Abstract
Ectopic expression of c-Myc (Myc) in most primary cell types results in programmed cell death, and malignant transformation cannot occur without additional mutations that block apoptosis. The development of Myc-induced lymphoid tumors has been well studied and supports this model. Myc can be upregulated in acute myeloid leukemia (AML), but its exact role in myeloid leukemogenesis is unclear. To study its role in AML, we used a murine stem cell virus (MSCV) retroviral gene transfer/transplantation system to broadly express Myc in the bone marrow of mice either alone or in combination with antiapoptotic mutations. Myc expression in the context either of Arf/Ink4a loss or Bcl-2 coexpression induced a mixture of acute myeloid and acute lymphoid leukemias (AML+ALL). In the absence of antiapoptotic mutations however, all mice transplanted with MSCV-Myc (100%, n = 110) developed AML exclusively. MSCV-Myc-induced AML was polyclonal, readily transplantable, possessed an intact Arf-p53 pathway, and did not display cytogenetic abnormalities by spectral karyotyping (SKY) analysis. Lastly, we found that Myc preferentially stimulated the growth of myeloid progenitor cells in methylcellulose. These data provide the first direct evidence that Myc is a critical downstream effector of myeloid leukemogenesis and suggest that myeloid progenitors are intrinsically resistant to Myc-induced apoptosis. (Blood. 2005;106: 2452-2461)
Introduction
Deregulation of the proto-oncogene c-Myc (MYC) is considered one of a series of oncogenic events required for mammalian tumorigenesis.1-4 MYC encodes a basic helix-loop-helix leucine zipper transcription factor that dimerizes with its partner, Max, and regulates multiple cellular functions including cell cycle, cell growth, differentiation, apoptosis, metabolism, and angiogenesis via transcription of downstream target genes.2 The Myc/Max dimer can also repress transcription of another set of target genes through a less well-understood mechanism.5,6 The c-Myc proto-oncogene is involved in transformation and cell proliferation in part via activation of the cyclin D2 promoter,7 but also induces programmed cell death, mediated by nuclear respiratory factor 1 (NRF-1)8 and the Arf-p53 pathway.9,10 Forced expression of Myc in primary cells is generally thought to induce growth arrest or apoptosis.11,12
The oncogenic function of c-Myc has been best studied in the Eμ-Myc transgenic mouse, in which c-Myc expression is targeted to the lymphoid compartment by the immunoglobulin heavy chain gene promoter and enhancer.13 In Eμ-Myc mice, expression of Myc is not sufficient to cause leukemia. A latency period of 4 to 6 months is required for the accumulation of cooperating mutations before lymphoma can develop.14 The majority of the Eμ-Myc tumors harbor mutations in the now canonical Arf-Mdm2-p53 pathway.14,15 Although the p53 pathway controls many functions including cell cycle, DNA damage response, and apoptosis, Bcl-2, or dominant-negative caspase 9 also cooperate with Myc, and alleviate the pressure to inactivate p53 during lymphomagenesis.15 Therefore, of the many functions of the Arf-Mdm2-p53 pathway, it is the ability to mediate apoptosis that is targeted by cooperating mutations in Myc-induced lymphomagenesis.
MYC dysregulation, via a variety of mechanisms, has also been associated with myeloid leukemias.16 Double minute chromosomes in patients with acute myeloid leukemia (AML) contain MYC amplifications.17,18 c-Myc expression is apparently required for the oncogenic effects of the Philadelphia chromosome product, BCL-ABL,19 and overexpression of c-Myc also complements the transformation defects of BCR-ABL mutants.20 A recent study showed that many important oncogenes in myeloid leukemogenesis including AML1-ETO, PML-RARA, and PLZF-RARA induce leukemogenesis by activating c-Myc,21 suggesting c-Myc is a downstream target of these oncogenes. Lastly, c-Myc is upregulated by activating mutations in the gene encoding the FLT3 receptor tyrosine kinase, found in nearly one-third of all patients with AML.22,23
There is a lack of useful animal models for studying the role of Myc in myeloid leukemia. AML infrequently develops in EμSR-Myc mice, in which a heterologous promoter inducibly drives Myc expression in bone marrow cells; however, most of these animals develop T-cell lymphomas,24 making it a cumbersome system to study myeloid disease. Retroviral transduction of c-Myc into p53-null bone marrow cells rapidly causes lymphoid leukemias, but not myeloid disease.25 Finally, v-Myc transduction can induce myeloid leukemias in a retroviral transduction/transplantation system,26,27 however v-Myc bears point mutations known to contribute synergistically to transforming potential measured in culture,28 so results obtained using v-Myc cannot be extrapolated to unmutated c-Myc.
Using the murine stem cell virus (MSCV) system to broadly express Myc in primary murine bone marrow cells, we compared the effects of ectopic Myc expression on myeloid and lymphoid progenitors in methylcellulose colony assays, and found a preferential increase in myeloid colonies. We used two different retroviral bone marrow transduction/transplantation systems to examine the contribution of antiapoptotic mutations to the development of Myc-induced disease in mice. In the setting of either Ink4a loss or Bcl-2 coexpression, Myc induced both myeloid and lymphoid leukemias. In contrast, rapidly fatal, disseminated myeloid leukemia developed independently of antiapoptotic mutations. We analyzed AML cells from MSCV-Myc mice extensively and found no evidence of spontaneous antiapoptotic mutations. Our results provide direct evidence of Myc's role in myeloid leukemogenesis and suggest that myeloid progenitor cells are relatively resistant to Myc-induced apoptosis.
Materials and methods
Plasmid constructs
To make the MSCV constructs that express c-Myc, the cDNAs encoding murine c-Myc (Myc; provided by Michael Cole, Dartmouth Medical School, Hanover, NH) were subcloned into the Xho site in the MSCV-internal ribosome entry site (IRES)-green fluorescent protein (GFP) vector (provided by Warren Pear, University of Pennsylvania). The MSCV-Bcl2 construct was made by subcloning the Xho1/EcoRI fragment of Bcl2 (provided by Carlo Croce, Thomas Jefferson University, Philadelphia, PA) into Xho-EcoRI sites of MSCV-IRES-GFP. To make the MSCV construct that expresses both Myc and Bcl-2, Bcl-2 was first amplified using primers with NcoI and Cla sites engineered. The amplicon was purified and cut with those enzymes and put into NcoI-ClaI-digested MSCV-GFP (replacing the GFP). c-Myc was then added into the XhoI site of this intermediate.
Ecotropic retroviral gene transfer and murine bone marrow transplants
Replication-incompetent supernatant was made by transiently transfecting 293T cells with each construct and ecotropic packaging sequence pIK6.1MCV.ecopac.UTD (Ecopac; M. Finer Cell Genosys, Redwood City, CA). Supernatants were harvested 48 hours after transfection, passed through a 0.45 μ filter, and kept in frozen aliquots at -80°C. Retroviral titers were determined by measuring percentage of GFP-positive 3T3 cells or quantitation of proviral DNA in infected cells using real-time polymerase chain reaction (PCR) specific for MSCV Psi-sequences (protocol available on request). Bone marrow cell transduction and transplantation were carried out according to the method previous reported.29 Briefly, mononuclear bone marrow cells were harvested from 6- to 7-week-old BALB/c, B6 129, or B6 129-Cdkn2atm1Rdp donor mice (Jackson Laboratories, Bar Harbor, ME) treated with 0.45 mg 5-flourouracil (5-FU; Sigma, Saint Louis, MO). Spinfection was performed twice on day -1 and day 0. Cells (1 × 106) were injected intravenously into lethally irradiated (on day 1) syngeneic Balb/c (800 cGy) or B6 129 (1200 cGy) mice. Secondary transplantations were performed by intravenously injecting unsorted mononuclear spleen cells isolated from moribund primary transplant recipients into secondary recipient mice treated with 500 cGy gamma irradiation.
Analysis of mice
Mice were monitored 3 times per week for disease by palpation and observation. Peripheral blood was obtained by either retro-orbital phlebotomy after adequate methoxyflurane (Springvale, Australia) anesthesia or femoral phlebotomy using heparinized capillary tubes (Fisher Scientific, Pittsburgh, PA). Mononuclear-cell suspensions of spleen, lymph nodes, or thymus were made by passing tissue through nylon mesh cell strainers (Falcon, Lincoln Park, NJ) wetted with phosphate-buffered saline (PBS). Tissues from the mice were stored in a 10% buffered formalin solution (Sigma). Kaplan Meier and significance analyses were performed using Statview software (SAS Institute, Cary, NC).
Cytospin and formalin-fixed tissue slides were stained with hematoxalin and eosin (H&E) and were imaged using an Olympus BX40 F4 microscope equipped with an oil-immersion 50×/0.90 or 100×/1.30 objective lens (Olympus Optical, Tokyo, Japan). Pictures were taken with an Olympus DP70 digital camera using DPController software.
Detection of provirus and clonality assays
Genomic DNA was isolated from mouse spleen and bone marrow mononuclear cells using the Puregene DNA Isolation Kit (Gentra, Minneapolis, MN) per the manufacturer's instructions. DNA (10 μg) was digested with XbaI (cuts once in each long terminal repeat [LTR] and releases a 4.2-kb provirus band). To assess clonality, genomic DNA was digested with BglII (cuts once in provirus). DNA was precipitated and subjected to agarose gel electrophoresis and transferred to a nylon membrane (Hybond-N Plus; Amersham Pharmacia, Piscataway, NJ). After a 2-hour prehybridization, the blot was hybridized for 24 hours at 65°C with an 800-bp GFP (HindIII-NcoI) probe labeled with α32P adenosine triphosphate (ATP) using the Random Primed DNA labeling kit (Roche Diagnostics, Indianapolis, IN) per the manufacturer's instructions. Membranes were washed with 2X standard saline citrate (SSC) twice for 5 minutes and 2X SSC and 1% sodium dodecyl sulfate (SDS) 3 times for 10 minutes at 65°C and exposed to radiographic film overnight or a week at -80°C. Blots were stripped by incubation in boiling 0.1% SDS, and incubating until SDS reached room temperature. The Ink4a exon1β probe was kindly provided by Ron DePinho and Ned Sharpless, Harvard Medical School, Boston, MA.
Immunophenotyping
One million freshly isolated mononuclear cells were suspended in 100 μL Flow buffer and incubated for 30 minutes on ice with appropriate antibodies, including phycoerythrin (PE)-conjugated Gr-1, PE-B220, PE-CD4, Biotin-Mac-1, Biotin-CD3, Biotin-CD8, Biotin-CD43, and Biotin-IgM (all from Becton Dickinson, San Diego, CA). Samples were then incubated with 1 μL streptavidin in 100 μL Flow buffer for Biotin samples for 30 minutes before being analyzed on a FACScan Flow Cytometer (BD Biosciences, San Jose, CA).
Sequence analysis
RNA was isolated from BALB/c and MSCV-Myc mice using the RNeasy kit (Qiagen, Valencia, CA) per the manufacturer's instructions. cDNA was made using a Superscript First Strand Synthesis System (Invitrogen, Carlsbad, CA) per the manufacturer's instructions. PCR reactions for P53 and Arf were performed using 2 μL cDNA, 2 mM final MgCl2, 10 μM final primer concentration, 10X PCR reaction buffer (Fisher Scientific, Pittsburgh, PA), and 1 U Taq DNA polymerase (Fisher Scientific). The thermal program was as follows: 94°C for 2 minutes; 39 cycles of 94°C for 30 seconds, 71.5°C for 1 minute, 72°C for 1 minute. Ink4a amplification was the same except for 1 mM final MgCl2 and annealing temp of 66°C. Primers were as follows: P53: TRP53F3 GCGGGGGTTGCTGGGATTG, TRP53R2 CCGCGGATCTTGAGGGTGAAATAC, TRP53F4 TGGCCCCTGTCATCTTTTGTCC; Arf: P19F1 GAGGCCGCCGCTGAGGGAGTA, P19R1 CTAAGAAGAAAAAGGCGGGCTGAG; Ink4a: INK4A 2F CGCCCAACGCCCCGAACTCT, INK4A 2R AAGGCGGGCTGAGGCGGATTTA. Products were gel purified using the QIAEX II Gel Extraction kit (Qiagen, Valencia, CA) per the manufacturer's instructions. Sequences were compared with normal BALB/c P53 (GI 200 198), p19Arf (accession no. nm_009 877), and p16Ink4a (GI 3 002 946). For Arf, 8 mice were sequenced. Data ranges from base pair 2 to 586 (1 = A from the ATG start codon). For Ink4a, 6 mice were sequenced. Data ranges from base pair 85 to 525. For P53, sequence data were collected from 8 mice. Data span base pair -113 to 347. At the C terminal end, 2 mice were sequenced, and data ranges from base pair 654 to 952.
Western blots
Protein lysates were made from sorted GFP-positive cells or unsorted bone marrow, spleen, and lymph node cells from affected and control animals with lysis buffer containing protease inhibitors (Cell Signaling, Beverly, MA) and 1 mM phenylmethylsulfonyl fluoride (PMSF; Sigma). Protein samples (20 μg) were resolved on a 9% SDS polyacrylamide resolving gel and transferred to a nitrocellulose membrane (Midwest Scientific, Saint Louis, MO). After incubated in 5% blocking solution (5% nonfat dried milk, 150 mM NaCl, 50 mM Tris, pH 8) at room temperature for 1 hour, membranes were incubated with primary antibody at 4°C overnight, washed 3 times for 15 minutes with TBST (10 mM Tris, pH 7.6, 150 mM NaCl, 0.05% Tween 20), and then incubated with secondary antibody for 40 minutes at room temperature. Blots were subsequently washed 3 times for 20 minutes with TBST. P53 (1:250) and P19 antibodies (1:500) were kindly provided by Jason Weber, Washington University (Saint Louis, MO). P21 (1:250), Bcl-2 (1:1000), and c-Myc (1:1000) were from Santa Cruz Biotechnology (Santa Cruz, CA). Bcl-xl (1:500) was from Cell Signaling Technologies. β-actin (1:5000) was from Santa Cruz Biotechnology, and actin (1:5000) was from Sigma. Horseradish peroxidase (HRP)-conjugated antirabbit (1:5000) antibody was from Upstate Biotechnology (Lake Placid, NY); HRP-conjugated antimouse (1:5000) was from Sigma. Blots were developed using the Pierce Enhanced Chemiluminescence system (Pierce, Rockford, IL) and exposed to Kodak Biomax film (SciMart, Saint Louis, MO).
Methylcellulose culture
B6 129 mice were transplanted with either MSCV-GFP- or MSCV-MycER-GFP-transfected bone marrow cells following the method described in “Ecotropic retroviral gene transfer and murine bone marrow transplants.” At 8 to 10 weeks later, GFP-positive bone marrow cells from the recipient mice were plated in methylcellulose media containing myeloid cytokines (stem cell factor [SCF], IL-3, IL-6, erythropoietin [Epo]), lymphoid cytokines (IL-7), or no cytokines (Stemcell Technologies). In each experiment, 4 × 104 cells/well for myeloid culture or 4 × 105 cells/well for lymphoid and noncytokine culture were plated in triplicate. Two hundred nM 4-hydroxy-tamoxifen (Sigma) or ethanol carrier was added when the culture was set up. Seven days later, the colony numbers were counted and the cells in each culture were subjected to immunophenotype analysis.
Results
Myc rapidly induces acute myeloid leukemia (AML), independent of Ink4a status
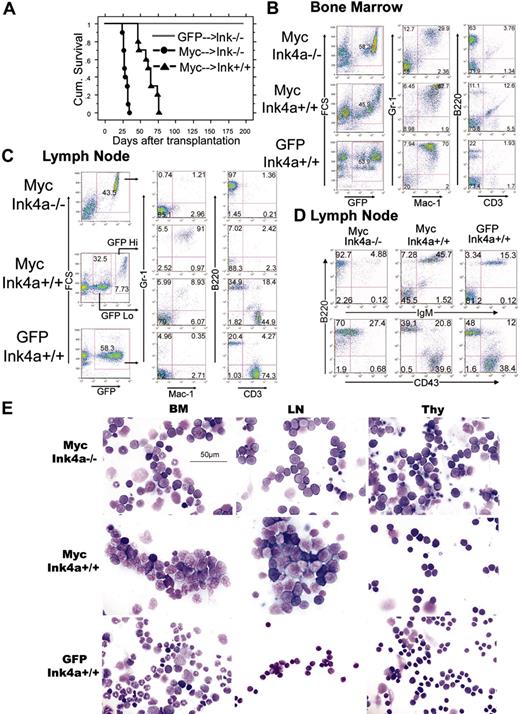
To characterize the effects of c-Myc on hematopoietic progenitor cells in vivo, we performed bone marrow transduction/transplantation experiments using an MSCV retrovirus encoding the wild-type c-Myc (MSCV-Myc). Unfractionated bone marrow was harvested from wild-type mice or mice bearing a targeted deletion in Ink4a exon 2A that results in the loss of expression of both p19Arf and p16Ink4a proteins (Ink4a-/-). MSCV-Myc mice that underwent transplantation using Ink4-/- bone marrow (MSCV-Myc→Ink4a-/-) developed a rapidly fatal disease characterized by significant peripheral leukocytosis with circulating myeloblasts and lymphoblasts, splenomegaly, lymphadenopathy, and hind-limb paralysis (Figure 1A and Table 1). To our surprise, MSCV-Myc mice transplanted with Ink4a+/+ bone marrow (MSCV-Myc→Ink4a+/+) also succumbed to disease rapidly (median survival, 61 days) with splenomegaly and hind-limb paralysis. Blood leukocytosis and lymphadenopathy was conspicuously absent/reduced (Figure 1A and Table 1). Notably, nonsaturating retroviral insertion with replication-incompetent MSCV-GFP virus did not cause disease either in wild-type or tumor-prone Ink4a-/- mice during a 200-day observation (Figure 1A).
Summary of primary bone marrow transplantation experiments
Construct . | Donor background . | No. . | Median survival, days . | GFP, % at 3 weeks* . | WBC count, × 103/μL . | Spleen weight, mg . | Phenotype . |
|---|---|---|---|---|---|---|---|
| Myc | B6 129 Ink−/− | 10 | 28 ± 1.1 | 79.5 ± 2.7 | 65.5 ± 20.7 | 214 ± 50 | AML + ALL |
| Myc | B6 129 Ink+/+ | 10 | 61 ± 3.7 | 56.2 ± 3.9 | 6.0 ± 1.4 | 380 ± 40 | AML |
| GFP | B6 129 Ink−/− | 10 | >200 | 40.5 ± 2.8 | 2.9 ± 0.4 | NA | NA |
| GFP | B6 129 Ink+/+ | 6 | >200 | 78.2 ± 5.4 | 6.2 ± 0.6 | NA | NA |
| Myc + Bcl2 | BALB/c | 33 | 16 ± 0.7 | NA† | 113.7 ± 18.8 | 355 ± 15 | AML + ALL |
| Myc | BALB/c | 35 | 47 ± 2.9 | 86.9 ± 0.6 | 12.3 ± 1.5 | 386 ± 30 | AML |
| Bcl-2 | BALB/c | 10 | >200 | 76.3 ± 3.2 | 9.9 ± 0.7 | NA | NA |
Construct . | Donor background . | No. . | Median survival, days . | GFP, % at 3 weeks* . | WBC count, × 103/μL . | Spleen weight, mg . | Phenotype . |
|---|---|---|---|---|---|---|---|
| Myc | B6 129 Ink−/− | 10 | 28 ± 1.1 | 79.5 ± 2.7 | 65.5 ± 20.7 | 214 ± 50 | AML + ALL |
| Myc | B6 129 Ink+/+ | 10 | 61 ± 3.7 | 56.2 ± 3.9 | 6.0 ± 1.4 | 380 ± 40 | AML |
| GFP | B6 129 Ink−/− | 10 | >200 | 40.5 ± 2.8 | 2.9 ± 0.4 | NA | NA |
| GFP | B6 129 Ink+/+ | 6 | >200 | 78.2 ± 5.4 | 6.2 ± 0.6 | NA | NA |
| Myc + Bcl2 | BALB/c | 33 | 16 ± 0.7 | NA† | 113.7 ± 18.8 | 355 ± 15 | AML + ALL |
| Myc | BALB/c | 35 | 47 ± 2.9 | 86.9 ± 0.6 | 12.3 ± 1.5 | 386 ± 30 | AML |
| Bcl-2 | BALB/c | 10 | >200 | 76.3 ± 3.2 | 9.9 ± 0.7 | NA | NA |
Numbers are shown as ± standard deviation.
NA indicates not applicable; WBC, white blood cell; AML, acute myeloid leukemia; ALL, acute lymphoid leukemia.
The percentage of GFP-positive peripheral-blood leukocytes measured by flow cytometry 3 weeks after marrow transplantation.
The MSCV-Myc + Bcl2 construct does not contain GFP.
We used flow cytometry to compare the immunophenotypes of GFP-positive cells from MSCV-Myc→Ink4a+/+ and MSCV-Myc→Ink4a-/- mice. MSCV-Myc→Ink4a-/- bone marrow was packed with 2 distinct populations of malignant cells: one with features of myeloblasts and another with features of lymphoblasts. The immunophenotypic studies confirmed the lineage of these two populations: one stained double-positive with Gr-1 and Mac-1, representing myeloid origin; and another stained positive with B220, representing B-cell lymphoid origin (Figure 1B). These two malignant populations composed over 90% of bone marrow. All of the MSCV-Myc→Ink4a-/- transplant recipients developed significant lymphadenopathy and the GFP-positive cells from their lymph nodes were exclusively immature CD43+IgM- B lymphocytes (Figure 1C-D). All of the MSCV-Myc→Ink4a-/- transplant recipients also developed thymus enlargement. Pathology revealed infiltration of the thymus with lymphoblasts and immunophenotyping showed these cells to be immature B-lineage lymphoblasts (Figure 1E). In summary, mice that underwent transplantation with Ink4a-/- bone marrow expressing Myc (MSCV-Myc→Ink4a-/-) developed myeloid and lymphoid leukemias simultaneously.
In contrast, 100% of the MSCV-Myc→Ink4a+/+ transplant recipients developed acute myeloid leukemia exclusively (Figure 1 and Table 1). The MSCV-Myc→Ink4a+/+ transplant recipients developed splenomegaly and hind-limb paralysis, but did not develop significant lymphadenopathy or peripheral leukocytosis to the same degree as Ink4a-/- mice (Table 1). Immunophenotypic studies of the GFP-positive bone marrow cells showed a dominant cell population that stained double-positive with Gr-1 and Mac-1, confirming their myeloid lineage, and a conspicuous reduction in the size of the B220+ lymphoid population (Figure 1B). Analysis of the lymph nodes revealed two distinct populations: a high GFP population stained positive with Gr-1 and Mac-1, representing myeloid leukemic cells infiltrating lymph nodes; and a group of GFP-low cells that expressed either B220 or CD3 and were indistinguishable from normal, mature lymphocyte populations (Figure 1C-E). Microscopic analysis of the tissues of MSCV-Myc→Ink4a+/+ mice revealed bone marrow packed with myeloblasts with abundant cytoplasm, fine chromatin, and prominent nucleoli (Figure 1F), and these leukemic cells infiltrated the spleen, liver, and nervous system, leading to hind-limb paralysis (Figure S1; see the Supplemental Figure link at the top of the online article, at the Blood website). Histologic examination of lymph nodes from MSCV-Myc mice revealed infiltration of large malignant cells with characteristic myeloblast morphology, normal-appearing lymphocytes, and an absence of transformed lymphoblasts (Figure 1F). Therefore, targeted disruption of the Ink4a locus cooperated with Myc in our model system to achieve malignant transformation of lymphoid cells. In the setting of wild-type Ink4a, MSCV-Myc mice lived 33 days longer (Table 1) and displayed no evidence of lymphoid malignancy. However, Myc-expressing wild-type Ink4a mice did succumb uniformly to AML.
Coexpression of Bcl-2 is required for Myc-induced lymphoid, but not myeloid, leukemia
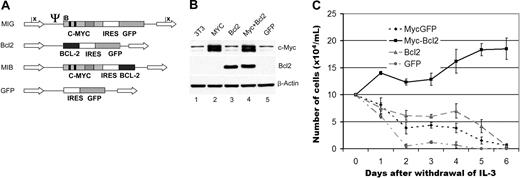
To further study the role of apoptosis in MSCV-Myc AML, we made additional retroviral constructs designed to coexpress Myc with the antiapoptotic protein Bcl-2 (Figure 2A). We confirmed that these constructs expressed Myc and Bcl-2 proteins (Figure 2B). Consistent with published data,30 coexpression of Myc and Bcl-2 together, but not either gene alone, transformed Ba/F3 cells to factor independence (Figure 2C).
MSCV-Myc induces myeloid and lymphoid disease phenotypes simultaneously in Ink4a-/-mice, and myeloid leukemia exclusively in Ink4a+/+mice. (A) Kaplan-Meier survival analysis of mice that underwent transplantation with Ink4a+/+ or Ink4a-/- bone marrow. GFP indicates MSCV-GFP; Myc, MSCV-Myc; Ink, Ink4a. Mice reconstituted with Ink4a-/- bone marrow transduced with GFP (GFP/Ink-/-) were free of disease. Median survival of Myc→Ink4a-/- reconstituted mice was 28 days, and that of Myc→Ink4a+/+ mice was 62 days. (B) Flow cytometric analysis of bone marrow cells from Myc→Ink4a+/+ and Myc→Ink4a-/- mice. Bone marrow data from GFP→Ink4a+/+ mice are shown as controls. GFP-positive cells from Ink4a-/- mice show two populations of leukemic cells: a Mac-1+/Gr-1+ myeloid population, and a B220+/CD3- lymphoid population. In Ink4a+/+ mice, there is only a single Mac-1+/Gr-1+ myeloid population. (C) Flow cytometric analysis of lymph node cells from Myc→Ink4a+/+ and Myc→Ink4a-/- mice. Lymph node cells from GFP→Ink4a+/+ mice are shown as controls. Myc→Ink4a-/- mice show a single GFP-positive population with a B-lymphoid immunophenotype (Mac-1-/Gr-1-, and B220+/CD3-). Lymph node cells from Myc→Ink4a+/+ mice show two populations of cells, a GFP-low and a GFP-high population. The GFP-high cells are Mac-1+/Gr-1+ myeloid cells, and the GFP-low cells are a mixture of B220+ B lymphocytes and CD3+ T lymphocytes similar to those seen in lymph nodes of control GFP→Ink4a+/+ mice. (D) Further immunophenotyping of lymph node cells from leukemic mice. IgM and CD43 staining reveals that lymphoid cells from Myc→Ink4a-/- mice are immature (IgM-/CD43-low) B-lineage cells, in contrast to lymph node cells from Myc→Ink4a+/+ mice and GFP→Ink4a+/+ mice, which are a mixture of mature IgM+ B cells and T cells that are CD3+. (E) Cell morphology. Myc→Ink4a-/- mice show a mixture of myeloid and lymphoid blast cells in the bone marrow (BM), and lymphoid blasts in the lymph nodes (LN) and thymus (Thy). Myc→Ink4a+/+ mice show only myeloid blasts in the bone marrow and lymph nodes, and normal-appearing cells in thymus.
MSCV-Myc induces myeloid and lymphoid disease phenotypes simultaneously in Ink4a-/-mice, and myeloid leukemia exclusively in Ink4a+/+mice. (A) Kaplan-Meier survival analysis of mice that underwent transplantation with Ink4a+/+ or Ink4a-/- bone marrow. GFP indicates MSCV-GFP; Myc, MSCV-Myc; Ink, Ink4a. Mice reconstituted with Ink4a-/- bone marrow transduced with GFP (GFP/Ink-/-) were free of disease. Median survival of Myc→Ink4a-/- reconstituted mice was 28 days, and that of Myc→Ink4a+/+ mice was 62 days. (B) Flow cytometric analysis of bone marrow cells from Myc→Ink4a+/+ and Myc→Ink4a-/- mice. Bone marrow data from GFP→Ink4a+/+ mice are shown as controls. GFP-positive cells from Ink4a-/- mice show two populations of leukemic cells: a Mac-1+/Gr-1+ myeloid population, and a B220+/CD3- lymphoid population. In Ink4a+/+ mice, there is only a single Mac-1+/Gr-1+ myeloid population. (C) Flow cytometric analysis of lymph node cells from Myc→Ink4a+/+ and Myc→Ink4a-/- mice. Lymph node cells from GFP→Ink4a+/+ mice are shown as controls. Myc→Ink4a-/- mice show a single GFP-positive population with a B-lymphoid immunophenotype (Mac-1-/Gr-1-, and B220+/CD3-). Lymph node cells from Myc→Ink4a+/+ mice show two populations of cells, a GFP-low and a GFP-high population. The GFP-high cells are Mac-1+/Gr-1+ myeloid cells, and the GFP-low cells are a mixture of B220+ B lymphocytes and CD3+ T lymphocytes similar to those seen in lymph nodes of control GFP→Ink4a+/+ mice. (D) Further immunophenotyping of lymph node cells from leukemic mice. IgM and CD43 staining reveals that lymphoid cells from Myc→Ink4a-/- mice are immature (IgM-/CD43-low) B-lineage cells, in contrast to lymph node cells from Myc→Ink4a+/+ mice and GFP→Ink4a+/+ mice, which are a mixture of mature IgM+ B cells and T cells that are CD3+. (E) Cell morphology. Myc→Ink4a-/- mice show a mixture of myeloid and lymphoid blast cells in the bone marrow (BM), and lymphoid blasts in the lymph nodes (LN) and thymus (Thy). Myc→Ink4a+/+ mice show only myeloid blasts in the bone marrow and lymph nodes, and normal-appearing cells in thymus.
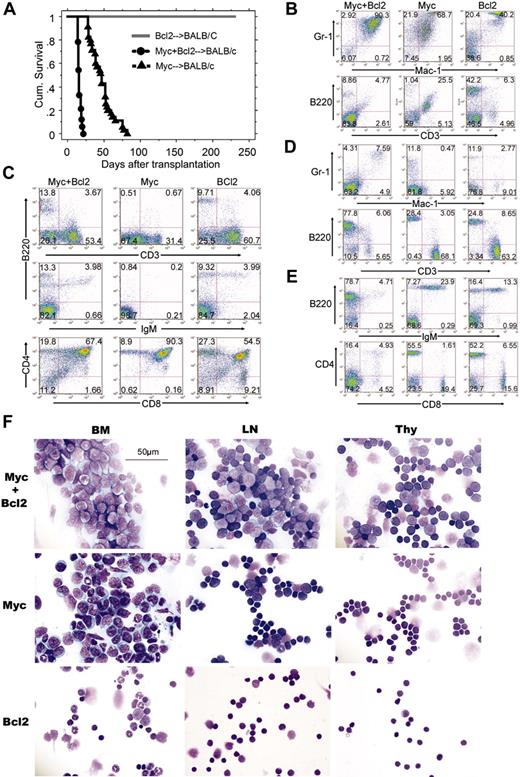
To assess the effect of apoptosis suppression on Myc-induced leukemogenesis, we performed bone marrow transduction/transplantation assays with Myc alone, Bcl2 alone, or both together. Successful retroviral transduction was confirmed in all experiments by flow cytometry of GFP-positive cells in peripheral blood 3 weeks after transplantation (with the exception of mice transplanted with the GFP-less MSCV-Myc+Bcl2 construct (Table 1). Notably, retroviral transduction of the Bcl2 proto-oncogene by itself was insufficient to induce disease in a single animal, consistent with published data.31 The analyses of bone marrow, lymph nodes, spleen, and thymus from MSCV-Bcl2 mice showed normal morphology microscopically and normal immunophenotype even several months after transplantation (Figure 3F). In sharp contrast, all MSCV-Myc+Bcl2 mice (n = 33, 100%) developed a rapidly fatal leukemia/lymphoma characterized by leukocytosis, hind-limb paralysis, splenomegaly, and lymphadenopathy and a median survival of 16 days (Figure 3A and Table 1). Flow cytometric analysis demonstrated that, in the bone marrow of MSCV-Myc+Bcl2 mice, Mac1/Gr-1 double-positive cells predominated (Figure 3B), but thymus and lymph node tissues were also heavily infiltrated with large B220+ lymphoblastic cells (Figure 3C-E). Further analysis of these B220+ cells showed that these represent immature B lymphocytes that stained negative for surface IgM (Figure 3E). Consistent with these data, examination of affected tissues from MSCV-Myc+Bcl2 mice demonstrated 2 morphologically distinct leukemic populations: the bone marrow of MSCV-Myc+Bcl2 mice was packed with myeloblasts, whereas the lymph nodes of these mice were packed with lymphoblasts (Figure 3F).
Design of retroviral constructs expressing Myc and Bcl-2. (A) Murine c-Myc was inserted into the MSCV-IRES-GFP backbone, which uses the long terminal repeat (LTR) sequence to drive high expression. Dark bars represent Myc Box I and Box II. Striped bar represents the basic helix-loop-helix leucine zipper motif. Arrows represent LTRs. Psi represents the viral packaging sequence. B indicates BamHI; E, EcoRI; n, NcoI; H, HindIII; X, Xba. The murine Bcl-2 coding region was inserted into the viral backbone in a similar fashion. A construct coexpressing both Myc and Bcl-2 was generated by replacing the GFP sequence in the Myc construct with the Bcl-2 coding region. MIG indicates MSCV-IRES-GFP; MIB; MSCV-Myc+Bcl 2 (B) Expression of Myc and Bcl-2 in transfected 3T3 cells. Cells in exponential growth phase that were transfected with either GFP or Bcl-2 express similar levels of c-Myc. The cells transfected with either Myc or Myc/Bcl-2 construct express significantly higher levels of c-Myc. Only the cells transfected with either Bcl-2 or c-Myc/Bcl-2 express Bcl-2 protein. (C) IL-3-independent growth of BaF3 cells. Only cells transfected with both Myc and Bcl-2 (black line), but not Myc or Bcl-2 alone (gray line and long dashed line), grow in the absence of IL-3. Cells transfected with GFP alone do not grow in the absence of IL-3 (dotted line).
Design of retroviral constructs expressing Myc and Bcl-2. (A) Murine c-Myc was inserted into the MSCV-IRES-GFP backbone, which uses the long terminal repeat (LTR) sequence to drive high expression. Dark bars represent Myc Box I and Box II. Striped bar represents the basic helix-loop-helix leucine zipper motif. Arrows represent LTRs. Psi represents the viral packaging sequence. B indicates BamHI; E, EcoRI; n, NcoI; H, HindIII; X, Xba. The murine Bcl-2 coding region was inserted into the viral backbone in a similar fashion. A construct coexpressing both Myc and Bcl-2 was generated by replacing the GFP sequence in the Myc construct with the Bcl-2 coding region. MIG indicates MSCV-IRES-GFP; MIB; MSCV-Myc+Bcl 2 (B) Expression of Myc and Bcl-2 in transfected 3T3 cells. Cells in exponential growth phase that were transfected with either GFP or Bcl-2 express similar levels of c-Myc. The cells transfected with either Myc or Myc/Bcl-2 construct express significantly higher levels of c-Myc. Only the cells transfected with either Bcl-2 or c-Myc/Bcl-2 express Bcl-2 protein. (C) IL-3-independent growth of BaF3 cells. Only cells transfected with both Myc and Bcl-2 (black line), but not Myc or Bcl-2 alone (gray line and long dashed line), grow in the absence of IL-3. Cells transfected with GFP alone do not grow in the absence of IL-3 (dotted line).
Myc and Bcl-2 expression cooperate to transform primary lymphoid cells30,32 ; however, even in the absence of transgenic Bcl-2 expression, MSCV-Myc mice universally (n = 35, 100%) developed ruffled fur, cacexia, splenomegaly, and hind-limb paralysis. Median survival of MSCV-Myc mice was 47 days following marrow transplantation (Figure 3A, Table 1). Immunophenotype studies showed that the majority of bone marrow cells from MSCV-Myc mice stained positive for the myeloid markers Mac-1 and Gr-1 (Figure 3B). Although the composition of the bone marrow of MSCV-Myc+Bcl2 and MSCV-Myc was similar (Figure 3B), analysis of the lymph nodes and thymus of leukemic animals revealed stark contrasts. The lymphoid organs of MSCV-Myc contained only myeloid blasts and immunophenotypically mature lymphoid cells. The immature B lymphoblasts dominant in MSCV-Myc+Bcl2 mice were not detected in MSCV-Myc mice (Figure 3C-E).
We did observe a small highly autofluorescent population in the bone marrow of MSCV-Myc mice (weakly B220 and CD3 double-positive) that stained strongly with annexin V and 7-amino actinomycin D (7-AAD), with cytoplasmic blebbing and nuclear fragmentation upon histologic examination (not shown). Despite this evidence of apoptosis, the bone marrow cavity of all MSCV-Myc mice analyzed was also packed with neoplastic myeloid blasts, with expansion of cells to the surrounding soft tissue and musculature (Figure 3F). Hind-limb paralysis was due to invasion by malignant cells from vertebral body marrow cavities into the spinal canal (Figure S1). Malignant cells were also found disrupting the architecture of the spleen, and in perivascular foci in the liver (Figure S1). Differential counts of May-Grunwald-stained bone marrow cytospins demonstrated elevated myeloid blast counts (> 30%) with promyelocytes or more mature neutrophils (at least 10% of nucleated bone marrow cells) characteristic of AML of the M2 subtype (AML with maturation, Figure 3F, and data not shown).
Moribund MSCV-Myc mice, in contrast to MSCV-Myc+Bcl2 mice, had no significant lymphadenopathy or leukocytosis (Table 1). Histologic examination of lymph node cells revealed small, mature-appearing lymphocytes with normal morphology (Figure 3F), consistent with the immunophenotypic analysis (Figure 3D-E). The thymuses from MSCV-Myc mice were of normal size and were found to contain normal lymphocytes predominantly (Figure 3D-E). However, histologic studies revealed a small population of cells with myeloid leukemic morphology (Figure 3F).
We performed secondary transplantation experiments to assess the self-renewal capacity of leukemic cells from our mice. We isolated mononuclear cells from the spleens of moribund primary MSCV-Myc or MSCV-Myc+Bcl2 animals and intravenously injected serial dilutions into sublethally irradiated syngeneic mice. Both MSCV-Myc+Bcl2 mixed lineage and MSCV-Myc myeloid leukemias were easily transplantable into secondary recipients. Secondary recipients succumbed to leukemias with phenotypes identical to the primary animals (data not shown), and the median survival of these mice was inversely correlated with the number of cells injected, allowing an estimation of the frequency of leukemia-initiating cells. The number of leukemia-initiating cells was more than 1 in 500 in MSCV-Myc+Bcl2 mice and between 1 in 5000 to 1 in 500 in MSCV-Myc mice (Figure 4). These findings demonstrate that leukemic cells from both MSCV-Myc+Bcl2 and MSCV-Myc mice have the capacity to self-renew, and suggest that differences in disease latency between MSCV-Myc mice and MSCV-Myc+Bcl2 mice (16 days vs 47 days) may be due in part to the presence of a greater number of transformed clones in the setting of antiapoptotic mutations (eg, lymphoid plus myeloid versus myeloid alone).
Inhibition of apoptosis with Bcl-2 is required for development of lymphoid but not myeloid leukemias. (A) Kaplan-Meier survival analysis of bone marrow transplants using MSCV-Bcl2, MSCV-Myc, and MSCV-Myc+Bcl2 using wild-type Balb/c donors and recipients. (B) Flow cytometric analysis of bone marrow cells from animals that underwent transplantation. Samples from MSCV-Myc and MSCV-Myc+Bcl2 mice were isolated from moribund animals. MSCV-Bcl2 mice never appeared ill and were killed as controls 5 months after marrow transplantation. Compared with MSCV-Bcl2 mice, both MSCV-Myc and MSCV-Myc+Bcl2 mice have a large Gr-1+/Mac1+ blast population. MSCV-Myc mice also show a population of cells pseudo-positive for both B220 and CD3. These cells were judged apoptotic cells as judged by annexin V and 7-AAD staining, morphology, and failure to give disease in secondary transplant recipients (not shown). (C) Flow cytometric analysis of thymocytes from the same leukemic and control animals. MSCV-Myc+Bcl2 mice show an increase in B220+/IgM- early B cells in the thymus whereas MSCV-Myc mice show a relative loss of mature single-positive thymocytes. (D, E) Flow cytometric analysis of lymph node cells isolated from MSCV-Myc+Bcl2, MSCV-Myc, and MSCV-Bcl2 animals. Both MSCV-Bcl2 and MSCV-Myc mice have a near-normal complement of B and T cells. (D) Immunophenotype of lymph node cells of mice that underwent transplantation. B220+ cells predominate in MSCV-Myc+Bcl2 mice at the expense of CD3+ T cells. (E) Lymph node cells in MSCV-Myc+Bcl2 mice are immature B lymphoblasts (CD43+, IgM-). In contrast, lymph node cells from MSCV-Myc and MSCV-Bcl2 mice are a mixture of mature B cells (CD43-, IgM+) and mature T cells (CD4 and CD8 single-postive cells). (F) Histopathologic analysis of hematopoietic tissues from MSCV-Myc+Bcl2, MSCV-Myc, and MSCV-Bcl2 mice. MSCV-Myc+Bcl2 mice have myeloid blast cells in the bone marrow and lymphoid blast cells in the lymph node and thymus. MSCV-Myc mice have myeloid blast cells in the bone marrow, mostly mature lymphocytes in the lymph nodes, with some invading myeloid blasts, and normal-appearing thymocytes in the thymus. MSCV-Bcl2 mice have mature-appearing, apparently normal cells in the bone marrow, lymph nodes, and thymus.
Inhibition of apoptosis with Bcl-2 is required for development of lymphoid but not myeloid leukemias. (A) Kaplan-Meier survival analysis of bone marrow transplants using MSCV-Bcl2, MSCV-Myc, and MSCV-Myc+Bcl2 using wild-type Balb/c donors and recipients. (B) Flow cytometric analysis of bone marrow cells from animals that underwent transplantation. Samples from MSCV-Myc and MSCV-Myc+Bcl2 mice were isolated from moribund animals. MSCV-Bcl2 mice never appeared ill and were killed as controls 5 months after marrow transplantation. Compared with MSCV-Bcl2 mice, both MSCV-Myc and MSCV-Myc+Bcl2 mice have a large Gr-1+/Mac1+ blast population. MSCV-Myc mice also show a population of cells pseudo-positive for both B220 and CD3. These cells were judged apoptotic cells as judged by annexin V and 7-AAD staining, morphology, and failure to give disease in secondary transplant recipients (not shown). (C) Flow cytometric analysis of thymocytes from the same leukemic and control animals. MSCV-Myc+Bcl2 mice show an increase in B220+/IgM- early B cells in the thymus whereas MSCV-Myc mice show a relative loss of mature single-positive thymocytes. (D, E) Flow cytometric analysis of lymph node cells isolated from MSCV-Myc+Bcl2, MSCV-Myc, and MSCV-Bcl2 animals. Both MSCV-Bcl2 and MSCV-Myc mice have a near-normal complement of B and T cells. (D) Immunophenotype of lymph node cells of mice that underwent transplantation. B220+ cells predominate in MSCV-Myc+Bcl2 mice at the expense of CD3+ T cells. (E) Lymph node cells in MSCV-Myc+Bcl2 mice are immature B lymphoblasts (CD43+, IgM-). In contrast, lymph node cells from MSCV-Myc and MSCV-Bcl2 mice are a mixture of mature B cells (CD43-, IgM+) and mature T cells (CD4 and CD8 single-postive cells). (F) Histopathologic analysis of hematopoietic tissues from MSCV-Myc+Bcl2, MSCV-Myc, and MSCV-Bcl2 mice. MSCV-Myc+Bcl2 mice have myeloid blast cells in the bone marrow and lymphoid blast cells in the lymph node and thymus. MSCV-Myc mice have myeloid blast cells in the bone marrow, mostly mature lymphocytes in the lymph nodes, with some invading myeloid blasts, and normal-appearing thymocytes in the thymus. MSCV-Bcl2 mice have mature-appearing, apparently normal cells in the bone marrow, lymph nodes, and thymus.
We also used secondary transplantation to functionally assess the normal-appearing cells detected in the lymphoid organs of MSCV-Myc mice. Unfractionated thymocytes or lymph node cells from moribund MSCV-Myc→Ink4a-/- or MSCV-Myc+Bcl2 mice gave rise to lymphoid leukemias in secondary recipients, whereas thymocytes or lymph node cells from MSCV-Myc mice either failed to cause disease in secondary recipients or gave rise to myeloid lineage leukemias due to contaminating myeloblasts (Table 2). Therefore, in contrast to malignant lymphoid cells transformed by Myc in the setting of antiapoptotic mutations, lymphocytes from mice expressing Myc alone, while potentially “premalignant,” were not able to cause disease in secondary recipients.
Summary of results from secondary transplantation experiments using lymphoid tissues: lymphoid cells from MSCV-Myc mice are not fully transformed
Donor leukemic mice . | Tissue . | Recipients affected . | Mean survival, days . | Disease phenotype . |
|---|---|---|---|---|
| Myc→Ink4a+/+ | Thymocytes | 3/3 | 34 | Myeloid leukemia |
| MIB→BALB/c | Thymocytes | 2/2 | 58 | Mixed-lineage leukemia |
| MIB→BALB/c | LN cells | 3/3 | 37 | Mixed-lineage leukemia |
| Myc→BALB/c | LN cells | 0/2 | NA | None |
Donor leukemic mice . | Tissue . | Recipients affected . | Mean survival, days . | Disease phenotype . |
|---|---|---|---|---|
| Myc→Ink4a+/+ | Thymocytes | 3/3 | 34 | Myeloid leukemia |
| MIB→BALB/c | Thymocytes | 2/2 | 58 | Mixed-lineage leukemia |
| MIB→BALB/c | LN cells | 3/3 | 37 | Mixed-lineage leukemia |
| Myc→BALB/c | LN cells | 0/2 | NA | None |
Single-cell suspensions containing 1 × 106 to 2 × 106 cells from indicated tissues of moribund primary animals were injected intravenously into sublethally irradiated syngeneic mice.
Myc indicates MSCV-Myc; MIB, MSCV-Myc + Bcl2; LN, lymph node; NA, not achieved.
Malignant cells from MSCV-Myc and MSCV-Myc+Bcl-2 mice are readily transplantable into secondary recipients. Kaplan-Meier survival analysis of secondary recipients of (A) MSCV-Myc and (B) MSCV-Myc+Bcl2 cells. Viable mononuclear cells isolated from the spleens of moribund primary animals were counted, serially diluted, and injected into sublethally irradiated syngeneic mice. Number of mononuclear cells injected per mouse is shown. Comparison of panels A and B shows a higher frequency of leukemia-initiating cells in MSCV-Myc+Bcl2 mice.
Malignant cells from MSCV-Myc and MSCV-Myc+Bcl-2 mice are readily transplantable into secondary recipients. Kaplan-Meier survival analysis of secondary recipients of (A) MSCV-Myc and (B) MSCV-Myc+Bcl2 cells. Viable mononuclear cells isolated from the spleens of moribund primary animals were counted, serially diluted, and injected into sublethally irradiated syngeneic mice. Number of mononuclear cells injected per mouse is shown. Comparison of panels A and B shows a higher frequency of leukemia-initiating cells in MSCV-Myc+Bcl2 mice.
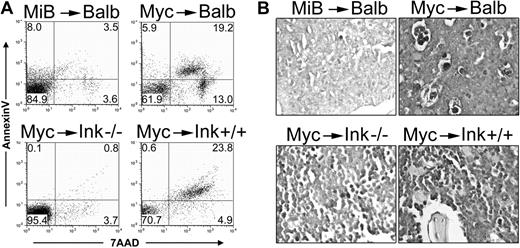
We assessed apoptosis in our leukemic mice and found robust Myc-induced apoptosis in the bone marrow of moribund MSCV-Myc mice with AML. Apoptosis was notably reduced in the marrow of leukemic MSCV-Myc+Bcl2 and MSCV-Myc→Ink4a-/- mice (Figure 5A). Similar results were obtained using transferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) staining of diseased tissues (Figure 5B). Taken together, our data demonstrate that while transformation of lymphoid cells requires the active inhibition of apoptosis, Myc is able to transform immature myeloid cells, and induce fatal AML, without suppression of apoptosis by genetic manipulation.
Myc-induced myeloid leukemias are characterized by increased bone marrow apoptosis. (A) Apoptotic cells in the bone marrow of moribund leukemic mice were assessed by annexin V and 7-AAD staining and followed by flow cytometry. Numbers represent the percentage of events in each quadrant. MSCV-Myc+Bcl2 (MiB)/Balb/c showed 15% annexin V- and/or 7-AAD-positive cells compared with 38% dying cells in matched stain mice expressing Myc but not Bcl-2 (Myc/Balb, top panels). Bone marrow from leukemic Myc/Ink4a-/- mice are 5% apoptotic compared with 29% apoptotic cells in the marrow of wild-type mice expressing Myc (Myc/Ink4a+/+, bottom panels). A representative of 3 independent analyses is shown. (B) Bone marrow sections from moribund leukemic mice were prepared and stained with terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL). Myc induces AML and apoptosis, whereas Myc+Bcl2 induces AML and ALL. Expressing Myc in bone marrow with targeted disruption of the Ink4a gene (Ink4a-/-) yields results similar to Myc+Bcl2. MiB, MSCV-Myc+Bcl2; Myc, MSCV-Myc; Ink, Ink4a.
Myc-induced myeloid leukemias are characterized by increased bone marrow apoptosis. (A) Apoptotic cells in the bone marrow of moribund leukemic mice were assessed by annexin V and 7-AAD staining and followed by flow cytometry. Numbers represent the percentage of events in each quadrant. MSCV-Myc+Bcl2 (MiB)/Balb/c showed 15% annexin V- and/or 7-AAD-positive cells compared with 38% dying cells in matched stain mice expressing Myc but not Bcl-2 (Myc/Balb, top panels). Bone marrow from leukemic Myc/Ink4a-/- mice are 5% apoptotic compared with 29% apoptotic cells in the marrow of wild-type mice expressing Myc (Myc/Ink4a+/+, bottom panels). A representative of 3 independent analyses is shown. (B) Bone marrow sections from moribund leukemic mice were prepared and stained with terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL). Myc induces AML and apoptosis, whereas Myc+Bcl2 induces AML and ALL. Expressing Myc in bone marrow with targeted disruption of the Ink4a gene (Ink4a-/-) yields results similar to Myc+Bcl2. MiB, MSCV-Myc+Bcl2; Myc, MSCV-Myc; Ink, Ink4a.
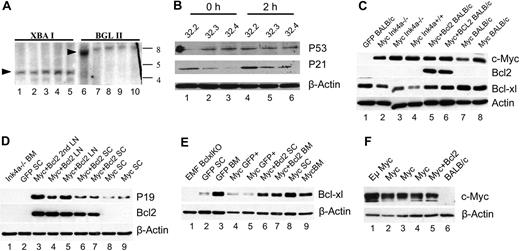
MSCV-Myc tumors display normal karyotypes and are polyclonal. (A) Spectral karyotyping of malignant spleen cells from 5 MSCV-Myc mice did not reveal any clonal abnormalities. A representative karyotype, 40, XX[10], is shown. (B) Analysis of proviral integration by Southern blot hybridization using an MSCV-specific (Psi region) probe. Digestion of spleen genomic DNA (gDNA) with an enzyme (XbaI) that cuts twice in the provirus releases a single 4.2-kb proviral band in MSCV-Myc mice (arrow, lanes 2-8), demonstrating the presence of provirus in affected tissues but not in Balb/c control (lane 1). Digestion of genomic DNA with an enzyme (BglII) that cuts once in the provirus reveals polyclonal smearing and oligoclonal bands. More than 15 individual murine leukemias were analyzed and no clonal tumors were found. A representative blot is shown. Lanes 2, 3, 4, 7, and 8 are from primary MSCV-Myc/Balb/c mice. Lanes 5 and 13 are from primary MSCV-Myc/ Black6 spleens and lanes 6 and 14 are gDNA from an isolated granulocytic sarcoma. Lanes 1 and 9 are Balb/c control splenocytes.
MSCV-Myc tumors display normal karyotypes and are polyclonal. (A) Spectral karyotyping of malignant spleen cells from 5 MSCV-Myc mice did not reveal any clonal abnormalities. A representative karyotype, 40, XX[10], is shown. (B) Analysis of proviral integration by Southern blot hybridization using an MSCV-specific (Psi region) probe. Digestion of spleen genomic DNA (gDNA) with an enzyme (XbaI) that cuts twice in the provirus releases a single 4.2-kb proviral band in MSCV-Myc mice (arrow, lanes 2-8), demonstrating the presence of provirus in affected tissues but not in Balb/c control (lane 1). Digestion of genomic DNA with an enzyme (BglII) that cuts once in the provirus reveals polyclonal smearing and oligoclonal bands. More than 15 individual murine leukemias were analyzed and no clonal tumors were found. A representative blot is shown. Lanes 2, 3, 4, 7, and 8 are from primary MSCV-Myc/Balb/c mice. Lanes 5 and 13 are from primary MSCV-Myc/ Black6 spleens and lanes 6 and 14 are gDNA from an isolated granulocytic sarcoma. Lanes 1 and 9 are Balb/c control splenocytes.
Myc-induced AML is polyclonal and maintains an intact Arf-p53 pathway
Myc expression can elicit genomic instability, raising the possibility that Myc may induce tumorigenesis by facilitating the accumulation of mutagenic chromosomal abnormalities.33 To examine the MSCV-Myc myeloid leukemia genome for chromosomal abnormalities, we examined metaphase spreads from the spleen tumors of 5 animals by spectral karyotyping (SKY). No clonal chromosomal abnormalities were identified in any of the tumors examined (Figure 6A). Analysis of proviral integration was also performed to determine the clonality of MSCV-Myc tumors. We examined genomic DNA isolated from the splenocytes for the presence of proviral sequences by Southern hybridization using a provirus-specific probe. Using a restriction endonuclease that cuts twice in the provirus, genomic DNA isolated from spleen mononuclear cells was subjected to Southern blot analysis using an enhanced GFP (EGFP) probe. A band of the expected size was found in all affected tissues from MSCV-Myc mice, but not in genomic DNA from control animals (Figure 6B, left panel). Using an enzyme that cuts only once in the provirus, analysis of the same genomic DNAs revealed that none of the tumors analyzed (n > 20) were clonally derived. Instead, repeated analysis of multiple tumors demonstrated a heterogeneous pattern of faint bands at varying molecular weights, suggesting an polyclonal or oligoclonal tumor-cell population (Figure 6B, right panel).
MSCV-Myc leukemias contain intact Arf-p53 pathways. (A) Germline DNA from spleens of leukemic mice was analyzed by Southern hybridization using an Ink4a exon 1β probe. Lanes 1 and 6 are Balb/c control mice and demonstrate the germline configuration of the locus. Arrowheads indicate size of germline fragments. Lanes 2 to 5 and 7 to 10 are MSCV-Myc spleen cells. Lanes 1 to 5 are spleen genomic DNA samples digested with XbaI and lanes 6 to 10 are digested with BglII. The band in lane 6 is due to inadvertent DNA overloading noted on ethidium-stained gel (data not shown). In all leukemias examined, the Ink4 locus was found to be in germline configuration. (B) Western blot analysis of p53 protein and the p53 target p21 in isolated leukemia cells before and after γ-irradiation. Leukemia cells were purified from spleens of moribund MSCV-Myc mice by FACS sorting on GFP and forward/side scatter profiles. Lysates were isolated at time zero (lanes 1-3) and from parallel cell aliquots 2 hours after irradiation with 500 Gy (lanes 4-6). All lanes have detectable p53 and the 2-hour samples reveal increased p21 levels, consistent with expected up-regulation by intact p53 response. β-actin levels indicated equivalent protein loading in all lanes. (C) Western analysis of proteins from tissues of leukemic and control animals that underwent transplantation. Myc protein is expressed in all MSCV-Myc and MSCV-Myc+Bcl2 mice. Bcl-2 is only expressed in MSCV-Myc+Bcl2 mice. Bcl-XL is expressed in normal bone marrow as well as unfractionated leukemia samples due to normal cell contamination (see panel E). (D) Western analysis of p19Arf and Bcl-2 proteins in animals that underwent transplantation. Arf is expressed in all tumors analyzed. Arf-null mouse embryonic fibroblasts (MEFs) are shown as negative control. Bcl-2 is expressed only in MSCV-Myc+Bcl2 samples. (E) Comparison of Bcl-XL protein levels in unfractionated and purified tumor-cell populations. Unfractionated tumor tissues show abundant Bcl-XL protein due to contamination with normal cells. Bcl-XL levels in purified leukemia cells are much lower (see panel B). (F) Comparison of c-Myc expression in Eμ-Myc lymphoma and MSCV-Myc leukemia. Whole-cell lysates were prepared from tumor-bearing lymph nodes from an Eμ-Myc mouse, and spleen cells from MSCV-Myc (Myc), MSCV-Myc+Bcl2 (Myc+Bcl2), and normal Balb/c control (BALB/c) mice. Expression levels appear equivalent in samples from retroviral and transgenic models.
MSCV-Myc leukemias contain intact Arf-p53 pathways. (A) Germline DNA from spleens of leukemic mice was analyzed by Southern hybridization using an Ink4a exon 1β probe. Lanes 1 and 6 are Balb/c control mice and demonstrate the germline configuration of the locus. Arrowheads indicate size of germline fragments. Lanes 2 to 5 and 7 to 10 are MSCV-Myc spleen cells. Lanes 1 to 5 are spleen genomic DNA samples digested with XbaI and lanes 6 to 10 are digested with BglII. The band in lane 6 is due to inadvertent DNA overloading noted on ethidium-stained gel (data not shown). In all leukemias examined, the Ink4 locus was found to be in germline configuration. (B) Western blot analysis of p53 protein and the p53 target p21 in isolated leukemia cells before and after γ-irradiation. Leukemia cells were purified from spleens of moribund MSCV-Myc mice by FACS sorting on GFP and forward/side scatter profiles. Lysates were isolated at time zero (lanes 1-3) and from parallel cell aliquots 2 hours after irradiation with 500 Gy (lanes 4-6). All lanes have detectable p53 and the 2-hour samples reveal increased p21 levels, consistent with expected up-regulation by intact p53 response. β-actin levels indicated equivalent protein loading in all lanes. (C) Western analysis of proteins from tissues of leukemic and control animals that underwent transplantation. Myc protein is expressed in all MSCV-Myc and MSCV-Myc+Bcl2 mice. Bcl-2 is only expressed in MSCV-Myc+Bcl2 mice. Bcl-XL is expressed in normal bone marrow as well as unfractionated leukemia samples due to normal cell contamination (see panel E). (D) Western analysis of p19Arf and Bcl-2 proteins in animals that underwent transplantation. Arf is expressed in all tumors analyzed. Arf-null mouse embryonic fibroblasts (MEFs) are shown as negative control. Bcl-2 is expressed only in MSCV-Myc+Bcl2 samples. (E) Comparison of Bcl-XL protein levels in unfractionated and purified tumor-cell populations. Unfractionated tumor tissues show abundant Bcl-XL protein due to contamination with normal cells. Bcl-XL levels in purified leukemia cells are much lower (see panel B). (F) Comparison of c-Myc expression in Eμ-Myc lymphoma and MSCV-Myc leukemia. Whole-cell lysates were prepared from tumor-bearing lymph nodes from an Eμ-Myc mouse, and spleen cells from MSCV-Myc (Myc), MSCV-Myc+Bcl2 (Myc+Bcl2), and normal Balb/c control (BALB/c) mice. Expression levels appear equivalent in samples from retroviral and transgenic models.
In Eμ-Myc lymphomas, loss of p19Arf expression occurs frequently due to biallelic deletion of the Ink4aArf locus. To characterize the Ink4aArf locus in our MSCV-Myc leukemias, we subjected gDNA from leukemic tissues to Southern analysis using a probe specific for Ink4a exon 1β. We found the Ink4aArf locus to be in the germ line configuration in all tumors tested (n = 6 primary, Figure 7A; n = 4 secondary, data not shown). Although we detected no large-scale deletions in the Ink4a locus, smaller deletions or point mutations affecting Ink4a or Arf would also be expected to contribute to tumorigenesis. To address this possibility, we generated cDNA using bone marrow RNA from MSCV-Myc transplants and sequenced the coding regions of both Arf (n = 8) and Ink4a (n = 6) genes. In every case, PCR amplification generated amplicons of the expected size, and no mutations were found in transcripts encoding p16Ink4a or p19Arf (data not shown). We also sequenced p53 from cDNA amplified from MSCV-Myc tumors, and again, in every tumor tested (n = 8), we found amplicons of the expected size. In 4 of 4 mice, we detected a silent C358T that likely represents an unreported polymorphism of our Balb/c substrain, but no base changes that would result in amino acid substitution or premature stop. Although we found no somatic mutations in p53, Arf, or Ink4a in any of our MSCV-Myc tumors, the wild-type germ line Balb/c Ink4a allele has been linked to plasmacytoma susceptibility and encodes a hypomorphic p16Ink4a protein.34 We excluded this as an explanation for the phenotype we observed in Balb/c mice by repeating MSCV-Myc bone marrow transduction/transplantation using bone marrow from Black6 mice, whose Ink4a allele encodes a fully functional protein. Black6 mice succumbed to an AML phenotype identical to that seen with Balb/c mice with similar median survival and 100% penetrance (data not shown).
We next examined protein extracts from MSCV-Myc and other tumors for expression of Myc, p19Arf, p16Ink4a, p53, p21, Bcl-2, and Bcl-XL proteins (Figure 7B-F). Mutations in modifier genes, such as Dmp1, or methylation of Arf may functionally impair the Arf/Mdm/p53 pathway. To exclude the possibility that other genetic or epigenetic changes could be affecting p53 levels or function, Myc-expressing tumor cells from moribund MSCV-Myc animals were purified using high-speed flow cytometry on the basis of GFP expression and forward and side scatter profiles to eliminate contamination from normal splenocytes. We subjected these purified MSCV-Myc tumor cells to 500 cGy gamma-irradiation and analyzed levels of p53 and the p53 target gene p21waf/cip. p53 levels were low, but uniformly detectable, in purified MSCV-Myc tumor cells both before and 2 hours after gamma irradiation (Figure 7B). Notably, we did not see supra-physiologic expression of p53 in any of our samples, as has been reported in association with inactivating mutations of p53.14 In all samples tested, p21waf/cip levels were increased after irradiation compared with lysates harvested before irradiation (Figure 7B), demonstrating a functional p53 response. P19Arf protein was also detected in all tumors tested, albeit at somewhat reduced levels in myeloid compared with lymphoid tumors (Figure 7D). Thus, despite detailed examination of the Arf-p53 pathway at the DNA, RNA, and protein levels, no lesions in this tumor suppressor pathway were found in any of the tumors tested.
Examination of leukemic tissues by Western analysis also showed that the antiapoptotic protein Bcl-2 was not expressed at detectable levels except in MSCV-Myc+Bcl2 tumors (Figure 7C-D). The Bcl-2 family member Bcl-xL was broadly expressed in MSCV-GFP control bone marrow, as well as in unfractionated leukemia samples from MSCV-Myc and MSCV-Myc+Bcl2 mice (Figure 7C). However, analysis of FACS-purified MSCV-Myc tumor cells revealed much lower levels of Bcl-XL protein (Figure 7E). Therefore, the Bcl-xL signal we observed in leukemic tissues is likely due to contaminating normal marrow elements. Finally, to determine if the AML phenotype we observed in MSCV-Myc mice was an artifact of extremely high expression from the MSCV LTR, we compared the level of c-Myc protein in our MSCV-Myc AML tumor lysates with lysates made from an Eμ-Myc transgenic tumor. We found that our MSCV-Myc system does not express Myc more highly than the well-studied Eμ-Myc system. If anything, the Eμ-Myc tumor expressed Myc to a higher level than did our MSCV-Myc AML samples (Figure 7F).
c-Myc preferentially expands myeloid, but not lymphoid, progenitor cells ex vivo
Lastly, we used methylcellulose colony assays to compare the effects of forced Myc expression on progenitor cells of myeloid and lymphoid origin. We used FACS-purified, GFP-positive cells from mice that had undergone transplantation with bone marrow mononuclear cells expressing an inducible Myc allele (MSCV-MycER) or GFP alone (MSCV-GFP). Myc induction caused a significant increase in the number of myeloid progenitor colonies, and in the number of cytokine-independent progenitor colonies, but not in the number of lymphoid colonies (Table 3). To confirm the lineage of these colonies, we analyzed cells in aggregate by flow cytometry. Cells from colonies plated in myeloid cytokines or no cytokines were predominantly Gr-1-positive, whereas cells plated in the lymphoid cytokine IL-7 were predominantly B220+ (data not shown). These data support the model that myeloid progenitor cells are preferentially and cell-autonomously expanded by Myc induction.
Bone marrow transduction with tamoxifen-inducible Myc stimulates myeloid colony growth in methylcellulose colony assays
Construct . | 4-OH-T . | Myeloid cytokine colonies, no. . | Lymphoid cytokine colonies, no. . | Colonies with no cytokines, no. . |
|---|---|---|---|---|
| MycER | + | 930 ± 50 | 57 ± 3 | 207 ± 18 |
| MycER | − | 390 ± 50 | 64 ± 6 | 41 ± 5 |
| GFP | + | 410 ± 40 | 34 ± 5 | 5 ± 1 |
| GFP | − | 440 ± 30 | 35 ± 3 | 8 ± 1 |
Construct . | 4-OH-T . | Myeloid cytokine colonies, no. . | Lymphoid cytokine colonies, no. . | Colonies with no cytokines, no. . |
|---|---|---|---|---|
| MycER | + | 930 ± 50 | 57 ± 3 | 207 ± 18 |
| MycER | − | 390 ± 50 | 64 ± 6 | 41 ± 5 |
| GFP | + | 410 ± 40 | 34 ± 5 | 5 ± 1 |
| GFP | − | 440 ± 30 | 35 ± 3 | 8 ± 1 |
Bone marrow from mice that underwent transplantation with the indicated retroviruses was sorted for GFP and plated in methylcellulose media containing myeloid cytokines (SCF, IL-3, IL-6, Epo; 5 × 104 cells/well), lymphoid cytokines (IL-7; 4 × 105 cells/well), or no cytokines (4 × 105 cells/well). Cells were plated in triplicate in each experiment. Myeloid colony numbers were multiplied by 10 to normalize for number of cells plated. Colony numbers are ± SD. A representative of 4 independent experiments is shown.
4-OH-T indicates 4-hydroxy-tamoxifen.
Discussion
We used a murine bone marrow transduction/transplantation system to broadly express Myc in the bone marrow of mice both in the presence and absence of antiapoptotic mutations. When-apoptosis was blocked, either by the absence of Ink4a, or by coexpression of Bcl-2, Myc induced a “biphenotypic” leukemia comprised of two distinct myeloid and lymphoid malignant-cell populations (AML+ALL). In contrast, ectopic Myc expression in all mouse strains tested (Balb/c, Black6, and B6/129S) uniformly caused a rapidly fatal AML phenotype with 100% penetrance even in the absence of antiapoptotic mutations. In humans with myeloid leukemias, MYC expression is dysregulated via multiple mechanisms including gene amplification, transcriptional regulation by fusion oncoproteins such as AML/ETO, and by activating mutations in receptor and nonreceptor tyrosine kinases.19,21,22 Our results provide the first direct evidence of Myc's central role as a downstream mediator of myeloid leukemogenesis.
Our data are consistent with the recent findings that the cellular effects of Myc are dependent on the developmental context in which it is expressed.35-37 Two groups have also recently published data in regards to the role of Myc in modulating hematopoietic stem-cell function, albeit with different results. One group demonstrates that Myc expression promotes self-renewal of stem-cells,38 and the other proposes that Myc expression promotes differentiation at the expense of self-renewal.39 Our data support the idea that Myc promotes self-renewal; however, additional experiments are required to exclude the possibility that Myc in our system is transforming a cell that already possesses self-renewal capability.
Hematopoietic stem and progenitor cells may avoid Myc-induced apoptosis by the expression of endogenous antiapoptosis genes. The Bcl-2 family member Mcl-1 is expressed in developmentally immature hematopoietic cells and is critical for maintenance of stem and progenitor cells.40 The transcriptional repressor Slug is also expressed preferentially in immature hematopoietic cells and is required for protection of these cells from radiation-induced, p53-mediated apoptosis.41 Our model is that developmentally immature hematopoietic stem cells and/or myeloid progenitor cells are relatively apoptosis-resistant and therefore are especially susceptible to Myc-induced transformation. A more detailed understanding of the mechanisms by which immature bone marrow progenitor cells resist apoptosis and are transformed by Myc may allow us to identify novel, broadly applicable therapeutic targets for patients with AML.
Prepublished online as Blood First Edition Paper, June 21, 2005; DOI 10.1182/blood-2005-02-0734.
Supported by the Mentors in Medicine Program of Washington University School of Medicine (Q.L.), the Pfizer-Washington University Biomedical Agreement, and the Leukemia and Lymphoma Society (M.H.T.).
H.L. and J.O. performed research; Q.L. analyzed data and wrote the paper; F.K. performed histopathologic analyses; M.M.L.B. contributed vital analytic tools; and M.H.T. designed research and wrote the paper.
H.L., Q.L., and J.O. contributed equally to this report.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Bill Eades, Tim Graubert, and the Siteman Cancer Center Flow Cytometry Core for assistance with MoFlo cell sorting; Jan Nolta, Jesper Bonde, and David Hess for valuable reagents and help analyzing flow cytometry data; Steve Weintraub, Jason Weber, Troy Baudino, John Cleveland, and Michael D. Cole for valuable reagents; Wanghai Zhang for help with TUNEL staining; and Dan Link, Tim Graubert, Tim Ley, and Katherine Weilbaecher for valuable discussion and critical reading of the manuscript.


![Figure 6. MSCV-Myc tumors display normal karyotypes and are polyclonal. (A) Spectral karyotyping of malignant spleen cells from 5 MSCV-Myc mice did not reveal any clonal abnormalities. A representative karyotype, 40, XX[10], is shown. (B) Analysis of proviral integration by Southern blot hybridization using an MSCV-specific (Psi region) probe. Digestion of spleen genomic DNA (gDNA) with an enzyme (XbaI) that cuts twice in the provirus releases a single 4.2-kb proviral band in MSCV-Myc mice (arrow, lanes 2-8), demonstrating the presence of provirus in affected tissues but not in Balb/c control (lane 1). Digestion of genomic DNA with an enzyme (BglII) that cuts once in the provirus reveals polyclonal smearing and oligoclonal bands. More than 15 individual murine leukemias were analyzed and no clonal tumors were found. A representative blot is shown. Lanes 2, 3, 4, 7, and 8 are from primary MSCV-Myc/Balb/c mice. Lanes 5 and 13 are from primary MSCV-Myc/ Black6 spleens and lanes 6 and 14 are gDNA from an isolated granulocytic sarcoma. Lanes 1 and 9 are Balb/c control splenocytes.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/7/10.1182_blood-2005-02-0734/6/m_zh80190584620006.jpeg?Expires=1769293666&Signature=JRQYb0h0vHqNis3ubk2VqtOW~cSLmkHWFDXoF9Q4SDZZLcqPBcv4DodEXjF8uL18O23OnlgNWxdUS4XtevlcKkMfQMnQQwQ5BpmuSrfvKYs-t2aqNvHPkSvWOijm5Y46CnVjivxChmgp5TRTyN-zCNSrOcnwg7FclI1PmnAndqsBEImOX2NivbjZJfSS3Y4mDJxAnEu8Tb-ea7A8Vv-8D6bjVRwT7W0m89hWfB4Oi8vAbT0k63leGnfkzIOrkDkeMC2kG1nr4o8-lbVz8iXxThE1QExcufFdBpCxQf5aLEbmk9veK7WoDZhxRj7mGvZh6dzPgP2n4ItXC5Yuim~bmQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal