Abstract
Hemophilia A (HA) is caused by partial or total deficiency of F8 protein activity. In a small group, about 1.8% of patients with HA, no mutation is found in the F8 gene. Among this group, we report here on one patient with severe HA in whom no mRNA of the F8 gene was detected. Using 2 common polymorphisms in F8 exon 14, we were able to show that the same allele shared by the patient, his mother, and his sister was not detected by reverse transcription–polymerase chain reaction (RT-PCR) from total blood mRNA. Skewed X-chromosome inactivation in both the mother and the sister was excluded by studying the methylation profile of the androgen receptor gene (HUMARA locus). These findings strongly suggest that the cause of HA in this patient is either absence or rapid degradation of the F8 mRNA, which points to a novel mechanism leading to HA.
Introduction
Hemophilia A (HA) (OMIM, 306700) is caused by impaired F8 protein activity, resulting from a broad spectrum of mutations within the F8 gene (GenBank mRNA accession number, M14113).1 Various point mutations leading to either altered or truncated F8 protein and the 2 major inversion hot spots, intron 22 and intron 1 inversions, have been reported.2,3 Most interestingly, in about 1.8% (15 of 860) of the HA cases we were unable to detect intron 1 or 22 inversions or point mutations or deletions and insertions, even after sequencing the whole F8 exons and their flanking intronic sequences.4 In such cases in which the coding sequence looks normal on the DNA level, few reasons could explain the absence or the decrease in the activity of the corresponding gene product. The first is an absence of mRNA, which includes the rapid degradation or even the complete lack of transcription; the second possibility is an inefficient or failure in translation of the mRNA or even errors in the secretion process. The third, which is applicable to the F8 protein, is a rapid or accelerated clearance from blood as in the case of von Willebrand disease type 2 Normand.
Therefore, to further investigate the reason for HA in this small number of patients we have recently analyzed the F8 mRNA, isolated from total blood, without finding any rearrangements (in 11 unrelated patients).5 However, in one patient with severe HA that we are reporting here we were unable to detect a reverse transcription–polymerase chain reaction (RT-PCR) product from the F8 gene. We followed the segregation of this allele on the DNA level by 2 exon 14 polymorphisms in the family members, mainly his mother and his sister; the same allele present in the patient and his mother was also transmitted to his sister. In all 3 individuals we were unable to get an RT-PCR product corresponding to this allele. Our data strongly suggest that the cause of HA in this patient is either absence or rapid degradation of the F8 mRNA, thus pointing to a novel mechanism leading to hemophilia A.
Patients, materials, and methods
Patients
The index patient is a 13-year-old boy with severe HA. He was on a prophylactic treatment regime from his second year of life. No inhibitor has been reported during more than 1000 exposure days. The very first F8 activity test done at age 8 months prior to F8 substitution was below 1% (both with a chromogenic assay and a one-stage assay). Sensitivity of both assays cannot distinguish values between 0% and 1%. Moreover, the time of 152 minutes in the thromboblastogram (normal, 8-12 minutes) strongly suggests that there is absolutely no functional active F8 protein. He is the first hemophiliac in this family. The coding regions of the F8 gene in this patient were sequenced 3 times by 2 independent centers (the covered sequence relative to the F8 coding region is summarized in Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article); both intron 1 and 22 inversions were also excluded. Both the mother and the sister are suggested to be carriers because of significantly reduced FVIII:C of 33% and 42%, respectively, whereas the VWF:Ag levels were in the normal range (mother, 109%; sister, 139%). The family gave informed consent to participate in the study in accordance with the Declaration of Helsinki.
RT-PCR
Blood (3 mL) was collected in PAXgene blood RNA tubes (PreAnalytiX, Hombrechtikon, Switzerland). Total cellular RNA was extracted using the PAXgene Blood RNA kit according to the manufacturer's procedure (PreAnalytiX). RNA analysis by RT-PCR was performed as described earlier5,6 (Figure 1A). Briefly, for the reverse transcription F8 cDNA was divided into the following 4 different regions that cover all the splicing sites: region A, exons 1 to 8; region B, exons 8 to 14; region C, exons 14 to 22; and region D, exons 19 to 26. Reverse transcription was performed in a 20-μL total volume, using the Ominiscript reverse transcriptase (Qiagen, Hilden, Germany) according to the manufacturer's protocol. An internal control from the human porphobilinogen deaminase gene (PBGD) (GenBank accession number, NM_000190) was also included in the RT to exclude any failure in the RT-PCR that could be due to failure in the RT reaction; a PBGD-specific PCR was positive from each RT.
To increase the sensitivity of detection we performed the RT amplifications in 2 rounds of PCR using a nested approach. Each PCR consisted of 35 cycles. For the first PCR we used 5 μL (for a total of 50-μL PCR reaction) from the reverse transcription. We divided each of the 4 F8 regions (A-D) into 2 regions (total of 8 overlapping regions) that were amplified, by a nested PCR approach, using 5μL (in total of 50 μL) from the first RT-PCR as a template. Primer sequences and the conditions for amplifications are given in Table 1.
Primers used in this study
. | Primers 5′ to 3′ . | Annealing temperature, °C . | Product size, bp . |
|---|---|---|---|
| PBGD control primers | |||
| Outer | |||
| PBGD_hum-F | acc aag gag ctt gaa cat gc | 55 | 784 |
| PBGD_hum-R | gaa aga caa cag cat cat gag | ||
| Nested | |||
| PBGD-N-F | aac atg ccc tgg aga | 47 | 144 |
| PBGD-N-R | atg agg gtt ttc ccg | ||
| F8 exon 14-15 RT-PCR | |||
| Outer | |||
| F1 | aag ggt gaa ttt aca aag gac gt | 52 | 1867 |
| R1 | tga aag tta cca tga tat tat ctt | ||
| Nested | |||
| F2 | aga tgg ttt ttc caa gca gca | 54 | 1731 |
| R2 | agc cat cag taa att cct gga a | ||
| SNuPE | |||
| SN-F8Ex14-1241 | gta gaa ggt tca tat ga | 28 | |
| SN-F8Ex14-1269 | cac aca gct cat ttc tc | 28 | |
| F8B-specific RT-PCR | |||
| RT | |||
| Ex19-26/4* | — | — | — |
| Outer | |||
| F8B-Ex1-F1 | cgt atg caa atc gag ggt ct | 52 | 770 |
| Ex19-26/3* | — | — | — |
| Nested | |||
| F8B-Ex1-F2 | gat cca aga ccc tgg gaa g | 53 | 657 |
| Ex23/26-R | agt ggc cac cct cag tag ag | ||
| F8 promoter region | |||
| F8P-F1 | gga gtg aca gga ctc gct tt | 58 | 695 |
| F8P-R1 | aat cgc aaa agg cac aga aa | ||
| F8P-F2 | ctc tcc cta ata aac att aac ctg a | 58 | 699 |
| F8P-R2 | cac tgc tgc cag tat gag ga | ||
| F8P-F3 | ggc tgt agg caa gcc att ta | 58 | 688 |
| F8P-R3 | cat cag aat gta gcc atg gtg | ||
| F8P-F4 | gga gcc ctt taa cag tgt gc | 58 | 844 |
| F8P-R4 | aga aat ccc agt tcc caa cc | ||
| F8P-F5 | tga aaa ttt ggt aat ggg gtc t | 58 | 785 |
| F8P-R5 | gga tct gtg ggc ttt gag aa | ||
| F8P-F6 | cct tca aca gaa aat cca gca | 58 | 828 |
| F8P-R6 | gca aca aga gcg aga ctc tg | ||
| F8P-F7 | gta gag gat ctg cgc ggt ag | 58 | 838 |
| F8P-R7 | cct atg gtg cca caa aat cc | ||
| F8P-F8 | gat ggc tct gtt tag caa tgg | 58 | 802 |
| F8P-R8 | cac gct gag ctc tgt gat gt | ||
| F8 3′ untranslated region | |||
| F8-3′ end-F1 | gcg agg cac agg acc tct a | 58 | 481 |
| F8-3′ end-R1 | tgc ttt gtt cca tgc ttg at | ||
| F8-3′ end-F2 | gga gaa acc tgc atg aaa gc | 58 | 596 |
| F8-3′ end-R2 | ttg gcc atc aca aat ttc aa | ||
| F8-3′ end-F3 | tga gaa tta tag atg ggg ttc aag a | 58 | 593 |
| F8-3′ end-R3 | ggc aga tgg aag gag cag ta | ||
| F8-3′ end-F4 | ccg tga ctg aaa act aga gtc c | 58 | 576 |
| F8-3′ end-R4 | tgg ggt taa ctc ctc tac cg | ||
| F8-3′ end-F5 | tcc tgc ttg acc ctt atc tga | 58 | 689 |
| F8-3′ end-R5 | ctc cca aag tgc cag gat ta | ||
| Methylation analysis | |||
| Outer | |||
| Bi-F8-PN-F1: | ttt aaa att aat aat aaa gta gtt g | 48 | 210 |
| Bi-F8-PN-R1: | acc taa cca ccc taa aat atc | ||
| Nested | |||
| Bi-F8-PN-F2: | ggg agg tgt ttt gta tat aga atg a | 57 | 195 |
| Bi-F8-PN-R2: | tct cca ata aaa ctc acc caa cct t | ||
| SNuPE | |||
| SN-F8-P-1: | ata aag aag tta gat | 28 | |
| HUMARA assay | |||
| HUMARA-Nor-F | 6-FAM-tcc aga atc tgt tcc aga gcg tgc | 60 | 231 |
| HUMARA-Nor-R | ctc tac gat ggg ctt ggg gag aac | ||
| F8 mRNA analysis | — | — | — |
. | Primers 5′ to 3′ . | Annealing temperature, °C . | Product size, bp . |
|---|---|---|---|
| PBGD control primers | |||
| Outer | |||
| PBGD_hum-F | acc aag gag ctt gaa cat gc | 55 | 784 |
| PBGD_hum-R | gaa aga caa cag cat cat gag | ||
| Nested | |||
| PBGD-N-F | aac atg ccc tgg aga | 47 | 144 |
| PBGD-N-R | atg agg gtt ttc ccg | ||
| F8 exon 14-15 RT-PCR | |||
| Outer | |||
| F1 | aag ggt gaa ttt aca aag gac gt | 52 | 1867 |
| R1 | tga aag tta cca tga tat tat ctt | ||
| Nested | |||
| F2 | aga tgg ttt ttc caa gca gca | 54 | 1731 |
| R2 | agc cat cag taa att cct gga a | ||
| SNuPE | |||
| SN-F8Ex14-1241 | gta gaa ggt tca tat ga | 28 | |
| SN-F8Ex14-1269 | cac aca gct cat ttc tc | 28 | |
| F8B-specific RT-PCR | |||
| RT | |||
| Ex19-26/4* | — | — | — |
| Outer | |||
| F8B-Ex1-F1 | cgt atg caa atc gag ggt ct | 52 | 770 |
| Ex19-26/3* | — | — | — |
| Nested | |||
| F8B-Ex1-F2 | gat cca aga ccc tgg gaa g | 53 | 657 |
| Ex23/26-R | agt ggc cac cct cag tag ag | ||
| F8 promoter region | |||
| F8P-F1 | gga gtg aca gga ctc gct tt | 58 | 695 |
| F8P-R1 | aat cgc aaa agg cac aga aa | ||
| F8P-F2 | ctc tcc cta ata aac att aac ctg a | 58 | 699 |
| F8P-R2 | cac tgc tgc cag tat gag ga | ||
| F8P-F3 | ggc tgt agg caa gcc att ta | 58 | 688 |
| F8P-R3 | cat cag aat gta gcc atg gtg | ||
| F8P-F4 | gga gcc ctt taa cag tgt gc | 58 | 844 |
| F8P-R4 | aga aat ccc agt tcc caa cc | ||
| F8P-F5 | tga aaa ttt ggt aat ggg gtc t | 58 | 785 |
| F8P-R5 | gga tct gtg ggc ttt gag aa | ||
| F8P-F6 | cct tca aca gaa aat cca gca | 58 | 828 |
| F8P-R6 | gca aca aga gcg aga ctc tg | ||
| F8P-F7 | gta gag gat ctg cgc ggt ag | 58 | 838 |
| F8P-R7 | cct atg gtg cca caa aat cc | ||
| F8P-F8 | gat ggc tct gtt tag caa tgg | 58 | 802 |
| F8P-R8 | cac gct gag ctc tgt gat gt | ||
| F8 3′ untranslated region | |||
| F8-3′ end-F1 | gcg agg cac agg acc tct a | 58 | 481 |
| F8-3′ end-R1 | tgc ttt gtt cca tgc ttg at | ||
| F8-3′ end-F2 | gga gaa acc tgc atg aaa gc | 58 | 596 |
| F8-3′ end-R2 | ttg gcc atc aca aat ttc aa | ||
| F8-3′ end-F3 | tga gaa tta tag atg ggg ttc aag a | 58 | 593 |
| F8-3′ end-R3 | ggc aga tgg aag gag cag ta | ||
| F8-3′ end-F4 | ccg tga ctg aaa act aga gtc c | 58 | 576 |
| F8-3′ end-R4 | tgg ggt taa ctc ctc tac cg | ||
| F8-3′ end-F5 | tcc tgc ttg acc ctt atc tga | 58 | 689 |
| F8-3′ end-R5 | ctc cca aag tgc cag gat ta | ||
| Methylation analysis | |||
| Outer | |||
| Bi-F8-PN-F1: | ttt aaa att aat aat aaa gta gtt g | 48 | 210 |
| Bi-F8-PN-R1: | acc taa cca ccc taa aat atc | ||
| Nested | |||
| Bi-F8-PN-F2: | ggg agg tgt ttt gta tat aga atg a | 57 | 195 |
| Bi-F8-PN-R2: | tct cca ata aaa ctc acc caa cct t | ||
| SNuPE | |||
| SN-F8-P-1: | ata aag aag tta gat | 28 | |
| HUMARA assay | |||
| HUMARA-Nor-F | 6-FAM-tcc aga atc tgt tcc aga gcg tgc | 60 | 231 |
| HUMARA-Nor-R | ctc tac gat ggg ctt ggg gag aac | ||
| F8 mRNA analysis | — | — | — |
PBGD indicates porphobilinogen deaminase gene; SNP, single nucleotide polymorphism; SNuPE, single nucleotide primer extension; SIRPH, SNuPE-Ion Pair Reverse Phase HPLC (high-performance liquid chromatography); and —, as described by El-Maarri et al.5
Search for informative markers
Informative markers (single nucleotide polymorphisms, SNP) in exon 14 of the factor VIII gene at codon 1241 (nucleotide 3780, C>G) and codon 1269 (nucleotide 3864, C>A), which are known to harbor 2 frequent SNPs, were analyzed by direct sequencing in both the mother and the sister of the index patient. To obtain an RNA-specific RT-PCR (including these 2 polymorphisms) that is not contaminated by genomic DNA we used an outer pair of primers that amplified part of exon 14 to exon 16, F1/R1 (Figure 1B; Table 1). This was followed by a nested PCR that would increase the yield and the sensitivity of the RT-PCR, F2/R2 (Figure 1B; Table 1).
Allele quantification
To distinguish the presence of 1 or 2 alleles in the RT-PCR product and to quantify the amount of different alleles, we used linear amplification using single nucleotide primer extension assay (SNuPE). In such an assay a primer whose 3′ end is just flanking the polymorphic site is extended by one of either ddNTPs corresponding to the base changes (ie, ddCTP or ddGTP for polymorphism at F8 nucleotide 3780). Separation and detection of the products is done by high performance liquid chromatography (HPLC) as described by El-Maarri et al7 (Figure 2) (see also Table 1, SIRPH analysis).
X-chromosome inactivation
X-chromosome inactivation test at the human androgen receptor gene (HUMARA) locus was done as described by Allen et al8 (Figure 3). Briefly, every DNA sample to be tested was digested with HpaII whose activity is inhibited by methylation; therefore, it will not cut the inactive (methylated) X chromosome. In a normal, randomly inactivated X chromosome, about 50% of each X chromosome is active and the other 50% is inactive; therefore, after digestion with HpaII, both copies of X chromosomes should still be represented as uncut DNA. The highly polymorphic cag repeats in the human androgen receptor are a useful polymorphic marker to identify the 2 X chromosomes. From each case to be tested both digested and undigested DNA are used separately for amplification using fluorescently labeled primer (HUMARA-Nor-(6-FAM)-F; Table 1). To separate the 2 alleles, the PCR products were run on an ABI sequencer machine (ABI Instruments, Foster City, CA); PCR product(s) derived from the undigested DNA would give 2 bands if the 2 alleles have different numbers of cag repeats. In the case of normal random X-chromosome inactivation, about 50% of each X allele is unmethylated and active; therefore, the same 2 alleles should also be detected after HpaII digestion (Figure 3).
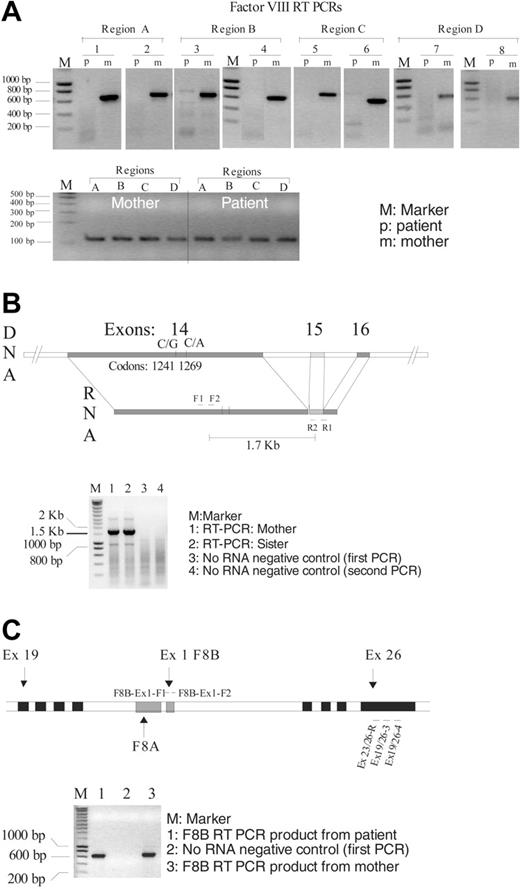
RT-PCR analysis in F8. (A) RT-PCR picture in F8 from both the patient (p; showing no product) and his mother (m; with positive product). Different RT-PCR regions of the F8 are indicated by Reg A (exons 1-8), B (exons 8-14), C (exons 14-19), and D (exons 19-26), whereas the nested PCRs are labeled 1 to 8. An internal control for the success of the reverse transcription, in the PBGD gene, is also shown (A, bottom panel). The PBGD control primer used for the RT was included in each of the RT reactions of the F8 gene. The PCRs from the control PBGD gene and the F8 gene were done separately (regions A, B, C, and D correspond to the same regions in the top part of the figure). (B) A schematic diagram showing the relative positions of the informative markers in F8 exon 14, a picture of the 1.7 kb (kilobase) RNA-specific RT-PCR product that covers exon 15 and the polymorphic markers in exon 14 is also shown. (C) Schematic diagram of the intron 22 region showing the 2 nested genes F8A and F8B. The position of the primers used for the reverse transcription (Ex 19/26-4) and the 2 subsequent PCRs are also shown (first PCR, F8B-Ex1-F1/Ex 19/26-3; second PCR, F8B-Ex1-F2/Ex 23/26-R). A representative picture of the RT-PCR product of the F8B is also shown. Primers are listed in Table 1.
RT-PCR analysis in F8. (A) RT-PCR picture in F8 from both the patient (p; showing no product) and his mother (m; with positive product). Different RT-PCR regions of the F8 are indicated by Reg A (exons 1-8), B (exons 8-14), C (exons 14-19), and D (exons 19-26), whereas the nested PCRs are labeled 1 to 8. An internal control for the success of the reverse transcription, in the PBGD gene, is also shown (A, bottom panel). The PBGD control primer used for the RT was included in each of the RT reactions of the F8 gene. The PCRs from the control PBGD gene and the F8 gene were done separately (regions A, B, C, and D correspond to the same regions in the top part of the figure). (B) A schematic diagram showing the relative positions of the informative markers in F8 exon 14, a picture of the 1.7 kb (kilobase) RNA-specific RT-PCR product that covers exon 15 and the polymorphic markers in exon 14 is also shown. (C) Schematic diagram of the intron 22 region showing the 2 nested genes F8A and F8B. The position of the primers used for the reverse transcription (Ex 19/26-4) and the 2 subsequent PCRs are also shown (first PCR, F8B-Ex1-F1/Ex 19/26-3; second PCR, F8B-Ex1-F2/Ex 23/26-R). A representative picture of the RT-PCR product of the F8B is also shown. Primers are listed in Table 1.
HPLC chromatograms of the single nucleotide primer extension product to distinguish the 2 alleles in exon 14 to 15 RT-PCRs. The presence of 2 peaks in the DNA-PCR (C and G at codon 1241 and C and A at codon 1269, respectively) indicate that this is an informative marker in females. The analysis of the DNA-PCR product that showed 2 alleles (2 peaks) and the RT-PCR product that showed the presence of only 1 allele (1 peak) indicate a monoallelic expression.
HPLC chromatograms of the single nucleotide primer extension product to distinguish the 2 alleles in exon 14 to 15 RT-PCRs. The presence of 2 peaks in the DNA-PCR (C and G at codon 1241 and C and A at codon 1269, respectively) indicate that this is an informative marker in females. The analysis of the DNA-PCR product that showed 2 alleles (2 peaks) and the RT-PCR product that showed the presence of only 1 allele (1 peak) indicate a monoallelic expression.
Methylation analysis
Bisulfite treatment of the DNA was done as previously described.9 Briefly, 50 to 100 ng DNA were digested with EcoRI (total volume of 21 μL). DNA was denatured by mixing it with 4 μL of 2 M NaOH followed by incubation at 50°C for 15 minutes. After mixing with hot 2% low-melting agarose, the DNA-Agarose beads were pipetted in 10 μL volumes in ice-cold heavy mineral oil (Sigma, Steimheim, Germany). After the solidification of the beads, 500 μL of a 2.5 M sodium metabisulfite (Merck, Darmstadt, Germany) and 125 mM hydroquinone (Sigma) pH 5.0 were added. The tubes were incubated for 30 minutes on ice then for 3 hours 30 minutes at 50°C in the dark. The beads were washed 4 times in 1 × TE (pH 8.0), treated twice with 0.2 M NaOH for 15 minutes, and washed twice with 1 × TE for 10 minutes each. Prior to PCR amplification, the beads were washed twice with H2O for 10 minutes each, and melted at 70°C for 10 minutes. Five microliters of this melted agarose were used in each 50-μL PCR reaction. PCR was performed in 2 rounds of amplification to increase the specificity for the amplification of fully bisulfite-converted templates.
X-chromosome inactivation patterns at the HUMARA locus. Two controls of random inactivation (the presence of both alleles 1r and 2r before and after HpaII digestion) and skewed inactivation (presence of 2 alleles 1k and 2k before and 1 allele 2k after HpaII digestion) are included. The male patient has 1 allele 1p corresponding to his active X chromosome; the same allele is absent after HpaII digestion, indicating its normal active status. Two alleles were detected for the mother (1m and 2m) and the sister (1s and 2s) before and after HpaII digestion, indicating a normal random X-Chr inactivation in both. In most of the diagrams, peaks appear in duplicate, which is the result of unspecific flanking dATP added by the taq polymerase (for some of the product) at the 3′ end of the PCR product. The small peaks to the left side of each major peak are a result of slippage of the taq polymerase during the PCR reaction that results in amplifications of n-1 of the cag repeats.
X-chromosome inactivation patterns at the HUMARA locus. Two controls of random inactivation (the presence of both alleles 1r and 2r before and after HpaII digestion) and skewed inactivation (presence of 2 alleles 1k and 2k before and 1 allele 2k after HpaII digestion) are included. The male patient has 1 allele 1p corresponding to his active X chromosome; the same allele is absent after HpaII digestion, indicating its normal active status. Two alleles were detected for the mother (1m and 2m) and the sister (1s and 2s) before and after HpaII digestion, indicating a normal random X-Chr inactivation in both. In most of the diagrams, peaks appear in duplicate, which is the result of unspecific flanking dATP added by the taq polymerase (for some of the product) at the 3′ end of the PCR product. The small peaks to the left side of each major peak are a result of slippage of the taq polymerase during the PCR reaction that results in amplifications of n-1 of the cag repeats.
SIRPH analysis for quantitative methylation
The SNuPE-IP RP HPLC assay (SIRPH) was performed as previously described.7 Briefly, to remove the unreacted PCR oligonucleotides and excess of dNTPs, the PCR product(s) were purified using QIAquick gel extraction kit (Qiagen). The SNuPE reaction was prepared by mixing 100 ng PCR product, 12 pmole SNuPE oligonucleotides, 50 μM of each ddCTP and ddTTP (Amersham, Uppsala, Sweden), 3 units thermosequenase (Amersham) in a total volume of 20 μL. This mixture was then incubated for 60 cycles as follows: 15 seconds at 92°C, 15 seconds at 30°C, and 15 seconds at 60°C. Then, the product was directly loaded on the dHPLC machine (Wave; Transgenomics, San Jose, CA) to separate the extended products and quantitatively measure their corresponding peaks.
Results
Absence of detectable mRNA from the index patient and monoallelic expression in the mother and the sister
As a result of previous extensive work to identify causative mutations in patients with HA, a small group of patients (about 1.8%; 15 of 860) was identified as having no detectable DNA alteration in the F8 coding region.4 In one of these patients we were unable to detect any RT-PCR product from the F8 mRNA from 3 different RNA preparations. A total of 24 RT-PCRs (8 from each RNA extraction) failed to give any product corresponding to the F8 mRNA (Figure 1). In contrast to the index patient, we were able to obtain all 8 RT-PCR products from his mother (his sister was not investigated except for the exon 14 region).
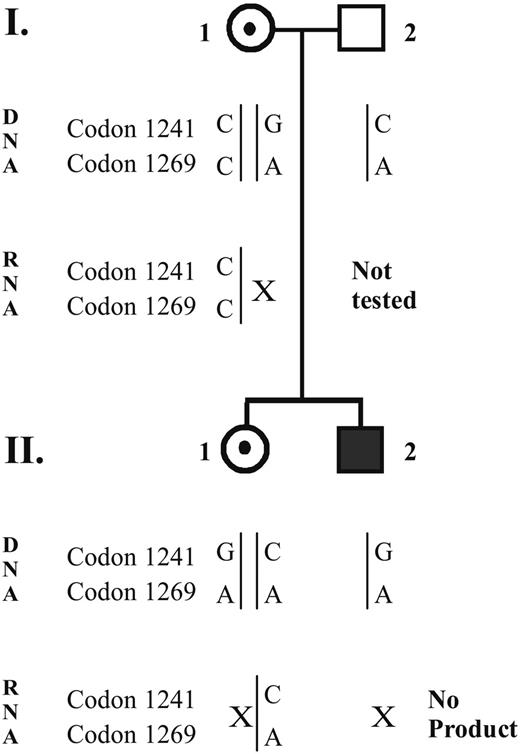
The absence of the RT-PCR product from this patient led us to investigate whether the same allele is expressed in the mother and the sister, which are both likely to be carriers because of their low FVIII:C values. Therefore, to distinguish the 2 alleles, we looked for the 2 common polymorphisms in F8 exon 14 region (codons 1241, nucleotide 3780C>G, and 1269, nucleotide 3864C>A) in both the mother and the sister. Both were informative with the mother having alleles CC/GA (allele 1 nt3780-nt3864/allele 2 nt3780-nt3864); the daughter has CA/GA allele combination (Figure 2). The GA allele is shared by the index patient, his mother, and his sister. We than looked at the RT-PCR products that cover these 2 polymorphisms in both the mother and the sister (Figure 1B). We were able to detect only the CC (mother) and CA (sister) alleles but not the GA allele (Figure 2). This observation would suggest either a monoallelic expression or a rapid degradation of the GA allele on the mRNA level.
To investigate whether this lack of mRNA in this patient is specific for the F8 gene or includes a broader region, we looked for the expression of other neighboring transcripts in the F8 region. The F8 locus is known to contain 2 nested genes: Factor 8A (F8A) and Factor 8B (F8B).10,11 The F8B transcript consists of exons 23 to 26 of the F8 gene and additionally has a unique small exon (64 bp) located in F8 intron 22. A bidirectional promoter is shared with the F8A gene that is transcribed in the opposite direction to both F8 and F8B. For the F8B-specific RT-PCR we used a forward primer located in the first exon of F8B and a reverse primer in exon 26 of F8, followed by a second PCR with nested primers (Figure 1C). A clear RT-PCR product corresponding to part of the F8B transcript was obtained from both the mother and the patient (the sister was not investigated for this transcript). Moreover, a specific RT-PCR product was also obtained from HCBP6 (hepatitis C virus core-binding protein 6; gene bank accession number, AY032594) a neighboring gene about 5 kb upstream of F8, in both the patient and the mother (data not shown).
X-chromosome inactivation
The absence of detectable expression of the GA allele in the lymphocyte-derived RNA in both the mother and the sister could theoretically also be the result of skewed X-chromosome inactivation that is leaving only the allele from the active X chromosome to be actively expressed. However, analysis of the HUMARA locus proved a normal random X-chromosome inactivation in both the mother and the sister, demonstrated by the presence of both alleles before and after HpaII digestion (Figure 3). Therefore, a biased or skewed X-chromosome inactivation does not explain this observed monoallelic expression.
Methylation analysis
The promoter region of the F8 gene contains no CpG island. However, about 5 kb upstream of the transcription starting site a CpG-rich region was identified (GenBank accession number, AL645722; nucleotides, 14701-15601). Because high level of methylation in the gene promoter region is known to be associated with gene silencing, we investigated the possibility that this region could be more methylated in the silenced allele (ie, in the patient). Therefore, the methylation pattern of this region was analyzed using bisulfite-based DNA treatment followed by SIRPH analysis as a quantitative approach to determine the level of methylation7,9 (Figure 4). We included for this study the 3 family members plus 13 healthy males and females. The observed level of methylation at the studied CpG site in the patient was 9%, which falls in the normal range of healthy male controls, 8% to 13%. Therefore, it is unlikely that a methylation-related silencing mechanism could explain this absence of F8 RT-PCR from the patient. The mother and the sister have 26% and 30%, respectively; healthy female controls varied between 18% and 25%. The differences between the male and female group of controls suggest that this region is subject to some slight differential methylation associated with the X-Chr inactivation process.
Quantitative methylation analysis. The methylation levels obtained at 1 CpG site (nucleotide 14779 in AL645722) in a CpG-rich region 5 kb upstream of the F8 transcription starting site.
Quantitative methylation analysis. The methylation levels obtained at 1 CpG site (nucleotide 14779 in AL645722) in a CpG-rich region 5 kb upstream of the F8 transcription starting site.
Sequencing of F8 promoter and 3′-untranslated regions
To exclude any DNA mutations in either the promoter region or the 3′-untranslated region, we sequenced about 3 kb upstream of the translation starting site and about 2 kb downstream from the stop codon. Both regions were normal (Table S1).
Integrity of the X chromosome and the F8 promoter region
To exclude also gross chromosomal rearrangements that could influence the F8 expression because of a position effect we performed karyotype analysis (G-banding at the 500 band level); no abnormality was observed (data not shown). Moreover, the integrity of the promoter region was further assessed by Southern blot analysis using F8 exon 1 as a probe; DNA from the patient, his mother, his sister, and healthy control subjects were digested with the following enzymes: EcoRI, BglI, or HpaI (each separately). This covers a region that extends from 3.3 kb upstream of exon 1 to 11.6 kb downstream of the same exon. All samples gave expected band sizes as healthy controls (data not shown).
Discussion
In the present study, we report on a patient with hemophilia A, in whom we were unable to detect RT-PCR product specific for the F8 gene from 3 separate mRNA preparations (from total blood) and 8 different regions of the F8 gene (Figure 1A). To investigate whether this finding is specific only for this allele in the patient's genetic background we studied his mother and his sister, both of whom were suggested to be carriers from the laboratory measurements (mother, FVIII:C 33% and VWF:Ag 109%; sister, FVIII:C 42% and VWF:Ag 139%).
The 2 alleles in both the mother and the sister were distinguished by the analysis of 2 known polymorphisms in exon 14 of the F8 gene, C3780G (codon 1241) and C3864A (codon 1269). The mother was informative for both SNPs, whereas the sister was informative for only the C3780G polymorphism (Figures 2,5). RT-PCR revealed that the same GA allele that was not detected in the patient was also absent from the RT-PCR products of both the mother and the sister in 3 different experiments. In a control RNA sample both the CA and GA alleles were detected by RT-PCR (Figure 2).
The F8 gene main expression sites are the sinusoidal endothelial cells and Copper cells in the liver as studied in mice.12 Therefore, this absence of detectable expression in this patient could be because the studied mRNA are isolated from total blood that expresses only low levels of F8. Two reasons argue against this: (1) in humans an ectopic expression of F8 in total blood can be detected, and this is widely used to screen for mutations or rearrangements in the F8 cDNA13-15 ; (2) the results obtained in control subjects and the reproducibility and consistency of the experiments in the patient, his mother, and his sister should exclude any bias being specific for ectopic lymphocyte RT-PCR.
Pedigree of the family with a summary of the results. The observed alleles obtained from genomic DNA or from the RT-PCR products are shown as vertical lines, labeled with C, A, or G for the corresponding polymorphisms at codons 1241 and 1269 (nucleotides 3780 and 3864, respectively). A given allele that was present in DNA, but not observed in the RT-PCR product, is indicated by X.
Pedigree of the family with a summary of the results. The observed alleles obtained from genomic DNA or from the RT-PCR products are shown as vertical lines, labeled with C, A, or G for the corresponding polymorphisms at codons 1241 and 1269 (nucleotides 3780 and 3864, respectively). A given allele that was present in DNA, but not observed in the RT-PCR product, is indicated by X.
The absence of detectable expression of the GA allele in the lymphocyte-derived RNA in both the mother and the sister could theoretically also be the result of skewed X-chromosome inactivation that is leaving only the allele from the active X chromosome to be actively expressed. However, analysis of the HUMARA locus showed a normal random X-chromosome inactivation in both the mother and the sister (Figure 3).
Another possibility of our finding could be a local abnormal “condensed” chromatin structure that would affect the expression of the F8 gene among other neighboring genes. The F8B transcript, which is completely nested within the F8 locus, represents a good candidate to further elucidate this possibility. An RT-PCR product specific for the F8B transcript could be obtained from the patient RNA (Figure 1C). Similar results were obtained from HCBP6 locus located 5 kb upstream of the F8 exon 1. Furthermore, the level of DNA methylation at a CpG-rich region located about 5 kb upstream from the transcription start site was not different from male control subjects (Figure 4), thus further arguing against any global regional silencing because of epigenetic factors.
A second nested gene in the F8 locus is the F8A (located in a 9.1-kb region called intron 22 homologous region 1; int22h-1) (GenBank accession number, NM_012151) which is expressed in antisense direction to F8 and is present in 2 additional copies some 500 to 580 kb downstream of the F8 gene. Antisense transcription is suggested to play a role in gene regulation through the degradation in the sense transcript as observed for RNA interference. However, a correlation of expression between sense and antisense is not always observed; Katayama et al16 studied 15 sense and antisense pairs of expressed genes in bone marrow–derived macrophages after induction with bacterial lipopolysaccharide (LPS). Seven pairs showed variation in expression after LPS induction; 3 have coregulation in expression, 2 reciprocal correlation, and a further 2 showed no correlation. However, it is unlikely that a misexpression (ie, overexpression) of antisense F8A mRNA could cause the silencing of the F8 allele in this patient. Such a down-regulation because of up-regulation of expression in the F8A should affect both F8 alleles in the females and not only one allele as we observed in both the mother and the sister.
Furthermore, we could exclude DNA mutation in the promoter region or the 3′-untranslated regions by direct sequencing of about 3 kb upstream of the translation start site and about 2 kb downstream of the stop codon. Alternatively, a defect in the 3′-untranslated region could be excluded because that F8B gene, which shares the last F8 4 exons, was normally expressed.
Most autosomal genes are biallelically expressed; however, some normal monoallelic expressions are known in mammals. Among these are imprinted genes that are expressed, nonrandomly, from only maternal or paternal copy, and various random monoallelic expressions in autosomal genes like immunoglobulins, T-cell receptors, interleukins, odorant receptors, and the protocadherin genes.17 In addition to these genes, pathologic monoallelic expression in disease-related genes were also reported.18-20 For example, congenital hypothyroidism is a recessive disease caused by mutations in thyroid peroxidase (TPO); patients are usually homozygous or compound heterozygous for gene mutations. In some 17% of the cases there is only one mutated allele; in one of these cases Fugazzola et al18 showed that the intact allele on the DNA level is unexpressed and undetectable on the RNA level. The exact molecular mechanism leading to such absence or decrease in expression is not known. In the case of F8, because of the X-linked inheritance of the F8 gene, the absence of mRNA in a single allele with apparently normal DNA sequence is sufficient to cause hemophilia A in males. Reasons that could explain the absence of RT-PCR product from a single allele could be a mutation lying either deep in one of the large F8 introns that affects the stability or induces the rapid degradation of the mRNA or a mutation located in a still unknown control region that enhances in cis the expression of the gene.
Factors affecting genomewide variation in gene expression over long distances are known to exist, most of them were found to work in trans; however, some are also known to work in cis.21 Such factors are found to act over the long range, because they could be located in large noncoding DNA regions, called gene deserts, which eventually make any mutation mechanism that neutralizes them very difficult to detect.22 An example of such long-range cis-acting regulator was observed in preaxial polydactyly (PPD), a common limb malformation in humans, whereby the disruption of a region that resides about 1 Mb (megabase) away causes the misexpression of the Sonic Hedgehog (Shh) gene.23 Currently, no hints are available for the existence of such a cis-acting regulator that acts on the F8 gene; however, the presence of a long-range acting element cannot be excluded.
In conclusion we are presenting a unique case of HA, where we provided strong evidence that the absence of expression of the F8 and/or the rapid degradation of the F8 mRNA is the cause of HA. This finding points to a novel mechanism leading to HA; however, the molecular basis behind this phenomenon is still to be elucidated.
Prepublished online as Blood First Edition Paper, December 8, 2005; DOI 10.1182/blood-2005-09-3702.
Supported by the German Human Genome Project (BMBF/DLR 01KW0305 and BMBF/DLR-01GS0424) (J.O.) and (DHGP 01KW9905) (H.H.B., J.G., J.O., R.S., and W.S.).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the family members for their cooperation and their great interest during this study. Special thanks are due to Judith Schwalbach and Alexandra Schmitt for excellent technical support.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal