Abstract
Human T-cell leukemia virus type I (HTLV-1) causes adult T-cell leukemia (ATL), a fatal T-cell leukemia resistant to chemotherapy, after more than 50 years of clinical latency from transmission through breast-feeding. Polyclonal expansion of virus-infected T cells predisposes them to transformation. Constitutive activation of nuclear factor-κB (NF-κB) in the leukemic cells is essential for their growth and survival. Blocking NF-κB has been shown to be a potential strategy to treat ATL. We tested this approach using a novel NF-κB inhibitor, dehydroxymethylepoxyquinomicin (DHMEQ), and also examined its application to chemoprevention by selective purging of the HTLV-1-infected cells. DHMEQ inhibited NF-κB activation in primary ATL cells and cell lines derived from them and induced apoptotic cell death. NF-κB inhibition down-regulated expression of genes involved in antiapoptosis or cell-cycle progression. DHMEQ protected severe combined immunodeficiency (SCID) mice inoculated with HTLV-1-transformed cells from death. In addition, DHMEQ selectively targeted HTLV-1-infected cells in the peripheral blood of virus carriers in vitro, resulting in a decreased number of infected cells. We conclude that NF-κB is a potential molecular target for treatment and prevention of ATL. As a potent NF-κB inhibitor, DHMEQ is a promising compound allowing the translation of this strategy into clinical medicine. (Blood. 2005;106:2462-2471)
Introduction
Adult T-cell leukemia (ATL) is a fatal T-cell neoplasm caused by infection of human T lymphocytes by human T-cell leukemia virus type I (HTLV-1).1,2 HTLV-1 is transmitted to children mainly through breast-feeding; however, ATL usually manifests clinically in middle- or older-age individuals. The lifetime incidence of ATL among carriers is estimated to be 2.6% in women and 4.5% in men. Almost 1000 cases of ATL are diagnosed each year in Japan, where HTLV-1 is endemic.2 ATL has a poor prognosis with a mean survival time of 13 months, being refractory to currently available combination chemotherapy.3 Thus, it is critical to develop a novel treatment strategy, particularly a molecularly targeted therapy.
Although the precise mechanisms of ATL leukemogenesis remain to be clarified, transformation of virus-infected T cells in vivo is a multistage process in which at least 5 genetic events are involved.4,5 In virus-expressing cells, a virus-encoded regulatory protein, Tax, plays a critical role in the growth and survival of the infected T cells by perturbing normal regulatory mechanisms including transcription, signal transduction, and cell-cycle progression and resulting in uncontrolled cell growth and polyclonal expansion.6 Because HTLV-1-infected T cells can be immortalized in vitro, it is assumed that a similar immortalization occurs in vivo probably through the multiple functions of Tax. However, viral gene expression is silenced in ATL cells in vivo by deletion and/or mutation of the provirus genome7,8 as well as heavy CpG methylation of the 5′ region of the genome.9 Thus, in addition to direct growth promotion early in infection, Tax also may endow infected T cells with capacities that assist in the progression to a transformed phenotype.
In this context, induction of a mutator phenotype by Tax in infected cells appears to be important. Tax impairs the cell's ability to repair DNA damage. The mechanism appears to involve both suppression of base excision repair (BER) through repression of human DNA polymerase-β10,11 and also the inhibition of cellular nucleotide excision repair (NER) by suppression of DNA polymerase-δ activity.12 Induction of Bcl-xL by Tax may also contribute to suppression of homologous recombination through inhibition of the RAD51 recombination pathway.13,14 Furthermore, functional inactivation of p53 by Tax15,16 allows HTLV-1-infected cells to survive and proliferate in the presence of unrepaired genomic damage. Thus, Tax not only stimulates proliferation of cells but also induces DNA damage in HTLV-1-infected cells, hence rendering a mutator phenotype5 that may constitute a basis for multistep leukemogenesis. In other words, polyclonal expansion of HTLV-1-infected cells is an early step in the development of leukemogenesis in ATL, supporting the idea that a high proviral load (ie, an increased number of HTLV-1-infected cells in the peripheral blood) is one of the risk factors for developing ATL.2,17 Thus, suppression of clonal expansion of HTLV-1-infected cells is a logical target of preventive intervention.
Nuclear factor-κB (NF-κB) is activated in HTLV-1-infected cells by transcriptional regulator Tax. Tax activates NF-κB by stimulating the activity of the IκB kinase (IKK),18 which in turn leads to phosphorylation and degradation of the NF-κB inhibitor, IκBα. Tax activates NF-κB by binding to IKKγ, a component of IKK complex, inducing constitutive activation of IKK.19 However, it is well accepted that NF-κB is activated in the absence of viral gene expression in primary ATL cells20 and some ATL cell-derived cell lines.21 Thus, the mechanisms of NF-κB activation in ATL cells are still unclear. In particular, it remains to be elucidated whether NF-κB is activated in HTLV-1-infected but untransformed T cells in asymptomatic virus carriers, where viral genes are not expressed.
As in normal T cells,22 NF-κB activation is considered to prevent ATL cells from undergoing apoptosis.23 Thus, constitutively active NF-κB appears to be a molecular basis for aberrant growth and cytokine gene expression in ATL cells, thereby providing a target for molecular therapy of ATL. If untransformed HTLV-1-infected cells have constitutive NF-κB activation, these cells also can be targeted as a preventive intervention.
Dehydroxymethylepoxyquinomicin (DHMEQ) is a new NF-κB inhibitor that is a 5-dehydroxymethyl derivative of epoxyquinomicin C.24 Epoxyquinomicin C is a novel compound isolated from Amicolatopsis sp. MK299-95F4 as antibiotics and anti-inflammatory agents having a 4-hydroxy-5,6-epoxycyclohexenone structure like panepoxydone.25 Panepoxydone alone had been found to inhibit tumor necrosis factor-α (TNF-α)-induced activation of NF-κB.26 Therefore, we have designed and synthesized an NF-κB inhibitor, DHMEQ, as a 5-hydroxymethyl derivative of epoxyquinomicin C.24 DHMEQ inhibits nuclear translocation of p65, a component of NF-κB.27
To develop a new strategy for prevention and treatment of ATL, in the current study we have investigated the effects of DHMEQ on ATL tumor cells, asked whether NF-κB is activated in untransformed primary HTLV-1-infected cells in virus carriers, and examined the effects of DHMEQ on virus infected cells. The results show clear inhibition of NF-κB activation and efficient induction of apoptosis in primary ATL cells and cell lines derived from ATL, prolonging survival of ATL xenografted mice. We demonstrate that NF-κB is constitutively activated in untransformed HTLV-1-infected cells in the peripheral blood of virus carriers. Subsequent DHMEQ treatment of HTLV-1 carriers' peripheral blood mononuclear cells (PBMCs) ex vivo resulted in selective purging of HTLV-1-infected cells without significant toxicity of normal cells. These results provide the first evidence of effective chemoprevention of ATL as well as targeted therapy for established ATL using an NF-κB inhibitor.
Materials and methods
Cells
Jurkat and K562 cells were obtained from the Japanese Cancer Research Resources Bank (Tokyo, Japan). MT-1 and MT-2 cell lines are gifts from Dr I. Miyoshi, Kochi University; TL-Om1 from Dr K. Sugamura, Tohoku University; and ST-1 and KK-1 from Dr Y. Yamada, Nagasaki University. MT-1 and TL-Om1 cell lines were derived from leukemic cells of ATL patients and do not express viral genes, whereas the MT-2 cell line was established from cord blood lymphocytes cocultured with peripheral blood mononuclear cells from an ATL patient and constitutively expresses viral genes.28-30 PBMCs were obtained from healthy volunteers, asymptomatic carriers of HTLV-1, and patients with ATL after obtaining informed consent per the Declaration of Helsinki. Approval was obtained from the University of Tokyo institutional review board for these studies. In all experiments to test effects of DHMEQ treatment, PBMCs from healthy volunteers, HTLV-1 carriers, and ATL patients were cultured in RPMI 1640 supplemented with 10% self serum and antibiotics to avoid stimulation of lymphocytes. To prepare T-cell-enriched PBMCs, RosetteSep T Cell Enrichment (StemCell Technologies, Vancouver, BC, Canada) was used according to the protocol of the manufacturer.
Array experiments
Gene expression analysis was performed essentially as described in the manual of CodeLink Expression Bioarray System (Amersham Biosciences, Freiburg, Germany). CodeLink UniSet Human 20K I Bioarray was used. Total RNA was harvested from both DHMEQ (10 μg/mL)-treated and vehicle control populations after 12 hours. Fluorescence intensities were captured with a GenePix 4000B Array Scanner. A comparative analysis between DHMEQ treatment and control samples was performed with CodeLink Expression Analysis v2.3 software.
Growth inhibition assay and cell-cycle analysis
The effect of DHMEQ on cell growth was assayed by the (3-(4,5-dimethylthiazol-2-yl)-2,5diphenyltetrazolium (MTT) method as described.31 Briefly, 5 × 104 cells treated by MTT solution were measured by a microplate reader (Bio-Rad, Hercules, CA) at a reference wavelength of 570 nm and test wavelength of 450 nm. Cell viability was measured as a percentage of the control. In dose-response experiments, cells were treated with 2 μg/mL, 5 μg/mL, and 10 μg/mL for 48 hours. In time-course experiments, cells were treated with 10 μg/mL DHMEQ for indicated periods.
To analyze cell-cycle distribution, cells were collected after 48 hours of incubation, washed with phosphate-buffered saline (PBS), fixed with cold 70% ethanol overnight and treated with RNase and propidium iodide (PI) (50 μg/mL), and then analyzed by flow cytometry using BD-LSR II (Becton Dickinson, Franklin Lakes, NJ). The resulting DNA histograms were interpreted using the Cell Quest Pro software (Becton Dickinson).
Apoptosis and pathways analysis
To quantify apoptosis, cells were labeled with fluorescein isothiocyanate (FITC)-conjugated annexin V (BD Biosciences, Palo Alto, CA) followed by flow cytometric analysis. For analysis of morphologic changes of nuclei, cells were stained by 10 μM Hoechst 33342 and photographed through an ultraviolet filter using an Olympus BX50F microscope (Olympus, Tokyo, Japan). To study apoptosis pathways, detection of cleaved caspases by immunoblotting was done as well as inhibition of apoptosis by specific inhibitors. Primary antibodies used for immunoblotting are as follows: mouse monoclonal antibodies for caspase-3/CPP32 (BD Biociences), cleaved caspase-8 (Asp384) (Cell Signaling Technology, Beverly, MA), and rabbit polyclonal antibody for cleaved caspase-9 (Asp330) (Cell Signaling Technology). Caspase inhibitors used and the final concentrations are as follows: caspase-3 inhibitor Z-Asp-Glu-Val-Asp (DEVD)-FMK (20 μM), caspase-8 inhibitor Z-Ile-Glu-Thr-Asp (IETD)-FMK (20 μM), and caspase-9 inhibitor z-Leu-Glu (OMe)-His-Asp (OMe) (LEHD)-FMK (20 μM) (all from Calbiochem, San Diego, CA).
Immunohistochemistry
Immunohistochemical analysis was done as described,31 using a confocal microscope (Radiance 2000; Bio-Rad). Numerical aperture of the objective was 1.4. Images were analyzed using NIH-Image (Research Services Branch, National Institutes of Mental Health, National Institute of Neurological Disorders and Strokes, Bemesda, MD) and Adobe Photoshop (Adobe Systems, San Jose, CA). Primary antibodies used are as follows: goat polyclonal antibody for NF-κB p50 (C-19), rabbit polyclonal antibody for NF-κB p65 (C-20) (both from Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal antibody for activated NF-κB p65 subunit (Chemicon International, Temecula, CA), and rabbit polyclonal antibody for interleukin-2 receptor-α (IL-2Rα). Santa Cruz Biotechnology Secondary antibodies used in these studies are as follows: FITC-labeled antirabbit goat antibody (Santa Cruz Biotechnology); FITC-labeled antigoat donkey antibody (Santa Cruz Biotechnology). HTLV-1 p19 antigen expression in paraffin-embedded specimen was detected using anti-HTLV-I p19 monoclonal antibody (Chemicon International) as primary antibody and FITC-conjugated anti-mouse immunoglobulin G (IgG) (Cappel, Durham, NC) as secondary antibody.
Immunoblotting
Immunoblot analysis was done as described.31,32 Antibodies used were as follows: cyclin D1 (HD11) mouse monoclonal antibody, Bcl-xL (H-62) rabbit polyclonal antibody, c-myc (9E10) mouse monoclonal antibody, FLICE inhibitory protein, long and short isoforms (FLIPS/L) (H-202) rabbit polyclonal antibody, antitubulin (TU-02) mouse IgM monoclonal antibody (all from Santa Cruz Biotechnology), phospho-Rb (Ser795) rabbit polyclonal antibody, phospho-Rb (Ser807/811) rabbit polyclonal antibody, phospho-p53 (Ser15) mouse monoclonal antibody (all from Cell Signaling Technology). Alkaline phosphatase-conjugated secondary antibodies are as follows: anti-mouse IgG (H&L) antibody, anti-rabbit IgG (Fc) antibody (both from Promega, Madison, WI, and anti-mouse IgM antibody (Vector Laboratories, Burlingame, CA).
Reporter gene assays
For reporter assays, a κB site-dependent luciferase vector, p[κB]6-Luc, and an AP-1 site-dependent vector, AP-1-Luc, (Stratagene, La Jolla, CA) were used.31,32 Using 2 × 105 cells, transfection was done with Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA). After culturing 16 hours, DHMEQ was added to the medium at the concentration of 10 μg/mL. After 6 hours of DHMEQ treatment, cells were harvested and luciferase activities were measured by Dual Luciferase assay kit (Promega). Assays were performed in triplicate.
Electrophoretic mobility shift analysis (EMSA)
Double-stranded oligonucleotides containing the AP-1 consensus sequence (5′-CGCTTGATGAGTCAGCCGGAA-3′) and OCT1 consensus sequence (5′-TGTCGAATGCAAATCACTAGAA-3′) were purchased from Promega. For detecting NF-κB binding, a double-stranded oligonucleotide containing the κB site of the promoter for the mouse H-2Kb class I major histocompatibility antigen gene was used as a probe.33 The nucleotide sequence is 5′-GATCCGGCTGGGAATCCCCGCTGGGAATCCCCATCTA-3′. Nuclear extracts were prepared basically as described.34 DNA-protein complexes were resolved on 6% polyacrylamide gels in 0.5 × TBE (tris(hydroxymethyl)aminomethane-borate-ethylenediaminetetraacetic acid) buffer. The gels were then dried and autoradiographed.
In vivo therapeutic effect of DHMEQ
Male C.B17-scid/scid (severe combined immunodeficiency [SCID]) mice were obtained from Charles River Japan (Tokyo, Japan). Mice at 5 weeks old were treated intraperitoneally with 1 mg anti-IL-2Rβ monoclonal antibody TM-β1 (a gift from Dr M. Miyasaka, Osaka University) in PBS for 3 to 5 days before inoculation of ATL cells.35,36 Then, mice were injected with 5 × 107 MT-2 cells intraperitoneally. In the treatment group, mice injected with MT-2 cells received an intraperitoneal injection of DHMEQ (4 mg/kg or 12 mg/kg) dissolved in 0.5% carboxymethyl cellulose (CMC) (Sigma, St Louis, MO) solution, on day 0 and 3 times a week thereafter until a month after inoculation with the cells. In control groups, mice were treated with 0.5% CMC solution by the same procedure. Survival was calculated by the method of Kaplan and Meier. Statistical significance was examined by Cox-Mantel test.
Real-time PCR of HTLV-1 provirus
Provirus copy numbers (ie, provirus load) of PBMC samples derived from asymptomatic carriers were measured by real-time polymerase chain reaction (PCR) using ABI PRISM 7000 Sequence Detection System (Applied Biosystems Japan, Tokyo, Japan). Cells were treated with DHMEQ or solvent dimethyl sulfoxide (DMSO) alone for 72 hours, followed by removal of dead cells using a DEAD Cell Removal Kit (MACS; Miltenyi Biotec, Auburn, CA). Collected cells were subjected to DNA isolation by a genomic DNA purification kit (PUREGENE; Gentra Systems, Minneapolis, MN). As previously reported,37 quantitative real-time PCR was done by multiplex PCR using 2 set of primers, for HTLV-1 provirus and a single copy gene, RNaseP. The primers and the probe for RNaseP were purchased from Applied Biosystems (Foster City, CA). The primers and the probe for the pX region of HTLV-1 provirus are as follows: forward primer pX2-S 5′-CGGATACCCAGTCTACGTGTT-3′, reverse primer pX2-AS 5′-CAGTAGGGCGTGACGATGTA-3′, and carboxyfluorescein (FAM)-labeled pX2 probe 5′-CTGTGTACAAGGCGACTGGTGCC-3′. For a control experiment, equal number of cells from K562 and TL-Om1 cell lines were mixed and treated with DHMEQ and subjected to real-time PCR analysis for quantitation of provirus copy numbers. Results were presented as reduction rates by comparing copy numbers between DHMEQ-treated and mock-treated samples.
Results
DHMEQ blocks constitutive NF-κB activity in ATL-derived cell lines
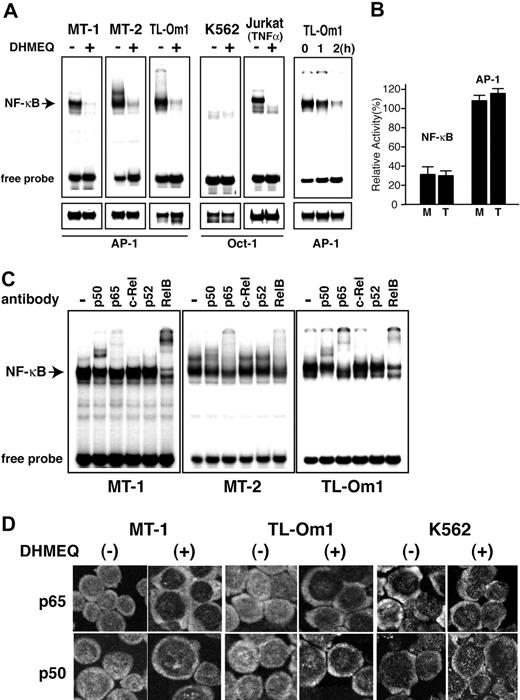
We first examined effects of DHMEQ on constitutive NF-κB activity in 2 ATL-derived cell lines (MT-1 and TL-Om1) and the MT-2 cell line that was transformed by HTLV-1 in vitro. Electrophoretic mobility shift analysis (EMSA) with an NF-κB probe showed that DHMEQ efficiently abrogates constitutive NF-κB binding activity in the 3 ATL-derived cell lines (Figure 1A, left upper 3 panels). Inhibition appeared specific to NF-κB and not due to cell death, because no significant change in binding activity of AP-1 was observed after treatment of cells with DHMEQ at a concentration of 10 μg/mL (Figure 1A, left lower 3 panels). When K562 cells, which do not show constitutive NF-κB activation, were treated by DHMEQ (10 μg/mL), no change in binding activity of OCT1 was observed (Figure 1A, fourth panels). To confirm NF-κB inhibitory activity of DHMEQ after TNF-α stimulation, NF-κB binding activity was tested with TNF-α-stimulated Jurkat cells with or without DHMEQ treatment. Results clearly showed selective inhibition of NF-κB binding and almost no change in OCT1 binding (Figure 1A, upper and lower fifth panels). Next we tested the time course of NF-κB inhibition by DHMEQ using TL-Om1 cells. NF-κB binding was significantly inhibited after 2-hour treatment with DHMEQ without significant changes in the binding activity of AP-1 (Figure 1A, upper and lower rightmost panels).
DHMEQ inhibits constitutive NF-κB activity in HTLV-1-transformed and ATL-derived cell lines. (A) EMSA of NF-κB. Inhibition of constitutive NF-κB binding activity by DHMEQ in HTLV-1-transformed and ATL-derived cell lines (left 3 panels). A myeloid leukemia cell line K562 without HTLV-1 infection was used as a control. Cells were cultured with or without 10 μg/mL DHMEQ for 16 hours. Nuclear extracts from Jurkat cells treated with TNF-α served as a control. Upper panels show inhibition of NF-κB binding activity by DHMEQ. Lower panels show results of EMSA with control probes, AP-1 and OCT1. The OCT1 probe was used for HTLV-1-uninfected cells that do not show constitutive activation of AP-1. (B) Inhibition of NF-κB transcription activities in ATL-derived cell lines by DHMEQ. Relative levels of luciferase activities are shown in percentages compared with the levels of untreated cells. M indicates MT-1 cells; T, TL-Om1 cells; NF-κB, NF-κB-driven luciferase construct; AP-1, AP-1-driven luciferase construct. Renilla luciferase vector (pRL-TK) was used to standardize the transfection efficiency. (C) Supershift analysis of the NF-κB components in HTLV-1-transformed and ATL-derived cell lines. Antibodies used are indicated on top. The position of a shifted band corresponding to NF-κB is indicated on the left. (D) Inhibition of nuclear translocation of NF-κB by DHMEQ. Representative results of confocal immunofluorescence analysis using antibodies against NF-κB p65 or NF-κB p50. Original magnification ×600. Images captured with a60×/1.4 objective lens.
DHMEQ inhibits constitutive NF-κB activity in HTLV-1-transformed and ATL-derived cell lines. (A) EMSA of NF-κB. Inhibition of constitutive NF-κB binding activity by DHMEQ in HTLV-1-transformed and ATL-derived cell lines (left 3 panels). A myeloid leukemia cell line K562 without HTLV-1 infection was used as a control. Cells were cultured with or without 10 μg/mL DHMEQ for 16 hours. Nuclear extracts from Jurkat cells treated with TNF-α served as a control. Upper panels show inhibition of NF-κB binding activity by DHMEQ. Lower panels show results of EMSA with control probes, AP-1 and OCT1. The OCT1 probe was used for HTLV-1-uninfected cells that do not show constitutive activation of AP-1. (B) Inhibition of NF-κB transcription activities in ATL-derived cell lines by DHMEQ. Relative levels of luciferase activities are shown in percentages compared with the levels of untreated cells. M indicates MT-1 cells; T, TL-Om1 cells; NF-κB, NF-κB-driven luciferase construct; AP-1, AP-1-driven luciferase construct. Renilla luciferase vector (pRL-TK) was used to standardize the transfection efficiency. (C) Supershift analysis of the NF-κB components in HTLV-1-transformed and ATL-derived cell lines. Antibodies used are indicated on top. The position of a shifted band corresponding to NF-κB is indicated on the left. (D) Inhibition of nuclear translocation of NF-κB by DHMEQ. Representative results of confocal immunofluorescence analysis using antibodies against NF-κB p65 or NF-κB p50. Original magnification ×600. Images captured with a60×/1.4 objective lens.
DHMEQ treatment significantly suppressed NF-κB transcription activity of 2 ATL-derived cell lines, MT-1 and TL-Om1, in transient reporter gene assays using an NF-κB-driven luciferase construct (Figure 1B). However, DHMEQ treatment did not affect transcription activity of AP-1 when an AP-1-driven luciferase construct was used in the assay (Figure 1B). These results suggest that DHMEQ specifically inhibits NF-κB activation in ATL-derived cell lines.
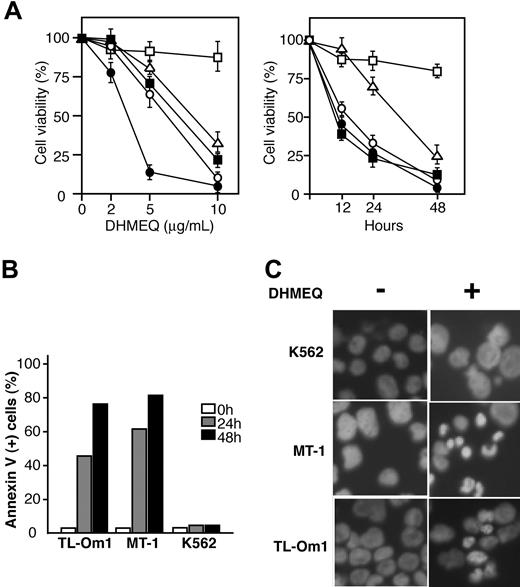
DHMEQ induces apoptosis in ATL-derived cell lines. (A) Results of dose-response and time-course experiments. Relative levels of cell viability of DHMEQ-treated ATL-derived cell lines compared with those treated with DMSO. Mean and SD of triplicate experiments are presented. TL-Om1, MT-1, KK-1, and ST-1 are ATL-derived cell lines; K562, an uninfected cell line used as a control. □ indicates K562;▵, TL-Om1; ▪, MT-1; ○, KK-1; •, ST-1. (B) Induction of apoptosis by DHMEQ. Cells were treated with 10 μg/mL DHMEQ for indicated time periods and binding of FITC-conjugated annexin V was analyzed by flow cytometry. Representative results of 3 independent experiments are shown. (C) Hoechst 33342 staining of the cells. Cells were treated with DHMEQ (10 μg/mL) or DMSO (0.1%) for 24 hours and stained by Hoechst 33342. Original magnification, × 600. Objective lens = 60×/1.4.
DHMEQ induces apoptosis in ATL-derived cell lines. (A) Results of dose-response and time-course experiments. Relative levels of cell viability of DHMEQ-treated ATL-derived cell lines compared with those treated with DMSO. Mean and SD of triplicate experiments are presented. TL-Om1, MT-1, KK-1, and ST-1 are ATL-derived cell lines; K562, an uninfected cell line used as a control. □ indicates K562;▵, TL-Om1; ▪, MT-1; ○, KK-1; •, ST-1. (B) Induction of apoptosis by DHMEQ. Cells were treated with 10 μg/mL DHMEQ for indicated time periods and binding of FITC-conjugated annexin V was analyzed by flow cytometry. Representative results of 3 independent experiments are shown. (C) Hoechst 33342 staining of the cells. Cells were treated with DHMEQ (10 μg/mL) or DMSO (0.1%) for 24 hours and stained by Hoechst 33342. Original magnification, × 600. Objective lens = 60×/1.4.
We then analyzed the composition of constitutively activated NF-κB subcomponents in MT-1, MT-2, and TL-Om1 cell lines by supershift EMSA. The results showed that constitutively activated NF-κB contains p65, p50, and RelB (Figure 1C). DHMEQ efficiently blocked binding, resulting in a significant reduction in the intensities of most shifted bands (Figure 1A).
We next examined the effects of DHMEQ on the cellular distribution of NF-κB components using immunofluorescence confocal microscopy. DHMEQ blocked nuclear localization of NF-κB p65 as well as p50 in MT-1 and TL-Om1 cells in which there is constitutive nuclear distribution of these proteins (in the absence of DHMEQ) (Figure 1D). DHMEQ treatment of K562 cells, which lack NF-κB DNA binding (Figure 1A), did not induce any change in localization of p50 and p65 (Figure 1D). Thus, these results indicate that DHMEQ can inhibit nuclear translocation of several NF-κB components in tumor cells with constitutively activated NF-κB as was reported in cells with TNF receptor-mediated activation of NF-κB.27
DHMEQ selectively induces apoptosis of ATL-derived cell lines
To evaluate the role of constitutively activated NF-κB in the cell growth of ATL-derived cell lines, we studied the in vitro effects of DHMEQ on these cells. Results of MTT assays showed that DHMEQ significantly reduced the viability of all cell lines in a dose- and time-dependent manner, whereas the effect was not significant in a control cell line K562 without constitutive NF-κB activity, even at higher concentrations (Figure 2A). To examine whether the reduced viability of ATL-derived cell lines is due to induction of apoptosis, we examined the population of apoptotic cells by annexin V-FITC staining and fluorescence-activated cell sorter (FACS) analysis. In MT-1 and TL-Om1 cell lines, annexin V-positive cells were significantly increased after 24 to 48 hours of DHMEQ treatment (Figure 2B). After 48 hours of DHMEQ treatment, Hoechst 33342 staining of nuclei showed apoptotic changes in the ATL-derived cell lines but not in K562 cells (Figure 2C). Taken together, these data suggest that DHMEQ specifically targets ATL-derived cell lines with constitutively activated NF-κB and induces apoptosis of these cells.
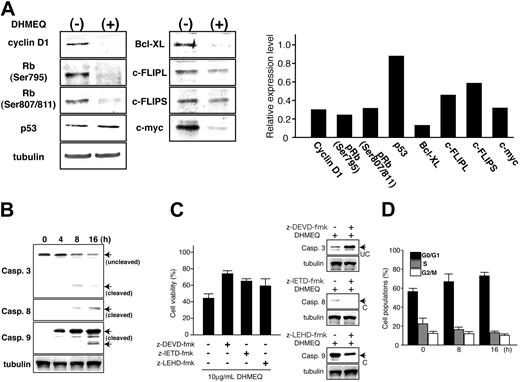
Expression of genes involved in cell-cycle progression and antiapoptosis after DHMEQ treatment. (A) Immunoblot analysis of DHMEQ-treated MT-1 cells (left panels). Cells were treated with 10 μg/mL DHMEQ for 16 hours. Total cell lysates were subjected to analysis. Antibodies used for detection are presented on the left. Levels of tubulin expression are used to confirm equal amounts of total cell lysates in each lane. (Right panel) Results of densitometric analysis of detected bands. Results are expressed as relative levels compared with those of untreated samples. Tubulin bands were used to normalize densitometric measurement. (B) Activation of caspases. Cleavage of caspase-3, -8, and -9 was examined by immunoblot analysis. Samples were prepared at the indicated time points after DHMEQ treatment. Arrows indicate the position of cleaved or uncleaved fragments. (C) Inhibition of apoptosis pathways. Effects of caspase inhibitors on DHMEQ-induced apoptosis were studied using specific inhibitors for caspase-3 (Z-DEVD-FMK) and caspase-8 (Z-IETD-FMK) as well as caspase-9 inhibitor (z-LEHD-FMK). Inhibitors were added to the culture media 1 hour prior to addition of DHMEQ. (Left panel) Cell viabilities after DHMEQ treatment in the presence of specific caspase inhibitors. (Right panel) Immunoblot analysis of caspase cleavage. Protein expression levels of cleaved or uncleaved caspases are shown. UC indicates uncleaved; C, cleaved. (D) Effects of DHMEQ on cell-cycle progression. MT-1 cells were treated with DHMEQ (10 μg/mL) for indicated periods followed by PI staining and subjected to cell-cycle analysis by flow cytometry.
Expression of genes involved in cell-cycle progression and antiapoptosis after DHMEQ treatment. (A) Immunoblot analysis of DHMEQ-treated MT-1 cells (left panels). Cells were treated with 10 μg/mL DHMEQ for 16 hours. Total cell lysates were subjected to analysis. Antibodies used for detection are presented on the left. Levels of tubulin expression are used to confirm equal amounts of total cell lysates in each lane. (Right panel) Results of densitometric analysis of detected bands. Results are expressed as relative levels compared with those of untreated samples. Tubulin bands were used to normalize densitometric measurement. (B) Activation of caspases. Cleavage of caspase-3, -8, and -9 was examined by immunoblot analysis. Samples were prepared at the indicated time points after DHMEQ treatment. Arrows indicate the position of cleaved or uncleaved fragments. (C) Inhibition of apoptosis pathways. Effects of caspase inhibitors on DHMEQ-induced apoptosis were studied using specific inhibitors for caspase-3 (Z-DEVD-FMK) and caspase-8 (Z-IETD-FMK) as well as caspase-9 inhibitor (z-LEHD-FMK). Inhibitors were added to the culture media 1 hour prior to addition of DHMEQ. (Left panel) Cell viabilities after DHMEQ treatment in the presence of specific caspase inhibitors. (Right panel) Immunoblot analysis of caspase cleavage. Protein expression levels of cleaved or uncleaved caspases are shown. UC indicates uncleaved; C, cleaved. (D) Effects of DHMEQ on cell-cycle progression. MT-1 cells were treated with DHMEQ (10 μg/mL) for indicated periods followed by PI staining and subjected to cell-cycle analysis by flow cytometry.
DHMEQ down-regulates expression of genes involved in antiapoptosis and cell-cycle progression
To investigate the molecular mechanism(s) through which DHMEQ induces apoptosis in ATL cells, we next examined the global changes in transcriptional profiles caused by DHMEQ treatment using the CodeLink Expression Bioarray System. The results demonstrated induction of proapoptotic genes and repression of antiapoptotic ones, respectively. Furthermore, the results revealed the antiproliferative sequelae with down-regulation of genes that promote cell-cycle progression (Table S1; see the Supplemental Table link at the top of the online article, at the Blood website). Down-regulation was observed in Bcl-xL, Bcl-2, c-myc, cyclin D1, Rb, and p53, whereas proapoptotic genes such as caspase-3, -8, and -9 showed up-regulation of transcripts (Table 1). Immunoblot analysis showed down-regulation of protein expression levels of these genes as well as decreased levels of phosphorylated retinoblastoma (Rb) protein (Ser 795, Ser807/811) expression. DHMEQ did not alter the expression of p53 protein (Figure 3A, left panels). Densitometric analysis of the detected bands using those of tubulin as controls confirmed the changes in expression levels of these proteins (Figure 3A, right panels). Thus, DHMEQ-induced apoptosis of ATL-derived cell lines is accompanied by a down-regulation of genes involved in antiapoptosis or cell-cycle progression and induction of proapoptotic genes.
Relative levels of transcripts after DHMEQ treatment
Gene, by family name . | Ratio . |
|---|---|
| Caspase | |
| Caspase-3 | 1.61 |
| Caspase-8 | 1.21 |
| Caspase-9 | 2.01 |
| Bcl | |
| Bcl-xL | −1.43 |
| Bcl-2 | −1.40 |
| Bad | −1.12 |
| BAK | 1.79 |
| Bid | 1.10 |
| CARD | |
| clAP-1 | −1.91 |
| clAP-2 | −2.78 |
| Apaf-1 | 1.12 |
| Bcl10 | 1.20 |
| TRAF | |
| TRAF2 | −1.14 |
| TRAF5 | −1.03 |
| TRAF6 | 1.00 |
| TNF/TNFR | |
| TNF-α | −1.16 |
| TNFR2 | −1.30 |
| FAS | −2.01 |
| Rel and related | |
| NFKB1 | −1.97 |
| NFKB2 | −1.43 |
| IKBα | −2.34 |
| IKBβ | −1.04 |
| IKKα | −1.07 |
| IKKβ | −1.94 |
| IKKγ | 1.30 |
| Cyclin | |
| Cyclin A1 | 1.29 |
| Cyclin B1 | 1.05 |
| Cyclin B2 | −1.31 |
| Cyclin C | −1.08 |
| Cyclin D1 | −1.60 |
| Cyclin D2 | −6.34 |
| Cyclin E1 | −3.00 |
| Others | |
| Myc | −1.48 |
| TP53 | −1.20 |
| RBI | −3.79 |
Gene, by family name . | Ratio . |
|---|---|
| Caspase | |
| Caspase-3 | 1.61 |
| Caspase-8 | 1.21 |
| Caspase-9 | 2.01 |
| Bcl | |
| Bcl-xL | −1.43 |
| Bcl-2 | −1.40 |
| Bad | −1.12 |
| BAK | 1.79 |
| Bid | 1.10 |
| CARD | |
| clAP-1 | −1.91 |
| clAP-2 | −2.78 |
| Apaf-1 | 1.12 |
| Bcl10 | 1.20 |
| TRAF | |
| TRAF2 | −1.14 |
| TRAF5 | −1.03 |
| TRAF6 | 1.00 |
| TNF/TNFR | |
| TNF-α | −1.16 |
| TNFR2 | −1.30 |
| FAS | −2.01 |
| Rel and related | |
| NFKB1 | −1.97 |
| NFKB2 | −1.43 |
| IKBα | −2.34 |
| IKBβ | −1.04 |
| IKKα | −1.07 |
| IKKβ | −1.94 |
| IKKγ | 1.30 |
| Cyclin | |
| Cyclin A1 | 1.29 |
| Cyclin B1 | 1.05 |
| Cyclin B2 | −1.31 |
| Cyclin C | −1.08 |
| Cyclin D1 | −1.60 |
| Cyclin D2 | −6.34 |
| Cyclin E1 | −3.00 |
| Others | |
| Myc | −1.48 |
| TP53 | −1.20 |
| RBI | −3.79 |
CARD indicates caspase recruitment domain; TRAF, TNF receptor (TNFR)-associated factor.
To distinguish between use of the extrinsic and intrinsic pathways of apoptosis induced by DHMEQ, we studied activation of caspase-3, -8, and -9. Cleavage of these caspases was examined by immunoblot analyses of cells treated by DHMEQ for 4, 8, and 16 hours. Results demonstrated that caspase-3, -8, and -9 are activated after 4 and 8 hours of treatment, suggesting both the extrinsic and intrinsic pathways are activated by DHMEQ (Figure 3B). Furthermore, all inhibitors specific to caspase-3, -8, and -9 showed significant antiapoptotic effects on DHMEQ treatment (Figure 3C, left panel), with inhibition of cleavage of respective caspases (Figure 3C, right panels), confirming involvement of both apoptotic pathways in DHMEQ-induced cell death.
Antiproliferative effects of DHMEQ were examined by cell-cycle analysis of DHMEQ-treated cells. There was a time-dependent increase of cells in G0/G1 phase after DHMEQ treatment, demonstrating antiproliferative effects of DHMEQ (Figure 3D).
Effects of DHMEQ on fresh primary ATL cells
Because NF-κB is constitutively activated in ATL-derived cell lines as well as in primary ATL cells,23 we next examined the effects of DHMEQ on freshly isolated ATL cells. Treatment of ATL cells from 3 independent patients with 10 μg/mL DHMEQ significantly blocked NF-κB DNA binding activities in each instance without changing binding activity of AP-1 (Figure 4A, left 3 panels). Supershift analysis of these 3 samples confirmed that NF-κB in primary ATL cells contains p50 and p65 but not RelB or c-Rel as previously described23 (Figure 4A, right 3 panels). EMSA of a control T cell-enriched PBMC sample revealed constitutive NF-κB DNA binding activity that was resistant to DHMEQ treatment, showing a clear contrast to ATL cells (Figure 4B, left panel). The NF-κB components in normal T cell-enriched PBMCs were found to consist mainly of p50, but not of p65 or RelB (Figure 4B, right panel), which may explain the difference from ATL cells in the sensitivity to DHMEQ.
Results of MTT assays demonstrated that DHMEQ significantly reduced the viability of primary ATL cells but not of control PBMCs (Figure 4C). Primary ATL cells showed a significant increase in annexin V-positive cells after 24 and 48 hours of DHMEQ treatment, indicating that DHMEQ induced apoptosis of primary ATL cells, whereas little increase in annexin V-positive cells was observed in control PBMC samples (Figure 4D). Hoechst 33342 staining of DHMEQ-treated ATL cells revealed apoptotic characteristics of the nuclei, such as nuclear condensation, whereas no change in staining was observed in PBMCs of healthy controls (Figure 4E). In sum, DHMEQ specifically blocks NF-κB activation, which induces apoptosis of primary ATL cells.
In vivo effects of DHMEQ on SCID mice inoculated with HTLV-1-transformed cell line
Because the DHMEQ-induced apoptosis of ATL cells suggests that DHMEQ can be an effective agent to treat ATL, we next examined the in vivo effects of DHMEQ in a SCID mouse model. Mice were inoculated intraperitoneally with 5 × 107 MT-2 cells, followed by intraperitoneal injection of 4 mg/kg or 12 mg/kg DHMEQ in 0.5% CMC, or 0.5% CMC alone. DHMEQ or CMC was then injected 3 times weekly until 1 month after tumor cell inoculation. In the experiment using 4 mg/kg DHMEQ, 4 of 6 mice treated with DHMEQ were alive after 6 months, whereas none survived in the untreated control group. In the experiment using 12 mg/kg DHMEQ, only 1 of 6 mice died in the treated group 35 days after inoculation, whereas 5 of 6 died in the untreated control group. Thus, at both 4 mg/kg and 12 mg/kg doses, a significant increase in the survival rate was observed in the mice treated with DHMEQ compared with controls (Cox-Mantel test; P < .05; Figure 5A, upper and lower panels, respectively).
DHMEQ inhibits constitutive NF-κB activity and induces apoptosis of primary ATL cells. (A) EMSA and supershift analyses. (Left upper 3 panels) DHMEQ inhibition of constitutively activated NF-κB in 3 samples of primary ATL cells. Cells were treated with or without 10 μg/mL DHMEQ for 16 hours. Positions of shifted bands and free probes are indicated on the left. (Left lower 3 panels) Results of EMSA with AP-1 probe. (Right 3 panels) Supershift analysis of NF-κB components in 3 samples of primary ATL cells. Nuclear extracts of ATL samples were subjected to supershift analysis with antibodies specific for p50, p65, c-Rel, p52, and RelB. (B) EMSA of DHMEQ effects on T cell-enriched normal PBMCs and supershift analysis of NF-κB components. (Left panel) Nuclear extracts were prepared after 16 hours of treatment with or without 10 μg/mL DHMEQ. Results of control EMSA with an OCT1 probe are shown at the bottom. (Right panel) Supershift analysis of NF-κB components using antibodies indicated above the gel. The position of shifted bands is indicated on the left. Antibodies used are indicated on the top. (C) Effects of DHMEQ on the viability of primary ATL cells. Cells were treated with 10 μg/mL DHMEQ for 48 hours. Cell viability was measured by MTT assay, and the relative levels compared with those of DMSO-treated cells are presented. Data represent the mean ± SD of 3 independent experiments. (D) Detection of apoptosis by annexin V. Three ATL samples and normal PBMCs were treated with 10 μg/mL DHMEQ, and binding of FITC-conjugated annexin V was analyzed by flow cytometry after 24 or 48 hours. Data represent the mean ± SD of 3 independent experiments. (E) Changes in the nuclear morphology by DHMEQ treatment. Primary ATL cells and control PBMCs were treated with 10 μg/mL DHMEQ for 24 hours and stained with Hoechst 33342. Original magnification ×400. Objective lens, 60×/4.4.
DHMEQ inhibits constitutive NF-κB activity and induces apoptosis of primary ATL cells. (A) EMSA and supershift analyses. (Left upper 3 panels) DHMEQ inhibition of constitutively activated NF-κB in 3 samples of primary ATL cells. Cells were treated with or without 10 μg/mL DHMEQ for 16 hours. Positions of shifted bands and free probes are indicated on the left. (Left lower 3 panels) Results of EMSA with AP-1 probe. (Right 3 panels) Supershift analysis of NF-κB components in 3 samples of primary ATL cells. Nuclear extracts of ATL samples were subjected to supershift analysis with antibodies specific for p50, p65, c-Rel, p52, and RelB. (B) EMSA of DHMEQ effects on T cell-enriched normal PBMCs and supershift analysis of NF-κB components. (Left panel) Nuclear extracts were prepared after 16 hours of treatment with or without 10 μg/mL DHMEQ. Results of control EMSA with an OCT1 probe are shown at the bottom. (Right panel) Supershift analysis of NF-κB components using antibodies indicated above the gel. The position of shifted bands is indicated on the left. Antibodies used are indicated on the top. (C) Effects of DHMEQ on the viability of primary ATL cells. Cells were treated with 10 μg/mL DHMEQ for 48 hours. Cell viability was measured by MTT assay, and the relative levels compared with those of DMSO-treated cells are presented. Data represent the mean ± SD of 3 independent experiments. (D) Detection of apoptosis by annexin V. Three ATL samples and normal PBMCs were treated with 10 μg/mL DHMEQ, and binding of FITC-conjugated annexin V was analyzed by flow cytometry after 24 or 48 hours. Data represent the mean ± SD of 3 independent experiments. (E) Changes in the nuclear morphology by DHMEQ treatment. Primary ATL cells and control PBMCs were treated with 10 μg/mL DHMEQ for 24 hours and stained with Hoechst 33342. Original magnification ×400. Objective lens, 60×/4.4.
Mice injected with more than 5 × 107 MT-2 cells developed gross tumors, the sizes of which were usually less than 1 × 1 cm. To confirm that these tumors were derived from the inoculated cells, PCR was performed to detect the HTLV-I Tax and human β-globin genes (data not shown). All tumors found in mice were positive for both genes. All tumors reacted with anti-HTLV-I p19 (Figure 5B, top panel) and anti-HLA-DR monoclonal antibodies in immunofluorescence assays (data not shown). Histologic examination of hematoxylin and eosin (H&E)-stained sections revealed that the tumor masses were lymphomas or consisted of fibrosis tissues and lymphoid cells (data not shown). After DHMEQ treatment, no HTLV-1 p19-positive cells were detected in residual tumors (Figure 5B, bottom panel). DHMEQ at these treatment dosages was well tolerated, and there were no adverse findings such as weight loss in the treated mice compared with the controls. We confirmed that DHMEQ blocked constitutive NF-κB binding activity of MT-2 cells in vitro (Figure 1A) and induced cell death (Figure 5C). These results suggest that DHMEQ can rescue SCID mice inoculated with HTLV-1-transformed T cells, probably through induction of apoptosis by abrogation of constitutive NF-κB activity.
DHMEQ can rescue SCID mice inoculated with MT-2 cells. (A) Survival curves of the SCID mice injected with MT-2 cells. DHMEQ was used at a dose of 4 mg or 12 mg/kg body weight. The differences are statistically significant (both P < .05 by Cox-Mantel test). (B) Reduction of MT-2 cells in the peritoneal mass after DHMEQ treatment. MT-2 cells were detected by immunohistochemistry using anti-p19 antibody. Original magnification ×100. Objective lens, 10×/0.4. (C) Effects of DHMEQ on MT-2 cells. DHMEQ cytotoxicity on MT-2 cells. Cells were treated with 10 μg/mL DHMEQ for 48 hours, and viabilities were studied by MTT assay. K562 cells were used as a control. Data represent the mean ± SD of 3 independent experiments. M indicates MT-2; K, K562.
DHMEQ can rescue SCID mice inoculated with MT-2 cells. (A) Survival curves of the SCID mice injected with MT-2 cells. DHMEQ was used at a dose of 4 mg or 12 mg/kg body weight. The differences are statistically significant (both P < .05 by Cox-Mantel test). (B) Reduction of MT-2 cells in the peritoneal mass after DHMEQ treatment. MT-2 cells were detected by immunohistochemistry using anti-p19 antibody. Original magnification ×100. Objective lens, 10×/0.4. (C) Effects of DHMEQ on MT-2 cells. DHMEQ cytotoxicity on MT-2 cells. Cells were treated with 10 μg/mL DHMEQ for 48 hours, and viabilities were studied by MTT assay. K562 cells were used as a control. Data represent the mean ± SD of 3 independent experiments. M indicates MT-2; K, K562.
DHMEQ-mediated reduction of HTLV-1 provirus load in the PBMCs of carriers
It has been suggested that an increased provirus load in the PBMCs is one of the risk factors for developing HTLV-1-associated diseases, including ATL.38 Increase of the provirus load (ie, the number of HTLV-1-infected cells) has been shown to result from clonal expansion of HTLV-1-infected cells, although the exact mechanisms remain to be clarified.39 Because HTLV-1-immortalized cells as well as ATL cells show constitutive activation of NF-κB,18 it is conceivable that NF-κB is also activated in untransformed HTLV-1-infected T cells in vivo, in which DHMEQ can induce apoptosis. Therefore, we first examined whether HTLV-1-infected cells in the PBMCs show activation of NF-κB. However, it is not possible to identify HTLV-1-infected cells in vivo, because viral genes are not expressed in vivo and in situ hybridization to detect integrated provirus is not sensitive. On the other hand, it is well known that HTLV-1-infected cells regularly express IL-2Rα on the cell surface irrespective of viral gene expression. Thus, we attempted to identify lymphocytes positive for both IL-2Rα and activated p65 by confocal immunofluorescence microscopy. We could identify lymphocytes positive for both IL-2Rα and p65 in each sample from HTLV-1 carriers (n = 5), whereas no control PBMC sample showed such cells (n = 3). The percentage of the positive cells ranged from a few to more than 10% in the carriers' PBMCs. A representative result is shown in Figure 6A. These results indicate that NF-κB is activated in untransformed HTLV-1-infected cells in PBMCs of virus carriers.
Thus, we next tested whether inhibition of NF-κB by DHMEQ can reduce the number of HTLV-1-infected cells (proviral load) in PBMCs of carriers. The experimental protocol is summarized in Figure 6B. PBMC samples were cultured in RPMI 1640 medium supplemented with 10% of patients' own sera and antibiotics in the presence of DHMEQ (10 μg/mL) or DMSO (0.1%) for 72 hours. Dead cells were then removed using DEAD Cell Removal Kit (MACS), and viable cells were collected. Genomic DNA samples prepared from viable cells were subjected to measurement of HTLV-1 proviral copies by quantitative PCR analysis. When equal numbers of cells of 2 cell lines, K562 and TL-Om1, were mixed and treated by DHMEQ as a control experiment, the provirus copy number was reduced by about 40% after 72 hours. When PBMC samples of HTLV-1 carriers were treated by DHMEQ for 72 hours, the cells were shown to have a reduced number of proviral copies compared with those treated by DMSO alone. The reduction rate ranged from about 10% to more than 80% (mean ± SD, 65.6% ± 25.8%) (Figure 6C). These results suggest that DHMEQ can purge PBMCs of HTLV-1-infected cells while uninfected cells are not affected.
Discussion
Strong and constitutive activation of NF-κB is a hallmark of primary ATL cells and ATL-derived cell lines and is thought to contribute to their aberrant growth and cytokine gene expression characteristic of ATL cells.20,23 ATL is characterized by dysregulation of various signaling pathways,23,40-42 and blockade of constitutive NF-κB activity is predicted to disrupt cell survival. In this study we provide evidence for NF-κB activation in untransformed HTLV-1-infected cells in vivo (Figure 6A). Thus, the NF-κB pathway appears to be a potential target for treatment and prevention of ATL. To translate this hypothesis into an effective ATL therapy, targeting NF-κB with low-molecular-weight compounds is an appropriate strategy. Thus, we tested whether a novel NF-κB inhibitor, DHMEQ, could be a potential agent for these purposes. Indeed, the results of our study showed that DHMEQ is an effective agent that blocks constitutive NF-κB activity and induces apoptosis in ATL-derived cell lines as well as in primary ATL cells. DHMEQ did not significantly affect the viability of control PBMCs or inhibit basal NF-κB binding activity. Administration of DHMEQ rescued SCID mice from death when they were inoculated with HTLV-1-transformed MT-2 cells. Furthermore, DHMEQ treatment of the carriers' PBMCs reduced the HTLV-1 provirus copy number, suggesting selective cytotoxicity against HTLV-1-infected cells. Taken together, our results suggest that DHMEQ appears to be a promising compound targeting NF-κB to overcome resistance of ATL to conventional chemotherapy and also to reduce the risk of ATL development by purging carriers of HTLV-1-infected cells as a preventive intervention.
Reduction of HTLV-1 provirus load of PBMCs by DHMEQ. (A) Detection of NF-κB-activated cells with IL-2R expression in PBMCs of asymptomatic HTLV-1 carriers. Confocal immunofluorescence microscopy of PBMC samples (upper panels) and control cell lines (lower panels). Primary antibodies used are indicated on the top as well as the agent used for nuclear staining. Original magnification ×600. Objective lens, 60×/1.4. (B) The experimental protocol for measuring changes in the provirus copies after DHMEQ treatment of PBMCs. (C) Reduction of HTLV-1 provirus load in PBMCs by DHMEQ. Reduction rates of HTLV-1 proviral copies of DHMEQ-treated PBMCs of virus carriers are presented. Mixture of equal number of K562 and TL-Om1 cells served as a control (C).
Reduction of HTLV-1 provirus load of PBMCs by DHMEQ. (A) Detection of NF-κB-activated cells with IL-2R expression in PBMCs of asymptomatic HTLV-1 carriers. Confocal immunofluorescence microscopy of PBMC samples (upper panels) and control cell lines (lower panels). Primary antibodies used are indicated on the top as well as the agent used for nuclear staining. Original magnification ×600. Objective lens, 60×/1.4. (B) The experimental protocol for measuring changes in the provirus copies after DHMEQ treatment of PBMCs. (C) Reduction of HTLV-1 provirus load in PBMCs by DHMEQ. Reduction rates of HTLV-1 proviral copies of DHMEQ-treated PBMCs of virus carriers are presented. Mixture of equal number of K562 and TL-Om1 cells served as a control (C).
NF-κB inhibitors, which have a potential to target the NF-κB pathway, can be categorized into several groups.43 Representative of these groups are inhibitors of (1) IκBα phosphorylation, (2) proteasome or protease, (3) translocation of NF-κB into the nucleus, (4) binding of NF-κB to DNA, or (5) interactions with basal transcription machinery. DHMEQ could be added to the third group as a specific inhibitor of NF-κB translocation into the nucleus. Although various NF-κB inhibitors have been reported so far,43 only a limited number of them are under consideration for clinical translation.44 One of the reasons for this may be their relatively lower specificities that result in unexpected side effects at required treatment doses.
We and others recently reported that blockade of constitutive NF-κB activation in tumor cells was sufficient to induce apoptosis. We showed induction of apoptosis of Hodgkin and Reed-Sternberg cell lines by blockade of constitutive NF-κB activity by adenovirus-mediated transduction of an IκBα mutant.31 Results in the present study showed that this strategy is also effective in ATL, another lymphoid malignancy associated with strong and constitutive NF-κB activity.
The effect of several NF-κB inhibitors upon constitutive NF-κB activity was examined recently.44,45 The compounds include IκB phosphorylation inhibitors such as aspirin, sodium salicylate, and Bay-11-7082 and nuclear translocation inhibitors such as SN50 and caffeic acid phenethyl ester (CAPE). Among them, Bay 11-7082 was reported to block constitutive NF-κB activation and induce apoptosis in Kaposi sarcoma-associated herpesvirus (KSHV)-infected B cells and ATL cells.45,46 Compared with the results of Bay-11-7082 described in these reports, results of the present study suggest a higher specificity of DHMEQ that efficiently induces apoptosis of ATL cells by completely blocking NF-κB activation at a concentration that does not affect viability of normal PBMCs. These different specificities may depend on the differences in the mechanisms of function. Furthermore, a published paper47 that is referred to in the Sigma Aldrich catalog of this compound reports that in addition to inhibition of IκBα phosphorylation, Bay-11-7082 shows other activities, including stimulation of the stress-activated protein kinases, p38 and Jun N-terminal kinase-1 (JNK-1), and activation of tyrosine phosphorylation of 130 to 140 kDa protein of unknown origin. These data indicate that Bay-11-7082 has nonspecific activities unrelated to inhibition of IKK activity.
As to the proteasome inhibitor PS-341 and ATL, several reports have been published recently.48-50 NF-κB inhibition by PS-341 was not consistent in these reports, showing almost no inhibition at comparable levels to those of sodium salicylate. Also, antiproliferative effects against ATL cells were not consistent, showing no or little effects in 2 reports48,50 or potent inhibition in one49 by using PS-341 alone. On the other hand, DHMEQ targets translocation of p65 into the nucleus, which is located downstream of IKKS that are the targets of Bay-11-7082. Furthermore, blockade of NF-κB by DHMEQ is almost complete (Figures 1 and 4) and appears to be transient and reversible (M.W., R.H., and T.W.; unpublished observation, April 2004), whereas that of Bay-11-7082 is irreversible. Thus, our data also suggest that permanent inhibition of NF-κB activity is not required for induction of apoptosis in ATL cells. Moreover, the transient activity of DHMEQ might help to minimize unexpected toxicity on the normal tissues.
DHMEQ showed little effect on the DNA binding activity of NF-κB in control T cell-enriched PBMC samples at a concentration that completely blocked NF-κB activity in ATL cells (Figure 4B). These results appear to relate to the differences in NF-κB components between normal PBMCs and ATL cells. Constitutive activity found in normal T cell-enriched PBMCs appears to be due to the presence of p50 homodimers (Figure 4B), whereas activity in ATL cells is due to p50/p65 heterodimers (Figure 4A and Mori et al45 ). The residual binding in DHMEQ-treated ATL cells is observed as faint fast-migrating bands that may correspond to or contain the p50 homodimer (Figure 4A). Therefore, the data suggest that either primary ATL cells contain small amounts of p50 homodimer or ATL samples used in the current study contained a small number of contaminating normal T lymphocytes. Thus, DHMEQ may function as an inhibitor of NF-κB complex except for p50 homodimers, although this hypothesis remains to be proven.
After administration of DHMEQ, blockade of NF-κB is evident at 1 hour and until 24 hours (part of the data is shown in Figure 1A, and unpublished observation). On the other hand, apoptosis appears to be induced at a relatively late phase after DHMEQ treatment (Figure 2B and Figure 4D), suggesting that apoptosis induction in ATL cells by DHMEQ may be indirect and mediated by altered levels of gene expression. This notion is supported by DHMEQ-induced down-regulation of expression of antiapoptotic and proliferative genes in ATL-derived cell lines (Figure 3A) and confirmed in the expression profile analysis using a commercial system (CodeLink Expression Bioarray System) (Table 1). Decreased levels of Rb protein phosphorylation may be due to down-regulation of cyclin D1, whose expression level directly affects the phosphorylation status of Rb protein. Because phosphorylation of Rb results in the release of E2F that determines G0/G1-S phase progression, DHMEQ may induce G1 cell-cycle arrest in ATL cells. Results of cell-cycle analysis shown in Figure 3D support this idea. Collectively, these results suggest that constitutive NF-κB activation appears to support survival of ATL cells through induction of proteins involved in antiapoptosis and cell-cycle progression.
DHMEQ treatment contributes to survival of mice injected with HTLV-1-transformed MT-2 cells. We used 2 doses of DHMEQ in these experiments, 4 mg/kg and 12 mg/kg, administered 3 times a week. These doses are far less than the median lethal dose (LD50) for DHMEQ 180 mg/kg (Naoki Matsumoto, K.U.; unpublished observation, July 1999). Thus, DHMEQ appears to be a feasible and less toxic candidate as a novel anti-ATL agent as well as for chemoprevention of ATL, although the pharmacokinetics is not yet fully elucidated.
It is difficult to identify HTLV-1-infected cells in vivo, because they are latently infected.9,51 In the immunohistochemical studies, we showed that some cells are positive both for IL-2Rα and activated p65 in the PBMC samples from HTLV-1 carriers but not in those from healthy control PBMC samples (Figure 6A). Because ATL cells express IL-2Rα and show NF-κB activation in the absence of viral gene expression, it is very likely that the cells with IL-2Rα and activated p65 are HTLV-1-infected cells. The number of those cells is correlated with the proviral load measured by real-time PCR (data not shown), which provides supportive evidence for the concept that IL-2R expression and NF-κB activation characterize HTLV-1-infected cells in vivo. We provide the first supportive evidence for constitutive activation of NF-κBin untransformed HTLV-1-infected cells in vivo (Figure 6A). We also demonstrate that DHMEQ can selectively reduce the number of HTLV-1-infected cells in the PBMCs (Figure 6C). These findings can provide a rationale for prevention of HTLV-1-associated diseases by treatment with a small-molecular-weight compound targeting a specific molecule. In this context, DHMEQ appears to be a good candidate agent because of its apparently low toxicity against normal PBMCs.
In conclusion, our study indicates that a novel and unique NF-κB inhibitor, DHMEQ, is a promising compound that targets constitutive activation of NF-κB in HTLV-1-infected untransformed T cells and ATL cells. Further analysis of the action of DHMEQ as well as its pharmacokinetics may provide a rationale for future clinical trials in ATL patients.
Prepublished online as Blood First Edition Paper, June 14, 2005; DOI 10.1182/blood-2004-09-3646.
Supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology and Japan Society of Promotion of Science (T.W., R.H.); a grant from Integrative Research Program of the Graduate School of Medical Sciences, Kitasato University (R.H.); The Mochida Memorial Foundation for Medical and Pharmaceutical Research (R.H.); a Grant-in-Aid for Scientific Research (T.W.); and a Grant-in-Aid for Cancer Research from the Ministry of Education, Culture, Sports Science, and Technology, Japan (T.W., K.Y.).
The online version of this article contains a data supplement
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The PBMC samples of carriers used in the present work were a part of those collected as a collaborative project of the Joint Study on Prognostic Factors of ATL Development (JSPFAD). The authors thank Drs Marshall Kadin and Lawrence Abraham for critical comments on the manuscript and helpful discussions and Ms Michiko Hirayama and Ms Yukiko Shimizu for their technical assistance.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal