Abstract
The interleukin-6 (IL-6) cytokine family plays an important role in regulating cellular responses during hematopoiesis. We report here that mice homozygous for a knock-in mutation in the IL-6 cytokine family receptor signaling subunit glycoprotein (gp) 130 (gp130Y757F/Y757F) that leads to gp130-dependent signal transducers and activators of transcription (STAT) 1/3 hyperactivation develop a broad spectrum of hematopoietic abnormalities, including splenomegaly, lymphadenopathy, and thrombocytosis. To determine whether STAT3 hyperactivation was responsible for the perturbed hematopoiesis in gp130Y757F/Y757F mice, we generated gp130Y757F/Y757F mice on a Stat3 heterozygous (Stat3+/-) background to specifically reduce gp130-dependent activation of STAT3, but not STAT1. Normal hematopoiesis was observed in gp130Y757F/Y757F:Stat3+/- bone marrow and spleen, with no evidence of the splenomegaly and thrombocytosis displayed by gp130Y757F/Y757F mice. The perturbed cellular composition of thymus and lymph nodes in gp130Y757F/Y757F mice was also alleviated in gp130Y757F/Y757F: Stat3+/- mice. Furthermore, we show that hematopoietic cells from gp130Y757F/Y757F mice exhibited increased survival and proliferation in response to IL-6 family cytokines. Collectively, these data provide genetic evidence that gp130-dependent STAT3 hyperactivation during hematopoiesis has pathological consequences affecting multiple organs, and therefore identify the threshold of STAT3 signaling elicited by IL-6 family cytokines as a critical determinant for hematopoietic homeostasis.
Introduction
The interleukin-6 (IL-6) cytokine family, in particular IL-6, IL-11, oncostatin M (OSM), and leukemia inhibitory factor (LIF), exhibit both functional redundancy and pleiotropy in regulating proliferation, differentiation, and functional maturation of cells belonging to multiple hematopoietic lineages.1,2 The importance of IL-6 family cytokines in regulating hematopoiesis in vivo has been demonstrated in mice by genetic modification of individual genes encoding members of this family, or their receptor subunits. For instance, mice lacking the common signal-transducing receptor β-subunit glycoprotein (gp) 130 exhibit severely impaired fetal hematopoiesis,3 and mice null for IL-6, LIF, or the LIF receptor (LIFR) β-subunit display hematopoietic defects characterized by a reduction in the numbers of primitive hematopoietic progenitor cells.4-6 Conversely, pathological consequences from the continuous activation of gp130 within the hematopoietic compartment have also been described, with mice overexpressing either LIF or both IL-6 and its soluble ligand-binding alpha subunit (sIL-6Rα) displaying hyperproliferative abnormalities, including splenomegaly and thrombocytosis.7,8
Functional redundancy of the IL-6 cytokine family results from the use of the common receptor subunit gp130,2 while the pleiotropic actions are associated with the coordinated and simultaneous activation of multiple signaling cascades which ultimately modulate gene expression and cellular responses. Ligand-induced gp130 homodimerization in response to IL-6 and IL-11,9,10 or heterodimerization of gp130 with either the LIFR11 or OSMR12 β-subunits in response to other IL-6 family cytokines, results in activation of Janus kinases (JAKs) and the subsequent tyrosine phosphorylation of the receptor cytoplasmic domain.13 The 4 C-terminal phospho-tyrosine residues (pY) in murine gp130 serve as binding sites for signal transducers and activators of transcription (STAT) 1 and STAT3,14 whereas the membrane-proximal pY757 (pY759 in human gp130) facilitates binding of both suppressor of cytokine signaling (SOCS) 3 and the src homology 2 (SH2) domain–containing tyrosine phosphatase SHP2,15 the latter of which when phosphorylated also mediates activation of the Rasmitogen–activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K) pathways.16
It has recently been shown that the magnitude and duration of gp130-dependent signaling pathways are tightly coordinated by their reciprocal negative regulation.17 For instance, while pY757 is required for transduction of SHP2-mediated signaling,18 it also enables negative regulation of gp130 signaling through the combined actions of the protein tyrosine phosphatase activity of SHP2,19 and allows SOCS3 to down-modulate JAK activity and target the receptor complex for proteasomal degradation.20 On the other hand, STAT3 activation induces expression of SOCS3 to create a negative feedback loop to terminate gp130-dependent signaling.21 The latter mechanism has also been shown to be critical for granulocyte colony-stimulating factor (G-CSF)–mediated granulopoiesis in mice, where specific deletion of STAT3 in hematopoietic progenitors leads to neutrophilia due to the absence of G-CSFR/STAT3-dependent induction of SOCS3 to limit neutrophil production.22
The physiologic importance of these negative regulatory mechanisms for gp130-dependent biological responses has been recently demonstrated by our laboratory17 and others23 through the generation of mice homozygous for knock-in mutations that abolish binding of specific signaling molecules to gp130. For instance, several abnormalities in gp130Y757F/Y757F mice, in which both SOCS3 and SHP2 are unable to bind to gp130, resemble those associated with continuous activation of gp130 in IL-6/sIL-6Rα or LIF transgenic mice, such as splenomegaly.7,8,17 At the molecular level, the combined loss of SHP2 and SOCS3 binding to gp130 in gp130Y757F/Y757F mice leads to exaggerated gp130-dependent STAT1 and STAT3 activation, which may account for, at least in part, the phenotype of these mice.24
The activation of STAT3 is critical for the regulation of hematopoietic cell survival and proliferation,22,25,26 and persistent STAT3 activation has also been observed in various hematologic diseases, including myeloproliferative and lymphoproliferative disorders.27 Our laboratory has previously demonstrated that gp130ΔSTAT/ΔSTAT mice unable to elicit gp130-dependent STAT1/3 activation display leukocytosis and thrombocytopenia,28 thus suggesting an important role for gp130-dependent STAT1 and/or STAT3 activation in regulating hematopoietic processes in vivo. Here, we report that gp130Y757F/Y757F mice develop multiple hematopoietic abnormalities characterized by an expansion in the numbers of immature and committed hematopoietic progenitor cells in the bone marrow and spleen, as well as an altered cellular distribution within the lymphoid organs, resulting in thrombocytosis, lymphadenopathy, and splenomegaly. Importantly, genetically reducing the activity of STAT3 in gp130Y757F/Y757F mice prevents the onset of the hematopoietic phenotype, thus providing genetic evidence that the level of STAT3 activation is crucial for gp130-controlled hematopoietic homeostasis in vivo.
Materials and methods
Mice and treatments
Mice homozygous for the gp130(Y757F) knock-in mutation (gp130Y757F/Y757F mice17 ), and mice heterozygous for a Stat3 null mutation (Stat3+/- mice29 ), both of which are on a mixed 129 × C57BL/6 background, were mated to generate gp130Y757F/Y757F:Stat3+/- mice. All experiments were fully approved by the Ludwig Institute for Cancer Research/Department of Surgery Animal Ethics Committee, and included wild-type littermate controls that were genetically matched. Animals were housed under specific pathogen-free conditions.
For biochemical analyses, mice were subjected to a single intraperitoneal injection with human (h) IL-6 (5 μg).
Cytokines and antibodies
Recombinant murine (m) stem cell factor (SCF) was a kind gift from N. Nicola (Walter and Eliza Hall Institute, Melbourne, Australia). Recombinant hIL-6 and hIL-11 were kindly provided by C. G. Begley (Amgen, Thousand Oaks, CA) and L. Robb (Walter and Eliza Hall Institute), respectively. Recombinant mIL-3 was purchased from PeproTech (Rocky Hill, NJ). Commercially available antibodies were purchased as follows: gp130, extracellular signal-regulated kinase 1/2 (ERK1/2), STAT1, and STAT3 antibodies from Santa Cruz Biotechnology (Santa Cruz, CA), phospho(Tyr701)STAT1, phospho(Tyr705)STAT3, and phospho(Ser727)STAT3 antibodies from Cell Signaling Technology (Beverly, MA), and immunoglobulin G (IgG) Fc receptor, fluoroscein isothiocyanate (FITC)–Gr1, FITC-B220, FITC-CD4, phycoerythrin (PE)–Thy1.2, and biotin-CD8a from Pharmingen (San Diego, CA).
Hematologic analyses
Blood from the retro-orbital plexus of mice was collected into EDTA (ethylenediaminetetraacetic acid)–coated tubes and analyzed on an Advia120 System quantitative hematology analyzer (Bayer Diagnostics, Tarrytown, NY). Manual differential cell counts were performed on May-Grunwald-Giemsa–stained blood smears and cytocentrifuge preparations of spleen and bone marrow single-cell suspensions.
Hematopoietic progenitor cell assays
Clonal cultures of hematopoietic progenitor cells were assayed in semisolid agar containing 0.3% agar, Dulbecco modified Eagle medium (DMEM), and 20% fetal bovine serum (FBS). Briefly, triplicate cultures of cells from bone marrow (2.5 × 104) or spleen (2.5 × 105) were stimulated with SCF (50 ng/mL) and IL-3 (10 ng/mL). Cultures were incubated for 7 days at 37°C in a humidified atmosphere of 10% CO2 in air. Colonies containing 50 or more cells were enumerated using a dissection microscope. Agar cultures were fixed with 2.5% glutaraldehyde and sequentially stained with acetylcholinesterase, Luxol Fast Blue, and hematoxylin for determination of the composition of each colony by light microscopy. Megakaryocyte colonies were defined as those containing 3 or more megakaryocytes.
Colony-forming unit–spleen assays
Colony-forming unit–spleen (CFU-S) assays were performed using 7.5 × 104 bone marrow or 3.75 × 105 spleen cells from wild-type or gp130Y757F/Y757F mice as described previously.28
Histologic analyses
Following dissection, spleen and femoral bone marrow specimens were fixed in 10% formalin solution and embedded in paraffin. Sections were stained with hematoxylin and eosin. Megakaryocytes were enumerated by light microscopy under × 200 (spleen) and × 400 (bone marrow) magnification. A minimum of 30 microscopic fields was scored.
Protein extraction and immunoblot analysis
Lysates were prepared from snap-frozen cell pellets and tissues in ice-cold lysis buffer.30 Lysates were cleared of debris by centrifugation, separated on 10% polyacrylamide gels, and then transferred to nitrocellulose membranes. Membranes were blocked in TBST (300 mM NaCl, 10 mM Tris [tris(hydroxymethyl)aminomethane]-HCl pH 7.5, 0.5% Tween 20) containing 1% bovine serum albumin (BSA) and incubated with primary antibodies according to the manufacturer's recommendations, after which they were incubated with the appropriate secondary antibodies (antimouse or -rabbit) coupled to horseradish peroxidase (Bio-Rad, Hercules, CA). Proteins were visualized using enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ).
Flow cytometry
Thymus and lymph node cell suspensions in phosphate-buffered saline (PBS) containing 2% FBS were initially incubated with the anti-IgG Fc receptor antibody on ice for 5 minutes, after which they were incubated with the various biotin-labeled, FITC-labeled, or PE-labeled antibodies on ice for 30 minutes. Cells stained with biotin-labeled antibody were washed once in PBS plus 2% FBS prior to 30 minutes incubation on ice with Streptavidin-PE. Cells were then washed twice in PBS plus 2% FBS, and stained cells were immediately analyzed on a FACScan flow cytometer (Becton Dickinson, San Jose, CA).
For cell sorting, bone marrow cells were flushed from femurs and collected in DMEM plus 2% FBS, following which they were stained with FITC-Gr1 antibody. Stained cells were washed twice, and Gr1+ cells were sorted on a FACSVantage flow cytometer (Becton Dickinson).
RNA isolation and expression analysis
Total RNA was extracted from frozen cell pellets and tissue using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's protocol. cDNA was prepared from 5 μg of total RNA using the SuperScript III First-Strand Synthesis System (Invitrogen Life Technologies) as per the manufacturer's instructions. For PCR, the following mouse primer sets were used: β-actin (190 base pair [bp]), forward 5′-AAGGTGACAGCATTGCTTCT-3′ and reverse 5′-GGCTGCCTCAACACCTCA-3′; Socs3 (450 bp), forward 5′-GGACCAGCGCCACTTCTTCAC-3′ and reverse 5′-TACTGGTCCAGGAACTCCCGA-3′; and Bcl-xL (685 bp), forward 5-AGCAACCGGGAGCTGGTGGTCGAC-3′ and reverse 5′-GACTGAAGAGTGAGCCCAGCAGA-3′. β-actin was used as an internal control for sample integrity and normalization. Amplification conditions were 94°C/45 seconds, 55°C/45 seconds, and 72°C/45 seconds, and reactions were performed over a range of cycles to determine the number of cycles yielding maximal linear amplification of PCR products, as follows: 24 cycles (liver β-actin); 25 cycles (liver Socs3); and 34 cycles (bone marrow β-actin, Socs3, and Bcl-xL). PCR products were resolved by electrophoresis on 2% agarose gels and visualized after ethidium bromide staining.
Proliferation assays
Unfractionated and Gr1+ bone marrow cells were added to 96-well plates at 105 cells per well in triplicate containing 100 μL of DMEM supplemented with 10% FBS with or without IL-6 (100 ng/mL). After 2 days of culture at 37°C, 0.037 MBq (1 μCi) of 3H-thymidine (ICN Biomedicals Australasia, Seven Hills, NSW, Australia) was pulsed for the last 12 hours of culture, and 3H-thymidine incorporation was measured in a TopCountNXT Microplate Scintillation Counter (Packard, Meriden, CT).
Cell viability and apoptosis assays
Bone marrow and spleen cells (5 × 106) were added to 6-well plates containing 2 mL DMEM supplemented with 1% FBS with or without IL-6 (100 ng/mL). After 5 days of culture at 37°C, viable cell numbers were determined by exclusion of trypan blue dye. To measure apoptosis, cells were stained with FITC-labeled annexin-V and propidium iodide (PI) according to the manufacturer's protocol (R&D Systems, Minneapolis, MN), and flow cytometric analysis was performed.
Statistical analyses
Comparisons between mean values were performed using analysis of variance (ANOVA) and Student t tests as appropriate. A P value of less than .05 was considered statistically significant.
Results
Thrombocytosis, splenomegaly, and lymphadenopathy in gp130Y757F/Y757F mice
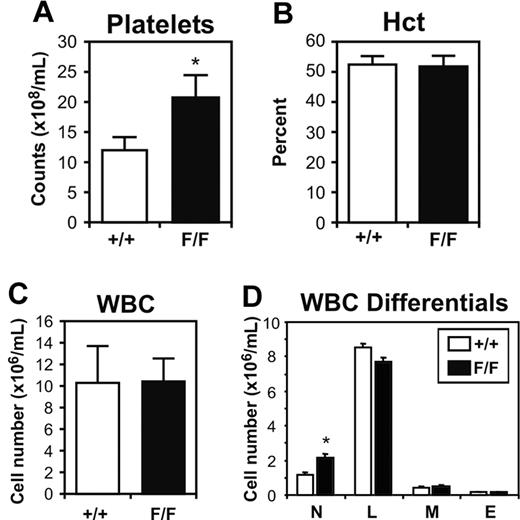
We have previously reported that gp130ΔSTAT/ΔSTAT mice, which express a truncated gp130 receptor unable to bind and activate STAT1 and STAT3, display leukocytosis and thrombocytopenia, thus demonstrating the requirement for gp130-dependent STAT1 and/or STAT3 signaling to maintain hematopoietic homeostasis in the adult mouse.28 To assess the importance of gp130-mediated SOCS3 and SHP2 binding and activation in hematopoiesis, we examined various hematologic parameters in 12-week-old gp130Y757F/Y757F mice on a mixed 129 × C57BL/6 background. Analysis of platelet numbers revealed that gp130Y757F/Y757F mice displayed thrombocytosis, with a circulating platelet count (20.52 ± 4.17 × 108/mL, mean ± SD, P < .05) 1.7-fold higher than gp130+/+ littermates (12.16 ± 1.78 × 108/mL; Figure 1A). While the hematocrit and total white blood cell count of gp130Y757F/Y757F mice were comparable to those of gp130+/+ controls (Figure 1B-C), there was a 1.8-fold increase in the number of circulating neutrophils (gp130+/+, 1.18 ± 0.45 × 106/mL versus gp130Y757F/Y757F, 2.14 ± 1.28 × 106/mL, mean ± SD, P < .05; Figure 1D).
Peripheral blood profile of gp130Y757F/Y757F (F/F) mice and gp130+/+ (wild-type) littermates. Platelet counts (A), hematocrit (B), white blood cell (WBC) counts (C), and WBC differentials (D). Data from 5 mice of each genotype are expressed as the mean ± SD. *P < .05 versus data from gp130+/+ mice. N indicates neutrophils; L, lymphocytes; M, monocytes; and E, eosinophils.
Peripheral blood profile of gp130Y757F/Y757F (F/F) mice and gp130+/+ (wild-type) littermates. Platelet counts (A), hematocrit (B), white blood cell (WBC) counts (C), and WBC differentials (D). Data from 5 mice of each genotype are expressed as the mean ± SD. *P < .05 versus data from gp130+/+ mice. N indicates neutrophils; L, lymphocytes; M, monocytes; and E, eosinophils.
At 12 weeks of age, the spleens of gp130Y757F/Y757F mice were noticeably larger in size compared with gp130+/+ littermates, with spleen weight and cellularity increased by 2.6-fold and 2.1-fold, respectively (Table 1). Splenomegaly was characterized by a 2.3-fold and 3-fold increase in the frequency of nucleated red blood cells and neutrophils, respectively (Table 1), corresponding to even greater increases in absolute numbers (4.9-fold for nucleated red blood cells, and 6.4-fold for neutrophils). Although the cellularity of gp130Y757F/Y757F femoral bone marrow was normal, analysis of the cellular composition of gp130Y757F/Y757F bone marrow revealed a 1.7-fold excess of neutrophils and myeloid precursors, which was accompanied by a 39% and 34% reduction in the frequency of nucleated red blood cells and lymphocytes, respectively (Table 1). Histologic sections of gp130Y757F/Y757F femoral bone marrow and spleen revealed a 1.5-fold and 7-fold increase, respectively, in the number of megakaryocytes compared with that seen in gp130+/+ bone marrow and spleen (Table 1).
Characterization of hematopoietic organs in gp130Y757F/Y757F mice
Parameter . | +/+, mean ± SD* . | F/F, mean ± SD* . |
|---|---|---|
| Bone marrow | ||
| Cellularity per femur, × 107 | 3.58 ± 0.26 | 3.50 ± 1.02 |
| Blasts, % | 3 + 1 | 4 + 2 |
| Promyelocytes/myelocytes, % | 7 ± 2 | 10 ± 2 |
| Metamyelocytes/neutrophils, % | 27 ± 4 | 46 ± 7§ |
| Lymphocytes, % | 32 ± 7 | 21 ± 8 |
| Monocytes, % | 5 ± 1 | 4 ± 1 |
| Eosinophils, % | 3 ± 2 | 2 ± 1 |
| Nucleated RBCs, % | 23 ± 6 | 14 ± 3 |
| Megakaryocytes, per 10 hpf† | 88 ± 10 | 132 ± 17§ |
| Spleen | ||
| Mass, mg | 79 ± 16 | 203 ± 63§ |
| Cellularity, × 108 | 1.70 ± 0.71 | 3.63 ± 1.22§ |
| Blasts, % | 0 ± 0 | 0 ± 0 |
| Promyelocytes/myelocytes, % | 0 ± 0 | 0 ± 0 |
| Metamyelocytes/neutrophils, % | 2 ± 1 | 6 ± 1§ |
| Lymphocytes, % | 89 ± 6 | 76 ± 7 |
| Monocytes, % | 2 ± 1 | 2 ± 1 |
| Eosinophils, % | 0 ± 0 | 0 ± 0 |
| Nucleated RBCs, % | 7 ± 5 | 16 ± 6§ |
| Megakaryocytes, per 10 hpf† | 11 ± 5 | 76 ± 2§ |
| Thymus | ||
| Mass, mg | 46 ± 9 | 74 ± 17§ |
| Cellularity, × 108 | 2.01 ± 0.48 | 2.69 ± 0.84 |
| CD4+ CD8+‡§ | 84.63 ± 2.53 | 77.52 ± 5.6§ |
| B220+‡§ | 2.34 ± 1.82 | 4.02 ± 3.1 |
| Lymph nodes | ||
| Mass, mg | 43 ± 10 | 69 ± 13§ |
| Cellularity, × 107 | 2.09 ± 0.45 | 4.59 ± 0.60§ |
| Thy 1.2+‡§ | 57.90 ± 7.39 | 42.88 ± 7.46§ |
| B220+‡ | 39.60 ± 8.38 | 54.5 ± 10.45§ |
Parameter . | +/+, mean ± SD* . | F/F, mean ± SD* . |
|---|---|---|
| Bone marrow | ||
| Cellularity per femur, × 107 | 3.58 ± 0.26 | 3.50 ± 1.02 |
| Blasts, % | 3 + 1 | 4 + 2 |
| Promyelocytes/myelocytes, % | 7 ± 2 | 10 ± 2 |
| Metamyelocytes/neutrophils, % | 27 ± 4 | 46 ± 7§ |
| Lymphocytes, % | 32 ± 7 | 21 ± 8 |
| Monocytes, % | 5 ± 1 | 4 ± 1 |
| Eosinophils, % | 3 ± 2 | 2 ± 1 |
| Nucleated RBCs, % | 23 ± 6 | 14 ± 3 |
| Megakaryocytes, per 10 hpf† | 88 ± 10 | 132 ± 17§ |
| Spleen | ||
| Mass, mg | 79 ± 16 | 203 ± 63§ |
| Cellularity, × 108 | 1.70 ± 0.71 | 3.63 ± 1.22§ |
| Blasts, % | 0 ± 0 | 0 ± 0 |
| Promyelocytes/myelocytes, % | 0 ± 0 | 0 ± 0 |
| Metamyelocytes/neutrophils, % | 2 ± 1 | 6 ± 1§ |
| Lymphocytes, % | 89 ± 6 | 76 ± 7 |
| Monocytes, % | 2 ± 1 | 2 ± 1 |
| Eosinophils, % | 0 ± 0 | 0 ± 0 |
| Nucleated RBCs, % | 7 ± 5 | 16 ± 6§ |
| Megakaryocytes, per 10 hpf† | 11 ± 5 | 76 ± 2§ |
| Thymus | ||
| Mass, mg | 46 ± 9 | 74 ± 17§ |
| Cellularity, × 108 | 2.01 ± 0.48 | 2.69 ± 0.84 |
| CD4+ CD8+‡§ | 84.63 ± 2.53 | 77.52 ± 5.6§ |
| B220+‡§ | 2.34 ± 1.82 | 4.02 ± 3.1 |
| Lymph nodes | ||
| Mass, mg | 43 ± 10 | 69 ± 13§ |
| Cellularity, × 107 | 2.09 ± 0.45 | 4.59 ± 0.60§ |
| Thy 1.2+‡§ | 57.90 ± 7.39 | 42.88 ± 7.46§ |
| B220+‡ | 39.60 ± 8.38 | 54.5 ± 10.45§ |
Values represent the mean ± SD from 5 mice of each genotype
Number of megakaryocytes counted at × 400 magnification (bone marrow) or × 200 magnification (spleen), with 30 high-power fields (hpf) counted per mouse
Percentage of CD4+/CD8+ cells, and B220 or Thy1.2 single-positive cells as determined by flow cytometric analysis of single cell suspensions of thymus and lymph nodes
P < .05 versus data from wild-type (gp 130+/+) mice. No other statistically significant differences were observed between genotypes
We also observed a marked increase in the weight and cellularity of thymus and lymph nodes of gp130Y757F/Y757F mice, and flow cytometric analysis with the monoclonal antibody B220 revealed a significantly higher percentage of immature and mature B-lineage cells in these organs (Table 1). In contrast, the percentage of Thy1.2+ T cells was reduced in the thymus and lymph nodes of gp130Y757F/Y757F mice, and this correlated with a reduction in the percentage of CD4+CD8+ T cells (Table 1; data not shown). Despite this observation, absolute T-cell numbers were higher in the lymph nodes and thymus of gp130Y757F/Y757F mice because of the increased cellularity of these organs.
To discount the possibility these differences observed on a mixed 129 × C57BL/6 background were a function of strain differences, these experiments were repeated with mice backcrossed onto a C57BL/6 background over 10 generations. Similar magnitudes of differences were observed (data not shown), and data presented in the remainder of this study were generated from mice of all genotypes on a mixed 129 × C57BL/6 background.
Increased numbers of hematopoietic progenitor cells in gp130Y757F/Y757F mice
To determine if the perturbed hematopoiesis in gp130Y757F/Y757F mice resulted from aberrant production of immature and committed hematopoietic progenitors, we assessed the frequency and lineage commitment of hematopoietic progenitors from bone marrow and spleen of gp130Y757F/Y757F mice in agar cultures containing SCF and IL-3. Indeed, the total number of colonies formed in gp130Y757F/Y757F bone marrow cultures was significantly higher, thus demonstrating an elevated frequency of colony-forming cells (CFCs) representing the various myeloid lineages in bone marrow of gp130Y757F/Y757F mice compared with gp130+/+ controls (Table 2). Examination of colony formation by bone marrow progenitors from both genotypes in response to various concentrations of cytokines revealed that gp130Y757F/Y757F progenitors were not hyperresponsive to either SCF or IL-3 (Figure S1, available on the Blood website; see the Supplemental Figures link at the top of the online article). Surprisingly, the frequency of megakaryocyte progenitors (Mk-CFCs) in gp130Y757F/Y757F bone marrow was not increased. However, substantial increases in the frequency of Mk-CFCs, as well as myeloid CFCs, were observed in spleens of gp130Y757F/Y757F mice, which corresponded to further increases in absolute numbers due to the increased gp130Y757F/Y757F splenic cellularity (Table 2).
Hematopoietic progenitor cell production in gp130Y757F/Y757F mice
Parameter . | +/+, mean ± SD* . | F/F, mean ± SD* . |
|---|---|---|
| Bone marrow | ||
| Total† | 54 ± 16 | 81 ± 20§ |
| Granulocyte | 16 ± 2 | 27 ± 2§ |
| Granulocyte/macrophage | 10 ± 3 | 15 ± 2§ |
| Macrophage | 17 ± 3 | 20 ± 4 |
| Eosinophil | 2 ± 1 | 11 ± 3§ |
| Blast | 5 ± 1 | 6 ± 2 |
| Megakaryocyte | 4 ± 2 | 2 ± 1 |
| CFU-Sd12‡§ | 4.5 ± 1.1 | 9.1 ± 1.6§ |
| Spleen | ||
| Total† | 5 ± 3 | 36 ± 28§ |
| Granulocyte | 1 ± 1 | 7 ± 1§ |
| Granulocyte/macrophage | 1 ± 0 | 8 ± 4§ |
| Macrophage | 1 ± 1 | 7 ± 2§ |
| Blast | 1 ± 0 | 6 ± 4§ |
| Megakaryocyte | 2 ± 1 | 8 ± 6 |
| CFU-Sd12‡§ | 1.2 ± 0.2 | 5.7 ± 1.2§ |
Parameter . | +/+, mean ± SD* . | F/F, mean ± SD* . |
|---|---|---|
| Bone marrow | ||
| Total† | 54 ± 16 | 81 ± 20§ |
| Granulocyte | 16 ± 2 | 27 ± 2§ |
| Granulocyte/macrophage | 10 ± 3 | 15 ± 2§ |
| Macrophage | 17 ± 3 | 20 ± 4 |
| Eosinophil | 2 ± 1 | 11 ± 3§ |
| Blast | 5 ± 1 | 6 ± 2 |
| Megakaryocyte | 4 ± 2 | 2 ± 1 |
| CFU-Sd12‡§ | 4.5 ± 1.1 | 9.1 ± 1.6§ |
| Spleen | ||
| Total† | 5 ± 3 | 36 ± 28§ |
| Granulocyte | 1 ± 1 | 7 ± 1§ |
| Granulocyte/macrophage | 1 ± 0 | 8 ± 4§ |
| Macrophage | 1 ± 1 | 7 ± 2§ |
| Blast | 1 ± 0 | 6 ± 4§ |
| Megakaryocyte | 2 ± 1 | 8 ± 6 |
| CFU-Sd12‡§ | 1.2 ± 0.2 | 5.7 ± 1.2§ |
Values represent the mean ± SD from 5 mice for each genotype
Number of colonies in semisolid agar cultures containing 2.5 ± 104 bone marrow cells or 2.5 ± 105 spleen cells. Cells were stimulated with IL-3 and SCF
Number of colonies on spleens from irradiated wild-type mice injected with 7.5 × 104 bone marrow cells or 3.75 × 105 spleen cells from gp130+/+ or gp130Y757F/Y757F mice
P < .05 versus data from gp130+/+ mice. No other statistically significant differences were observed between genotypes
The frequency of day-12 CFU-S progenitor cells, which represent immature multipotential hematopoietic progenitors, was increased in the bone marrow and spleen of gp130Y757F/Y757F mice by approximately 2-fold and 5-fold, respectively, compared with gp130+/+ littermates (Table 2). Furthermore, we observed a similar 2-fold increase in the absolute numbers of CFU-S progenitors in gp130Y757F/Y757F bone marrow (4249 ± 218 per femur) compared with gp130+/+ bone marrow (2149 ± 38 per femur), and a striking 10-fold increase in absolute CFU-S numbers in the spleens of gp130Y757F/Y757F mice (gp130Y757F/Y757F, 5513 ± 391 versus gp130+/+, 544 ± 38).
Genetic reduction of gp130-dependent STAT3 activation in gp130Y757F/Y757F mice
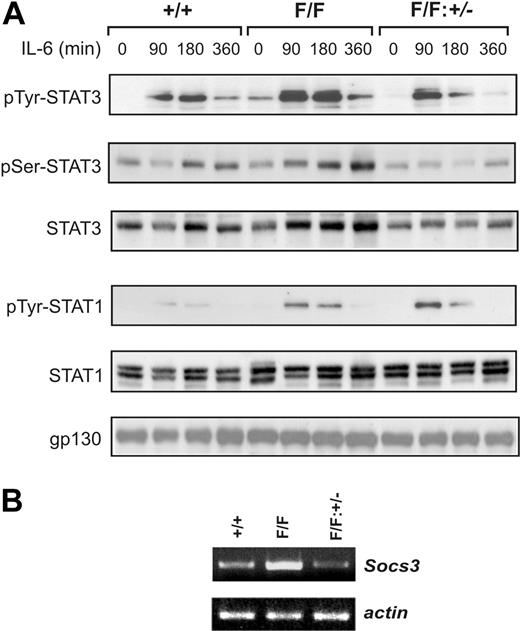
The hematopoietic abnormalities in heterozygous gp130+/Y757F mice were intermediate between those of gp130+/+ and gp130Y757F/Y757F mice (Figure S2), suggesting that the dosage of gp130-dependent signaling is critical to maintain normal hematopoiesis. In this respect, we have previously shown that the gp130(Y757F) mutation represents both a hypomorphic mutation abolishing gp130-dependent activation of the SHP2-Ras-MAPK pathway, as well as a hypermorphic mutation for gp130-dependent STAT1/3 hyperactivation.17 Since continuous gp130-dependent activation of both STAT1/3 and MAPK pathways in IL-6/sIL-6Rα or LIF transgenic mice also resulted in thrombocytosis and splenomegaly,7,8 we hypothesized that the gp130Y757F/Y757F phenotype may result from excessive STAT1/3 activation. To assess the specific contribution of gp130-dependent STAT3 hyperactivation to the hematopoietic phenotype, we crossed gp130Y757F/Y757F mice onto a Stat3 heterozygous (Stat3+/-) background to genetically reduce the cellular pool, and therefore activity, of STAT3. Indeed, immunoblot analyses of lysates from livers, the cellular composition of which is unaffected by the gp130(Y757F) mutation, from untreated and IL-6–treated mice revealed that the elevated level of STAT3 tyrosine phosphorylation in gp130Y757F/Y757F mice was reduced in gp130Y757F/Y757F: Stat3+/- mice to a level comparable to that observed in gp130+/+ mice (Figure 2A). In contrast, the level of IL-6-induced STAT3 serine phosphorylation, which is required for full transcriptional activation,31 relative to the total amount of STAT3 was similar among the genotypes, suggesting that the gp130(Y757F) mutation does not affect serine kinases that phosphorylate STAT3. This genetic intervention specifically affected STAT3 activation, since it neither altered the level of expression nor tyrosine phosphorylation of gp130 and STAT1 in gp130Y757F/Y757F:Stat3+/- mice (Figure 2A; data not shown). Furthermore, we confirmed enhanced expression of a representative IL-6-induced STAT3-response gene, Socs3,32 in gp130Y757F/Y757F mice compared with gp130Y757F/Y757F:Stat3+/-mice (Figure 2B).
Gp130-dependent STAT3 hyperactivation in gp130Y757F/Y757F mice is reduced on a Stat3 heterozygous (Stat3+/-) background. (A) Gp130+/+, gp130Y757F/Y757F, and gp130Y757F/Y757F:Stat3+/- mice were injected with 5 μg IL-6 for the times indicated. Liver lysates were run out on 10% polyacrylamide gel electrophoresis (PAGE) gels, transferred to nitrocellulose membranes, and then immunoblotted with the indicated antibodies. (B) Semiquantitative reverse transcriptase–polymerase chain reaction (RT-PCR) analysis of Socs3 expression was performed on cDNA derived from total RNA prepared from livers of untreated mice. Also shown is the RT-PCR for β-actin as an internal control for sample normalization and integrity.
Gp130-dependent STAT3 hyperactivation in gp130Y757F/Y757F mice is reduced on a Stat3 heterozygous (Stat3+/-) background. (A) Gp130+/+, gp130Y757F/Y757F, and gp130Y757F/Y757F:Stat3+/- mice were injected with 5 μg IL-6 for the times indicated. Liver lysates were run out on 10% polyacrylamide gel electrophoresis (PAGE) gels, transferred to nitrocellulose membranes, and then immunoblotted with the indicated antibodies. (B) Semiquantitative reverse transcriptase–polymerase chain reaction (RT-PCR) analysis of Socs3 expression was performed on cDNA derived from total RNA prepared from livers of untreated mice. Also shown is the RT-PCR for β-actin as an internal control for sample normalization and integrity.
Reduced gp130-dependent STAT3 activation in gp130Y757F/Y757F:Stat3+/- mice prevents abnormal hematopoiesis
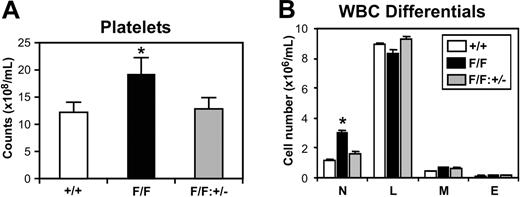
We next examined whether the reduction of exaggerated Stat3 signaling in gp130Y757F/Y757F:Stat3+/- mice would prevent the onset of the hematopoietic abnormalities observed in gp130Y757F/Y757F mice. Indeed, the circulating platelet counts of gp130Y757F/Y757F: Stat3+/- mice were 33% lower than gp130Y757F/Y757F littermates, and indistinguishable from gp130+/+ controls (Figure 3A). Similarly, the neutrophilia evident in gp130Y757F/Y757F mice was absent in gp130Y757F/Y757F:Stat3+/- mice (Figure 3B). All other peripheral blood parameters from gp130Y757F/Y757F:Stat3+/- mice were normal (data not shown).
Absence of peripheral blood abnormalities in gp130Y757F/Y757F:Stat3+/- mice. (A) Platelet counts and (B) WBC differentials on gp130+/+ (□), gp130Y757F/Y757F (▪), and gp130Y757F/Y757F:Stat3+/- (▦) mice. Data from 5 mice of each genotype are expressed as the mean ± SD. *P < .05 versus data from gp130+/+ mice. N indicates neutrophils; L, lymphocytes; M, monocytes; and E, eosinophils.
Absence of peripheral blood abnormalities in gp130Y757F/Y757F:Stat3+/- mice. (A) Platelet counts and (B) WBC differentials on gp130+/+ (□), gp130Y757F/Y757F (▪), and gp130Y757F/Y757F:Stat3+/- (▦) mice. Data from 5 mice of each genotype are expressed as the mean ± SD. *P < .05 versus data from gp130+/+ mice. N indicates neutrophils; L, lymphocytes; M, monocytes; and E, eosinophils.
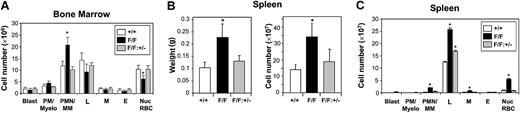
The cellular distribution in bone marrow from gp130Y757F/Y757F: Stat3+/- mice was similar to gp130+/+ bone marrow, with no evidence of the shift toward myelopoiesis observed in gp130Y757F/Y757F bone marrow (Figure 4A). At 12 weeks of age and beyond, gp130Y757F/Y757F: Stat3+/- mice showed no signs of the splenomegaly observed in age-matched gp130Y757F/Y757F mice, with the weight and cellularity of gp130Y757F/Y757F:Stat3+/- spleens comparable to those of gp130+/+ controls (Figure 4B). Except for a slight increase in lymphocyte numbers, the distribution of mature cells in gp130Y757F/Y757F:Stat3+/- spleens was also normal (Figure 4C). Clonal cultures of bone marrow and spleen cells revealed that the absolute numbers of myeloid progenitors in the femoral marrow and spleen of gp130Y757F/Y757F: Stat3+/- mice were reduced from those observed in gp130Y757F/Y757F mice back to wild-type levels (Figure 5). Similarly, the total number of blast-cell CFCs, which represent the most immature progenitor cell detectable in clonogenic assays, was normal. The absolute number of splenic Mk-CFCs was also normal in gp130Y757F/Y757F:Stat3+/- mice (Figure 5B).
Normal distribution of mature cells in the bone marrow and spleen of gp130Y757F/Y757F:Stat3+/- mice. (A) Bone marrow cells from gp130+/+ (□), gp130Y757F/Y757F (▪), and gp130Y757F/Y757F:Stat3+/- (▦) mice were cytocentrifuged and stained with May-Grunwald-Giemsa solution. (B) Weight and cellularity of spleens from the above mice. (C) Cytocentrifuge analysis of spleen cells as in panel A. Data from 5 mice of each genotype are expressed as the mean ± SD. *P < .05 versus data from gp130+/+ mice. PM/Myelo indicates myeloblast/myelocyte; PMN/MM, neutrophil/metamyelocyte; L, lymphocyte; M, monocyte; E, eosinophil; and Nuc RBC, nucleated red blood cell.
Normal distribution of mature cells in the bone marrow and spleen of gp130Y757F/Y757F:Stat3+/- mice. (A) Bone marrow cells from gp130+/+ (□), gp130Y757F/Y757F (▪), and gp130Y757F/Y757F:Stat3+/- (▦) mice were cytocentrifuged and stained with May-Grunwald-Giemsa solution. (B) Weight and cellularity of spleens from the above mice. (C) Cytocentrifuge analysis of spleen cells as in panel A. Data from 5 mice of each genotype are expressed as the mean ± SD. *P < .05 versus data from gp130+/+ mice. PM/Myelo indicates myeloblast/myelocyte; PMN/MM, neutrophil/metamyelocyte; L, lymphocyte; M, monocyte; E, eosinophil; and Nuc RBC, nucleated red blood cell.
Normalization of hematopoietic progenitor cell formation in gp130Y757F/Y757F: Stat3+/- mice. In vitro hematopoietic colony formation of (A) bone marrow cells and (B) spleen cells from gp130+/+ (□), gp130Y757F/Y757F (▪), and gp130Y757F/Y757F: Stat3+/- (▦) mice in semisolid agar containing SCF (50 ng/mL) and IL-3 (10 ng/mL). Data from 4 to 6 mice of each genotype are expressed as the mean ± SD. *P < .05 versus data from gp130+/+ mice. G indicates granulocyte; GM, granulocyte/macrophage; M, macrophage; Eo, eosinophil; and Meg, megakaryocyte.
Normalization of hematopoietic progenitor cell formation in gp130Y757F/Y757F: Stat3+/- mice. In vitro hematopoietic colony formation of (A) bone marrow cells and (B) spleen cells from gp130+/+ (□), gp130Y757F/Y757F (▪), and gp130Y757F/Y757F: Stat3+/- (▦) mice in semisolid agar containing SCF (50 ng/mL) and IL-3 (10 ng/mL). Data from 4 to 6 mice of each genotype are expressed as the mean ± SD. *P < .05 versus data from gp130+/+ mice. G indicates granulocyte; GM, granulocyte/macrophage; M, macrophage; Eo, eosinophil; and Meg, megakaryocyte.
We next determined whether the abnormalities observed in lymphoid organs of gp130Y757F/Y757F mice were rescued in gp130Y757F/Y757F: Stat3+/- mice. Thymic cellularity of gp130Y757F/Y757F:Stat3+/- mice was indistinguishable from that of gp130+/+ mice, and flow cytometric analyses also demonstrated that the perturbed distribution of B (B220+) and T (CD4+CD8+) cells in the thymus of gp130Y757F/Y757F mice was no longer apparent in gp130Y757F/Y757F:Stat3+/- thymi (Figure 6A-B). In contrast, the altered cellularity and distribution of B (B220+) and T (Thy1.2+) cells in lymph nodes from gp130Y757F/Y757F:Stat3+/- mice were only partially restored back to normal (Figure 6C-D). From these data, we conclude that removal of one Stat3 allele in gp130Y757F/Y757F mice reduces gp130-dependent STAT3 activation to a level that leads to rescue of the myeloid and thrombopoietic abnormalities, as well as amelioration of the perturbed lymphopoiesis.
Amelioration of perturbed lymphopoiesis in the thymus and lymph nodes of gp130Y757F/Y757F:Stat3+/- mice. Cellularity of (A) thymus and (C) lymph nodes from gp130+/+ (□), gp130Y757F/Y757F (▪), and gp130Y757F/Y757F:Stat3+/- (▦) mice. Data from 6 mice of each genotype are expressed as the mean ± SD. *P < .05 versus data from gp130+/+ mice. (B) Thymocytes and (D) lymph node cells were subjected to dual-color flow cytometric analysis with the indicated antibodies. For dot plots, x-axis represents FITC intensity, y-axis represents PE intensity, and numbers indicate the percent of positive cells in each quadrant. For histograms, x-axis represents either FITC or PE intensity, and y-axis represents relative cell number. The numbers indicate the percent of positive cells in the marked regions. These flow cytometric profiles are representative of at least 5 mice of each genotype.
Amelioration of perturbed lymphopoiesis in the thymus and lymph nodes of gp130Y757F/Y757F:Stat3+/- mice. Cellularity of (A) thymus and (C) lymph nodes from gp130+/+ (□), gp130Y757F/Y757F (▪), and gp130Y757F/Y757F:Stat3+/- (▦) mice. Data from 6 mice of each genotype are expressed as the mean ± SD. *P < .05 versus data from gp130+/+ mice. (B) Thymocytes and (D) lymph node cells were subjected to dual-color flow cytometric analysis with the indicated antibodies. For dot plots, x-axis represents FITC intensity, y-axis represents PE intensity, and numbers indicate the percent of positive cells in each quadrant. For histograms, x-axis represents either FITC or PE intensity, and y-axis represents relative cell number. The numbers indicate the percent of positive cells in the marked regions. These flow cytometric profiles are representative of at least 5 mice of each genotype.
Enhanced survival and proliferation of gp130Y757F/Y757F hematopoietic cells
Since STAT3 has been implicated as a crucial regulator of cell survival and proliferation,22,25,26 we next examined whether these responses were enhanced in cultured gp130Y757F/Y757F hematopoietic cells displaying STAT3 hyperactivation. Indeed, the number of viable bone marrow and spleen cells after 5 days in culture with or without IL-6 was higher in gp130Y757F/Y757F cultures compared with corresponding gp130+/+ and gp130Y757F/Y757F:Stat3+/- cultures (Figure 7A). Similar results were also obtained from cultures treated with IL-11 (Figure S3). This observation correlated with a lower frequency of apoptotic (annexin-V+) cells in IL-6–treated gp130Y757F/Y757F cultures (Figure 7B), thus demonstrating increased survival of gp130Y757F/Y757F hematopoietic cells. We confirmed elevated STAT3 tyrosine phosphorylation in IL-6–treated gp130Y757F/Y757F bone marrow cells (Figure 7C), as well as increased expression of Socs3 and the antiapoptotic Bcl-xL33 gene (Figure 7D). The proliferation of gp130Y757F/Y757F bone marrow cells in response to IL-6 was also increased (Figure 7E). Considering that these observations may reflect differences in the cellular composition of gp130+/+, gp130Y757F/Y757F, and gp130Y757F/Y757F:Stat3+/- bone marrow, we measured the proliferation of sorted Gr1+ bone marrow cells from each genotype. Similar to unfractionated bone marrow cells, 3H-thymidine incorporation by gp130Y757F/Y757F Gr1+ myeloid cells was enhanced in cultures containing IL-6 compared with IL-6–treated gp130+/+ and gp130Y757F/Y757F:Stat3+/- cells (Figure 7F), thus supporting the notion that gp130-dependent STAT3 hyperactivation augments biological responses from cells of various hematopoietic lineages.
Enhanced survival and proliferation of gp130Y757F/Y757F hematopoietic cells. (A) Survival of bone marrow and spleen cells from each genotype in response to IL-6. Equivalent cell numbers were cultured over 5 days, and the number of viable cells was counted. Data from 3 separate experiments were pooled and are presented as the mean ± SD. *P < .05 versus data from the corresponding gp130+/+ cultures. □ indicates gp130+/+; ▪, gp130Y757F/Y757F; and ▦, gp130Y757F/Y757F:Stat3+/- mice. (B) Cells from IL-6–treated bone marrow and spleen cultures were stained with FITC-labeled annexin-V and subjected to flow cytometric analysis. The x-axis represents FITC intensity, and y-axis represents relative cell number. The numbers indicate the percent of annexin-V- (M1 region) or annexin-V+ (M2 region) cells. (C,D) Bone marrow cells were cultured in the presence of IL-6 for 3 hours and 24 hours, after which whole-cell lysates were immunoblotted with the indicated antibodies (C), and semiquantitative RT-PCR analysis of Socs3 and Bcl-xL gene expression was performed on cDNA derived from total RNA (D). Also shown is the RT-PCR for β-actin as an internal control for sample normalization and integrity. Proliferation of unfractionated (E) and Gr1+ (F) bone marrow cells from gp130+/+, gp130Y757F/Y757F, and gp130Y757F/Y757F:Stat3+/- mice with or without IL-6 treatment over 48 hours prior to being pulsed with 3H-thymidine for 12 hours. Data are presented as the mean ± SD for triplicate cultures. *P < .05 versus data from the corresponding gp130+/+ cultures. No significant differences in 3H-thymidine incorporation were observed in cultures of Gr1+ bone marrow cells from each genotype treated with IL-3 (gp130+/+, 3740 ± 1101 counts per minute [cpm]; gp130Y757F/Y757F, 3612 ± 472 cpm; gp130Y757F/Y757F:Stat3+/-, 4227 ± 372 cpm).
Enhanced survival and proliferation of gp130Y757F/Y757F hematopoietic cells. (A) Survival of bone marrow and spleen cells from each genotype in response to IL-6. Equivalent cell numbers were cultured over 5 days, and the number of viable cells was counted. Data from 3 separate experiments were pooled and are presented as the mean ± SD. *P < .05 versus data from the corresponding gp130+/+ cultures. □ indicates gp130+/+; ▪, gp130Y757F/Y757F; and ▦, gp130Y757F/Y757F:Stat3+/- mice. (B) Cells from IL-6–treated bone marrow and spleen cultures were stained with FITC-labeled annexin-V and subjected to flow cytometric analysis. The x-axis represents FITC intensity, and y-axis represents relative cell number. The numbers indicate the percent of annexin-V- (M1 region) or annexin-V+ (M2 region) cells. (C,D) Bone marrow cells were cultured in the presence of IL-6 for 3 hours and 24 hours, after which whole-cell lysates were immunoblotted with the indicated antibodies (C), and semiquantitative RT-PCR analysis of Socs3 and Bcl-xL gene expression was performed on cDNA derived from total RNA (D). Also shown is the RT-PCR for β-actin as an internal control for sample normalization and integrity. Proliferation of unfractionated (E) and Gr1+ (F) bone marrow cells from gp130+/+, gp130Y757F/Y757F, and gp130Y757F/Y757F:Stat3+/- mice with or without IL-6 treatment over 48 hours prior to being pulsed with 3H-thymidine for 12 hours. Data are presented as the mean ± SD for triplicate cultures. *P < .05 versus data from the corresponding gp130+/+ cultures. No significant differences in 3H-thymidine incorporation were observed in cultures of Gr1+ bone marrow cells from each genotype treated with IL-3 (gp130+/+, 3740 ± 1101 counts per minute [cpm]; gp130Y757F/Y757F, 3612 ± 472 cpm; gp130Y757F/Y757F:Stat3+/-, 4227 ± 372 cpm).
Discussion
In this paper, we report that gp130Y757F/Y757F mice expressing a gp130 receptor unable to bind to SOCS3 and SHP2 display multiple hyperproliferative hematopoietic abnormalities, namely thrombocytosis, lymphadenopathy, and splenomegaly. This phenotype contrasts the loss-of-function phenotype characteristic of mice carrying null mutations in genes for IL-6 family cytokines, or their receptors.3-6 Rather, the thrombocytosis and splenomegaly displayed by gp130Y757F/Y757F mice are shared features with LIF single-7 or IL-6/sIL-6Rα double-transgenic mice8 in which gp130 is persistently activated. Significantly, we and others have demonstrated that functional inactivation of the Y757/759 residue leads to gp130-dependent STAT1 and STAT3 hyperactivation as a consequence of impaired negative regulation of gp130 signaling.17,23 Indeed, the striking finding of our present study was that the thrombocytosis, splenomegaly and, to a lesser extent, lymphadenopathy in gp130Y757F/Y757F mice were eliminated by genetically reducing the cellular pool of STAT3 protein and activity, thus identifying STAT3 hyperactivation, rather than loss of SHP2-MAPK activation, via gp130 as a major mechanism triggering the hematopoietic abnormalities. Furthermore, these abnormalities are intrinsic to the hematopoietic progenitors themselves, since wild-type mice reconstituted with bone marrow from gp130Y757F/Y757F mice also displayed thrombocytosis and splenomegaly (data not shown). The partial restoration of abnormalities in the lymph nodes of gp130Y757F/Y757F:Stat3+/- mice may be indicative of a STAT3 signal threshold requirement for homeostasis in this secondary lymphoid organ that is different to that in bone marrow, spleen and thymus.
Notably, our observations contrast with those relating to the previously described gp130F759/F759 mice, which express the gp130(Y759F) substitution in the SHP2/SOCS3 docking site of the human cDNA, and only partially resemble the gp130Y757F/Y757F hematopoietic phenotype, namely splenomegaly and lymphadenopathy.23 It was reported that cell-autonomous gp130-dependent signaling in gp130F759/F759 mice was largely dispensable for hematopoiesis, since colony-forming assays and reconstitution experiments using gp130 mutant fetal liver cells failed to identify any hematopoietic abnormalities.23 As discussed previously,24 it is likely that the more subtle hematopoietic phenotype of gp130F759/F759 mice is a consequence of a STAT3 signal that is below the threshold enabling the hyperresponsiveness of hematopoietic progenitor cells that is observed in gp130Y757F/Y757F mice.
With respect to the role of STAT3 during hematopoiesis, different strains of bone marrow–specific Stat3 null mice have been generated which display either neutrophilia22 or an inflammatory disease in the gastrointestinal tract.26 While the variation in these phenotypes is the result of differences in the Mx22 and Tie226 promoters driving Cre-mediated excision of STAT3 in bone marrow, both phenotypes were underscored by the hyperproliferation of myeloid cells in response to G-CSF or macrophage (M)–CSF. Moreover, this hyperproliferation correlated with the failure of G-CSF to transcriptionally induce Socs3 in STAT3-deficient bone marrow cells, thus implying a regulatory role for STAT3 to suppress myelopoiesis through induction of Socs3 expression.22 This notion was directly supported by a recent study in which hematopoietic-specific deletion of Socs3 in Socs3-/ΔVav mice also resulted in neutrophilia and a complex inflammatory disease that was characterized by the hyperresponsiveness of myeloid cells to G-CSF, most likely as a consequence of exaggerated G-CSF–dependent STAT3 activation.34 However, it was noted in this latter study that the pathology observed in older diseased Socs3-/ΔVav mice was unlikely caused by dysregulated G-CSF signaling alone. In this respect, SOCS3-deficient macrophages from Socs3-/ΔLysM mice display increased IL-6–induced STAT3 activation and are also hyperresponsive to IL-6,35 which is consistent with the emerging view that SOCS3 is a critical physiologic regulator of gp130-dependent signaling, and suggests that IL-6 family cytokines may contribute to disease development in conditional Socs3-/ΔVav mice. Importantly, these mice develop hematopoietic abnormalities which strikingly resemble those observed in the gp130Y757F/Y757F mice reported here, including splenomegaly characterized by increased myelopoiesis and erythropoiesis, and increased myelopoiesis at the expense of erythropoiesis in the bone marrow.34 Therefore, it is tempting to speculate that at least some of the shared pathologies between the conditional Socs3-/ΔVav and gp130Y757F/Y757F mice might be a consequence of increased STAT3 activation from IL-6 family cytokines. However, we cannot rule out the possibility that some of the abnormalities in gp130Y757F/Y757F mice might be due to the observed increase in Socs3 expression which, although unable to negatively regulate signaling from the gp130(Y757F) receptor, may inappropriately trans–down-modulate signaling from other cytokines affected by SOCS3 activity, such as G-CSF.34
A mechanism highly sensitive to the extent of STAT3 signaling throughput in megakaryocytes and their progenitors is likely to play a crucial role in regulating platelet production. We have previously demonstrated that gp130ΔSTAT/ΔSTAT mice displayed mild thrombocytopenia caused by impaired IL-6 and IL-11 induced megakaryocyte maturation, suggesting that megakaryocyte maturation induced by IL-6 family cytokines is dependent upon STAT1/3 activation.28 In contrast, the thrombocytosis in gp130Y757F/Y757F mice resulted from increased numbers of megakaryocytes rather than enhanced megakaryocyte maturation, since the size of megakaryocytes in gp130Y757F/Y757F bone marrow and spleen sections appeared normal (data not shown), an observation similar to that observed in thrombocytotic IL-6/sIL-6Rα or LIF transgenic mice.7,8 Previous studies have shown that long-term administration of IL-636 or IL-1137,38 to wild-type mice elevated in a dose-dependent manner the circulating platelet count, which was associated with an increase in both megakaryocyte size (maturation) and number. Since we have observed a dose-dependent relationship between the amount of IL-6 or IL-11 administered and the extent of STAT3 phosphorylation in gp130+/+ and gp130Y757F/Y757F mice (data not shown), we speculate that long-term treatment of wild-type mice with high doses of IL-6 family cytokines is likely to induce a stronger STAT3 signal than under “steady-state” conditions in IL-6/sIL-6Rα transgenic, LIF transgenic, or gp130Y757F/Y757F mice. Furthermore, the administration of exogenous IL-6 or IL-11 would then be likely to exceed the STAT3 signaling threshold required to promote both megakaryocyte production and maturation. Considering that the levels of circulating platelets in gp130ΔSTAT/ΔSTAT mice28 (reduced), gp130Y757F/Y757F mice (elevated), and gp130Y757F/Y757F:Stat3+/- mice (normal) directly correlate with the extent of STAT3 activation, we propose that the thrombopoietic activity of IL-6 family cytokines is elicited by STAT3-dependent mechanisms. A lower STAT3 signaling threshold (surpassed in IL-6/sIL-6Rα transgenic, LIF transgenic and gp130Y757F/Y757F mice) directs the expansion of megakaryocyte precursors from immature progenitor cells, whereas a higher STAT3 threshold (reached by long-term IL-6 or IL-11 administration) promotes megakaryocyte maturation.
The notion that the magnitude/intensity of gp130 signals can influence the fate of a hematopoietic cell is supported by recent studies based on the well-documented synergistic actions of IL-6 family cytokines with other hematopoietic growth factors such as stem-cell factor and flt3-ligand to promote the expansion of primitive hematopoietic cells.39,40 These studies suggest that the decision of hematopoietic stem cells (HSCs) to either self-renew or differentiate is dependent upon a signaling threshold effect determined by the intensity of gp130 signaling, possibly mediated by STAT3.41 Furthermore, STAT3 expression was greatest in the most primitive hematopoietic progenitors, and the blockade of STAT3 activation impaired the repopulating capacity of HSCs, suggesting that STAT3 is a crucial regulator of HSC amplification.42 Such findings are reminiscent of the importance of gp130-dependent STAT3 activation for maintaining totipotency of embryonic stem cells.43 Therefore, the increased number of immature CFU-S progenitors, which are derived from the more primitive HSCs, in the bone marrow and spleen of gp130Y757F/Y757F mice might reflect an expanded stem cell compartment as a result of gp130-dependent STAT3 hyperactivation.
In summary, we have provided genetic evidence that the level of gp130-dependent STAT3 activation is a critical determinant for pathophysiologic responses during hematopoiesis. The correlation between gp130-dependent STAT3 hyperactivation and enhanced proliferative/antiapoptotic responses of gp130Y757F/Y757F hematopoietic cells provides a molecular explanation for the expansion of hematopoietic cells that underlie the hematopoietic phenotype of gp130Y757F/Y757F mice. Compared with tissue-specific knockouts of, for instance, STAT3, hypo/hypermorphic signaling module mutants such as gp130Y757F/Y757F mice provide 2 distinct advantages in correlating molecular pathways with complex biological responses. First, they restrict alterations to the throughput of a widely used signaling cascade (eg, STAT3) to a specific receptor, and second, they enable the investigation of its global requirement rather than limiting it by conditional gene modification to a predetermined temporal or spatial cellular compartment. This latter point is particularly pertinent to variations in the phenotype of existing conditional knock-out mouse models examining the role of STAT3 in hematopoiesis, which have predominantly focused on the regulation of myelopoiesis by STAT3 as a consequence of STAT3 deletion being restricted to the bone marrow compartment.22,26 Finally, the wide availability of mice with null alleles for many components of the gp130 signaling network will enable the further genetic dissection of molecular mechanisms upstream and downstream of the gp130 receptor subunit, which ultimately restrict the extensive pleiotropic activities of this cytokine family to the appropriate physiologic response during adult development and tissue homeostasis.
Prepublished online as Blood First Edition Paper, January 13, 2005; DOI 10.1182/blood-2004-09-3751.
Supported by the Cancer Council of Victoria Sir Edward Dunlop Clinical Research Fellowship (A.W.R.) and by a research grant from the National Health and Medical Research Council (NHMRC) of Australia (B.J.J., M.E.).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank K. Takeda (University of Osaka, Japan) for generously providing Stat3+/- mice, and E. Richardson and M. Inglese for expert technical assistance. Additional thanks also to P. Hertzog, J. Cohen, M. Arnold, T. Helman, and T. Thorne. We are grateful to T. Burgess for critical reading of the manuscript.



![Figure 7. Enhanced survival and proliferation of gp130Y757F/Y757F hematopoietic cells. (A) Survival of bone marrow and spleen cells from each genotype in response to IL-6. Equivalent cell numbers were cultured over 5 days, and the number of viable cells was counted. Data from 3 separate experiments were pooled and are presented as the mean ± SD. *P < .05 versus data from the corresponding gp130+/+ cultures. □ indicates gp130+/+; ▪, gp130Y757F/Y757F; and ▦, gp130Y757F/Y757F:Stat3+/- mice. (B) Cells from IL-6–treated bone marrow and spleen cultures were stained with FITC-labeled annexin-V and subjected to flow cytometric analysis. The x-axis represents FITC intensity, and y-axis represents relative cell number. The numbers indicate the percent of annexin-V- (M1 region) or annexin-V+ (M2 region) cells. (C,D) Bone marrow cells were cultured in the presence of IL-6 for 3 hours and 24 hours, after which whole-cell lysates were immunoblotted with the indicated antibodies (C), and semiquantitative RT-PCR analysis of Socs3 and Bcl-xL gene expression was performed on cDNA derived from total RNA (D). Also shown is the RT-PCR for β-actin as an internal control for sample normalization and integrity. Proliferation of unfractionated (E) and Gr1+ (F) bone marrow cells from gp130+/+, gp130Y757F/Y757F, and gp130Y757F/Y757F:Stat3+/- mice with or without IL-6 treatment over 48 hours prior to being pulsed with 3H-thymidine for 12 hours. Data are presented as the mean ± SD for triplicate cultures. *P < .05 versus data from the corresponding gp130+/+ cultures. No significant differences in 3H-thymidine incorporation were observed in cultures of Gr1+ bone marrow cells from each genotype treated with IL-3 (gp130+/+, 3740 ± 1101 counts per minute [cpm]; gp130Y757F/Y757F, 3612 ± 472 cpm; gp130Y757F/Y757F:Stat3+/-, 4227 ± 372 cpm).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/9/10.1182_blood-2004-09-3751/6/m_zh80090577650007.jpeg?Expires=1765885713&Signature=P3EQJmOAWZDY8Ja65Q8R4ZN5mbnsShVNE2TU3S99hCvclLu8zbiLgF9YI-bOmXrtGuAw01BWnIZAVpGB~F4v~OX11f-t58Vl7nDwT4F1T16xdkYgxrwDckeO5gWEbqoi9FV~Y00~WLOhjcrvS95T9PJ-gvkNSXfyqyJgrUDD50msLfLZ146u3epeivtEHQqDOcT3YSpi-i~fd8yNMkjjh0cssihiXWH3xIiYBomeeo1c65pvTnotlfVcx3u5AR5GRZ8ySylIRR3Cc7DJWbaPy2MhWoyFq7byUMriIwJsAra4~n8HYoaUdcb8yXCyv3uaPg6ErEI3NqS1PCTFjFk7ng__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal