The JAK2V617F mutation is present in most patients with polycythemia vera (PV) and in some patients with essential thrombocythemia (ET) and myeloid metaplasia/myelofibrosis (MMM). We sought to investigate the relationship between granulocyte clonality and JAK2V617F allelic ratio. A total of 168 of 190 female patients were informative for a clonality assay at the HUMARA locus; 80% of MMM, 75% of PV, and 67% of ET patients demonstrated clonal granulopoiesis. The JAK2V617F allele was detected by quantitative real-time polymerase chain reaction (PCR) in 99% of PV, 72% of ET, and 39% of MMM. A correlation between clonality and JAK2V617F allelic ratio was demonstrated for PV (P < .001) but not for ET or MMM (both P > .6). These data suggest that acquisition of the JAK2V617F mutation may be sufficient for the development of PV, but additional genetic events are necessary in ET and MMM. In addition, some ET and MMM patients with clonal granulopoiesis have somatic mutations other than JAK2V617F.
Introduction
In 1951, Dr William Dameshek classified polycythemia vera (PV), essential thrombocythemia (ET), myeloid metaplasia/myelofibrosis (MMM), and chronic myelogenous leukemia (CML) as phenotypically related myeloproliferative disorders (MPDs).1 The molecular pathogenesis of BCR-ABL–negative MPDs was poorly understood until the recent identification of the JAK2V617F activating mutation.2-8 JAK2V617F confers factor-independent growth and erythropoietin hypersensitivity to hematopoietic cells,2,3 and expression of JAK2V617F in a murine bone marrow transplantation model results in erythrocytosis.3 These data suggest that acquisition of JAK2V617F is an important pathogenetic event in JAK2V617F-positive MPDs. Nonetheless, some PV, ET, and MMM patients are JAK2V617F negative, as assessed by DNA resequencing. There are several potential explanations for this observation, including mutations in other genes that phenocopy JAK2V617F. Granulocytes from some patients with PV and ET are polyclonal,9,10 and thus sequence analysis of granulocyte DNA may underestimate the true frequency of the JAK2V617F allele. Although granulocyte clonality and JAK2V617F mutational status were reported in ET,11 no study has investigated the relationship between the acquisition of JAK2V617F and the emergence of clonal hematopoiesis in PV, ET, and MMM. We therefore investigated the relationship between clonality and JAK2V617F allelic ratio in MPDs.
Study design
Harvard Myeloproliferative Disorders Study
The Harvard Myeloproliferative Disorders Study enrolled patients with PV, ET, and MMM to collect clinical information and biologic samples.2 All subjects provided informed consent. Approval for the studies was obtained from the Dana-Farber Cancer Institutional Review Board.
HUMARA clonality assay and X-inactivation ratio determination
Polymerase chain reaction (PCR) amplification of the polymorphic CAG repeat at the HUMARA locus12-14 was performed in tandem on undigested (6-FAM–labeled primer) and on HpaII-digested (HEX-labeled primer) DNA. PCR products were analyzed on an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA) to determine the area under the curve (AUC) for each allele. If the alleles are 1 or 2 repeat lengths different in size, shadow banding from the higher molecular weight allele affects the measured AUC of the lower molecular weight allele. To compensate for this, homozygous samples and samples with more than 5 repeat lengths between the 2 alleles were used to measure the percentage of shadow banding for each repeat length, and this percentage was subtracted from the lower molecular weight allele.
The ratio between the X-linked alleles was expressed as the degree of skewing (DS). To determine DS, the proportion of the superior allele (Psup) is calculated as follows:
A and a represent the AUC for the upper and lower alleles from the digested sample; A′ and a′ represent the AUC for the upper and lower alleles in the undigested sample. DS = |Psup – 0.5|. Allelic skewing consistent with clonal granulopoiesis was defined as a 3:1 ratio between X-linked alleles, which is equivalent to DS at least 0.25.12,15 Patients with “clonal granulopoiesis” have at least 50% clonally derived granulocytes but may have 50% or fewer admixed polyclonal cells; conversely, patients with “polyclonal granulopoiesis” may have 50% or fewer admixed clonal granulocytes.
Real-time quantitative PCR assay for JAK2V617F
PCR primers and probes are listed in Table 1. PCR amplification and detection were performed on an ABI Prism 7000 analyzer (Applied Biosystems) with an initial step of 10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. DNA from a healthy control homozygous for JAK2-WT and from a PV erythroid colony homozygous for JAK2V617F were mixed in various proportions to generate a standard curve for JAK2V617F/JAK2total against ΔCt (CtJAK2V617F – CtJAK2WT). All samples were measured in triplicate, and the mean ΔCt was used to calculate JAK2V617F /JAK2total.
Primer and probe sequences used for JAK2V617F and clonality assays
Primer . | Probe sequence . |
|---|---|
| HUMARA-Primer-F | GCTGTGAAGGTTGCTGTTCCTCAT |
| HUMARA-Primer-R | TCCAGAATCTGTTCCAGAGCGTGC |
| JAK2-PCR-Primer-F | AAGCTTTCTCACAAGCATTTGGTTT |
| JAK2-PCR-Primer-R | AGAAAGGCATTAGAAAGCCTGTAGTT |
| JAK2-Probe-WT; VIC | TCTCCACAGACACATAC |
| JAK2-Probe-V617F; FAM | TCCACAGAAACATAC |
Primer . | Probe sequence . |
|---|---|
| HUMARA-Primer-F | GCTGTGAAGGTTGCTGTTCCTCAT |
| HUMARA-Primer-R | TCCAGAATCTGTTCCAGAGCGTGC |
| JAK2-PCR-Primer-F | AAGCTTTCTCACAAGCATTTGGTTT |
| JAK2-PCR-Primer-R | AGAAAGGCATTAGAAAGCCTGTAGTT |
| JAK2-Probe-WT; VIC | TCTCCACAGACACATAC |
| JAK2-Probe-V617F; FAM | TCCACAGAAACATAC |
Biostatistical analysis
Results and discussion
A total of 168 of 190 (88%) female subjects were heterozygous at the HUMARA locus. Of this subset, 121 patients (72%) had allelic skewing (DS at least 0.25) consistent with clonal granulopoiesis. The proportion of patients with clonal granulopoiesis was highest for MMM (80%), followed by PV (75%), and then ET (67%); these data are consistent with previous reports.9,10,18 We and others have reported that excessive skewing can occur in granulocytes from some healthy females and that this phenomenon increases with age.13,19 However, in our cohort there was no difference in age between MPD patients with clonal and polyclonal granulopoiesis (P = .1). The proportion of MPD patients 60 years old or younger with clonal granulocytes (68%) did not differ significantly from the entire cohort (P = .5) but was significantly higher than age-matched controls (P < .001). In addition, the mean DS value for all patients (0.36 versus 0.22, P < .001) and for patients 60 years or younger (0.34 versus 0.197, P < .001) was significantly higher in MPD patients as compared with age-matched controls. These data indicate that the skewing observed in MPD patients cannot primarily be attributed to differences in age.
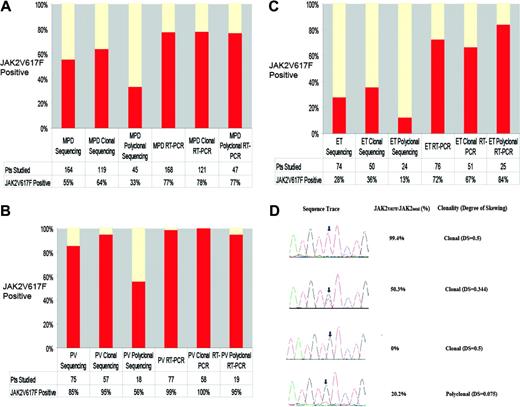
We then compared the clonality results with sequence analysis for JAK2V617F (Figure 1A-C).2 JAK2V617F mutations were identified in 64% of patients with clonal granulocytes versus only 33% of patients with polyclonal granulocytes (P < .001). JAK2V617F mutations were present in 95% of PV patients with clonal granulocytes and in 56% of PV patients with polyclonal granulocytes (P < .004), whereas JAK2V617F mutations were identified in 36% of ET patients with clonal granulocytes and in only 13% of ET patients with polyclonal granulocytes (P < .05). These data suggest that DNA resequencing cannot reliably detect JAK2V617F in a small population of clonally derived granulocytes.
We consequently developed a highly sensitive, quantitative, real-time PCR assay for JAK2V617F (Figure 1D). We did not detect JAK2V617F in 50 healthy controls, and analysis of 32 MPD samples in 2 independent experiments demonstrated that the assay is reproducible (mean difference of 2 measurements, 0.16%; standard deviation, 2.1%). We detected JAK2V617F at similar frequencies in patients with clonal (78%) and polyclonal (77%) granulocytes, due to the ability of the assay to detect JAK2V617F when JAK2V617F/JAK2total is 25% or less (Figure 1D). The frequency of JAK2V617F-positive PV (99%) by real-time PCR is similar to allele-specific PCR (97%)4 and is higher than DNA resequencing (65% to 81%).2-8 The frequency of JAK2V617F-positive ET by real-time PCR (72%) is higher than DNA resequencing (23% to 32%)2,3,6 or allele-specific PCR (57%).4 The frequency of JAK2V617F (39%) in the small number of MMM patients in our series is similar to previous reports.2,3,6
If acquisition of the JAK2V617F mutation is sufficient to cause an MPD, there should be a correlation between JAK2V617F/JAK2total and granulocyte clonality. A significant correlation between JAK2V617F/JAK2total and clonality was observed in PV (rho = 0.55; P < .001) but not ET or MMM (both P > .60). Analysis of patients 60 years old or younger yielded identical results. These results suggest that acquisition of the JAK2V617F allele is temporally related to acquisition of the PV phenotype, but additional genetic events are necessary in ET and MMM. Current efforts are focused on identification of genetic events that cooperate with JAK2V617F in ET and MMM.
Some patients with ET (23%) and PV (3%) had clonal granulopoiesis and a JAK2V617F/JAK2total of 2% to 25%, suggesting that development of a clonal MPD may precede acquisition of JAK2V617F. Alternatively, these patients may exhibit age-associated granulocyte skewing, and the low JAK2V617F/JAK2total ratio reflects the true proportion of clonal granulocytes. Serial assessment will be required to document acquisition and/or emergence of JAK2V617F. Most importantly, these data identify a subset of patients with ET (22%) and MMM (53%), but not PV, with a JAK2V617F-negative clonal MPDs. Genomic approaches similar to those used to identify JAK2V617F are warranted to search for mutations in JAK2V617F-negative ET and MMM.
Sequence analysis, JAK2V617F/JAK2total, and clonality results for patients with MPDs. (A) The frequency of the JAK2V617F allele as determined by sequencing and the real-time PCR (RT-PCR) assay in MPD patients according to granulocyte clonality, demonstrating that DNA resequencing more frequently identifies JAK2V617F mutations in MPD patients with clonal granulocytes but that RT-PCR is able to detect JAK2V617F mutations in patients with clonal and polyclonal granulocytes. (B) The frequency of the JAK2V617F allele as determined by sequencing and by RT-PCR in PV patients according to granulocyte clonality, demonstrating that DNA resequencing more frequently identifies JAK2V617F mutations in PV patients with clonal granulocytes but that RT-PCR is able to detect JAK2V617F mutations in PV patients with clonal and polyclonal granulocytes. (C) The frequency of the JAK2V617F allele as determined by sequencing and by the real-time PCR assay in ET patients according to granulocyte clonality, demonstrating that DNA resequencing more frequently identifies JAK2V617F mutations in ET patients with clonal granulocytes but that RT-PCR is able to detect JAK2V617F mutations in ET patients with clonal and polyclonal granulocytes. (D) Sequence traces of JAK2 exon 14 along with results from quantitative real-time assays for JAK2V617F and clonality results. The top trace shows a sequence trace from a patient with PV and a homozygous JAK2V617F mutation; the second trace shows a sequence trace from a patient with PV and a heterozygous JAK2V617F mutation; and the third trace shows a sequence trace from a patient with ET who was negative for the JAK2V617F mutation by sequence analysis and RT-PCR; each of these 3 patients had allele skewing consistent with clonal granulopoiesis. The bottom trace shows a sequence trace from a patient with PV who was scored as negative for the JAK2V617F mutation by DNA resequencing; there is a small peak (arrow) suggesting that the JAK2V617F mutation may be present in a subpopulation of cells. RT-PCR confirms that JAK2V617F is present in a minor population of cells, and clonality analysis demonstrates a lack of allele skewing consistent with polyclonal granulopoiesis.
Sequence analysis, JAK2V617F/JAK2total, and clonality results for patients with MPDs. (A) The frequency of the JAK2V617F allele as determined by sequencing and the real-time PCR (RT-PCR) assay in MPD patients according to granulocyte clonality, demonstrating that DNA resequencing more frequently identifies JAK2V617F mutations in MPD patients with clonal granulocytes but that RT-PCR is able to detect JAK2V617F mutations in patients with clonal and polyclonal granulocytes. (B) The frequency of the JAK2V617F allele as determined by sequencing and by RT-PCR in PV patients according to granulocyte clonality, demonstrating that DNA resequencing more frequently identifies JAK2V617F mutations in PV patients with clonal granulocytes but that RT-PCR is able to detect JAK2V617F mutations in PV patients with clonal and polyclonal granulocytes. (C) The frequency of the JAK2V617F allele as determined by sequencing and by the real-time PCR assay in ET patients according to granulocyte clonality, demonstrating that DNA resequencing more frequently identifies JAK2V617F mutations in ET patients with clonal granulocytes but that RT-PCR is able to detect JAK2V617F mutations in ET patients with clonal and polyclonal granulocytes. (D) Sequence traces of JAK2 exon 14 along with results from quantitative real-time assays for JAK2V617F and clonality results. The top trace shows a sequence trace from a patient with PV and a homozygous JAK2V617F mutation; the second trace shows a sequence trace from a patient with PV and a heterozygous JAK2V617F mutation; and the third trace shows a sequence trace from a patient with ET who was negative for the JAK2V617F mutation by sequence analysis and RT-PCR; each of these 3 patients had allele skewing consistent with clonal granulopoiesis. The bottom trace shows a sequence trace from a patient with PV who was scored as negative for the JAK2V617F mutation by DNA resequencing; there is a small peak (arrow) suggesting that the JAK2V617F mutation may be present in a subpopulation of cells. RT-PCR confirms that JAK2V617F is present in a minor population of cells, and clonality analysis demonstrates a lack of allele skewing consistent with polyclonal granulopoiesis.
Supported in part by the fond de la recherche en santé du Québec (FRSQ), National Institutes of Health grants CA66996 and DK50654 (D.G.G.), the Howard Hughes Medical Institute (D.G.G.), the Leukemia and Lymphoma Society, the Doris Duke Charitable Foundation (D.G.G.), and an American Society of Clinical Oncology Young Investigator Award (M.W.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Prepublished online as Blood First Edition Paper, January 24, 2006; DOI 10.1182/blood-2005-09-3900.
We thank the myeloproliferative disease patients who participated in this study and Joyce Niblack, JD, for support and access to the mpdinfo.org website. We are indebted to Jennifer Adelsperger, Sandra Moore, Sarah Cohen, Allison Coburn, Rachel Okabe, Elizabeth McDowell, Dana Cullen, Gulnar Pothiawala, and Claudia Tenen for assistance with sample processing, to Tarrah Kirk-patrick and Arnold Gonzales for assistance with the MPD study, and to Alexis Bywater for technical and administrative assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal