Abstract
Although the prognosis of pediatric leukemias has improved considerably, many patients still have relapses. Tipifarnib, a farnesyl transferase inhibitor (FTI), was developed to target malignancies with activated RAS, including leukemia. We tested 52 pediatric acute myeloid leukemia (AML) and 36 pediatric acute lymphoblastic leukemia (ALL) samples for in vitro sensitivity to tipifarnib using a total cell-kill assay and compared these results to those obtained with normal bone marrow (N BM) samples (n = 25). AML samples were significantly more sensitive to tipifarnib compared to B-cell precursor ALL (BCP ALL) or N BM samples. Within AML, French-American-British (FAB) M5 samples were most sensitive to tipifarnib. T-cell ALL samples were significantly more sensitive than BCP ALL and N BM samples. In AML there was a marked correlation between tipifarnib resistance and daunorubicin or etoposide resistance, but not to cytarabine or 6-thioguanine. RAS mutations were present in 32% of AML and 18% of ALL samples, but there was no correlation between RAS mutational status and sensitivity to tipifarnib. Future studies are needed to identify biomarkers predictive of tipifarnib sensitivity. In addition, clinical studies, especially in T-cell ALL, seem warranted.
Introduction
Although the prognosis of pediatric acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) has improved considerably over the past 20 years, many patients still have relapses. Therefore, further improvements in treatment are important. The success of imatinib in the treatment of chronic myelogenous leukemia has led to enthusiasm for the development of novel agents that target signaling abnormalities found in leukemic cells. One such agent is tipifarnib (Zarnestra), an orally available nonpeptidomimetic farnesyl transferase inhibitor (FTI), specifically developed to target RAS-driven malignancies.1
The RAS gene family consists of 3 G-proteins: neuroblastoma RAS (NRAS), Kirsten RAS (KRAS), and Harvey RAS (HRAS).2 The RAS proteins are important in relaying proliferation and survival signals from cell-membrane receptors to intracellular signal-transduction pathways. Mutations in exon 1 and 2 of RAS family genes induce constitutive activation of RAS-mitogen-activated protein kinase (MAPK) signaling and have been identified in numerous malignancies, including hematologic malignancies such as AML.2-4 Mutations of NRAS or KRAS are found in 13% to 20% of cases of adult AML.5-7 In addition, RAS mutations occur in approximately 15% of pediatric patients with AML and ALL.8-15 Mutations in HRAS have never been detected in pediatric AML or ALL samples.8,14-16 In addition to mutated RAS, signaling from other leukemia-associated oncoproteins such as KIT or fms-related tyrosine kinase 3 (FLT3) are dependent, in part, on signaling via the RAS-MAPK pathway. Therefore, FTIs might be effective in inhibiting proliferation and survival of leukemic cells with oncogenic events upstream of RAS or directly involving RAS family members.
RAS is synthesized as a precursor protein that needs several posttranslational modifications to become functional, including prenylation.1 Prenylation is catalyzed by 3 different enzymes, farnesyl protein transferase (FPTase), geranylgeranyl transferase type I (GGTase I), and geranylgeranyl transferase type II (GGTase II). Farnesylation is the dominant class of posttranslational modification required for proper RAS function.17 Notably, the addition of a farnesyl group is essential for proper intracellular localization of RAS to the inner surface of the cell membrane. Nonprenylated RAS cannot function in intracellular signaling. FTIs were developed to inhibit this process and thus interfere with the function of RAS. One such FTI is tipifarnib, which was previously known as R115777.
Tipifarnib is currently being evaluated in phase 2 and 3 clinical studies for many different malignancies, including hematologic malignancies. In the original phase 1 study in high-risk leukemias (AML, ALL, and chronic myeloid leukemia [CML]), 29% of patients responded, including 2 of 34 patients who reached complete response (CR) and toxicities were acceptable with neurotoxicity as the dose-limiting toxicity.18 None of the patients included in the phase 1 trial harbored a RAS mutation. In a large phase 2 study in untreated elderly patients with AML, CR was obtained in 18%, and an additional 16% of patients obtained a partial response (PR).19
In this study, we show that pediatric AML and T-cell ALL (T-ALL) cells have a similar sensitivity to tipifarnib, and both are more sensitive to tipifarnib in vitro than pediatric B-cell precursor (BCP) ALL or normal bone marrow (N BM) mononuclear cells. Within AML, French-American-British (FAB) M5 samples were most sensitive. Patient samples that were resistant to tipifarnib were also resistant to etoposide or daunorubicin, but not to cytarabine or 6-thioguanine. However, tipifarnib sensitivity did not correlate with the RAS mutational status of the samples. In fact, blasts with a RAS mutation may either be sensitive or resistant to tipifarnib. Future studies are needed to identify biomarkers predictive of tipifarnib sensitivity.
Patients, materials, and methods
Patient samples
BM or peripheral-blood (PB) samples from 113 children (≤ 18 years of age) were successfully tested for tipifarnib sensitivity using the methylthiazol-tetrazolium (MTT) assay. This group consisted of 52 samples from patients with newly diagnosed AML (35 BM and 17 PB samples), 36 with newly diagnosed ALL (27 BM and 9 PB samples), and 25 healthy children (all BM samples). The patient characteristics are presented in Table 1. The samples from healthy children were from children who underwent elective anesthesia for an ophthalmologic procedure with informed consent according to the Declaration of Helsinki. The study was approved by the Medical Ethical Committee of our hospital as well as by the Dutch Central Committee for Medical Research in Humans. Three collaborative groups participated in this study: the AML-Berlin-Frankfurt-Münster Study Group (AML-BFM-SG; Münster, Germany), the MRC Childhood Leukaemia Working Party (Glasgow, United Kingdom), and the Dutch Childhood Oncology Group (DCOG; The Hague, The Netherlands). All study groups performed central review of the diagnosis, classification, and clinical follow-up of the patients.
Clinical characteristics of the patients included in this study
. | AML . | ALL . | N BM . |
|---|---|---|---|
| No. of patients | 52 | 36 | 25 |
| Sex, % male | 62 | 66 | 56 |
| Age, y, median (p25-p75) | 9.6 (3.2-12.5) | 5.4 (2.6-8.4) | 8.1 (6.3-11.8) |
| WBC count, × 109/L, median (p25-p75) | 58.0 (19.0-157.0) | 55.5 (6.4-193.0) | — |
| FAB classification, no. | |||
| M0 | 2 | — | — |
| M1 | 4 | — | — |
| M2 | 8 | — | — |
| M3 | 4 | — | — |
| M4 | 18 | — | — |
| M5 | 11 | — | — |
| M7 | 1 | — | — |
| Unavailable | 4 | — | — |
| Immunophenotype | |||
| BCP ALL | 25 | — | |
| T-cell ALL | 11 | — |
. | AML . | ALL . | N BM . |
|---|---|---|---|
| No. of patients | 52 | 36 | 25 |
| Sex, % male | 62 | 66 | 56 |
| Age, y, median (p25-p75) | 9.6 (3.2-12.5) | 5.4 (2.6-8.4) | 8.1 (6.3-11.8) |
| WBC count, × 109/L, median (p25-p75) | 58.0 (19.0-157.0) | 55.5 (6.4-193.0) | — |
| FAB classification, no. | |||
| M0 | 2 | — | — |
| M1 | 4 | — | — |
| M2 | 8 | — | — |
| M3 | 4 | — | — |
| M4 | 18 | — | — |
| M5 | 11 | — | — |
| M7 | 1 | — | — |
| Unavailable | 4 | — | — |
| Immunophenotype | |||
| BCP ALL | 25 | — | |
| T-cell ALL | 11 | — |
The p25-p75 indicates 25th to 75th percentile. WBC indicates white blood cell; FAB, French-American-British morphology classification; BCP, B-cell precursor.
— indicates not applicable.
Drug resistance testing
Mononuclear cells were isolated by density gradient centrifugation with Ficoll Isopaque (both for the leukemic as well as the N BM samples). In the leukemic samples, when the blast percentage was low (< 80%) as determined by May-Grünwald-Giemsa (MGG) staining, contaminating lymphocytes were removed using immunomagnetic beads.20 Cellular drug resistance was measured using the MTT assay, a 4-day total cell-kill assay, as described before.21,22 Results from BM and PB samples were evaluated together because this does not influence the results of in vitro drug-resistance testing.21,23 Tipifarnib (kindly provided by Johnson & Johnson R&D, Titusville, NY) was dissolved in dimethyl sulfoxide (DMSO) and diluted further using culture medium (maximum final DMSO concentration 0.1%). Tipifarnib was tested in 6 different concentrations, in duplicate, with a concentration range of 0.008 to 25 μg/mL. The concentration range was established empirically, providing the best dose-response curves. The median coefficient of variation for the duplicates was below 10%. In the AML samples a panel of 4 different drugs was tested in addition to tipifarnib, including those frequently used in AML treatment (range of concentrations): etoposide (0.05-50 μg/mL), cytarabine (0.002-2.5 μg/mL), 6-thioguanine (1.56-50 μg/mL), and daunorubicin (0.002-2 μg/mL). For ALL samples prednisolone (0.008-250 μg/mL), vincristine (0.05-50 μg/mL), and l-asparaginase (0.003-10 IU/mL), commonly used drugs in the treatment of ALL, were tested in addition to tipifarnib.
Isolated cells (AML, 0.8-1.0 × 106/well; ALL, 2.0 × 106/well) were exposed to the selected drugs. Four wells contained only culture medium and 6 wells contained culture medium with cells to determine the control cell survival (CCS). After 4 days of culture, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (Sigma, Saint Louis, MO) was added and cells were incubated for another 6 hours. After 6 hours, formed formazan crystals were dissolved using acidified isopropanol. The colored product was measured spectrophotometrically at 560 and 720 nm. The optic density (OD) is linearly related to the number of viable cells. Cytotoxicity was calculated at each drug concentration by the equation: (OD treated well/mean OD control wells) × 100% after correction for the background OD of the wells with culture medium only. Results were considered evaluable only if (1) the control wells contained equal to or more than 70% leukemic cells after 4 days of culture (determined by morphology after MGG staining); (2) if the mean OD, after correction for background, at day 4 exceeded 0.05 arbitrary units; and (3) when the LC50 duplicates of one patient were within the same dilution step. The LC50 value, the drug concentration that kills 50% of the leukemic cells, was used as a measure of resistance.
Mutation detection
After isolation of the blasts, cytospin slides were prepared and stored at -20°C. After thawing, gDNA was extracted by rehydrating the slides and purifying the DNA using the QiaAmp DNA Mini Kit (Qiagen, Valencia, CA). Mutational analysis of NRAS and KRAS was possible in 44 newly diagnosed AML samples and 22 newly diagnosed ALL samples. The other samples could not be tested due to lack of cells.
For NRAS and KRAS mutation detection, the purified DNA was subjected to 45 cycles of polymerase chain reaction (PCR) using the High-Fidelity PCR System (no. 1732078; Roche, Basel, Switzerland). A nested PCR was performed in cases of low amplification yield. Primers and conditions were described in detail before.11,24 For the nested PCR, 1 μLof the initial PCR product was used as template for the nested PCR and amplified an additional 25 cycles. Negative controls were included in every set of amplifications. For nested PCRs, 1 μL of the negative control (water only) from the first-stage PCR was amplified as an additional negative control. Aliquots of the final PCR were screened for mutations on a Transgenomic WAVE high-performance liquid chromatography (HPLC) system (D-HPLC; Transgenomic, Omaha, NE) by running an HPLC under nondenaturing conditions and partially denaturing conditions. D-HPLC–detected mutations were confirmed by 2 methods: (1) reamplification of the exon and repeat D-HPLC analysis on a different day, and (2) bidirectional sequence analysis on an ABI 377 sequencer using the BigDye terminator kit (Applied Biosystems, Foster City, CA).
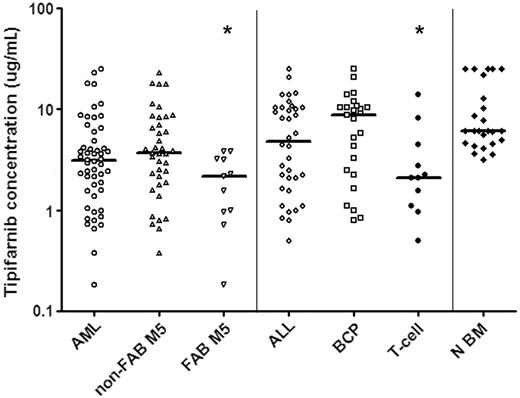
In vitro tipifarnib sensitivity of untreated ALL cells and N BM cells as measured with a total cell-kill assay. Results are depicted in LC50 values (μg/mL), the concentration of tipifarnib needed to kill 50% of the cells. Each symbol represents the LC50 value of an individual sample; the horizontal line depicts the median LC50 value. Within AML, samples classified as FAB M5 AML are more sensitive to tipifarnib in vitro than other AML samples (P = .021). Within ALL T-ALL samples are more sensitive to tipifarnib in vitro than BCP ALL samples (P = .029). BCP ALL and N BM cells are relatively resistant to tipifarnib in vitro when compared to AML blasts (BCP ALL versus AML P = .01, N BM versus AML, P < .001). *Statistically significant at P < .05 level. Non-FAB M5 indicates all FAB types other than FAB M5.
In vitro tipifarnib sensitivity of untreated ALL cells and N BM cells as measured with a total cell-kill assay. Results are depicted in LC50 values (μg/mL), the concentration of tipifarnib needed to kill 50% of the cells. Each symbol represents the LC50 value of an individual sample; the horizontal line depicts the median LC50 value. Within AML, samples classified as FAB M5 AML are more sensitive to tipifarnib in vitro than other AML samples (P = .021). Within ALL T-ALL samples are more sensitive to tipifarnib in vitro than BCP ALL samples (P = .029). BCP ALL and N BM cells are relatively resistant to tipifarnib in vitro when compared to AML blasts (BCP ALL versus AML P = .01, N BM versus AML, P < .001). *Statistically significant at P < .05 level. Non-FAB M5 indicates all FAB types other than FAB M5.
Statistical analysis
To assess differences in the distribution of continuous data, the nonparametric Mann-Whitney U test was used. P ≤ .05 was considered statistically significant (2-tailed test). The correlation between resistance to tipifarnib and resistance to other frequently used cytotoxic drugs was analyzed using the Spearman correlation coefficient (rho). P ≤ .05 was considered statistically significant (2-tailed test).
Results
Sensitivity to tipifarnib
A total of 88 pediatric acute leukemia samples were successfully tested using the MTT assay to determine sensitivity to tipifarnib. Tipifarnib cytotoxicity was dose-dependent in the concentration range tested. There were large interindividual differences in tipifarnib sensitivity, that is, LC50 values ranged from 0.18 to more than 25 μg/mL (140-fold). The tipifarnib median LC50 values and ranges in the different subgroups are depicted in Figure 1.
AML samples were more sensitive to tipifarnib compared to BCP ALL samples (2.8-fold, median LC50, 3.1 versus 8.8 μg/mL; P = .01; Figure 1). Within AML, FAB M5 samples were more sensitive to tipifarnib than samples with other FAB types (1.7-fold, median LC50, 2.2 versus 3.7 μg/mL; P = .021; Figure 1). Within ALL, T-ALL samples were more sensitive to tipifarnib than BCP ALL (4.2-fold, median LC50, 2.1 versus 8.8 μg/mL; P = .029; Figure 1). There was no statistically significant difference in sensitivity to tipifarnib between T-ALL samples and AML samples (median LC50, 2.1 versus 3.1 μg/mL; P = .36).
Bone marrow mononuclear cells from 25 healthy children were tested and these samples were more resistant to tipifarnib when compared to AML samples (2-fold, median LC50, 6.1 versus 3.1 μg/mL; P < .001) and T-ALL samples (2.9-fold, median LC50, 6.1 versus 2.1 μg/mL; P = .001). There was no statistically significant difference in sensitivity to tipifarnib between N BM and BCP ALL samples (median LC50, 6.1 versus 8.8 μg/mL; P = .49).
Is tipifarnib resistance associated with resistance to conventional cytotoxic drugs?
In addition to the novel agent tipifarnib, most samples were also tested for sensitivity to conventional cytotoxic drugs. In samples in which both tipifarnib and the conventional drug were tested, we analyzed whether tipifarnib resistance was correlated to resistance to other drugs, using the Spearman rho correlation coefficient. Within the AML samples tested, there was a marked correlation between tipifarnib resistance and resistance to etoposide (ρ = 0.53, P < .001) or daunorubicin (ρ = 0.62, P < .001), whereas there was only a weak correlation between resistance to tipifarnib and resistance to cytarabine (ρ = 0.36, P = .01) or 6-thioguanine (ρ = 0.32, P = .03; Table 2).
Correlation between in vitro intrinsic resistance to tipifarnib and resistance to other cytotoxic drugs in untreated pediatric AML and ALL samples
. | Median LC50 value, μg/mL (p25-p75) . | Spearman ρ* . | P . | No. . |
|---|---|---|---|---|
| AML | ||||
| Tipifarnib | 3.1 (1.58-5.45) | — | — | 52 |
| Daunorubicin | 0.17 (0.10-0.30) | 0.62 | < .001 | 50 |
| Etoposide | 2.99 (1.64-8.0) | 0.53 | < .001 | 49 |
| Cytarabine | 0.37 (0.13-0.67) | 0.36 | .01 | 50 |
| 6-Thioguanine | 5.0 (2.94-7.65) | 0.32 | .03 | 49 |
| ALL | ||||
| Tipifarnib | 4.8 (1.87-10.4) | — | — | 36 |
| Prednisolone | 0.32 (0.05-6.2) | –0.35 | .05 | 32 |
| Vincristine | 0.70 (0.13-6.7) | 0.009 | .96 | 32 |
| l-Asparaginase | 0.14 (0.03-0.94) | 0.003 | .99 | 32 |
. | Median LC50 value, μg/mL (p25-p75) . | Spearman ρ* . | P . | No. . |
|---|---|---|---|---|
| AML | ||||
| Tipifarnib | 3.1 (1.58-5.45) | — | — | 52 |
| Daunorubicin | 0.17 (0.10-0.30) | 0.62 | < .001 | 50 |
| Etoposide | 2.99 (1.64-8.0) | 0.53 | < .001 | 49 |
| Cytarabine | 0.37 (0.13-0.67) | 0.36 | .01 | 50 |
| 6-Thioguanine | 5.0 (2.94-7.65) | 0.32 | .03 | 49 |
| ALL | ||||
| Tipifarnib | 4.8 (1.87-10.4) | — | — | 36 |
| Prednisolone | 0.32 (0.05-6.2) | –0.35 | .05 | 32 |
| Vincristine | 0.70 (0.13-6.7) | 0.009 | .96 | 32 |
| l-Asparaginase | 0.14 (0.03-0.94) | 0.003 | .99 | 32 |
— indicates not analyzed.
Spearman ρ correlation coefficient for the correlation between tipifarnib resistance and resistance to the other tested drug
Within ALL there was no correlation between resistance to tipifarnib and resistance to vincristine (ρ = 0.009, P = .96), l-asparaginase (ρ = 0.003, P = .99), or prednisolone (ρ = -0.35, P = .05; Table 2). Among samples with T-ALL or BCP ALL, there was no correlation between resistance to tipifarnib and resistance to vincristine, l-asparaginase, or prednisolone (data not shown).
RAS mutations and tipifarnib sensitivity
We were able to examine NRAS and KRAS mutations in 44 newly diagnosed AML samples and 22 newly diagnosed ALL samples. Overall, 14 of 44 AML (32%) and 4 of 22 ALL (18.2%) samples had a RAS mutation. NRAS mutations (9 of 44, 20.5%) were more frequent than KRAS mutations (5 of 44, 11.4%) in AML, whereas there was no difference in the incidence of NRAS (2 of 22, 9.1%) and KRAS (2 of 22, 9.1%) mutations in ALL. We hypothesized that tipifarnib might have greater activity against leukemic cells with mutated NRAS or KRAS. We therefore compared the in vitro sensitivity to tipifarnib in patient samples with and without RAS mutations. The median LC50 values and ranges for AML samples with and without RAS mutations are depicted in Figure 2A. There were no statistically significant differences in the in vitro sensitivity to tipifarnib between AML samples with and without RAS mutations (median LC50, 3.5 versus 2.4 μg/mL, P = .24). Not only was there no difference in the median LC50 value, but both RAS mutated and wild-type subgroups contained samples that were relatively sensitive or resistant compared with the median. There was no difference in tipifarnib sensitivity between NRAS and KRAS mutated AML blasts (3.2 versus 4.9 μg/mL; P = .24).
The relationship between in vitro tipifarnib sensitivity and RAS mutational status in pediatric untreated AML and ALL samples. In vitro tipifarnib test results are depicted in LC50 values (μg/mL), the concentration of tipifarnib needed to kill 50% of the cells. Each symbol represents the LC50 value of an individual sample; the horizontal line depicts the median LC50 values. RAS mutational status is stated as either absence (wild-type RAS) or presence (RAS mutated). RAS mutated samples comprise mutations in either NRAS or KRAS, which are also depicted separately. (A) There were no differences in in vitro tipifarnib sensitivity between pediatric AML patients with and without RAS mutations (P = .20). In addition, there was no difference in sensitivity to tipifarnib between NRAS and KRAS mutated AML samples (P = .24). (B) There was no difference in in vitro tipifarnib sensitivity between RAS mutated ALL samples and wild-type RAS ALL samples. There were only 2 patients in both the NRAS and KRAS mutated subgroups.
The relationship between in vitro tipifarnib sensitivity and RAS mutational status in pediatric untreated AML and ALL samples. In vitro tipifarnib test results are depicted in LC50 values (μg/mL), the concentration of tipifarnib needed to kill 50% of the cells. Each symbol represents the LC50 value of an individual sample; the horizontal line depicts the median LC50 values. RAS mutational status is stated as either absence (wild-type RAS) or presence (RAS mutated). RAS mutated samples comprise mutations in either NRAS or KRAS, which are also depicted separately. (A) There were no differences in in vitro tipifarnib sensitivity between pediatric AML patients with and without RAS mutations (P = .20). In addition, there was no difference in sensitivity to tipifarnib between NRAS and KRAS mutated AML samples (P = .24). (B) There was no difference in in vitro tipifarnib sensitivity between RAS mutated ALL samples and wild-type RAS ALL samples. There were only 2 patients in both the NRAS and KRAS mutated subgroups.
Only 4 samples in the ALL cohort screened had an RAS mutation (Figure 2B). The 4 ALL samples with RAS mutations appeared more sensitive to tipifarnib than the 18 ALL samples without a RAS mutation (median LC50, 3.8 versus 8.5 μg/mL), but a meaningful statistical analysis was not possible because of the low number of samples with RAS mutations. Because KRAS or NRAS mutations were each found in only 2 patients, we cannot draw conclusions concerning differences in sensitivity between NRAS or KRAS mutated ALL samples and ALL samples with wild-type NRAS and KRAS genes.
Discussion
In this study, we determined the tipifarnib sensitivity of pediatric leukemic samples and N BM mononuclear cells. Tipifarnib was initially developed as a targeted drug, interfering with the function of RAS. RAS is involved in the development and progress of many malignancies including leukemia. RAS acts as a cellular switch that normally is activated in response to other molecular events such as ligand-dependent activation of growth factor receptors (eg, FLT3). In human cancer cells, RAS can either be activated by overexpression of these receptors or by oncogenic gain-of-function tyrosine kinases (eg, BCR-ABL) and RAS mutations.13,25,26 Thus, a drug targeting activated RAS could potentially benefit a large number of pediatric leukemia patients.
We found large, 140-fold, interindividual differences in the in vitro sensitivity of AML and ALL blasts to tipifarnib. The concentration range tested includes concentrations achieved in the adult phase 1 study, where at a dose level of 600 mg twice daily tipifarnib peak plasma concentrations were 1.8 μg/mL.18
Pediatric AML samples were more sensitive to tipifarnib in vitro than pediatric BCP ALL samples. This result is congruent with the results from a phase 1 trial conducted in patients with poor-risk leukemias that included both AML and ALL cases.18 In this study, none of the 6 patients with ALL responded, whereas 32% of patients with AML showed a response when treated with tipifarnib. In the published adult clinical trial of tipifarnib for poor-risk leukemias, no patients with T-ALL were included.18 Within AML, FAB M5 samples were most sensitive to tipifarnib. T-ALL samples were significantly more sensitive to tipifarnib than BCP ALL samples and in the same concentration range as AML samples. This is remarkable because we showed previously that pediatric T-ALL samples usually are more resistant to cytotoxic drugs in vitro, as were the T-ALL samples tested for vincristine, prednisolone, and l-asparaginase in this study (data not shown).27 These results suggest that further preclinical and clinical studies of tipifarnib in T-ALL are needed. The N BM samples were more resistant to tipifarnib than the AML and T-ALL samples, reflecting the therapeutic index. The BCP ALL samples were as resistant to tipifarnib as the N BM samples.
In AML, a moderate to strong correlation was found between intrinsic resistance to tipifarnib and resistance to daunorubicin or etoposide, but only a weak correlation between resistance to cytarabine or 6-thioguanine and resistance to tipifarnib. In the ALL samples there was no correlation between resistance to tipifarnib and resistance to prednisolone, vincristine, or l-asparaginase. These data suggest that combining tipifarnib with cytarabine or 6-thioguanine may be more efficacious than combining it with daunorubicin or etoposide. Notably, this resistance profile was determined using cells from untreated patients and therefore reflects intrinsic resistance at diagnosis. Currently, 2 clinical trials are evaluating the toxicity and efficacy of tipifarnib combined with conventional cytotoxic drugs. One study combines tipifarnib with idarubicin and cytarabine and the other combines tipifarnib and etoposide. One possible explanation for the correlation between in vitro resistance to tipifarnib and in vitro resistance to daunorubicin or etoposide could be their recognized status as P-glycoprotein substrates. There are conflicting data on the status of tipifarnib as a P-glycoprotein substrate.28-30 However, P-glycoprotein is expressed in less than 10% of newly diagnosed pediatric AML samples and is therefore unlikely to be the cause of the intrinsic drug resistance observed in the samples we tested.31 Another explanation for the observed correlation could be that these drugs share a common mechanism of action.
The causes of clinical resistance to FTIs, intrinsic or acquired, are not known yet. To study intrinsic tipifarnib resistance, cells from patients treated in clinical trials with tipifarnib are being analyzed using microarray analysis to determine what differences are found in responsive versus unresponsive patients.32 In the tipifarnib monotherapy phase 1 trial in heavily pretreated relapsed and refractory leukemia, the CR rate in AML was only 8%. However, in an ongoing clinical study of monotherapy with tipifarnib in untreated elderly patients with AML, a CR rate of 18% has been reported with an overall response rate of 34%.18,19 This difference in clinical response to tipifarnib between untreated and pretreated AML could be caused by additional, acquired, resistance between tipifarnib and drugs used in first-line treatment of AML such as daunorubicin and etoposide. In addition, in an in vitro study, cells with certain mutations in FTase β were resistant to treatment with the FTI lonafarnib and in 2 patients who developed clinical FTI resistance identical mutations were described.33
Because tipifarnib was originally developed to target malignancies with RAS mutations, we compared the RAS genotype with the in vitro sensitivity to tipifarnib and found that 32% of the AML and 18% of the samples from patients with ALL had a mutation in either NRAS or KRAS. However, there was no correlation of RAS genotype with in vitro sensitivity to tipifarnib in either ALL or AML. Patients with RAS mutations could be either relatively sensitive or relatively resistant to tipifarnib. Previous reports already indicated that malignant cells with wild-type RAS could be sensitive to tipifarnib, but we are the first to describe that leukemic blasts with RAS mutations can be resistant in vitro to the cytotoxic effects of tipifarnib.18,34 One explanation for the lack of correlation between RAS mutational status and sensitivity to tipifarnib may be that farnesylation is only one form of prenylation, and RAS proteins are also subject to other types of prenylation. Notably, HRAS undergoes farnesylation but not geranylgeranylation, whereas NRAS and KRAS can undergo farnesylation or geranylgeranylation.17 NRAS and KRAS become subject to geranylgeranylation when cells are treated with FTIs, allowing attachment to the cell membrane and participation in signal transduction. Therefore, geranylgeranylation of RAS could overcome the inhibitory effects of tipifarnib on RAS farnesylation and explain the lack of correlation between RAS mutations and in vitro sensitivity to tipifarnib.
Tipifarnib is a general inhibitor of farnesylation and thus not only affects RAS prenylation, but also interferes with the function of many other proteins that need farnesylation for normal function. The mediator of the cytotoxic effects of tipifarnib is thought to be one of these proteins. Cell-cycle–regulatory proteins that require farnesylation, such as RhoB and CENP-E, would be likely candidates for mediating the effects of tipifarnib. There are reports that an increase in geranylgeranylated RhoB (resulting from inhibition of farnesylation) inhibits proliferation and induces apoptosis.35,36 Treatment of cells with an FTI alters the interaction between CENP-E and the microtubules, resulting in an accumulation of cells prior to metaphase.37 Recently, a study identified Rab geranylgeranyl transferase, in addition to farnesyl transferase, as a target of many FTIs.38 Because the target for tipifarnib induced cytotoxicity is unknown, further investigation is warranted.
It should be noted that the differences in tipifarnib sensitivity we observed were larger within the subgroups than between the subgroups. This phenomenon has also been described for other drugs that we have previously reported on. For instance, it is well known that pediatric AML is clinically much more resistant to chemotherapy than pediatric ALL. Notably, we previously found that AML was 2.6-fold more resistant to daunorubicin and 4.9-fold more resistant to etoposide in vitro than ALL.22 Therefore, we believe that the differences described in this study are clinically relevant. Moreover, we have previously reported that in vitro drug resistance testing has prognostic significance both in ALL (where a combined prednisolone-vincristine-asparaginase score was most predictive) and AML (resistance to cytarabine).21,39
In conclusion, pediatric AML and T-ALL samples are more sensitive to tipifarnib than N BM and BCP ALL samples. We identified AML FAB M5 and T-cell ALL as the most tipifarnib-sensitive subsets of pediatric acute leukemia. These results suggest that tipifarnib might be an active agent in the treatment of T-ALL. Further preclinical studies are needed to confirm this observation. In AML we observed a marked correlation between resistance to tipifarnib and resistance to daunorubicin or etoposide. In contrast, there was only a weak correlation between resistance to tipifarnib and resistance to cytarabine or 6-thioguanine. These results suggest that combining tipifarnib and cytarabine might be efficacious in AML. There was no correlation between tipifarnib resistance and resistance to commonly used ALL drugs. Interestingly, sensitivity to tipifarnib was independent of RAS mutational status. Patients without RAS mutations can be sensitive to tipifarnib and patients with a RAS mutation can be resistant. Further studies are needed to identify robust biomarkers that are predictive of clinical response to tipifarnib.
Prepublished online as Blood First Edition Paper, July 28, 2005; DOI 10.1182/blood-2005-04-1640.
Supported by a VA Merit Review Grant (M.C.H.) and funding from the Doris Duke Charitable Foundation (M.C.H.); both are unrestricted grants. Tipifarnib was obtained from Johnson & Johnson R&D (Titusville, NY) free of charge.
B.F.G., C.M.Z., A.H., and G.J.L.K wrote the paper; C.M.Z., M.C.H., and G.J.L.K. designed the study; B.F.G. analyzed the results that were obtained by A.H., A.H.L., B.E.S.G., K.H., D.R., and U.C.; and B.E.S.G., K.H., D.R., U.C., M.C.H., and G.J.L.K. provided essential patient samples and characteristics and/or analytic tools.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank all the hospitals and clinicians participating in the AML-BFM Study Group, the MRC Childhood Leukaemia Working Party, and the Dutch Childhood Oncology Group, as well as their reference laboratories, who provided us with the patient samples and the clinical and cell-biologic data. The technicians of the research laboratory of Pediatric Oncology of the VU university medical center handled all samples.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal