1.515 | 5.230 | ||
| CD44* | 1423760 | 0.357 | 1.330 | 2.813 | ||
| CD38* | 1433741 | 0.472 | 1.309 | 3.771 | ||
| CD3ζ† | 1426396 | 1.181 | 0.613 | 0.586 | ||
| Transcription factors | ||||||
| T-bet* | 1449361 | 0.754 | 1.316 | 2.114 | ||
| TCF7 (TCF-1)† | 1433471 | 1.406 | 0.569 | 0.375 | ||
. | . | Developmental stage . | . | . | ||
|---|---|---|---|---|---|---|
| Protein encoded by gene . | Probe ID . | CD44- NK1.1- . | CD44+ NK1.1- . | CD44+ NK1.1+ . | ||
| Cytokines and receptors | ||||||
| IFNγ* | 1425947 | 0.460 | 1.557 | 8.114 | ||
| IL-13† | 1420802 | 2.466 | 0.459 | 0.126 | ||
| IL-10 | 1450330 | 1.103 | 1.135 | 0.559 | ||
| IL-4 | 1449864 | 1.276 | 1.090 | 0.863 | ||
| IL-21† | 1450334 | 1.243 | 1.104 | 0.195 | ||
| CD122* | 1448759 | 0.675 | 1.222 | 3.986 | ||
| Cytotoxicity | ||||||
| Granzyme B* | 1419060 | 0.327 | 1.700 | 11.41 | ||
| Perforin | 1451862 | 0.585 | 1.095 | 1.238 | ||
| FasL* | 1449235 | 0.285 | 1.695 | 9.186 | ||
| Chemokines and receptors | ||||||
| CxCL2 (MIP2α) | 1449984 | 0.354 | 1.250 | 1.440 | ||
| CxCL10 (IP10)* | 1418930 | 0.758 | 1.318 | 2.187 | ||
| XCL1 (lymphotactin)* | 1419412 | 0.764 | 2.327 | 16.53 | ||
| CCL4 (MIP1β) | 1421578 | 0.576 | 1.285 | 1.326 | ||
| CCL5 (RANTES)* | 1418126 | 0.438 | 2.275 | 40.82 | ||
| CCR1 | 1419610 | 0.151 | 2.087 | 0.482 | ||
| CCR5* | 1424727 | 0.847 | 0.999 | 2.833 | ||
| CCR7† | 1423466 | 1.494 | 0.317 | 0.068 | ||
| CCR9† | 1421920 | 2.591 | 0.703 | 0.124 | ||
| CxCR3* | 1449925 | 0.749 | 1.549 | 3.937 | ||
| CxCR6 | 1422812 | 0.551 | 1.442 | 1.399 | ||
| NK receptors | ||||||
| Klb1c (NK1.1)* | 1449570 | 0.388 | 1.601 | 9.780 | ||
| Klrk1 (NKG2D)* | 1450495 | 0.659 | 1.504 | 4.387 | ||
| Klrc2/3 (NKG2C)* | 1425005 | 0.107 | 3.291 | 24.33 | ||
| Klrd1 (CD94)* | 1460245 | 0.262 | 1.919 | 6.639 | ||
| Klra3 (Ly49C)* | 1425436 | 0.254 | 1.778 | 3.674 | ||
| Klra5 (Ly49E)* | 1420789 | 0.408 | 1.935 | 24.39 | ||
| Klra7 (Ly49G2)* | 1426171 | 0.264 | 1.822 | 8.525 | ||
| Klra20 (Ly49T)* | 1451942 | 0.160 | 1.566 | 2.644 | ||
| gp49b | 1420394 | 0.561 | 1.924 | 1.507 | ||
| CD160* | 1420066 | 0.836 | 1.455 | 2.572 | ||
| CD2* | 1418770 | 0.979 | 0.935 | 2.252 | ||
| Slamf4 (2B4)* | 1449991 | 0.316 | 1.525 | 12.83 | ||
| Stat proteins | ||||||
| Stat4* | 1448713 | 0.192 | 1.357 | 12.53 | ||
| Stat5b | 1422103 | 1.354 | 0.452 | 0.615 | ||
| Activation/memory | ||||||
| Ly6C* | 1421571 | 0.742 | 1.515 | 5.230 | ||
| CD44* | 1423760 | 0.357 | 1.330 | 2.813 | ||
| CD38* | 1433741 | 0.472 | 1.309 | 3.771 | ||
| CD3ζ† | 1426396 | 1.181 | 0.613 | 0.586 | ||
| Transcription factors | ||||||
| T-bet* | 1449361 | 0.754 | 1.316 | 2.114 | ||
| TCF7 (TCF-1)† | 1433471 | 1.406 | 0.569 | 0.375 | ||
Genes up-regulated in stage 2 or stage 3 relative to stage 1.
Genes down-regulated in stage 2 or stage 3 relative to stage 1.
These data confirmed that multiple cellular processes are involved in the maturation of Vα14i NKT cells. Like NK cells,24 the effector function of Vα14i NKT cells—namely, the granule-exocytosis pathway, the expression of Fas ligand, and the production of effector cytokines such as IFNγ—appeared to be closely linked to lineage maturation.
Vα14i NKT-cell maturation is arrested in the absence of T-bet
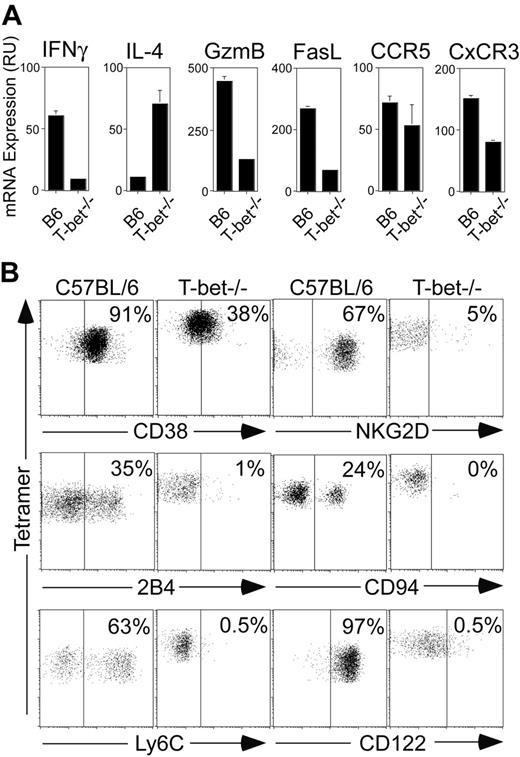
T-bet is up-regulated during the development of Vα14i NKT cells (Table 1 and Figure 1C), and the final maturation of these cells is impaired in the absence of T-bet.16 However, it is not clear whether T-bet controls the expression of only a limited set of genes or whether it enforces an entire genetic program driving the final maturation of Vα14i NKT cells. Therefore, we sought to examine in greater detail the phenotype of Vα14i NKT cells in the absence of T-bet. Figure 2A shows that mRNA levels for several genes expressed in mature Vα14i NKT cells (Table 1), including IFNγ, CD122, granzyme B, FasL, CCR5, and CxCR3 were decreased in the absence of T-bet. In contrast, the quantity of IL-4 mRNAin T-bet–deficient Vα14i NKT cells was greater than that detected in mature wild-type NKT cells. We also examined the expression of several other surface markers for which monoclonal antibodies were commercially available. We confirmed that C57BL/6-derived Vα14i NKT cells express CD38, NKG2D, 2B4, CD94, and Ly6C. However, the expression of these markers by T-bet–deficient Vα14i NKT cells was decreased and/or absent (Figure 2B). Altogether, these results suggest that T-bet plays a more profound role in the development of Vα14i NKT cells than originally anticipated.16 In the absence of T-bet, the entire genetic program associated with the final maturation of Vα14i NKT is arrested.
Expression of T-bet in T-bet–deficient and immature wild-type Vα14i NKT cells in vitro
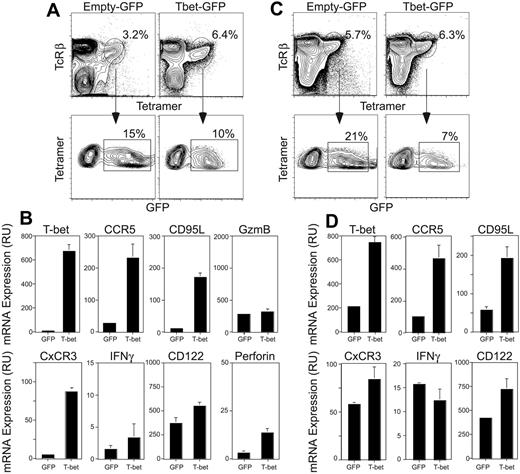
We next assessed the extent to which T-bet expression was sufficient to restore the gene-expression profile of mature Vα14i NKT cells. Wild-type immature and T-bet–deficient Vα14i NKT cells proliferate in the presence of IL-7.9,16 We took advantage of this cytokine-induced proliferation to introduce T-bet into T-bet–/– CD8-depleted thymocytes with a bicistronic retroviral vector allowing for coexpression of GFP. This strategy permits the transfection of Vα14i NKT cells, as well as a few conventional T cells, in absence of TCR signaling. To focus on direct or more immediate effects of T-bet, RNA was isolated from the cells within 48 hours after the second transduction and no effector restimulation was provided. GFP+ CD1d-tetramer+ cells were sorted and gene expression analyzed by quantitative PCR (Figure 3). In these conditions, expression of T-bet induced the up-regulation of CCR5, CxCR3, CD122, and FasL (CD95L) mRNA levels but only marginally modulated the mRNA expression levels of granzyme B, IFNγ, and perforin (Figure 3B). Similarly, we did not observe any significant change in mRNA expression levels for the other genes associated with the final maturation of Vα14i NKT cells, including NK receptors (NK1.1, Ly49s, NKG2/CD94), SLAM family members (2B4), and activation markers (Ly6C, CD38) (data not shown).
Modulation of gene expression during Vα14i NKT-cell development. (A) C57BL/6 thymocytes were stained for CD1d tetramer, TcRβ, NK1.1, and CD44. CD1d tetramer+ TcRβ+ cells were subdivided into 3 populations based on CD44 and NK1.1 expression and sorted. CD44low NK1.1– cells were defined as stage 1, CD44high NK1.1– as stage 2, and CD44high NK1.1+ as stage 3. (B) Gene tree clustering using similarity measure standard correlation. Genes were selected for analysis if their average expression level deviated from that of Stage 1 by at least a factor of 1.8 in 2 out of 2 experiments (597 genes met this criteria). Each column represents data from the indicated developmental stage. Each row represents a gene significantly induced (red) or repressed (green) during the development of Vα14i NKT cells. A color bar shows the magnitude of gene expression changes as a ratio of expression in Stage 1 versus Stage 2 and Stage 3. Genes names, probe ID, and normalized values for each gene can be found in Table S1. (C) Total RNA was prepared from Vα14i NKT cells at each of the 3 defined stages of development and quantitative real-time PCR was performed using primers and probes specific for granzyme B (GzmB), Fas ligand (CD95L), CCR5, IFNγ, CxCR3, CD122, CD38, T-bet, RANTES, and perforin. The amount of each transcript in the 3 developmental stages was determined by quantitative PCR with normalization to the amount of HPRT mRNA in each sample. Results are shown as mean ± standard deviation. Data are representative of at least 3 independent experiments.
Modulation of gene expression during Vα14i NKT-cell development. (A) C57BL/6 thymocytes were stained for CD1d tetramer, TcRβ, NK1.1, and CD44. CD1d tetramer+ TcRβ+ cells were subdivided into 3 populations based on CD44 and NK1.1 expression and sorted. CD44low NK1.1– cells were defined as stage 1, CD44high NK1.1– as stage 2, and CD44high NK1.1+ as stage 3. (B) Gene tree clustering using similarity measure standard correlation. Genes were selected for analysis if their average expression level deviated from that of Stage 1 by at least a factor of 1.8 in 2 out of 2 experiments (597 genes met this criteria). Each column represents data from the indicated developmental stage. Each row represents a gene significantly induced (red) or repressed (green) during the development of Vα14i NKT cells. A color bar shows the magnitude of gene expression changes as a ratio of expression in Stage 1 versus Stage 2 and Stage 3. Genes names, probe ID, and normalized values for each gene can be found in Table S1. (C) Total RNA was prepared from Vα14i NKT cells at each of the 3 defined stages of development and quantitative real-time PCR was performed using primers and probes specific for granzyme B (GzmB), Fas ligand (CD95L), CCR5, IFNγ, CxCR3, CD122, CD38, T-bet, RANTES, and perforin. The amount of each transcript in the 3 developmental stages was determined by quantitative PCR with normalization to the amount of HPRT mRNA in each sample. Results are shown as mean ± standard deviation. Data are representative of at least 3 independent experiments.
Since T-bet–deficient Vα14i NKT cells may not be equivalent to wild-type immature Vα14i NKT cells, we also assessed whether the expression of T-bet in these cells resulted in a similar outcome. Mature Vα14i NKT cells in C57BL/6 mice uniformly express the NK1.1 marker (Table 1, Figure 1). Therefore, thymocytes from C57BL/6 mice were depleted of CD8+ and NK1.1+ cells to enrich for immature NK1.1-Vα14i NKT cells. The remaining cells were then placed in culture with IL-7 and transduced to express T-bet (Figure 3C). As was observed when T-bet was introduced into T-bet–/– Vα14i NKT cells, introduction of T-bet into wild-type immature Vα14i NKT cells induced the up-regulation of CCR5, CxCR3, FasL, and CD122 mRNA levels, while the expression of IFNγ and granzyme B mRNA were unchanged (Figure 3D). Therefore, expression of T-bet in immature NK1.1-Vα14i NKT cells, either from wild-type or T-bet–deficient mice, can induce the expression of CCR5, CxCR3, FasL, and CD122.
The absence of induction of IFNγ and granzyme B mRNA was unexpected given the importance of T-bet in regulating the expression of these genes in conventional T cells.17,22 Indeed, ectopic expression of T-bet in Th2 or T cytotoxic 2 (Tc2) cells is sufficient to induce IFNγ and granzyme B gene transcription.17,22 However, one major difference between our experiments and those performed with cytokine-producing T-helper cells concerns how the cells are infected with retroviruses. T-helper cells are usually stimulated through their TCR to induce the proliferation necessary for retroviral infection. In our studies, Vα14i NKT cells were infected in absence of TCR triggering.
Analysis of expression of several markers on tetramer+ thymocytes from wild-type and T-bet–/– mice. (A) CD1d tetramer+ TcRβ+ thymocytes from wild-type and T-bet–/– mice were sorted and total RNA prepared. Expression levels of IFNγ, IL-4, CD122, granzyme B, Fas ligand, CCR5, and CxCR3 between the 2 cell populations was measured by quantitative PCR using specific primers and probes. The amount of transcript was normalized to the amount of HPRT in each sample. Results are shown as mean ± standard deviation. (B) C57BL/6 and T-bet–/– thymocytes were stained with CD1d tetramer, TcRβ, and diverse markers. CD1d tetramer+ TcRβ+ cells were gated and analyzed for the expression of CD38, NKG2D, 2B4, CD94, Ly6C, and CD122. Percentages of positive cells are indicated.
Analysis of expression of several markers on tetramer+ thymocytes from wild-type and T-bet–/– mice. (A) CD1d tetramer+ TcRβ+ thymocytes from wild-type and T-bet–/– mice were sorted and total RNA prepared. Expression levels of IFNγ, IL-4, CD122, granzyme B, Fas ligand, CCR5, and CxCR3 between the 2 cell populations was measured by quantitative PCR using specific primers and probes. The amount of transcript was normalized to the amount of HPRT in each sample. Results are shown as mean ± standard deviation. (B) C57BL/6 and T-bet–/– thymocytes were stained with CD1d tetramer, TcRβ, and diverse markers. CD1d tetramer+ TcRβ+ cells were gated and analyzed for the expression of CD38, NKG2D, 2B4, CD94, Ly6C, and CD122. Percentages of positive cells are indicated.
Expression of T-bet in immature Vα14i NKT cells. (A) T-bet–/– thymocytes were depleted of CD8+ cells using anti-CD8 coated magnetic beads. The cells were placed in culture in complete media with 10 ng/mL IL-7. On days 2 and 3 the cells were spin-infected using a control retrovirus (empty GFP) or a T-bet–encoding retrovirus (Tbet-GFP) and replaced in culture with IL-7 for 2 to 3 more days. At the end of the culture, the cells were harvested and stained with the CD1d tetramer and anti-TcRβ mAb. CD1d tetramer+ TcRβ+ GFP+ cells were sorted and total RNA prepared. Percentages of tetramer-positive, TCRβ+ cells are indicated. (B) Expression levels of T-bet, CCR5, Fas ligand, CxCR3, IFNγ, and CD122 in T-bet–/– Vα14i NKT cells infected with the control virus or the T-bet–encoding virus were analyzed by quantitative PCR using specific primers and probes. The amount of transcript in each sample was normalized to the amount of HPRT. Results are shown as mean ± standard deviation. Data are representative of 3 independent experiments. (C) C57BL/6 thymocytes were depleted of CD8+ and NK1.1+ cells using anti-CD8 and anti-NK1.1 magnetic beads. Depletion of NK1.1+ cells was 90% effective. The cells were placed in culture with rmIL-7 and infected and sorted as in panel A. Percentages of tetramer-positive, TCRβ+ cells are indicated. (D) Gene-expression level analysis was performed as in panel B. Results are shown as mean ± standard deviation.
Expression of T-bet in immature Vα14i NKT cells. (A) T-bet–/– thymocytes were depleted of CD8+ cells using anti-CD8 coated magnetic beads. The cells were placed in culture in complete media with 10 ng/mL IL-7. On days 2 and 3 the cells were spin-infected using a control retrovirus (empty GFP) or a T-bet–encoding retrovirus (Tbet-GFP) and replaced in culture with IL-7 for 2 to 3 more days. At the end of the culture, the cells were harvested and stained with the CD1d tetramer and anti-TcRβ mAb. CD1d tetramer+ TcRβ+ GFP+ cells were sorted and total RNA prepared. Percentages of tetramer-positive, TCRβ+ cells are indicated. (B) Expression levels of T-bet, CCR5, Fas ligand, CxCR3, IFNγ, and CD122 in T-bet–/– Vα14i NKT cells infected with the control virus or the T-bet–encoding virus were analyzed by quantitative PCR using specific primers and probes. The amount of transcript in each sample was normalized to the amount of HPRT. Results are shown as mean ± standard deviation. Data are representative of 3 independent experiments. (C) C57BL/6 thymocytes were depleted of CD8+ and NK1.1+ cells using anti-CD8 and anti-NK1.1 magnetic beads. Depletion of NK1.1+ cells was 90% effective. The cells were placed in culture with rmIL-7 and infected and sorted as in panel A. Percentages of tetramer-positive, TCRβ+ cells are indicated. (D) Gene-expression level analysis was performed as in panel B. Results are shown as mean ± standard deviation.
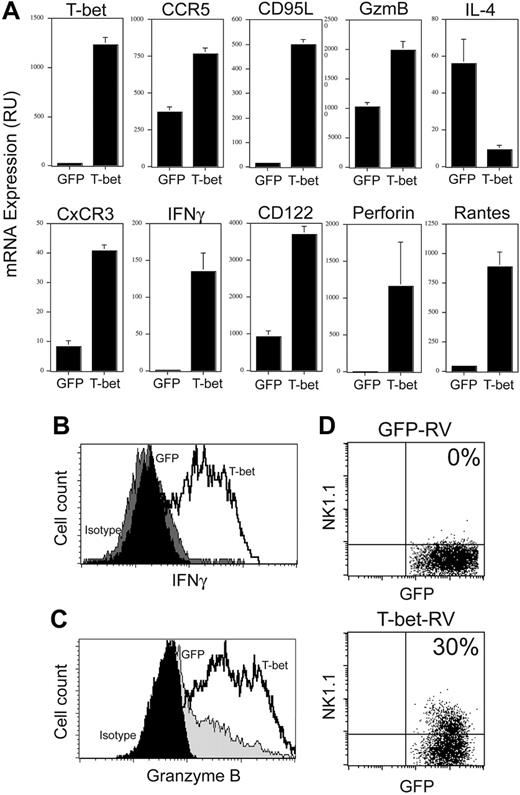
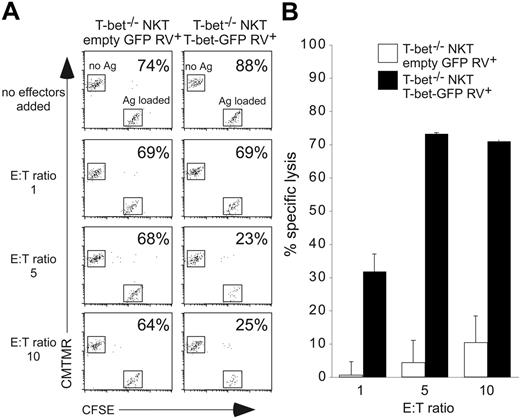
To assess whether TCR signaling could account for these differences, T-bet–/– Vα14i NKT cells were infected as previously, sorted, placed on anti-CD3/CD28–coated plates for 2 days, and expanded in IL-2 for several days. Under these in vitro experimental conditions, CxCR3, CCR5, CD122, and CD95L mRNA levels were up-regulated in the T-bet–expressing cells, in agreement with our previous results. In addition, IFNγ, granzymeB, perforin, and RANTES mRNA expression levels were also up-regulated in T-bet–expressing cells (Figure 4A). Expression of NK1.1, Ly6C, and 2B4, but not Ly49 markers, was also partly restored (Figure 4D and data not shown). The cells were then restimulated and their cytokine production assessed. T-bet–/– Vα14i NKT cells infected with T-bet retrovirus (RV) had reduced IL-4 production (data not shown) and had restored IFNγ expression (Figure 4B). In addition to the secretion of IFNγ, overexpression of T-bet in T-bet–/– Vα14i NKT cells was sufficient to induce granzyme B protein (Figure 4C) and the capacity to kill antigen-loaded CD1d+ target cells (Figure 5).
Altogether, these results demonstrate that under these conditions, T-bet is able to fully establish the Th1 program and partially represses the production of IL-4 in synergy with TCR/IL-2 signaling in Vα14i NKT cells.
Effects of a dominant-negative T-bet on the Vα14i NKT cell identity
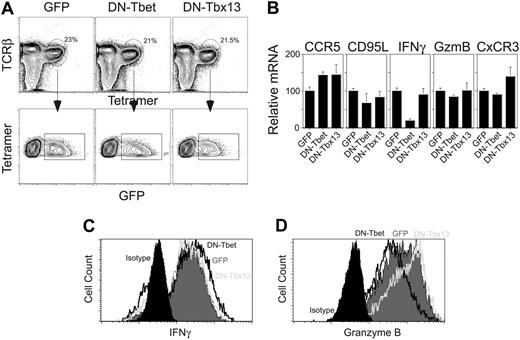
We next assessed the importance of continuing T-bet activity on the phenotype of mature Vα14i NKT cells. In agreement with the up-regulation of CD122 during Vα14i NKT-cell development, mature cells proliferate in vitro in response to IL-15.14 We took advantage of this cytokine-induced proliferation to introduce a dominant-negative form of T-bet21 with a bicistronic retroviral vector that contained the GFP marker into wild-type Vα14i NKT cells (Figure 6A). As a control for specificity, the cells were also infected with a retrovirus encoding for a dominant-negative form of Tbx13,22 a T-box transcription factor that does not belong to the T-bet subfamily.25
Antagonizing T-bet activity with DN T-bet in Vα14i NKT cells inhibited their constitutive expression of IFNγ mRNA but did not change significantly their expression of CCR5, CD95L, granzyme B, and CxCR3 (Figure 6B). Analysis of cytokine secretion and granzyme B expression following restimulation of the cells demonstrated that the introduction of the DN T-bet construct had only a modest effect on the secretion of IFNγ (Figure 6C) and on expression of granzyme B (Figure 6D). By contrast, a similar level of expression of the DN T-bet construct, as evaluated by GFP expression level, antagonized IFNγ secretion by Th1 cells by 70% to 80% (data not shown) and granzyme B expression in CD8+ T cells.22
Discussion
Large amounts of data support a stepwise model of T-cell and B-cell development with well-characterized and distinct stages of differentiation. Here we show that similar steps can be identified in Vα14i NKT-cell development. We have defined the gene-expression profile of developing Vα14i NKT cells and identified 597 genes with changes in expression levels during NKT-cell maturation in the thymus. As part of their developmental program, mature Vα14i NKT cells acquired an effector Th1-type phenotype characterized by the expression of certain cytokines, chemokines, chemokine receptors, and molecules involved in cytolysis. The transcription factor T-bet was necessary to drive this developmental program and mechanistically united the development and the effector functions of Vα14i NKT cells.
T-bet mediates IFNγ and granzyme B induction in Vα14i NKT cells. T-bet–/– Vα14i NKT cells were infected as described in Figure 3 and sorted. The cells were placed on anti-CD3/anti-CD28–coated plates for 48 hours, washed, and expanded in IL-2 (40 U/mL) for a week. (A) Expression levels of T-bet, CCR5, Fas ligand, CxCR3, IFNγ, IL-4, CD122, perforin, and RANTES were analyzed by quantitative PCR. Results are shown as mean ± standard deviation. (B) The cells were then stimulated with PMA/ionomycin for a total of 4 hours with the last 3 hours of the stimulation in the presence of golgi block, and analyzed by intracellular staining. At the end of the activation period, the cells were washed in phosphate-buffered saline (PBS)/0.1% bovine serum albumin (BSA) and surface-stained with CD1d tetramers. The cells were then fixed and stained for intracellular IFNγ. (C) The cells were treated as in panel B, and were analyzed for granzyme B expression. (D) NK1.1 expression on T-bet–/– Vα14i NKT cells infected with the control virus or the T-bet–encoding virus. Data are representative of 3 independent experiments. Percentages of positive cells are indicated.
T-bet mediates IFNγ and granzyme B induction in Vα14i NKT cells. T-bet–/– Vα14i NKT cells were infected as described in Figure 3 and sorted. The cells were placed on anti-CD3/anti-CD28–coated plates for 48 hours, washed, and expanded in IL-2 (40 U/mL) for a week. (A) Expression levels of T-bet, CCR5, Fas ligand, CxCR3, IFNγ, IL-4, CD122, perforin, and RANTES were analyzed by quantitative PCR. Results are shown as mean ± standard deviation. (B) The cells were then stimulated with PMA/ionomycin for a total of 4 hours with the last 3 hours of the stimulation in the presence of golgi block, and analyzed by intracellular staining. At the end of the activation period, the cells were washed in phosphate-buffered saline (PBS)/0.1% bovine serum albumin (BSA) and surface-stained with CD1d tetramers. The cells were then fixed and stained for intracellular IFNγ. (C) The cells were treated as in panel B, and were analyzed for granzyme B expression. (D) NK1.1 expression on T-bet–/– Vα14i NKT cells infected with the control virus or the T-bet–encoding virus. Data are representative of 3 independent experiments. Percentages of positive cells are indicated.
The Th1-type effector and cytolytic functions of Vα14i NKT cells are developmentally linked
Our results demonstrated that during development Vα14i NKT cells acquire their trademark features, including a predominantly Th1 cytokine profile, the ability to migrate to inflammatory sites, and cytolytic function.
In resting Vα14i NKT cells, IFNγ mRNA levels increased during development, while mRNA levels of the Th2-associated cytokines IL-4, IL-13, IL-10, and IL-21 remained constant or decreased (Table 1 and data not shown) in agreement with previous findings.7-9 In addition to cytokines, Vα14i NKT cells also expressed several mRNAs encoding for chemokines upon maturation, including MIP1α, MIP2α, IFNγ-inducible protein 10 (IP-10), the Th1-associated lymphotactin, and RANTES. This finding is in agreement with the chemokine secretion pattern previously reported for Vα14i NKT cells.26,27 Unexpectedly, at least 3 of these chemokines were differentially expressed between immature and mature Vα14i NKT cells, with IP-10, lymphotactin, and RANTES mRNAs all more highly expressed in mature as compared with immature Vα14i NKT cells.
Not only are the cytokines and chemokines expressed by mature Vα14i NKT cells reflective of a Th1-type cell, the chemokine receptor profile of mature Vα14i NKT cells was also indicative of Th1-type inflammatory homing cells, with high expression of CCR5, CxCR3, and CxCR6. This is similar to recently reported data with human peripheral blood Vα24i NKT cells28 and is consistent with the preferential expression of these chemokine receptors on effector/memory T cells.29-31 We found that CCR5, CxCR3, and CxCR6 mRNA levels were up-regulated during the course of Vα14i NKT-cell maturation, suggesting that Vα14i NKT cells acquire the ability to migrate to sites of inflammation late in their maturation. By contrast, CCR7 and CCR9, which are required for the basal homing of T lymphocytes into lymph nodes and the homing to intestinal tissues, were highly expressed in immature NK1.1-Vα14i NKT cells and were undetectable in mature Vα14i NKT cells. This is in agreement with the observation that only immature NK1.1-Vα14i NKT cells, but not NK1.1+ Vα14i NKT cells, migrated in response to the CCR7 ligand SLC/CCL21.29 In addition, these results could explain the paucity of Vα14i NKT cells in lymph nodes and the intestinal mucosa32 and the rapid recruitment of this population in inflamed and infected tissues.33
Vα14i NKT cells are potently cytolytic through the release of perforin and granzymes, and the expression of membrane-bound members of the tumor necrosis factor (TNF) family, FasL and TRAIL.34-36 All of these genes were detected in mature Vα14i NKT cells. Interestingly, as the cells progressed in vivo through the 3 developmental stages, they up-regulated granzyme B and perforin as well as FasL mRNA levels. These results suggest that Vα14i NKT cells acquire their cytolytic capacity during development in parallel with the acquisition of the Th1 phenotype.
Cytolytic activity of T-bet–/– Vα14i NKT cells upon re-expression of T-bet. (A) T-bet–/– Vα14i NKT cells retrovirally transduced with either an empty GFP control retrovirus or the T-bet–GFP retrovirus were used as effector cells. They were added in the indicated ratios to wells containing both CFSE-labeled A20 mCD1 target cells loaded with 4-deoxyGalCer and CMTMR-labeled A20 mCD1 target cells without antigen. After a 2.5-hour incubation period, each well was independently analyzed by flow cytometry and the ratio of cells loaded with antigen (Ag+) to cells not loaded with antigen (Ag–) was calculated and the percentage is indicated in each plot. (B) Percentage of specific lysis was determined as described in “Materials and methods,” using the analysis of triplicate wells to determine standard error for each condition. Data are representative of 2 independent experiments.
Cytolytic activity of T-bet–/– Vα14i NKT cells upon re-expression of T-bet. (A) T-bet–/– Vα14i NKT cells retrovirally transduced with either an empty GFP control retrovirus or the T-bet–GFP retrovirus were used as effector cells. They were added in the indicated ratios to wells containing both CFSE-labeled A20 mCD1 target cells loaded with 4-deoxyGalCer and CMTMR-labeled A20 mCD1 target cells without antigen. After a 2.5-hour incubation period, each well was independently analyzed by flow cytometry and the ratio of cells loaded with antigen (Ag+) to cells not loaded with antigen (Ag–) was calculated and the percentage is indicated in each plot. (B) Percentage of specific lysis was determined as described in “Materials and methods,” using the analysis of triplicate wells to determine standard error for each condition. Data are representative of 2 independent experiments.
Expression of DN T-bet in mature Vα14i NKT cells. C57BL/6 thymocytes were depleted of CD8+ cells using anti-CD8 magnetic beads. The cells were then placed in culture in complete media supplemented with 50 ng/mL IL-15. On day 2 and 3 the cells were spin-infected using a control retrovirus (empty GFP), a dominant-negative T-bet–encoding retrovirus (DN T-bet), or a control virus encoding an irrelevant dominant-negative T-box gene (Tbx13) and replaced in culture with IL-15 for 2 to 3 more days. At the end of the culture period, the cells were harvested and stained with the CD1d tetramer and anti-TcRβ mAb. CD1d tetramer+ TcRβ+ GFP+ cells were sorted (A), placed on anti-CD3/anti-CD28–coated plates for 48 hours, washed, and further expanded in complete media with IL-2 for a week. Percentages of tetramer-positive, TCRβ+ cells are indicated. (B) Total RNA was prepared and expression levels of CCR5, FasL (CD95L), IFNγ, granzyme B, and CxCR3 were measured by quantitative PCR as in Figure 4. Results are shown as mean ± standard deviation. (C-D) Alternatively, the cells were restimulated with PMA/ionomycin and analyzed as described in Figure 4. Data are representative of 2 independent experiments.
Expression of DN T-bet in mature Vα14i NKT cells. C57BL/6 thymocytes were depleted of CD8+ cells using anti-CD8 magnetic beads. The cells were then placed in culture in complete media supplemented with 50 ng/mL IL-15. On day 2 and 3 the cells were spin-infected using a control retrovirus (empty GFP), a dominant-negative T-bet–encoding retrovirus (DN T-bet), or a control virus encoding an irrelevant dominant-negative T-box gene (Tbx13) and replaced in culture with IL-15 for 2 to 3 more days. At the end of the culture period, the cells were harvested and stained with the CD1d tetramer and anti-TcRβ mAb. CD1d tetramer+ TcRβ+ GFP+ cells were sorted (A), placed on anti-CD3/anti-CD28–coated plates for 48 hours, washed, and further expanded in complete media with IL-2 for a week. Percentages of tetramer-positive, TCRβ+ cells are indicated. (B) Total RNA was prepared and expression levels of CCR5, FasL (CD95L), IFNγ, granzyme B, and CxCR3 were measured by quantitative PCR as in Figure 4. Results are shown as mean ± standard deviation. (C-D) Alternatively, the cells were restimulated with PMA/ionomycin and analyzed as described in Figure 4. Data are representative of 2 independent experiments.
Taken together, our results confirm that a significant number of differentially expressed genes underlie the progression of Vα14i NKT cells from an immature to a mature state. The granule-exocytosis pathway, the expression of Fas ligand, the production of effector cytokines such as IFNγ, and the expression of several other genes associated with a Th1 signature profile were closely linked to lineage maturation. The overall switch from a Th2 to a Th1-like gene-expression profile during development demonstrated that the mature functions of Vα14i NKT cells are developmentally controlled. A similar progression of events to the one we described for Vα14i NKT cells has also been documented for the development of human and mouse NK cells15,37 and the differentiation of CD8 T cells into effector cells,38 further emphasizing common pathways and mechanisms of development among these lymphocyte populations.
T-bet is necessary to drive Vα14i NKT-cell maturation
The gene-expression profile changes that occurred during the maturation of Vα14i NKT cells were reminiscent of the molecular program underlying the acquisition of a Th1-like39 and CD8 effector/memory-like profile.38 The transcription factor T-bet has been implicated in both the Th1-cell lineage commitment of CD4+ T cells and the regulation of cytolytic effector mechanisms of CD8+ T cells. T-bet was also shown to suppress Th2 cytokine production.40 Although the ectopic expression of T-bet into T-bet–deficient CD8-depleted thymocytes does not preclude the possibility that additional effects of T-bet on cells other than NKT cells might indirectly aid NKT-cell development, we believe it likely that the observed effects of T-bet are intrinsic to Vα14i NKT cells. First, T-bet mRNA levels increase naturally in Vα14i NKT cells during their maturation. Second, results from bone marrow chimera experiments demonstrated an intrinsic role for T-bet in developing NKT cells.16 Third, GFP-negative Vα14i NKT cells from cultures retrovirally transduced with T-bet were unable to undergo final maturation, as measured by up-regulation of CD122,16 making a role for T-bet in trans highly unlikely. Altogether, these results identify T-bet as a master regulator necessary for the final maturation of Vα14i NKT cells. In agreement with our previous observation,16 the few Vα14i NKT cells remaining in T-bet–/– mice lacked attributes of mature cells. This was further established by DNA array analysis of T-bet–/– Vα14i NKT cells (data not shown), confirming that T-bet is essential to the generation of fully mature Vα14i NKT cells.
Two sets of genes were identified following expression of T-bet into T-bet–/– Vα14i NKT cells. The first set of genes included CCR5, CxCR3, FasL, and CD122, and were up-regulated in T-bet–/– Vα14i NKT cells by the sole expression of T-bet. A second set of genes, including IFNγ, granzyme B, perforin, NK1.1, and RANTES, was also dependent upon the expression of T-bet, but the up-regulation of these genes became apparent only after TCR/IL-2 signaling. These results suggest a linear model of Vα14i NKT-cell maturation during which T-bet expression is absolutely necessary but defines only one required developmental step in an entire developmental process. Other signaling pathway(s) are then required for the acquisition of the full mature phenotype. TCR and/or IL-2 (and perhaps IL-15) signaling, associated to T-bet expression, are likely candidates of signaling pathways that are required to further drive the maturation of Vα14i NKT cells. This is the first demonstration that immature Vα14i NKT cells can be further differentiated to their mature phenotype in vitro. T-bet expression associated to TCR/IL-2 signaling was necessary and sufficient to achieve this in vitro maturation. Whether this sequence of events actually mimics the maturation of Vα14i NKT cells in vivo remains to be determined. However, based on the current knowledge of Vα14i NKT-cell development, we would argue that it is likely to reflect the in vivo situation. First, T-bet–/– Vα14i NKT cells are clearly blocked at the CD44+ NK1.1– stage of differentiation, and TCR/IL-2 signaling in vitro was not sufficient to induce their maturation. Second, recent findings suggest that the transition of immature (CD44+ NK1.1–)Vα14i NKT cells to the mature “NK stage” in vivo, as evaluated by expression of the NK1.1 marker, necessitate further interactions with CD1d-expressing antigen-presenting cells, most likely mediated through their TCR.41,42 Further in vitro and in vivo experiments with T-bet–transfected immature Vα14i NKT cells will help elucidate the precise sequence of events required for the final maturation of the cells.
Two nonexclusive models of differentiation mediated through T-bet can be envisaged. First, T-bet could be upstream of some membrane receptors required for further extrinsic signals in a T-bet–dependent or –independent manner. For example, expression of T-bet in immature Vα14i NKT cells induced the up-regulation of CD122, imparting the cells with IL-15 responsiveness. IL-15 is an important factor for the survival and homeostasis of Vα14i NKT cells in vivo14,43 and was shown to induce the expression of several chemokines and chemokine receptors in T lymphocytes.44 IL-15 signaling pathway has also been involved in the induction of Ly49 molecules on developing NK cells.45 Therefore, it is possible that the main role of T-bet is to induce CD122 on the surface of immature Vα14i NKT cells and that additional signaling by IL-15 through CD122 subsequently drives the acquisition of other markers associated with fully mature Vα14i NKT cells. Future experiments aimed at expressing CD122 on T-bet–/– Vα14i NKT cells should help identify those maturation events that are CD122 dependent.
Second, T-bet might have distinct intrinsic activities, some of them revealed only when associated with subsequent signaling by TCR and/or IL-2. This might reflect 2 layers of gene-expression control by T-bet. T-bet has been shown to directly transactivate transcription and to induce transcriptionally favorable histone modifications to promote locus accessibility.21,46,47 It is possible that T-bet regulates gene transcription directly but also remodels some gene loci during the development of Vα14i NKT cells. T-bet might directly transactivate the CD122, CxCR3, CCR5, and FasL promoters, which would explain why we detected up-regulation of these transcripts following the expression of T-bet alone. Alternatively, the observed effects in these conditions could be due to nonphysiologic levels of T-bet. Overexpression of T-bet using the retrovirus leads to an amount of T-bet about 3 times higher than the levels found in mature Vα14i NKT cells expanded in IL-15, as measured by quantitative PCR (data not shown). In these conditions, it is possible that the role of T-bet in regulating the expression of these genes is not by directly transactivating their promoter but has more to do with remodeling of their chromatin loci (see the next paragraph). However, the overexpression of T-bet might permit some transcription to occur due to some leakiness of the system. Further experiments aimed at testing whether T-bet can bind and directly transactivate these promoters are warranted.
T-bet might also remodel the chromatin structure of several other gene loci but acute transcription and therefore detection of the transcripts encoding for these genes only becomes apparent following TCR and/or IL-2 signaling. This might be the case for IFNγ, granzyme B, perforin, and RANTES. Consistent with this hypothesis, differences in the chromatin structure of the IFNγ locus between resting Vα14i NKT cells, NK cells, and conventional T cells have been reported.48,49 In contrast with conventional T cells, remodeling of the IFNγ locus occurs during the early development of Vα14i NKT and NK cells.48 Until now it was not clear whether the same factors that are involved in remodeling of the IFNγ locus in T cells are also involved in this developmentally regulated process in Vα14i NKT and NK cells. Our data suggest that, in addition to conventional T cells, T-bet might also be responsible for the remodeling of the IFNγ locus in Vα14i NKT cells. However, the relative paucity of Vα14i NKT cells in wild-type and T-bet–/– mice currently precludes further analysis of the chromatin structure of the IFNγ locus in these cells.
Regardless of the mechanism, our findings clearly established that the expression of these genes is directly linked to the activity of T-bet in Vα14i NKT cells and it is conceivable that these findings might be directly relevant to the biology of other T-bet-expressing cell types. First, CCR5, CxCR3, CxCR6, RANTES, IP-10, lymphotactin, and FasL expression is strongly associated with the T-bet–expressing Th1 phenotype.50-52 Second, a recent report also showed that ectopic expression of T-bet in human Th2 cells strongly up-regulated the expression of FasL, CCR5, and CxCR3 independent of IFNγ secretion.53 Hence, it is likely that T-bet modulates the expression of these genes in the physiologic setting of T-bet–expressing cells, including mature Vα14i NKT cells. We believe that the up-regulation of such genes is driven, either directly or indirectly, by T-bet. Comparison of Vα14i NKT cells from wild-type and T-bet–/– mice revealed lower mRNA levels of granzyme B in T-bet–/– cells. In addition, we found that T-bet–/– NKT cells could only modestly kill antigen-expressing target cells. By contrast, when these same cells were transduced with a T-bet–expressing retrovirus, they gained a potent capacity to kill target cells.
Mature Vα14i NKT cells are fully differentiated cells
Introduction of a dominant-negative form of T-bet into mature Vα14i NKT cells had little effect on the secretion of either IL-4 or IFNγ. Similar results were found for the CCR5, CxCR3, CD95L, CD122, and granzyme B gene expression. This is consistent with the hypothesis that T-bet might not be a classical transcription factor for these genes, but instead behaves as a key inducer of competence to express these genes. Indeed, introduction of the same DN T-bet protein into a fully differentiated Th1 clone could not interfere with the secretion of IFNγ, in sharp contrast with developing Th1 cells.21 This was interpreted as evidence that fully differentiated Th1 clones have undergone stable epigenetic modifications, including chromatin remodeling, that are resistant to the effects of DN T-bet.21 In agreement with the ability of Vα14i NKT cells to rapidly reiterate effector functions, we propose that mature NK1.1+ Vα14i NKT cells might represent fully differentiated cells that have undergone stable epigenetic modifications that became resistant to the effects of DN T-bet. Conditional T-bet gene-deletion strategies will be essential to formally address this possibility.
Conclusions
We find that Vα14i NKT cells final maturation or development of the cells is tightly linked to the acquisition of their trademark characteristics, including their capacity to migrate to inflammatory sites, their ability to proliferate to IL-15, their cytolytic function, and their IFNγ-producing capacity. These various functions are controlled by multiple transcriptional pathways but appear to share a common dependency upon the transcription factor T-bet. T-bet clearly acts as a master regulator that mechanistically unites Vα14i NKT-cell development and effector capacity. Much work remains to be done to elucidate the network of transcription factors that operate in Vα14i NKT lineage cells. However, the identification of genes relevant to the biology of these cells can now provide us with a conceptual framework for additional studies and defines new cell-surface markers as surrogate indicators of Vα14i NKT-cell maturation. Further analysis of gene expression in Vα14i NKT-cell development will likely provide insight into the molecular events that define their maturation as well as those events that can disrupt this developmental process.
Prepublished online as Blood First Edition Paper, December 15, 2005; DOI 10.1182/blood-2005-08-3103.
Supported by grants from the Cancer League of Colorado (L.G.) and the National Institutes of Health (NIH) (AI057485 to L.G. and AI057519 to A.R.H.). Support was also provided by postdoctoral fellowship from NIH NRSA 5 T32AI00048 (J.L.M.).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Mitchell Kronenberg for generously providing the α-GalCer/CD1d tetramers used in some experiments; Drs Steve Reiner and Laurie Glimcher for retroviral constructs; and Dr Laurent Brossay for mCD1d-transfected A20 cells. Biotinylated mCD1d monomers were obtained through the NIH Tetramer facility. We are grateful for the thoughtful review of the manuscript by Drs Mitchell Kronenberg, Yosef Refaeli, James Hagman, Laurent Brossay, and Philippa Marrack. We would like to thank the Gene Expression Core Facility at the University of Colorado Health Science Center and the cell-sorting facility at the National Jewish Medical and Research Center for excellent services.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal