Abstract
Angiogenesis is critical to the growth and regeneration of tissue but is also a key component of tumor growth and chronic inflammatory disorders. Endoglin plays a key role in angiogenesis by modulating cellular responses to transforming growth factor-β (TGF-β) signaling and is upregulated in proliferating endothelial cells. To gain insights into the transcriptional hierarchies that govern endoglin expression, we used a combination of comparative genomic, biochemical, and transgenic approaches. Both the promoter and a region 8 kb upstream of exon 1 were active in transfection assays in endothelial cells. In transgenic mice, the promoter directed low-level expression to a subset of endothelial cells. By contrast, inclusion of the –8 enhancer resulted in robust endothelial activity with additional staining in developing ear mesenchyme. Subsequent molecular analysis demonstrated that both the –8 enhancer and the promoter depend on conserved Ets sites, which were bound in endothelial cells in vivo by Fli-1, Erg, and Elf-1. This study therefore establishes the transcriptional framework within which endoglin functions during angiogenesis.
Introduction
Endoglin is a transmembrane glycoprotein that is expressed predominantly in endothelial cells and is critical for the normal development of blood vessels.1 Endoglin-null mice die in utero with defects in vascular remodeling,2-4 and haploinsufficiency of the gene in humans is associated with type 1 hereditary hemorrhagic telangiectasia (HHT), an autosomal dominant disorder characterized by bleeding from vascular malformations in the skin, mucosa, and viscera (reviewed in Lebrin et al5 and Abdalla and Letarte6 ).
Endoglin functions as an accessory receptor for members of the transforming growth factor-β (TGF-β) superfamily of cytokines.7 The TGF-β superfamily includes the TGF-β isoforms, nodal, activins, bone morphogenetic proteins (BMPs), and other related factors, which serve diverse functions during embryonic development and tissue homeostasis in the adult (reviewed in Massague8 ). These cytokines assemble a primary receptor complex made up of type I and type II receptors on the surface of cells and control gene expression by receptor mediated phosphorylation of SMAD proteins. The type I receptors confer signal specificity by preferential phosphorylation of SMAD2/3 or SMAD1/5/8 isoforms, which broadly engage different binding partners and act on separate sets of target genes. Ancillary nonsignaling receptors such as endoglin regulate the pathway by selectively facilitating the binding of subsets of the TGF-β family members to the primary receptor complex.9 Endoglin can also influence the cellular response to a particular cytokine by its preferential interaction with certain type I receptors. ALK1 is one such receptor, and mutations in this gene result in type 2 HHT, a clinical disorder that closely resembles that caused by mutations in the endoglin gene.10
The endothelium is intimately involved in the development and progression of a number of pathologies, including tumor growth and metastasis, chronic inflammatory disorders such as atherosclerosis, and rheumatoid arthritis and thrombosis.11,12 Each of these disorders accounts for significant morbidity and mortality in the community. Endoglin expression is up-regulated on the surface of activated and proliferating endothelial cells.13 Antimitogenic cytokines such as the TGF-β isoforms normally signal through the ALK5 type I receptor and phosphorylate SMAD2/3.14 Endoglin facilitates phosphorylation of SMAD1/5/8 and can switch an antimitogenic TGF-β signal to a proliferative signal in endothelial cells. However, the underlying molecular mechanism is unclear with endoglin reported to promote signaling through ALK1 either by directly engaging the receptor and facilitating its activity15,16 or by down-regulating ALK5 levels and indirectly promoting ALK1 activity.17 Endoglin is up-regulated in tumor neoangiogenesis and is a marker of intratumoral microvessel density, which correlates with poor tumor prognosis.18,19 Endoglin is a hypoxia-inducible gene,20 and the hypoxic environment within tumors and inflammatory lesions may contribute to its up-regulation.21,22
Despite the critical role played by endoglin in maintaining vascular integrity and angiogenesis, little is known about the cis-regulatory elements and transcription factors that control the expression of endoglin in vivo. Prior work on the endoglin promoter has revealed that gene transcription is initiated from a stretch of DNA that has neither a TATA nor CAAT consensus sequence.23,24 This promoter region contains GC-rich regions and consensus motifs for a number of transcription factors including Sp1, Ets, GATA, AP2, nuclear factor κB (NFκB), and TGF-β, glucocorticoid-, vitamin D–, and estrogen-response elements. From this list, biological validation has been limited to Sp1 and its cooperative interaction with SMAD3 and KLF6, an early vascular injury response gene, and an Ets binding site at –415 from the translation start site.25,26 Furthermore, binding of these factors to the promoter region in vivo has not been demonstrated. An approximately 750-bp fragment from the promoter region is reported to target variable levels of gene expression to the endothelium of capillaries, but not large vessels in transgenic mice.27 This, together with the common use of distal regulatory elements to enhance tissue specific gene transcription,28,29 points to the existence of regions outside the promoter fragment that enhance endogenous endoglin expression.
In this study, we used comparative genomics to identify potential regulatory regions of endoglin. We show that conserved Ets binding sites are required for transcriptional activity of the promoter and identify Fli-1, Erg, and Elf-1 as 3 Ets family members that are bound to the promoter in endothelial cells in vivo. In transgenic mice, the promoter has specific but relatively weak and patchy activity in the endothelium of small vessels. We have also identified a conserved –8-kb region of approximately 300 bp that is also bound in vivo by Fli-1, Erg, and Elf-1. We show that this fragment strongly enhancers reporter gene expression of the promoter in transgenic mice, in a tissue specific manner that closely resembles endogenous endoglin expression.
Materials and methods
Sequence analysis
Genomic endoglin sequences and feature files were downloaded from the Ensembl genome browser (www.ensemld.org). Sequence alignments of the endoglin loci were performed with multi-Lagan30 and displayed using SynPlot.31 Putative transcription factor binding sites in the conserved motifs were predicted using transcription factor binding site (TFBS) search.32 To help with sequence alignment and subsequent analysis, the incomplete sequence in the promoter region of the dog endoglin locus was determined by polymerase chain reaction (PCR) amplification and sequencing of genomic DNA extracted from a canine cell line, IIIG7 (a gift from Dr R. Huss, Ludwig Maximilian University Institute of Pathology, Munich, Germany; details available on request).
Quantitative ChIP analysis
Chromatin immunoprecipitation (ChIP) assays were performed as detailed elsewhere.33 Briefly, MS1 and NIH3T3 cells were treated with 0.4% formaldehyde and the cross-linked chromatin was retrieved by nuclei isolation and lysis. The chromatin was sonicated to yield an average fragment size of approximately 500 bp, precleared with rabbit serum, and immunoprecipitated with anti-Ets1 (sc-350x), anti-Ets2 (sc-351x), anti–Fli-1 (sc-356x), anti-Erg (sc-354x), anti–Elf-1 (sc-631x), and anti-GATA2 (sc-9008x) from Santa Cruz Biotechnology (Santa Cruz, CA) to recover the DNA-bound transcription factors, and an anti–acetyl H3 antibody (06-599) from Upstate (Lake Placid, NY) to recover acetylated histones. Enrichment was measured by real-time PCR using Sybr Green (Stratagene, La Jolla, CA) as previously described33 with the forward and reverse primer sets designed using Beacon Designer 4.0 software (Premier Biosoft International, Palo Alto, CA; primer sequences are available on request). The levels of enrichment were normalized to that obtained with a control rabbit antibody and were calculated as a fold increase over that measured at a control region. The 3′ untranslated region (UTR) of endoglin and the promoter of alpha-fetoprotein were used as control regions for the enrichment of transcription factors and acetylated histones, respectively.
Reverse transcriptase–PCR and plasmid constructs
Endoglin expression in MS1 and NIH3T3 cells relative to βactin was quantified using SYBR-Green (Stratagene) real-time PCR. The expression profiles of 20 Ets family members and 6 GATA family members in MS1 and NIH3T3 cells were also established and details of RNA extraction and transcript amplification will be provided on request. DNA fragments corresponding to the conserved regions E1-E9 were PCR amplified from human genomic DNA (primer sequences are available on request). Luciferase reporter constructs were generated according to standard procedures. The promoter fragment E3 was cloned into the XhoI/HindIII site of the pGL2 basic luciferase vector (Promega, Madison, WI). The 5′ E1 and E2 fragments were cloned into the SacI/MluI and SacI/XhoI sites of the pGL2 promoter luciferase vector (Promega), respectively. The 3′ E4-E9 fragments were cloned into the BamH1/SalI sites of the pGL2 promoter luciferase vector. The fragments were also cloned into the pGEM-T Easy vector (Promega) to facilitate downstream applications including the generation of mutant constructs. Plasmids were purified using the Plasmid Maxi Kit (Qiagen, Valencia, CA) and verified by sequencing.
Cell culture and transfection assays
MS1 and NIH3T3 cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum and antibiotics. For stable transfections, 10 μg linearized plasmid DNA and either 1 μg linearized pGK puro (MS1) or pGK neo (NIH3T3) were coelectroporated. Transfected cells were selected at 24 hours by adding either 1 μg/mL puromycin (MS1) or 750 μg/mL G418 (NIH3T3), and assayed 2 to 3 weeks later for luciferase activity as previously described.34
Histology and transgenic analysis
LacZ reporter plasmids with E3 and E1/E3 were generated using standard procedures (details available on request). F0 transgenic mouse embryos were generated by pronuclear injection of linearized plasmids, as described previously.28 For whole-mount analysis, embryos were harvested at 11.5 days after coitus (E11.5), fixed, and stained with 5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside (X-Gal) as detailed previously.28 For histology, the embryos were embedded in paraffin, sectioned, and counterstained with brazilin. To assess endoglin expression, frozen sections were prepared following standard protocols and stained with a biotinylated goat anti–mouse endoglin antibody (BAF1320; R&D Systems, Minneapolis, MN) and visualized with streptavidin–horseradish peroxidase (HRP). The sections were counterstained with hematoxylin (Sigma, St Louis, MO). Whole-mount images were acquired using a Pixera Pro 150ES digital camera (Pixera, Los Gatos, CA) attached to a Nikon SM7800 microscope (Nikon, Kingston upon Thames, United Kingdom). Images of sections were acquired with the same camera attached to an Olympus BX51 microscope (Olympus, Southall, United Kingdom) using Olympus UPlanApo 40×/0.85 numeric aperture (NA) and 100×/1.35 NA objectives. ImagePro Express version 4.5 (Images Processing Solutions, North Reading, MA) was used for acquisition of both types of images. Digital images were processed using Adobe Photoshop (Adobe Systems, San Jose, CA).
Results
Long-range comparative sequence alignment of mammalian endoglin loci reveals multiple regions of conserved noncoding sequences
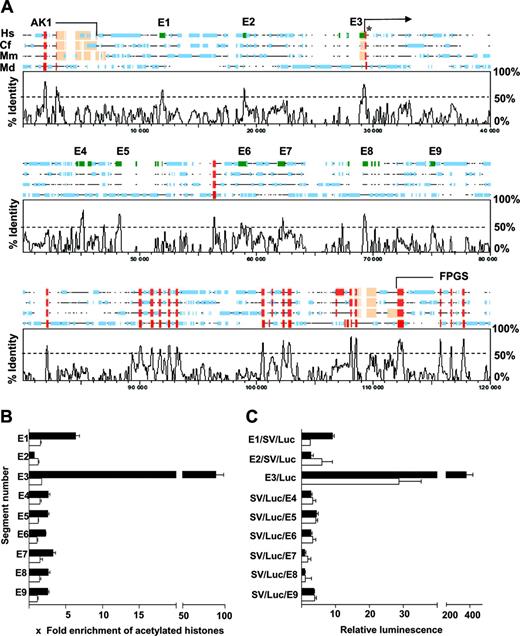
To identify candidate endoglin regulatory elements we compared the genomic loci of 4 mammalian species: human (Homo sapiens), dog (Canis familiaris), mouse (Mus musculus), and opossum (Monodelphis domestica) (Figure 1A). Multispecies sequence comparisons were performed with Lagan and homology plots constructed using SynPlot (“Materials and methods”). There were 9 regions with more than 50% cross-species sequence conservation marked E1-E9 along the human sequence in Figure 1A. The 5′ ends of human endoglin cDNA (listed at http://dbtss.bioinf.med.unigoettingen.de/) range from –357 to –292 with a majority initiated at –306 (the positions are relative to the translation start site of endoglin). This, together with published data,23,24 indicates that the conserved E3 region (–507 to –24) corresponds to the promoter region of endoglin. E1 and E2, the 5′ conserved regions, are approximately –8 and –4 kb from the translation start site of human endoglin and the 3′ conserved regions, E4, E5, E6, E7, E8, and E9, are at approximately +7.5, +9, +12.5, +15, +17.5, and +20 kb, respectively.
Survey of conserved noncoding sequences as potential regulatory regions of human endoglin. (A) Comparative sequence analysis reveals multiple conserved noncoding segments (E1-E9) both upstream and downstream of exon1 (asterisk) of endoglin. A SynPlot graphical representation of mammalian endoglin loci aligned using the multi-Lagan alignment program with sequences from Hs (Homo sapiens), Cf (Canis familiaris), Mm (Mus musculus), and Md (Monodelphis domestica). Segments within noncoding regions (green rectangles numbered E1-E9) with more than 50% sequence identity were selected for further study. The coding exons are represented as red rectangles along the sequence line and the untranslated regions and repetitive elements are marked with beige and blue rectangles, respectively. The base-pair numbering along the horizontal axis includes gaps introduced by the alignment program. The endoglin locus is flanked by AK1 (adenylate kinase 1) and FPGS (folylpolyglutamate synthetase) genes that are partially represented in the diagram. (B) Histone acetylation status determined by a chromatin immunoprecipitation assay comparing the endoglin expressing endothelial cell line, MS1 (▪), with nonexpressing fibroblasts, NIH3T3 (□). Histone acetylation status was used as a surrogate marker of chromatin accessibility and the results for each segment are expressed relative to acetylated histones around the promoter of a nonexpressed gene (alpha-fetoprotein). Segment E3 corresponds to the promoter region of endoglin and had the highest level of acetylation in MS1 cells. The enrichment of acetylated histones around segment E1 was second highest and more than 2-fold higher than any of the other segments. There was no significant enrichment in any segment in NIH3T3 cells. (C) Stable transfection assays in MS1 (▪) and NIH3T3 (□) cells to assess enhancer activity of the selected segments. E3, a 484-bp fragment that corresponds to the endoglin promoter, enhances luciferase activity over the promoterless vector, pGL2basic, by approximately 400-fold in MS1 cells and is significantly more active in MS1 cells than in NIH3T3 cells. E1, a 329-bp fragment, approximately 8-kb 5′ of exon 1, was the most active of the other segments with approximately 8-fold enhancement of luciferase activity over the pGL2 (SV) promoter vector in MS1 cells. Error bars indicate standard deviation
Survey of conserved noncoding sequences as potential regulatory regions of human endoglin. (A) Comparative sequence analysis reveals multiple conserved noncoding segments (E1-E9) both upstream and downstream of exon1 (asterisk) of endoglin. A SynPlot graphical representation of mammalian endoglin loci aligned using the multi-Lagan alignment program with sequences from Hs (Homo sapiens), Cf (Canis familiaris), Mm (Mus musculus), and Md (Monodelphis domestica). Segments within noncoding regions (green rectangles numbered E1-E9) with more than 50% sequence identity were selected for further study. The coding exons are represented as red rectangles along the sequence line and the untranslated regions and repetitive elements are marked with beige and blue rectangles, respectively. The base-pair numbering along the horizontal axis includes gaps introduced by the alignment program. The endoglin locus is flanked by AK1 (adenylate kinase 1) and FPGS (folylpolyglutamate synthetase) genes that are partially represented in the diagram. (B) Histone acetylation status determined by a chromatin immunoprecipitation assay comparing the endoglin expressing endothelial cell line, MS1 (▪), with nonexpressing fibroblasts, NIH3T3 (□). Histone acetylation status was used as a surrogate marker of chromatin accessibility and the results for each segment are expressed relative to acetylated histones around the promoter of a nonexpressed gene (alpha-fetoprotein). Segment E3 corresponds to the promoter region of endoglin and had the highest level of acetylation in MS1 cells. The enrichment of acetylated histones around segment E1 was second highest and more than 2-fold higher than any of the other segments. There was no significant enrichment in any segment in NIH3T3 cells. (C) Stable transfection assays in MS1 (▪) and NIH3T3 (□) cells to assess enhancer activity of the selected segments. E3, a 484-bp fragment that corresponds to the endoglin promoter, enhances luciferase activity over the promoterless vector, pGL2basic, by approximately 400-fold in MS1 cells and is significantly more active in MS1 cells than in NIH3T3 cells. E1, a 329-bp fragment, approximately 8-kb 5′ of exon 1, was the most active of the other segments with approximately 8-fold enhancement of luciferase activity over the pGL2 (SV) promoter vector in MS1 cells. Error bars indicate standard deviation
Histone acetylation and transcriptional activity of candidate regulatory elements
Histone acetylation can increase transcription factor access to chromatin35 by counteracting higher-order chromatin folding that may mask cis elements.36 To prioritize candidate regulatory elements predicted by the bioinformatic approach for transgenic analysis, we quantified the degree of histone acetylation at all 9 conserved regions of the endogenous endoglin locus in MS1 endothelial cells and in NIH3T3 fibroblasts. MS1 endothelial cells express, by quantitative reverse transcription PCR (RT-PCR), approximately 106 more endoglin transcripts than NIH3T3 fibroblasts. Histones in the promoter (E3) and –8 (E1) regions were highly acetylated in endoglin-expressing MS1 cells (filled bars in Figure 1B) relative to both the promoter region of alpha-fetoprotein (a gene that is not expressed in endothelial cells) and a 3′ UTR region of endoglin (data not shown). Acetylation at the other regions was lower than E1 and E3. By contrast, histone acetylation was not increased at any segment, in NIH3T3 cells (open bars in Figure 1B).
To assess the transcriptional activity of candidate regulatory regions, all 9 conserved sequence peaks were PCR amplified from human genomic DNA and subcloned into luciferase reporter plasmids. In stable transfection assays using the endothelial cell line MS1, the promoter construct E3/Luc had approximately 400-fold higher activity than a promoterless luciferase control plasmid (filled bars in Figure 1C corresponding to E3/Luc). The construct with the –8 region, E1/SV/Luc had approximately 8-fold higher activity than the simian virus (SV) promoter luciferase control plasmid, and was the most active of the distal candidate cis-regulatory elements (filled bars in Figure 1C corresponding to E1/SV/Luc). By contrast, the promoter construct, E3/Luc, and the –8 enhancer construct, E1/SV/Luc, had significantly less activity in NIH3T3 cells (compare open bars with filled bars in Figure 1C). The activity of the fragments was also verified by stable transfection of a second endothelial cell line, sEnd-1, which confirmed the results obtained with MS1 cells (data not shown). Taken together, these experiments established the –8 region and the endoglin promoter as key candidate endothelial regulatory elements.
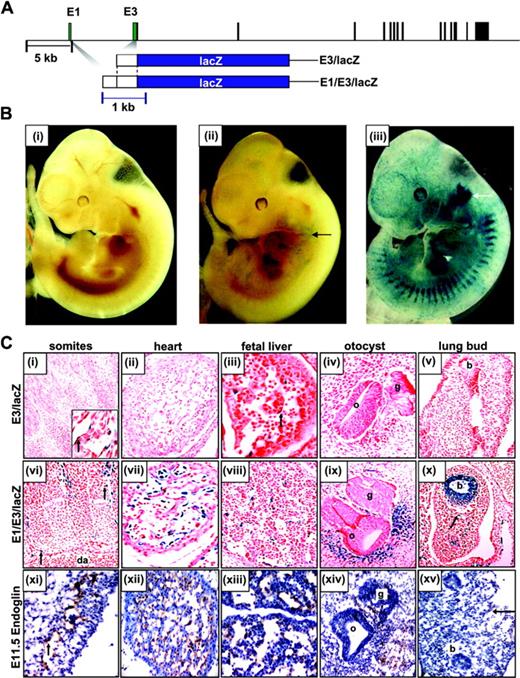
The –8 region (E1) enhances endothelial activity of the promoter region (E3) in transgenic mice, and also targets the developing ear and lung. (A) Transgenic reporter constructs in relation to the endoglin locus. The exons and enhancer fragments are drawn to scale and are represented as black and green rectangles, respectively. The 5-kb and 1-kb scale bars apply to the endoglin locus and the LacZ constructs, respectively. (B) Representative E11.5 embryos stained with X-Gal for LacZ expression (blue). (Bi) The wild-type embryo shows no staining. (Bii) A E3/LacZ transgenic embryo showing weak LacZ expression (blue) in the vascular network (black arrow). (Biii) A E1/E3/LacZ transgenic embryo showing strong LacZ expression in the vascular network and around the developing ear (white arrow). (C) Histologic analysis of tissue sections from X-Gal–stained transgenic embryos in panel B. (Ci-v) Tissue sections from E3/LacZ embryos. (Ci) Somites showing occasional endothelial staining (arrow in inset). (Cii) The heart shows no staining. (Ciii) The fetal liver showing occasional endothelial staining (arrow). (Civ) The developing otocyst and (Cv) lung bud show no staining. (Cvi-x) Tissue sections from E1/E3/LacZ embryos. (Cvi) The dorsal aorta and intersomitic vessels showing strong endothelial staining (arrows). (Cvii) The heart showing strong staining in the endocardium. (Cviii) The fetal liver showing staining in the endothelial cells lining the hepatic sinusoids. (Cix) The otocyst and facioacoustic ganglion showing strong staining in the mesenchyme adjacent to the otocyst. (Cx) The lung bud showing staining in the bronchial epithelium and parenchyma (arrow). (Cxi-xv) Frozen sections of an E11.5 embryo stained with an anti–mouse endoglin antibody. Endoglin-positive cells stain a brown color. (Cxi) Section through blood vessels showing endoglin expression in the endothelium (arrow). (Cxii) Section through the heart showing staining of the endocardium. (Cxiii) Section through the fetal liver showing staining of cells lining the hepatic sinusoids. (Cxiv) Section through the otocyst showing staining of cells in the surrounding mesenchyme. (Cxv) Section through the fetal lung bud shows no staining within the bronchial wall but staining in the endothelial lining of a blood vessel (arrow). b indicates bronchus; da, dorsal aorta; g, ganglion; and o, otocyst.
The –8 region (E1) enhances endothelial activity of the promoter region (E3) in transgenic mice, and also targets the developing ear and lung. (A) Transgenic reporter constructs in relation to the endoglin locus. The exons and enhancer fragments are drawn to scale and are represented as black and green rectangles, respectively. The 5-kb and 1-kb scale bars apply to the endoglin locus and the LacZ constructs, respectively. (B) Representative E11.5 embryos stained with X-Gal for LacZ expression (blue). (Bi) The wild-type embryo shows no staining. (Bii) A E3/LacZ transgenic embryo showing weak LacZ expression (blue) in the vascular network (black arrow). (Biii) A E1/E3/LacZ transgenic embryo showing strong LacZ expression in the vascular network and around the developing ear (white arrow). (C) Histologic analysis of tissue sections from X-Gal–stained transgenic embryos in panel B. (Ci-v) Tissue sections from E3/LacZ embryos. (Ci) Somites showing occasional endothelial staining (arrow in inset). (Cii) The heart shows no staining. (Ciii) The fetal liver showing occasional endothelial staining (arrow). (Civ) The developing otocyst and (Cv) lung bud show no staining. (Cvi-x) Tissue sections from E1/E3/LacZ embryos. (Cvi) The dorsal aorta and intersomitic vessels showing strong endothelial staining (arrows). (Cvii) The heart showing strong staining in the endocardium. (Cviii) The fetal liver showing staining in the endothelial cells lining the hepatic sinusoids. (Cix) The otocyst and facioacoustic ganglion showing strong staining in the mesenchyme adjacent to the otocyst. (Cx) The lung bud showing staining in the bronchial epithelium and parenchyma (arrow). (Cxi-xv) Frozen sections of an E11.5 embryo stained with an anti–mouse endoglin antibody. Endoglin-positive cells stain a brown color. (Cxi) Section through blood vessels showing endoglin expression in the endothelium (arrow). (Cxii) Section through the heart showing staining of the endocardium. (Cxiii) Section through the fetal liver showing staining of cells lining the hepatic sinusoids. (Cxiv) Section through the otocyst showing staining of cells in the surrounding mesenchyme. (Cxv) Section through the fetal lung bud shows no staining within the bronchial wall but staining in the endothelial lining of a blood vessel (arrow). b indicates bronchus; da, dorsal aorta; g, ganglion; and o, otocyst.
The in vivo endothelial activity of the endoglin promoter is enhanced by the –8 region, which also directs expression to the developing ear and lung
To evaluate the in vivo transcriptional activity of the conserved endoglin promoter, founder transgenic mouse embryos were generated using a construct with the –507/–24 promoter region cloned upstream of a promoterless lacZ reporter gene (E3/LacZ construct in Figure 2A). Reporter expression was analyzed by whole-mount X-Gal staining of E11.5 embryos. Four transgenic embryos were generated, and all 4 showed weak expression of the transgene, with staining that appeared to be localized to blood vessels (Figure 2B; compare Bii with Bi). Analysis of histological sections from these E11.5 F0 transgenic embryos confirmed that the staining was specific to the endothelium (Figure 2Ci). Nevertheless, the staining was weak and scattered and confined to subsets of capillaries and a minority of endothelial cells lining the hepatic sinusoids in the fetal liver. Furthermore, staining was absent in the endocardium, large vessels, and yolk sac vasculature where endoglin is expressed (Figure 2C; compare Ci-v with Cxi-xv; data not shown). These data suggested that sequences outside the promoter region (ie, an enhancer region) are required to recapitulate the full spectrum of endogenous endoglin expression in the endothelium.
To evaluate the role of the –8 region as a potential enhancer, we inserted a 329-bp fragment (–8179/–7851) containing the –8 enhancer upstream of the endoglin promoter (E1/E3/LacZ construct in Figure 2A) and assessed the activity of this construct in F0 transgenic embryos. Transgenic expression was evident in 4 of 6 transgenic embryos, 3 of which showed strong staining in blood vessels (Figure 2B; compare Biii with Bi-ii). Histological sections from these E11.5 F0 transgenic embryos confirmed that the extensive staining seen in whole-mount analyses reflected wide-spread endothelial expression in multiple tissues, including brain, somites, heart, fetal liver, and the dorsal aorta (Figure 2Cvi-viii; data not shown). Therefore, unlike the E3 promoter element alone, the E1 enhancer element together with the E3 promoter element recapitulates the expression profile of endogenous endoglin (Figure 2C; compare Cvi-ix with Cxi-xiv).
In E11.5 F0 whole-mount analysis, all 3 embryos expressing the E1/E3/LacZ transgene showed a prominent patch of staining around the developing ear that was absent in the 4 embryos expressing the E3/LacZ transgene (Figure 2B; compare region marked with a white arrow in Biii with the corresponding region in Bii). Analysis of histological sections through this region confirmed that the staining was in the mesenchyme surrounding the developing otocyst (Figure 2Cix). Tissue sections through the developing lung buds of 2 out of 3 E11.5 embryos expressing the E1/E3/LacZ transgene also showed staining within the bronchial epithelium, which was not seen in either the 4 E3/LacZ transgenic embryos (Figure 2C; compare Cx with Cv) or in embryos stained for endogenous endoglin expression (Figure 2C; compare Cx with Cxv).
Ets binding sites are required for transcriptional activity of the promoter
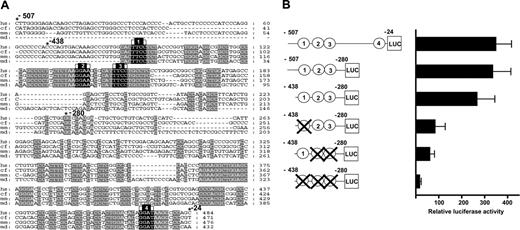
Prior analysis of the endoglin promoter in cell lines has shown that it is a TATA- and CAAT-less promoter with an Sp1 consensus site at –386 (Figure 3A; bp numbering is relative to the translation start site) that is required for basal transcription in vitro.25 There are 3 conserved Ets factor-binding sites (GGA A/T) at –415, –369, and –359 that flank this region and fall within a conserved sequence block, just proximal to the reported transcription initiation sites which range from –357 to –292. A fourth conserved Ets factor binding site (–37) and an overlapping GATA site (–36) lie outside this approximately 70-bp region from which transcription is initiated. To examine the relative importance of these Ets factor-binding sites, mutations were introduced individually and in combination (Figure 3B) and tested in stable transfections of MS1 endothelial cells. The activity of the wild-type and mutant constructs is expressed relative to transfection with the promoterless pGL2basic vector.
Deletion of a 256-bp fragment that includes the fourth Ets motif (and the overlapping GATA motif) resulted in little change to overall promoter activity (compare the –507/–24 construct with the –507/–280 construct in Figure 3B). Deletion of a 69-bp fragment consisting mostly of nonconserved sequences resulted in a relatively minor change (compare the –507/–280 construct with the –438/–280 construct in Figure 3B). In contrast, site-directed mutagenesis of the first Ets site more than halved the activity of the –438/–280 construct. Mutation of the second and third Ets sites in combination resulted in an even greater loss in activity. Mutation of all 3 Ets sites reduced activity by approximately 15-fold. Together, these results demonstrate that conserved Ets binding sites, which lie within a core block of conserved sequences, are critical for activity of the endoglin promoter in endothelial cells.
Conserved Ets transcription factor binding sites are required for promoter activity. (A) Nucleotide sequence alignment of the promoter region (E3) with conserved Ets binding sites marked in black (1-4) and other conserved sequence blocks in gray. (B) Shown on the left are the reporter constructs of human wild-type and mutated E3 fragments inserted upstream of the promoterless pGL2basic vector. The conserved Ets binding sites are represented as circles numbered 1-4 and are crossed out where mutated. Shown on the right are the results of stable transfection assays in MS1 cells corresponding to each construct. The luciferase activities are given as fold increases over the activity of the basic (pGL2B) vector. Each bar is the mean of the relative luciferase activity from at least 2 experiments performed in triplicate ± SD.
Conserved Ets transcription factor binding sites are required for promoter activity. (A) Nucleotide sequence alignment of the promoter region (E3) with conserved Ets binding sites marked in black (1-4) and other conserved sequence blocks in gray. (B) Shown on the left are the reporter constructs of human wild-type and mutated E3 fragments inserted upstream of the promoterless pGL2basic vector. The conserved Ets binding sites are represented as circles numbered 1-4 and are crossed out where mutated. Shown on the right are the results of stable transfection assays in MS1 cells corresponding to each construct. The luciferase activities are given as fold increases over the activity of the basic (pGL2B) vector. Each bar is the mean of the relative luciferase activity from at least 2 experiments performed in triplicate ± SD.
Ets binding sites are required for transcriptional activity of the –8 enhancer
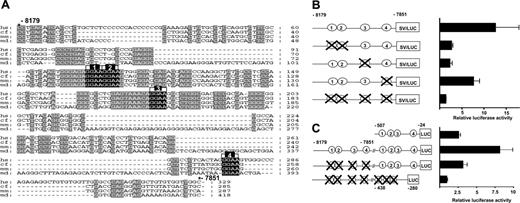
The –8 enhancer region of endoglin has several blocks of conserved sequences, including 4 conserved Ets motifs (Figure 4A). Therefore, to define functionally important Ets motifs within this region, mutations were introduced individually and in combination and the activity of the constructs tested in stable transfection of MS1 endothelial cells (Figure 4B). To avoid potential compensation by conserved Ets motifs in the endogenous promoter, the mutation analysis was first performed using the neutral SV40 promoter. The activity of the wild-type and mutant constructs was expressed relative to the pGL2promoter vector containing the SV minimal promoter. Site-directed mutagenesis of the double Ets motifs (1 and 2) resulted in approximately 75% loss of activity. A similar reduction in activity was seen with mutation of the third Ets motif. The fourth conserved Ets site lies outside the core block of conserved sequences and site-directed mutagenesis of this site resulted in comparatively little reduction in activity. Mutation of all 4 conserved Ets sites abolished enhancer activity within the fragment. Therefore, the activity of the –8 region is largely dependent on the first 3 Ets sites, which lie within a conserved block of sequences.
The in vitro activity of the –8 enhancer was also tested using the endogenous promoter (Figure 4C). The construct with the –8 enhancer (–8179/–7851) and the endogenous promoter fragment (–507/–24) had an approximately 3-fold higher activity than the construct with the promoter fragment alone. This increased activity was lost when the Ets binding sites in the –8 enhancer were mutated. Mutation of the Ets binding sites in the –8 enhancer and the promoter fragment (N1/N3/Luc) resulted in an approximately 8-fold loss of luciferase activity relative to the wild-type construct. The results are shown as a fold increase over the luciferase activity of the N1/N3/Luc construct. These results demonstrate that the activity of the endogenous promoter in endothelial cells is enhanced by the –8 region and is dependent on Ets binding sites. As shown in Figure 2, the –8 region also significantly enhances the in vivo endothelial activity of the endogenous promoter (compare Figure 2Biii with Bii).
Activity of the –8 core enhancer requires conserved Ets transcription factor binding sites. (A) Nucleotide sequence alignment of the –8 enhancer (E1) with conserved Ets binding sites marked in black (1-4) and other conserved sequence blocks in gray. (B) Shown on the left are the reporter constructs of human wild-type and mutated E1 fragments inserted upstream of the SV promoter of the pGL2p vector. As in Figure 3, the conserved Ets binding sites are represented as circles numbered 1-4 and are crossed out where mutated. The constructs were stably transfected into MS1 cells and the results are shown on the right. The luciferase activities are given as fold increases over the activity of the pGL2p vector and represent the mean results of at least 2 experiments performed in triplicate ± SD. (C) Shown on the left are reporter constructs of wild-type and mutated E1 fragments inserted upstream of the wild-type and mutated promoter (E3) fragment. The constructs were stably transfected into MS1 cells, and the results are shown on the right. The luciferase activities are given as fold increases over the activity of the construct with mutation of all Ets binding sites in E1 and E3.
Activity of the –8 core enhancer requires conserved Ets transcription factor binding sites. (A) Nucleotide sequence alignment of the –8 enhancer (E1) with conserved Ets binding sites marked in black (1-4) and other conserved sequence blocks in gray. (B) Shown on the left are the reporter constructs of human wild-type and mutated E1 fragments inserted upstream of the SV promoter of the pGL2p vector. As in Figure 3, the conserved Ets binding sites are represented as circles numbered 1-4 and are crossed out where mutated. The constructs were stably transfected into MS1 cells and the results are shown on the right. The luciferase activities are given as fold increases over the activity of the pGL2p vector and represent the mean results of at least 2 experiments performed in triplicate ± SD. (C) Shown on the left are reporter constructs of wild-type and mutated E1 fragments inserted upstream of the wild-type and mutated promoter (E3) fragment. The constructs were stably transfected into MS1 cells, and the results are shown on the right. The luciferase activities are given as fold increases over the activity of the construct with mutation of all Ets binding sites in E1 and E3.
The promoter region of endoglin and conserved Ets binding sites are required for the in vivo endothelial activity of the –8 enhancer
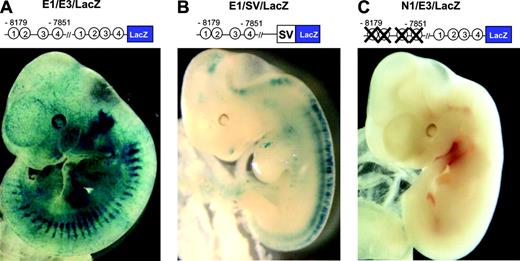
To evaluate the role of the endoglin promoter toward the in vivo endothelial activity of the –8 enhancer, mouse embryos were generated with the –8179/–7851 enhancer fragment cloned upstream of the SV minimal promoter (E1/SV/LacZ in Figure 5). Transgenic expression was seen in 4 of 5 transgenic embryos and was largely nonspecific (Figure 5B). In contrast with embryos generated with the E1/E3/LacZ construct, endothelial staining was minimal (compare Figure 5B with 5A). To evaluate the role of the Ets binding sites in the –8 enhancer toward in vivo endothelial activity, mouse embryos were generated with the –8179/–7851 enhancer fragment with targeted mutations in the conserved Ets motifs, cloned upstream of the endoglin promoter (N1/E3/LacZ in Figure 5). Six transgenic embryos were generated; 5 showed no staining and one showed ectopic staining in the retroperitoneum. In contrast with embryos generated with the E1/E3/LacZ construct, endothelial staining especially in capillaries was minimal (compare Figure 5C with 5A). Furthermore, unlike the E1/E3/LacZ transgenic embryos, neither the E1/SV/LacZ nor N1/E3/LacZ transgenic embryos showed staining around the otocyst or in the lung bud. These data show that the –8 enhancer acts in conjunction with the endoglin promoter in targeting not only the endothelium but also the mesenchyme surrounding the otocyst and the lung bud in vivo and requires Ets transcription factor binding sites for its activity.
The promoter region of endoglin and conserved Ets binding sites are required for the in vivo endothelial activity of the –8 enhancer. E11.5 F0 embryos were generated using either (A) E1/E3/LacZ (wild-type –8 enhancer with the endoglin promoter), (B) E1/SV/LacZ (wild-type –8 enhancer with the SV minimal promoter), or (C) N1/E3/LacZ (mutant –8 enhancer with the endoglin promoter). Representative F0 embryos are shown for each construct.
The promoter region of endoglin and conserved Ets binding sites are required for the in vivo endothelial activity of the –8 enhancer. E11.5 F0 embryos were generated using either (A) E1/E3/LacZ (wild-type –8 enhancer with the endoglin promoter), (B) E1/SV/LacZ (wild-type –8 enhancer with the SV minimal promoter), or (C) N1/E3/LacZ (mutant –8 enhancer with the endoglin promoter). Representative F0 embryos are shown for each construct.
Fli-1, Erg, and Elf-1 occupy the endoglin –8 enhancer and promoter in endothelial cells in vivo
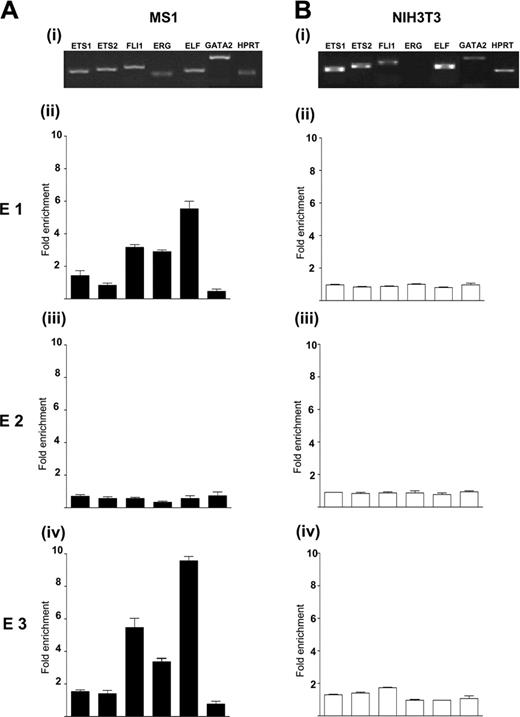
To determine the identity of Ets factors recruited to the endoglin promoter and –8 enhancer, we performed quantitative chromatin immunoprecipitation assays with antibodies to Ets1, Ets2, Fli-1, Erg, and Elf-1 in MS1 cells. These Ets family members were chosen, as they were known to be expressed in this cell line (Figure 6Ai and data not shown) and had been shown to regulate other genes in endothelial cells (reviewed in Lelievre et al37 ). The promoter region (E3) contains a conserved GATA motif (Figure 3) and GATA2 has been implicated in the regulation of endothelial genes.38-40 Because MS1 cells express GATA2 (Figure 6Ai), an antibody to GATA2 was added to this survey. With the exception of Erg, these Ets and GATA factors were also expressed in NIH3T3 fibroblasts (Figure 6Bi). Therefore, ChIP assays were also performed using these antibodies in NIH3T3 cells to compare transcription factor binding at the promoter and –8 enhancer in endoglin expressing cells with nonexpressing cells.
Fli-1, Erg, and Elf-1 bind the –8 enhancer and the promoter region of endoglin in MS1 cells but not NIH3T3 cells. (A) The expression profiles of selected Ets and GATA factors and their enrichment at E1-E3 segments in endoglin expressing MS1 endothelial cells. (Ai) Expression analysis by RT-PCR. (Aii) Chromatin immunoprecipitation assays were performed with anti-Ets1, anti-Ets2, anti–Fli-1, anti-Erg, anti–Elf-1, anti-GATA2, and control immunoglobulin G (IgG) antibodies. The DNA content of the immunoprecipitates was analyzed by real-time PCR for the conserved segments E1-E3. The level of enrichment with each antibody was normalized to the levels obtained with the control IgG and plotted as a fold increase over the level of enrichment at a control region (3′ UTR of endoglin). Fli-1, Erg, and Elf-1 are significantly enriched at E1 and E3 but not E2. (B) The expression profiles of selected Ets and GATA factors and their enrichment at E1-E3 segments in endoglin nonexpressing NIH3T3 fibroblasts. (Bi) RT-PCR expression analysis. Erg is not expressed in NIH3T3 cells. (Bii) Chromatin immunoprecipitation assay. None of the Ets and GATA factors were bound to E1-E3 segments in NIH3T3 cells. Error bars indicate standard deviation.
Fli-1, Erg, and Elf-1 bind the –8 enhancer and the promoter region of endoglin in MS1 cells but not NIH3T3 cells. (A) The expression profiles of selected Ets and GATA factors and their enrichment at E1-E3 segments in endoglin expressing MS1 endothelial cells. (Ai) Expression analysis by RT-PCR. (Aii) Chromatin immunoprecipitation assays were performed with anti-Ets1, anti-Ets2, anti–Fli-1, anti-Erg, anti–Elf-1, anti-GATA2, and control immunoglobulin G (IgG) antibodies. The DNA content of the immunoprecipitates was analyzed by real-time PCR for the conserved segments E1-E3. The level of enrichment with each antibody was normalized to the levels obtained with the control IgG and plotted as a fold increase over the level of enrichment at a control region (3′ UTR of endoglin). Fli-1, Erg, and Elf-1 are significantly enriched at E1 and E3 but not E2. (B) The expression profiles of selected Ets and GATA factors and their enrichment at E1-E3 segments in endoglin nonexpressing NIH3T3 fibroblasts. (Bi) RT-PCR expression analysis. Erg is not expressed in NIH3T3 cells. (Bii) Chromatin immunoprecipitation assay. None of the Ets and GATA factors were bound to E1-E3 segments in NIH3T3 cells. Error bars indicate standard deviation.
Immunoprecipitated chromatin samples were analyzed by quantitative real-time PCR. The levels of enrichment were normalized to that obtained with a control rabbit antibody and are plotted as a fold increase over that measured at a control region (3′UTR of endoglin). Very similar enrichments were obtained when plotted as a fold increase over measurements at a second control region, the alpha-fetoprotein promoter (data not shown). As shown in Figure 6Aii, there was specific, 3- to 5-fold enrichment of Fli-1, Erg, and Elf-1 at the –8 enhancer (E1) in MS1 cells. These same factors were enriched, 4- to 9-fold, at the promoter region (E3) (Figure 6Aiv). By contrast, there was no enrichment of any of the tested factors at the –4 region (E2) (Figure 6Aiii), which did not display increased acetylation or enhancer activity in vitro (Figure 1B-C). Furthermore, there was no enrichment of any of these factors at these segments in NIH3T3 cells (Figure 6Bii-iv). These data demonstrate that the Ets factors, Fli-1, Erg, and Elf-1, are recruited to the –8 enhancer and promoter of endoglin in endothelial cells in vivo. It is likely that the Ets factors bound to the –8 enhancer interact with those bound to the promoter in driving endoglin expression as evidenced by the drastic reduction of in vivo endothelial staining following the mutation of the conserved Ets motifs in the –8 enhancer (Figure 5). GATA2 was not enriched at these conserved regions in MS1 cells. This is consistent with the stable transfection data in MS1 endothelial cells where mutation of the conserved GATA motif did not lead to a loss of reporter activity (compare –507/–280 with –507/–24 in Figure 3B).
Discussion
We used a comparative genomics platform to identify conserved regulatory regions of endoglin. A conserved 484-bp region corresponding to the endoglin promoter was bound in vivo by the Ets family members Fli-1, Erg, and Elf-1, and directed reporter gene expression to the endothelium in transgenic mice. Another conserved 329-bp region –8 kb from exon 1 was also bound by these same Ets factors and strongly enhanced reporter gene expression in the endothelium in vivo. The endoglin promoter alone or in conjunction with the –8 enhancer did not direct expression to fetal liver blood progenitors when assessed by histological sections of E11.5 fetal livers. This contrasts with the regulatory elements of the transcription factors SCL,41 Fli-1, and PRH/HHEX.42 These genes, like endoglin, are expressed in blood stem cells and the endothelium but their regulatory elements target reporter gene expression to both tissues. This suggests that some genes coexpressed in endothelium and HSCs contain regulatory elements that are responsive to shared aspects of the 2 respective transcriptional environments, whereas others, such as endoglin, contain true endothelial specific regulatory elements.
Angiogenesis involves the generation of blood vessels from a preexisting vascular network and requires the integration of signals triggered by growth factors and adhesion events.12 Transcriptional control plays a key role in this dynamic process by adjusting gene expression patterns and/or levels in response to extracellular cues. The Ets family of transcription factors has previously been shown to play a crucial role in angiogenesis (reviewed in Carmeliet and Jain12 ). Ets transcription factors share a highly conserved winged helix-turn-helix domain, which binds to a consensus DNA sequence centered on the core GGA (A/T) motif. Fli-1 and Erg, 2 of the 3 Ets factors found in the current study to regulate endoglin expression in endothelial cells, have previously been shown to regulate genes implicated in endothelial cell injury, neovascularization, and tumor angiogenesis (Deramaudt et al43 and included references). They belong to a subfamily of Ets factors that share a highly conserved pointed domain, named after the Drosophila Ets factor Pointed.37 The pointed domain is a target for mitogen-activated protein (MAP) kinase–mediated phosphorylation and as such, permits modulation of Ets factor activity by extracellular signals (reviewed in Tootle and Rebay44 ). Elf-1, the third Ets factor found to occupy endoglin regulatory elements, is a regulator of vascular development and has recently been shown to play a critical role in tumor angiogenesis.45,46 Elf-1 is subjected to both phosphorylation and glycosylation, which promote nuclear localization, DNA binding, and target gene regulation.47 The data presented in this study therefore suggest that one or more of the Ets factors shown to regulate endoglin may play a role in integrating extracellular signals with the TGF-β pathway, of which endoglin is a component.
TGF-β plays a pivotal role in angiogenesis by regulating endothelial cell growth,16 vascular smooth muscle development,48 and the production and turnover of the extracellular matrix.49 TGF-β1 protein production is subjected to autocrine regulation50 and is defective in the absence of endoglin or the TGF-βR II receptor with which endoglin associates.48 The Ets family members Fli-1 and Erg, which we found in this study to occupy endoglin regulatory elements, also regulate the promoter of TGF-βR II.51 Therefore, the current study emphasizes the role of these Ets factors together with Elf-1 as potential regulators of TGF-β signaling by their role in regulating both endoglin and TGF-βR II. TGF-β induces growth of the extracellular matrix in part by inducing collagen synthesis and suppressing matrix metalloproteinases.51 The defective extracellular matrix in Fli-1–null mouse embryos52 is consistent with a role for this transcription factor in regulating components of the TGF-β autocrine loop. Partial compensation by Erg, which belongs to the same Ets subfamily and also regulates endoglin and TGF-βR II genes, could account for the relatively mild vascular phenotype in Fli-1–null mouse embryos in comparison with endoglin null embryos.
Interestingly, Ewing sarcoma and related primitive neuroectodermal tumors are associated with chromosomal translocations that fuse the EWS gene with Fli-1 and Erg (reviewed in Kovar53 ). One potential molecular explanation for the pathogenesis of this disease has been provided by studies demonstrating that expression of the antimitogenic TGF-βR II, which is normally regulated by Fli-1 and Erg,51 is repressed by the EWS-Fli-1 and EWS-Erg fusion genes.51,54 By analogy, expression of endoglin may also be repressed in Ewing sarcoma, which would result in further perturbation of TGF-β signaling and may thus contribute to the growth and development of these tumors. Erg is also overexpressed in the majority of prostate cancers55 as a result of a recurrent chromosomal rearrangement, which fuses the androgen responsive 5′ UTR of TMPRSS2 with Erg.56 It remains to be seen whether the N-terminal truncation of Erg that results from this gene fusion perturbs either TGF-βR II or endoglin expression.
Our transgenic studies revealed that the –8 region targets reporter gene expression to mesenchymal cells in the inner ear surrounding the developing otocyst, closely matching reporter gene expression in the endoglin LacZ knock-in57 and endogenous endoglin expression. BMP pathways are involved in otic capsule formation and epithelial-mesenchymal signaling in the developing ear.58 The expression of the transgene in this region could be consistent with a previously unsuspected role for endoglin as an accessory receptor in modulating inner ear development. The wild-type (E1) but not mutated (N1) –8 enhancer, in conjunction with the endoglin promoter, consistently targets expression to the bronchial epithelium. However, endoglin is not expressed in the bronchial epithelium. One possible explanation for this apparent discrepancy is the presence within the endogenous endoglin locus of suppressor elements, which are not included in the –8-enhancer/promoter construct.
In summary, we have established that the endoglin promoter is regulated in vivo by the Ets transcription factors Fli-1, Erg, and Elf-1. The promoter directs endothelial expression in transgenic mice but is insufficient to recapitulate the full spectrum of endothelial expression in vivo. We have identified a –8 enhancer region that is also bound by Fli-1, Erg, and Elf-1, and significantly enhances promoter activity in the endothelium in transgenic mice.
Prepublished online as Blood First Edition Paper, February 16, 2006; DOI 10.1182/blood-2005-12-4929.
Supported by grants from the Biotechnology and Biological Sciences Research Council, Leukaemia Research Fund, Wellcome Trust, and the Cambridge-MIT Institute, and a CJ Martin/RG Menzies Fellowship from the National Health and Medical Research Council of Australia (J.E.P).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Dr R. Huss for the IIIG7 canine cell line. We are grateful for the advice and assistance given us by Dr George Follows, Dr Juan Li, Dr Karen Steel, Dr Erika Bosman, Ms Sarah Kinston, and Ms Kathy Knezevic. We thank Paula Braker and Sandie Piltz for coordinating the animal husbandry.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal