The mutation status and usage of specific VH genes such as V3-21 and V1-69 are potentially independent pathogenic and prognostic factors in chronic lymphocytic leukemia (CLL). To investigate the role of antigenic stimulation, we analyzed the expression of genes involved in B-cell receptor (BCR) signaling/activation, cell cycle, and apoptosis control in CLL using these specific VH genes compared to VH mutated (VH-MUT) and VH unmutated (VH-UM) CLL not using these VH genes. V3-21 cases showed characteristic expression differences compared to VH-MUT (up: ZAP70 [or ZAP-70]; down: CCND2, P27) and VH-UM (down: PI3K, CCND2, P27, CDK4, BAX) involving several BCR-related genes. Similarly, there was a marked difference between VH unmutated cases using the V1-69 gene and VH-UM (up: FOS; down: BLNK, SYK, CDK4, TP53). Therefore, usage of specific VH genes appears to have a strong influence on the gene expression pattern pointing to antigen recognition and ongoing BCR stimulation as a pathogenic factor in these CLL subgroups.
Introduction
Chronic lymphocytic leukemia (CLL) with unmutated immunoglobulin variable heavy chain (VH) gene (VH-UM) displays a worse prognosis compared to VH-mutated CLL (VH-MUT).1,2 Higher expression of ZAP70 (or ZAP-70), a receptor-associated tyrosine kinase, was identified in VH-UM compared to VH-MUT CLL.3,4 B-cell receptor (BCR) cross-linking on ZAP70+ CLL cells led to increased tyrosine phosphorylation of p72 (SYK), indicating an increased activation after BCR stimulation as a potential mechanism accounting for the clinical differences of the VH mutation subgroups.5-7
CLL subsets with highly restricted BCR structure were identified, indicating a role for specific antigens in CLL pathogenesis.8-14 The V1-69 gene is the most common unmutated VH gene in CLL and is associated with a restricted VDJ structure that is distinct from the repertoire of normal B cells.13,14 Similarly, V3-21 gene usage comprises cases with a highly specific BCR structure as evidenced by homologous CDR3 sequences and a restricted VL gene usage.10 Moreover, these cases show a poor clinical outcome regardless of the VH mutation status,11 indicating an independent pathogenic and prognostic role of specific VH gene rearrangements.
To investigate the role of antigenic stimulation in the pathogenesis of CLL subgroups defined by specific VH genes, we analyzed the quantitative expression of 26 genes with central roles in BCR signaling, B-cell activation, cell cycle, and apoptosis control in cases defined by V3-21 and V1-69 usage as compared to VH-MUT and VH-UM CLL not using these VH genes.
Study design
Peripheral blood samples from patients with untreated CLL diagnosed according to established criteria were included after informed consent was obtained in accordance with the Declaration of Helsinki. These studies have been approved by the institutional review board of the University of Ulm. A non–CD19-purified cohort of 69 cases constituted the following subgroups: V3-21 usage, 16 cases (9 mutated, 7 unmutated); V1-69, 17 (all unmutated); VH-MUT (not using V3-21), 17; VH-UM (not using V3-21 or V1-69), 19. A CD19-purified cohort included 53 cases (30 overlapping with the nonpurified cohort): 8 using V3-21 (5 mutated, 3 unmutated); 12 V1-69, 12 were VH-MUT (not using V3-21); 21 VH-UM (not using V3-21 or V1-69). Distribution of age, sex, Binet stages, and the high-risk aberrations del11q22-23 and del17p13 within the cohorts and subgroups is detailed in Supplemental Table S1 (see the Supplemental Tables link at the top of the online article, at the Blood website). Preliminary data of our group implicate a specific biology for CLL using V3-23.15 However, these results were not reproduced independently and no specific VDJ configuration has been observed in these cases. Because of these uncertainties and to avoid potential interferences, we excluded V3-23 cases from the study. Fluorescence in situ hybridization (FISH) analysis and VDJ sequencing were performed as described.16,17 VH homology cut-off was 98%. CD19 purification was carried out using magnetically activated cell sorting (MACS) CD19 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany). Twenty-six candidate genes were analyzed (Table 1), of which ZAP70, LCK, FOS, E2F1, BCL2, and BCLXL showed an overexpression in the CD19- as compared to the CD19+ fraction of patient (n = 3) or healthy donor (n = 3) samples. These genes were therefore analyzed in the CD19-purified cohort. RNA extraction, cDNA synthesis, and real-time quantitative polymerase chain reaction (RQ-PCR) were performed as described18,19 with 3 housekeeping genes for expression normalization (PGK1, LMNB1, ACTB). Primers for SYBR Green detection were used or designed as described19 and as shown in Table S2. We used a closed testing procedure20 together with the step-down maxT method21 using nonparametric test statistics with bootstrap estimated null distributions to analyze gene expression differences between VH-MUT, VH-UM, and V3-21. For the analysis of VH unmutated CLL with versus without usage of the V1-69 gene adjusted P values were computed by the step-down maxT procedure using Wilcoxon rank-sum statistics. For both analyses, a global significance level of .025 was used to control an overall significance level of 5%. Hodges-Lehmann estimates of log2-transformed expression data served to compute fold-change estimates and their 95% confidence intervals (CIs). The purified and nonpurified cohorts were analyzed separately. All statistical computations were done using R, version 2.1.1.22
Comparison of candidate gene expression between the subgroups VH-MUT, VH-UM, and V3-21 and V1-69 and VH-UM
Gene . | VH-UM vs VH-MUT . | V3-21 vs VH-MUT . | V3-21 vs VH-UM . | V1-69 vs VH-UM . |
|---|---|---|---|---|
| Non–CD19-purified cohort | ||||
| AKT | 0.62 (0.45, 1.0) | 0.84 (0.6, 1.2) | 1.2 (0.8, 2.1) | 0.83 (0.58, 1.3) |
| ATM | 0.57 (0.33, 0.97) | 0.66 (0.47, 0.93) | 1.2 (0.69, 2.1) | 0.81 (0.44, 1.4) |
| BAX | 1.4 (1.0, 1.9)* | 0.75 (0.44, 1.1) | 0.55 (0.38, 0.73)* | 0.76 (0.57, 1.0) |
| BLNK | 1.0 (0.66, 1.5) | 0.77 (0.49, 1.2) | 0.8 (0.5, 1.2) | 0.51 (0.32, 0.8)* |
| CCND1 | 0.61 (0.4, 0.94) | 0.66 (0.43, 0.94) | 1.0 (0.69, 1.6) | 1.2 (0.72, 2.0) |
| CCND2 | 2.1 (1.3, 3.2)* | 0.58 (0.4, 0.8)* | 0.27 (0.17, 0.44)* | 0.8 (0.47, 1.4) |
| CCND3 | 1.4 (0.91, 1.9) | 0.7 (0.42, 1.2) | 0.51 (0.32, 0.95) | 0.72 (0.46, 1.0) |
| CDK4 | 1.6 (1.1, 3.3)* | 0.69 (0.42, 1.5) | 0.43 (0.27, 0.71)* | 0.32 (0.17, 0.62)* |
| FADD | 0.8 (0.55, 1.2) | 1.1 (0.74, 1.5) | 1.3 (0.88, 1.9) | 1.1 (0.71, 1.7) |
| JAK3 | 1.2 (0.86, 1.7) | 0.82 (0.56, 1.2) | 0.66 (0.49, 0.87) | 0.74 (0.57, 0.98) |
| LYN | 1.1 (0.7, 1.8) | 0.78 (0.56, 1.2) | 0.73 (0.42, 1.2) | 0.92 (0.58, 1.5) |
| MYC | 1.0 (0.6, 2.0) | 0.84 (0.46, 1.4) | 0.78 (0.43, 1.4) | 0.96 (0.46, 1.9) |
| NFKB | 1.4 (1.0, 2.4) | 0.87 (0.57, 1.1) | 0.58 (0.34, 0.88) | 1.3 (0.82, 2.0) |
| P27 | 0.98 (0.75, 1.2) | 0.63 (0.5, 0.83)* | 0.65 (0.51, 0.85)* | 1.1 (0.94, 1.5) |
| PI3K | 1.7 (1.2, 2.7)* | 0.77 (0.57, 1.1) | 0.44 (0.28, 0.66)* | 0.7 (0.47, 1.1) |
| PLCG2 | 0.75 (0.53, 1.1) | 0.7 (0.48, 0.98) | 0.93 (0.67, 1.4) | 0.78 (0.53, 1.1) |
| STAT6 | 0.95 (0.69, 1.4) | 0.96 (0.64, 1.4) | 0.98 (0.67, 1.4) | 0.64 (0.44, 1.0) |
| SYK | 1.1 (0.79, 1.4) | 0.69 (0.36, 1.1) | 0.63 (0.34, 1.0) | 0.57 (0.45, 0.8)* |
| TP53 | 1.3 (0.9, 1.9) | 0.77 (0.52, 1.2) | 0.57 (0.4, 0.82) | 0.49 (0.32, 0.8)* |
| TRAF3 | 0.64 (0.46, 0.95) | 0.87 (0.62, 1.3) | 1.4 (0.94, 2.1) | 1.3 (0.77, 2.1) |
| CD19-purified cohort | ||||
| BCL2 | 0.78 (0.51, 1.2) | 0.8 (0.29, 1.3) | 1.0 (0.36, 1.8) | 0.92 (0.51, 1.7) |
| BCLXL | 0.68 (0.49, 0.96) | 1 (0.45, 2.2) | 1.4 (0.63, 3.2) | 0.86 (0.59, 1.3) |
| E2F1 | 0.8 (0.24, 3.0) | 0.77 (0.37, 1.6) | 1.1 (0.58, 2.2) | 1.1 (0.61, 1.6) |
| FOS | 0.67 (0.43, 1.1) | 1.2 (0.48, 5.4) | 1.7 (0.56, 5.2) | 3.7 (1.5, 9.8)* |
| LCK | 0.92 (0.5, 2.1) | 1.4 (0.66, 3.3) | 1.3 (0.73, 2.9) | 0.65 (0.38, 1.3) |
| ZAP70 | 5.8 (2.6, 14)* | 9.3 (3.5, 25)* | 1.5 (0.75, 3.2) | 0.74 (0.36, 1.4) |
Gene . | VH-UM vs VH-MUT . | V3-21 vs VH-MUT . | V3-21 vs VH-UM . | V1-69 vs VH-UM . |
|---|---|---|---|---|
| Non–CD19-purified cohort | ||||
| AKT | 0.62 (0.45, 1.0) | 0.84 (0.6, 1.2) | 1.2 (0.8, 2.1) | 0.83 (0.58, 1.3) |
| ATM | 0.57 (0.33, 0.97) | 0.66 (0.47, 0.93) | 1.2 (0.69, 2.1) | 0.81 (0.44, 1.4) |
| BAX | 1.4 (1.0, 1.9)* | 0.75 (0.44, 1.1) | 0.55 (0.38, 0.73)* | 0.76 (0.57, 1.0) |
| BLNK | 1.0 (0.66, 1.5) | 0.77 (0.49, 1.2) | 0.8 (0.5, 1.2) | 0.51 (0.32, 0.8)* |
| CCND1 | 0.61 (0.4, 0.94) | 0.66 (0.43, 0.94) | 1.0 (0.69, 1.6) | 1.2 (0.72, 2.0) |
| CCND2 | 2.1 (1.3, 3.2)* | 0.58 (0.4, 0.8)* | 0.27 (0.17, 0.44)* | 0.8 (0.47, 1.4) |
| CCND3 | 1.4 (0.91, 1.9) | 0.7 (0.42, 1.2) | 0.51 (0.32, 0.95) | 0.72 (0.46, 1.0) |
| CDK4 | 1.6 (1.1, 3.3)* | 0.69 (0.42, 1.5) | 0.43 (0.27, 0.71)* | 0.32 (0.17, 0.62)* |
| FADD | 0.8 (0.55, 1.2) | 1.1 (0.74, 1.5) | 1.3 (0.88, 1.9) | 1.1 (0.71, 1.7) |
| JAK3 | 1.2 (0.86, 1.7) | 0.82 (0.56, 1.2) | 0.66 (0.49, 0.87) | 0.74 (0.57, 0.98) |
| LYN | 1.1 (0.7, 1.8) | 0.78 (0.56, 1.2) | 0.73 (0.42, 1.2) | 0.92 (0.58, 1.5) |
| MYC | 1.0 (0.6, 2.0) | 0.84 (0.46, 1.4) | 0.78 (0.43, 1.4) | 0.96 (0.46, 1.9) |
| NFKB | 1.4 (1.0, 2.4) | 0.87 (0.57, 1.1) | 0.58 (0.34, 0.88) | 1.3 (0.82, 2.0) |
| P27 | 0.98 (0.75, 1.2) | 0.63 (0.5, 0.83)* | 0.65 (0.51, 0.85)* | 1.1 (0.94, 1.5) |
| PI3K | 1.7 (1.2, 2.7)* | 0.77 (0.57, 1.1) | 0.44 (0.28, 0.66)* | 0.7 (0.47, 1.1) |
| PLCG2 | 0.75 (0.53, 1.1) | 0.7 (0.48, 0.98) | 0.93 (0.67, 1.4) | 0.78 (0.53, 1.1) |
| STAT6 | 0.95 (0.69, 1.4) | 0.96 (0.64, 1.4) | 0.98 (0.67, 1.4) | 0.64 (0.44, 1.0) |
| SYK | 1.1 (0.79, 1.4) | 0.69 (0.36, 1.1) | 0.63 (0.34, 1.0) | 0.57 (0.45, 0.8)* |
| TP53 | 1.3 (0.9, 1.9) | 0.77 (0.52, 1.2) | 0.57 (0.4, 0.82) | 0.49 (0.32, 0.8)* |
| TRAF3 | 0.64 (0.46, 0.95) | 0.87 (0.62, 1.3) | 1.4 (0.94, 2.1) | 1.3 (0.77, 2.1) |
| CD19-purified cohort | ||||
| BCL2 | 0.78 (0.51, 1.2) | 0.8 (0.29, 1.3) | 1.0 (0.36, 1.8) | 0.92 (0.51, 1.7) |
| BCLXL | 0.68 (0.49, 0.96) | 1 (0.45, 2.2) | 1.4 (0.63, 3.2) | 0.86 (0.59, 1.3) |
| E2F1 | 0.8 (0.24, 3.0) | 0.77 (0.37, 1.6) | 1.1 (0.58, 2.2) | 1.1 (0.61, 1.6) |
| FOS | 0.67 (0.43, 1.1) | 1.2 (0.48, 5.4) | 1.7 (0.56, 5.2) | 3.7 (1.5, 9.8)* |
| LCK | 0.92 (0.5, 2.1) | 1.4 (0.66, 3.3) | 1.3 (0.73, 2.9) | 0.65 (0.38, 1.3) |
| ZAP70 | 5.8 (2.6, 14)* | 9.3 (3.5, 25)* | 1.5 (0.75, 3.2) | 0.74 (0.36, 1.4) |
Comparison of candidate gene expression between the subgroups VH-MUT, VH-UM, and V3-21 (Kruskal-Wallis test for 3-group comparisons) and between V1-69 and VH-UM (Wilcoxon rank-sum test statistics for pair-wise comparisons). The numbers for the non–CD19-purified cohort are VH-UM versus VH-MUT, 19 and 17, respectively; for V3-21 versus VH-MUT, 16 versus 17; for V3-21 versus VH-UM, 16 versus 19; and for V1-69 versus VH-UM, 17 versus 19. For the CD19-purified cohort, the respective numbers are 21 versus 12, 8 versus 12, 8 versus 21, and 12 versus 21. Data presented are fold expression changes with the corresponding 95% CIs (lower and upper confidence limits).
Significant expression differences after P adjustment
Results and discussion
In line with previous studies,3,4 a higher ZAP70 expression was identified in VH-UM as compared to VH-MUT cases (Table 1; Figure 1). In addition, VH-UM showed a characteristic overexpression of PI3K and CCND2, for both of which a BCR-dependent up-regulation has been demonstrated experimentally.23,24 This finding is in line with Chen et al5 demonstrating that ZAP70 expression is associated with increased BCR signaling and reinforces the concept of ongoing BCR stimulation as a pathomechanism in VH-UM CLL.5-7
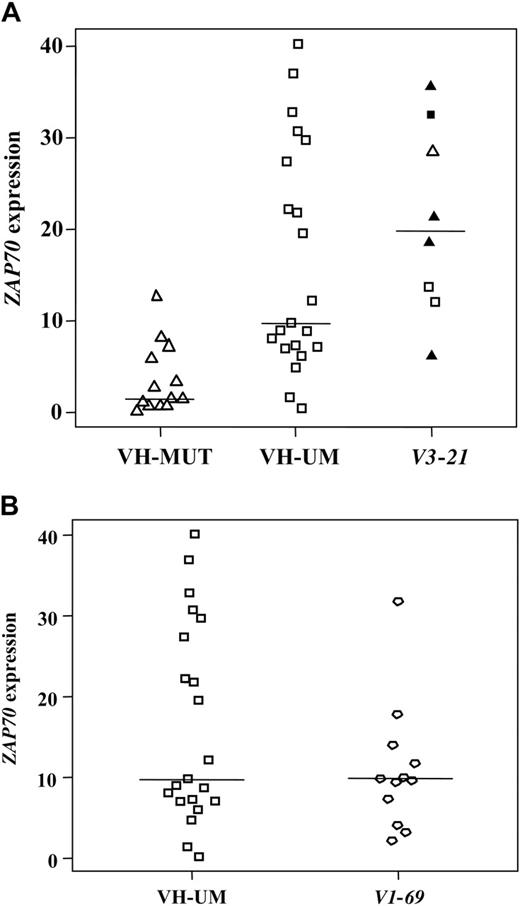
The V3-21 cases under study showed the characteristic distribution into VH mutated and VH unmutated cases with a median VH homology of 97.86%; half of them exhibited the characteristic CDR3 region of 7 amino acids of close homology (Table S3). V3-21 cases showed significantly higher ZAP70 levels as compared to VH-MUT with similar expression levels in the different V3-21 subsets (VH mutated/unmutated/7-amino acid CDR3; Table 1, Figure 1). ZAP70 expression in V3-21 cases was comparable to VH-UM cases, strengthening the role of ZAP70 as a prognostic marker recognizing unfavorable CLL subgroups and pointing to a common pathomechanism involving differential BCR signaling in these subgroups as recently proposed for VH unmutated CLL.6,7 However, marked differences occurred when comparing V3-21 cases with VH-UM CLL including a characteristic down-regulation of several genes in V3-21 cases (Table 1). The lower expression of PI3K and CCND2 in V3-21 cases implicates a down-regulation of these BCR target genes despite antigenic stimulation and points to alternative pathways. Indeed, the down-regulation of candidate genes such as BAX and P27 (Table 1) suggests apoptosis impairment and reduced cell cycle inhibition as additional pathomechanisms in V3-21 CLL, which is supported by a recent report.25 When comparing the mutated versus unmutated V3-21 subset, although restricted by low case numbers, no highly characteristic gene expression differences occurred (Table S4). Therefore, cases with V3-21 usage appear to represent a rather homogeneous biologic group independently of the VH mutation status.
ZAP70 expression. Results from the CD19-purified cohort. (A) VH-MUT, VH-UM, and V3-21 cases; (B) VH-UM and V1-69 cases. V3-21 subgroup: □, VH unmutated; ▵, VH mutated cases; ▴ and ▪, cases with the 7-amino acid CDR3.
ZAP70 expression. Results from the CD19-purified cohort. (A) VH-MUT, VH-UM, and V3-21 cases; (B) VH-UM and V1-69 cases. V3-21 subgroup: □, VH unmutated; ▵, VH mutated cases; ▴ and ▪, cases with the 7-amino acid CDR3.
V1-69 cases under study showed the previously described CLL-specific VDJ configuration,13,14 such as unmutated VH, long CDR3 lengths, and a biased usage of JH6 (Table S5) indicating antigen-specificity of the BCR. When comparing these cases with VH unmutated cases not using V1-69, a number of differentially expressed genes was detected despite the concordant VH mutation status including the BCR-related genes BLNK, SYK, and FOS (Table 1). This finding points to a distinct biology of the V1-69 subset among VH unmutated CLL, which might be explained by the involvement of a distinct antigen leading to antigen-specific BCR responses in V1-69 CLL cells. Additionally, V1-69 cases exhibited reduced levels of TP53, which was not due to a biased distribution of 17p cases (Table S1), and of CDK4 implicating cell-cycle deregulation and apoptosis impairment in these cases.
The concept of antigen-specific BCR stimulation is also supported by the characteristic gene expression differences that occurred between the V3-21 and V1-69 subgroups (Table S6).
Generally, the VH gene-specific gene expression pattern identified supports the concept of antigen selection, as already implicated by the highly restricted VDJ features of these cells, and a persisting antigen-dependence of the CLL cells with ongoing BCR-specific stimulation as a pathomechanism in these CLL subgroups.7-12 These findings strengthen the impact of VH gene usage on CLL pathogenesis and suggest a future role for epitope-specific therapeutic approaches in CLL.
Note added in proof. While this manuscript was under review, the results of high ZAP70 expression in V3-21 CLL could be confirmed by Kröber et al.26
Prepublished online as Blood First Edition Paper, December 1, 2005; DOI 10.1182/blood-2005-04-1483.
Supported by the Deutsche Forschungsgemeinschaft (DFG) (Sti 296/1-1) and the Sander-Stiftung (2001.004.2).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal