Abstract
Emerging evidence indicates that Notch receptors and their ligands play important roles in the development of T cells and B cells. However, little is known about their possible roles in the development of other lymphoid cells. Here we demonstrate that Jagged2, a Notch ligand, stimulates the development of natural killer (NK) cells from Lin- Sca-1+ c-kit+ hematopoietic stem cells. Our culture system supports NK cell development for 2 to 3 months, often leading to the establishment of continuous NK cell lines. The prototype of such cell lines is designated as KIL. KIL depends on interleukin-7 for survival and proliferation and is NK1.1+ CD3- TCRαβ- TCRδγ- CD4- CD8- CD19- CD25+ CD43+ CD45+ CD49b- CD51+ CD94+ NKG2D+ Mac-1-/low B220- c-kit+ perforin I+ granzyme B+ Notch-1+, and cytotoxic. Like normal natural killer cells, the T-cell receptor-β loci of KIL remain in the germ-line configuration. In response to interleukin-2, KIL proliferates extensively (increasing cell number by approximately 1010-fold) and terminally differentiates into adherent, hypergranular NK cells. Our findings indicate that Jagged2 stimulates the development of natural killer cells and the KIL cell line preserves most properties of the normal NK precursors. As such, KIL provides a valuable model system for NK cell research.
Introduction
Notch receptors and DSL (Delta-Serrate-Lag2) ligands assist in cell fate decisions during embryogenesis.1-5 Four Notch receptors (Notch-1, -2, -3, and -4)6-9 and 5 DSL ligands (Jagged1, Jagged2, Deltalike-1 or Dll-1, Dll-3, and Dll-4)10-15 have been identified in the murine system. After birth, mice continue to express Notch receptors and DSL ligands in many tissues. However, little is known about their functions in adult mice. Recently, reverse genetics and cell culture studies have begun to shed light on the functional roles of Notch and DSL ligands in T- and B-cell development.16-21 Conditional knockout of Notch-1 in the postnatal period abolishes T-cell development.17 The thymuses of these knockout mice lack a well-developed cortex. In tissue culture, Lin- Sca-1+ c-Kit+ murine hematopoietic stem cells (HSCs) stimulated by Flt3 ligand (Flt3L), interleukin-7 (IL-7), and OP-9 fibroblasts expressing ectopic Dll-1 undergo de novo T-cell development.20 Similar findings were made using progenitors derived from embryonic stem cells (ESCs).21 Subsequent studies indicate that continued presence of Dll-1 is required for T-cell commitment and maintenance at the double-negative 1 (DN1) and 2 (DN2) stages of thymocyte development. In its absence, the developing DN1 and DN2 thymocytes adopt the natural killer (NK) cell fate by default.22 More recently, it was reported that conditional knockout of Dll-1 blocks the development of marginal B cells but has no effect on T-cell development.23 This observation seems to contradict the findings of the Dll-1 studies.20,21 Nonetheless, the results of these studies underscore the importance of DSL ligands in lymphopoiesis.
In the current study, we used a slightly different coculture system to investigate the role of mouse Jagged2 (mJagged2),12 also a DSL ligand, in lymphoid development. In mouse embryos, mJagged2 expression is detected in thymus, dorsal aorta, paravertebral blood vessels, the basal layer of the entire epidermis, hair follicles, foregut, hindgut, brain, and dorsal root ganglia (S.T., unpublished data, June 1997).11,12,24,25 In newborn and adult mice, mJagged2 expression is detected in thymus, intestine, muscle, brain, testis, kidney, bone marrow, purified HSCs, hematopoietic progenitors, and endothelial cells.11,12,25 However, little is known about its role in lymphopoiesis except that mice homozygous for a mutated mJagged2 (Jagged2ΔDSL/ΔDSL) exhibit limb and craniofacial deformities, abnormalities in the thymic structure, and altered Tαβ/Tγδ ratios.26 Our coculture system differs from the one described in the cited Dll-1 studies in 3 ways. First, a slow-growing variant of OP-9,27 designated as OP-9S, was used to allow prolonged cocultivation and observation. Second, OP-9S was engineered to express mJagged2 rather than Dll-1. Third, interleukin-2 (IL-2) was added to half of the cocultures at a certain point to promote complete differentiation of T and NK cells. Our results indicate that mJagged2 has a strong stimulatory effect on the development of NK cells. Furthermore, NK precursors produced in the OP-9S/Jagged2 cocultures continue to proliferate for 2 to 3 months, often resulting in the establishment of permanent NK precursor cell lines, which have been extremely difficult to establish until now.
Materials and methods
Stromal cell lines and retroviral infection
A slow-growing derivative of the original OP-9 cell line,27 designated as OP-9S, was established in our laboratory and maintained in Dulbecco modified Eagle medium (DME; Gibco, Grand Island, NY)/30% fetal bovine serum (FBS). The original OP-9 was obtained from the laboratory of H. Kodama (Ohu University, Koriyama, Fukushima, Japan). OP-9S has a doubling time of 48 to 72 hours. W20 is a stromal fibroblast cell line derived from the bone marrow of a W+/+ mouse.12 It was maintained in DME/10% FBS. The construction of the retroviral vectors LXSN and LMJSN (expressing full-length mJagged2; GenBank accession no. AF038572) and the establishment of the corresponding retroviral producer cell lines have been described previously.12 OP-9S was infected with retroviral vectors LXSN and LMJSN and selected with G418 (0.75 mg/mL) for 12 days. The resultant cell lines are designated as OP-9S/LXSN and OP-9S/LMJSN, respectively.
Post-5-fluorouracil bone marrow and purification of Lin- Sca-1+ c-Kit+ HSCs
For post-5-fluorouracil (post-5-FU) bone marrow, 4- to 6-week-old C57BL/6 mice (Ly5.1) were injected intraperitoneally with 5-FU (150 mg/kg; SoloPak Laboratries, Elk Grove Village, IL). Bone marrows were harvested on day 4 and centrifuged over NycoPrep (1.077 mg/mL; Axis-Shield PoC, Oslo, Norway). Light-density mononuclear cells (MNCs) were collected from the interface and washed with Hanks balanced salt solution supplemented with 5% FBS. Lin- Sca-1+ c-Kit+ HSCs were purified as described.28 Briefly, red cells were lysed in an ammonium chloride solution. The unlysed cells were then stained with optimized concentrations of antibodies against lineage markers CD2, CD3, CD5, CD8, CD19, CD11b, CD45R, Ly-6G, and TER119. Lineage marker–positive cells were depleted by 2 successive incubations with sheep anti–rat Ig–coupled magnetic beads (Dynal AS, Oslo, Norway). The Lin- cells were then stained with phycoerythrin (PE)–Sca-1 antibody and sorted using a FACS Vantage sorter (Becton Dickinson, San Jose, CA) using the enrichment mode. Dead cells were excluded from sorting and all analyses by gating on forward scatter and propidium iodide (PI; Molecular Probes, Eugene, OR). The sorted Lin- Sca-1+ cells were restained with allophycocyanin-conjugated anti–c-Kit antibody (APC-c-Kit; BD Pharmingen) and resorted using the normal mode. Lin- Sca-1+ c-Kit+ cells were sorted into a tube containing FBS. An aliquot of each sorted population was reanalyzed to verify purity before use.
Bone marrow cocultures
Post-5-FU bone marrow MNCs or CD3-NK1.1- post-5-FU bone marrow MNCs (2.5 × 105 per well) or Lin- Sca-1+ c-Kit+ cells (104 per well) were cocultured with preformed monolayers of OP-9S/LXSN or OP-9S/LMJSN in 12-well plates in DME/30% FBS/5 × 10-5 M β-mercaptoethanol (β-ME), murine IL-7 (10 ng/mL; PeproTech, Rocky Hills, NJ), and human Flt3 ligand (15 ng/mL; R&D Systems, Minneapolis, MN). Half of the medium was changed every 2 to 4 days. To avoid crowding, one quarter to one half of the nonadherent cells were removed as needed at feeding.
Establishment and subcloning of KIL
KIL and 3 KIL-like cell lines were established from cocultures of post-5-FU bone marrow MNCs and OP-9S/LMJSN. These cultures were fed with the growth medium containing Flt3L and IL-7 only. No IL-2 was added at any point since it induced terminal NK differentiation and the eventual demise of the cocultures. NK precursors became the major cell type after about 6 weeks of coculturing. Once NK or NK precursors became the dominant (> 50%) cell type, other cells (mostly B progenitors and OP-9S) declined very rapidly, presumably due to the cytotoxicity and/or inhibitory cytokines of NK or NK precursors. KIL-like cell lines emerged after 3 months of continuous passaging in the same growth medium containing Flt3L and IL-7. Since OP-9S was the main source of stem cell factor (SCF) in the cocultures, we found it beneficial to provide exogenous SCF (10 ng/mL-20 ng/mL; R&D Systems) after most OP-9S cells had been destroyed. Once established, KIL was found to be unresponsive to Flt3L. It was then maintained in DME/30% FBS/5 × 10-5 M β-ME, murine IL-7 (25 ng/mL), and murine SCF (50 ng/mL) and subcultured at 1:2 to 1:8 ratios every 2 to 3 days. As an alternative, the conditioned medium (10%; vol/vol) of the BHK/MKL cell line29 can be used as a source of SCF. KIL was subcloned by limiting dilution at less than or equal to 0.2 clonogenic cells per well in 96-well plates. The clonal lines were designated KIL C.1-C.5. To induce the terminal differentiation of KIL and KIL C.2, human IL-2 (Chiron, Emeryville, CA) was added at a final concentration of 20 ng/mL (with or without IL-7 or SCF). The medium was changed as needed.
Monoclonal antibodies and flow cytometry
Biotinylated monoclonal antibodies against NK1.1, Ly5.1, CD3, CD25, CD4, CD8, CD19, Mac-1, CD43, CD49b (DX5), CD51, CD94, B220, Fc Block (anti-CD16/CD32), and PE-conjugated monoclonal antibodies against T-cell receptor β chain (TCRβ) and TCRγδ were purchased from BD Pharmingen (San Diego, CA). PE- or fluorescein isothiocyanate (FITC)–conjugated CD3, CD4, CD8, CD19, B220, Mac-1, Ly5.1 (clone A20), and Ly6G (clone RB6-8C5) were generous gifts from Gerald Spangrude (Division of Hematology, University of Utah). Antibodies against murine granzyme B, perforin I, and the carboxy terminus of human Notch-1 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal antibody against NKG2D (clone A10) was kindly provided by Wayne Yokoyama (Howard Hughes Medical Institute, Washington University School of Medicine, St Louis, MO). For flow cytometry, cells were incubated for 20 minutes with optimal concentrations of PE- or FITC-conjugated or biotinylated antibodies and PI. Biotinylated antibodies were secondarily stained with PE- or FITC- or peridinin chlorophyll–alpha protein (PerCP)–streptavidin (SAv; Biomedia, Foster City, CA). Cells were analyzed using a FACScan (Becton Dickinson).
Northern and Western analyses
Total RNAs (5 μg-10 μg) were resolved on 1% formaldehyde agarose gels, blotted onto Hybond-N (Amersham Pharmacia Biotech, Piscataway, NJ), and hybridized with P32-labeled probes at 65°C in Rapid-Hyb buffer (Amersham Pharmacia Biotech). Final washing was done in 0.1X standard saline citrate (SSC)/0.1% sodium dodecyl sulfate (SDS) at 65°C. For Western analyses, protein lysates (5 μg-10 μg) were separated by denaturing SDS–polyacrylamide (8%) gel electrophoresis, blotted onto Immobilon-P polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA), and visualized by enhanced chemiluminescence. The production of rabbit serum against the intracellular domain of mJagged2 has been previously described.12
Nested PCR and Southern hybridization
The sequences of polymerase chain reaction (PCR) primers used in detecting rearrangements in the TCRβ loci have been reported previously.30 In primary PCR, 0.5 μg genomic DNAs were amplified with the Dβ1.1ext/Dβ1.7ext or Dβ2.1ext/Dβ2.7ext primers for 30 cycles using the following cycling parameters: 94°C for 30 seconds, 59°C for 1 minute, 72°C for 3 minutes. A quantity of 2 μL of the primary PCR reactions was reamplified with the Dβ1.1int/Dβ1.7int or Dβ2.1int/Dβ2.7int primers for 30 cycles using the following cycling parameters: 94°C for 30 seconds, 63°C for 40 seconds, 72°C for 2 minutes. A quantity of 15 μL of the secondary PCR products were run on 2% agarose gels and stained with ethidium bromide. Southern blots of the PCR products were hybridized with P32-labeled, gel-purified Dβ1.1-Dβ1.7 or Dβ2.1-Dβ1.7 fragments, whose sequences had been verified by direct sequencing.
Cytotoxicity assay
OP-9S cells (or NIH3T3 cells that had been irradiated with 900 rad) were seeded in 12-well plates and grown to confluence. One confluent monolayer of OP-9S or NIH3T3 was dissociated by trypsin and the cell number was determined. KIL was added to the confluent stromal layers at 1:1 to 4:1 effector-to-target ratios and fed with growth medium containing IL-7 and SCF. After 24 to 48 hours, nonadherent cells were removed and the cultures were gently rinsed with phosphate-buffered saline, drained, and stained with Coomassie blue stain (BioRad, Hercules, CA) for 30 minutes. After staining, the cultures were rinsed with tap water and air-dried.
Microscopy
Photomicrographs were taken using a Nikon Eclipse TE300 microscope (Nikon, Tokyo, Japan) equipped with PlanFluor lenses (20 ×/0.45 and 40 ×/0.60) and a SPOT RT Slider digital camera (Diagnostic Instruments, Sterling Heights, MI) and SPORT RT software version 3.2 (Diagnostic Instruments). Image files were edited with Photoshop version 8.0 (Adobe Systems, San Jose, CA).
Results
Jagged2 stimulates NK development from post-5-fluorouracil marrow
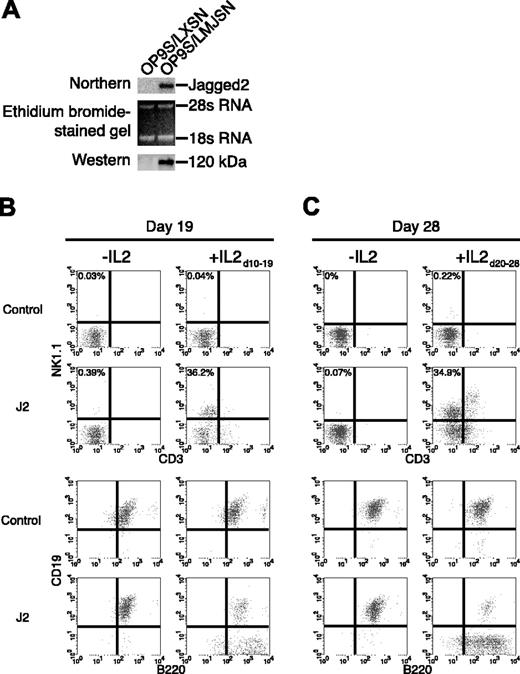
To study the effects of mJagged2 on NK cell development, we derived a slow-growing variant of the OP-9 cell line, designated as OP-9S, to minimize metabolic competition during prolonged cocultivation with bone marrow progenitors. The original OP-9 stromal cell line was derived from a B6C3F1 mouse with osteopetrosis.27 It does not express macrophage–colony-stimulating factor. OP-9S was transduced with retroviral vectors LXSN (negative control) and LMJSN (expressing full-length mJagged2)12 and selected with G418. The resultant cell lines are referred to as OP-9S/LXSN and OP-9S/LMJSN, respectively. Northern and Western analyses confirmed the expression of the full-length mJagged2 in OP-9S/LMJSN but not OP-9S/LXSN (Figure 1A).
Jagged2 stimulates the development of NK cells from post-5-FU bone marrow MNCs. (A) Retroviral vector–mediated expression of the full-length mJagged2 in OP-9S/LMJSN but not OP-9S/LXSN (negative control). The top panel is a Northern blot hybridized with an mJagged2 cDNA probe. The middle panel is the ethidium bromide–stained gel before blotting. The bottom panel is a Western blot probed with rabbit antiserum against the intracellular domain of mJagged2. The 120-kDa mJagged2 protein is indicated. (B) Expression of NK1.1 versus CD3 and CD19 versus B220 by 5-FU bone marrow MNCs cocultured with OP-9S/LXSN (negative control) or OP-9S/LMJSN (“J2”) in the presence of IL-7 and Flt3L for 19 days without or with IL-2 on days 10 to 19. The percent of CD3-NK1.1+ cells is indicated. (C) Expression of NK1.1 versus CD3 and CD19 versus B220 by post-5-FU bone marrow MNCs cocultured with OP-9S/LXSN or OP-9S/LMJSN (“J2”) in the presence of IL-7 and Flt3L for 28 days without or with IL-2 on days 20 to 28. The percent of CD3-NK1.1+ cells is indicated. Small numbers of CD3+NK1.1+ cells were present in OP-9S/LMJSN cocultures. They likely represented NKT or cytotoxic lymphocytes that had evolved from pre-existing T cells.
Jagged2 stimulates the development of NK cells from post-5-FU bone marrow MNCs. (A) Retroviral vector–mediated expression of the full-length mJagged2 in OP-9S/LMJSN but not OP-9S/LXSN (negative control). The top panel is a Northern blot hybridized with an mJagged2 cDNA probe. The middle panel is the ethidium bromide–stained gel before blotting. The bottom panel is a Western blot probed with rabbit antiserum against the intracellular domain of mJagged2. The 120-kDa mJagged2 protein is indicated. (B) Expression of NK1.1 versus CD3 and CD19 versus B220 by 5-FU bone marrow MNCs cocultured with OP-9S/LXSN (negative control) or OP-9S/LMJSN (“J2”) in the presence of IL-7 and Flt3L for 19 days without or with IL-2 on days 10 to 19. The percent of CD3-NK1.1+ cells is indicated. (C) Expression of NK1.1 versus CD3 and CD19 versus B220 by post-5-FU bone marrow MNCs cocultured with OP-9S/LXSN or OP-9S/LMJSN (“J2”) in the presence of IL-7 and Flt3L for 28 days without or with IL-2 on days 20 to 28. The percent of CD3-NK1.1+ cells is indicated. Small numbers of CD3+NK1.1+ cells were present in OP-9S/LMJSN cocultures. They likely represented NKT or cytotoxic lymphocytes that had evolved from pre-existing T cells.
To study the effects of mJagged2 on lymphoid development, light-density (< 1.077 g/mL) MNCs from post-5-FU murine bone marrows were cocultured with pre-established monolayers of OP-9S/LXSN and OP-9S/LMJSN in the presence of murine IL-7 (10 ng/mL) and Flt3L (15 ng/mL). To minimize contribution from mature NK or T cells, we used young C57BL/6 (Ly5.1) mice that were only 4 to 6 weeks old. In the first 6 to 9 days of coculturing, the proliferating cells were mostly small, round lymphoblasts. Around day 9, some racket-shaped cells began to appear in the OP-9S/LMJSN cocultures and to a much lesser extent in the control OP-9S/LXSN cocultures and correlated with the appearance of CD3-NK1.1+ cells. To stimulate further development of such cells, we added human IL-2 (25 ng/mL) to half of the cocultures on day 10. In the next 5 to 9 days, more racket-shaped cells emerged in the IL-2–stimulated OP-9S/LMJSN cocultures. Flow cytometric analyses on day 19 of cocultivation showed that many cells (∼36%) in the IL-2–treated OP-9S/LMJSN cocultures were CD3-NK1.1+ NK cells (Figure 1B). Such cells were rare (∼0.04%) in IL-2–stimulated OP-9S/LXSN cocultures (Figure 1B). Wright-Giemsa staining of sorted CD3-NK1.1+ cells revealed that they were large granular lymphocytes containing azurophilic granules (not shown).
Calculations based on the total cell numbers and the percentages of CD3-NK1.1+ cells showed that the absolute numbers of CD3-NK1.1+ NK cells in the OP-9S/LMJSN cocultures were 160 times greater than in the control OP-9S/LXSN cocultures when IL-2 was added on days 10 to 19. Without IL-2, the absolute numbers of CD3-NK1.1+ NK cells in the OP-9S/LMJSN cocultures were still 13 times greater than those in the negative control group. Taken together, our findings suggest that mJagged2 in combination with OP-9S, IL-7, and Flt3L stimulated the development of NK precursors, which underwent further proliferation and differentiation in response to IL-2.
To examine the effects of delayed IL-2 addition, we repeated the experiment but withheld the addition of IL-2 until day 20 of cocultivation. Flow cytometry was performed on day 28. The results again showed much higher frequencies (∼35%) of CD3-NK1.1+ NK cells in the OP-9S/LMJSN cocultures than the control OP-9S/LXSN cocultures (∼0.22%; Figure 1C). The remaining cells were mostly CD19+B220+ or CD19-B220+ B-lymphoid precursors (Figure 1C, bottom panels).
Jagged2 stimulates NK development from HSCs
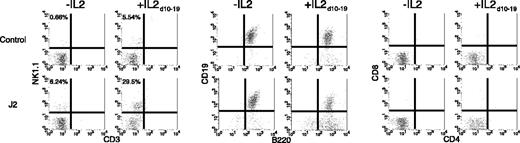
To examine the ability of mJagged2 to induce NK cell development from primitive hematopoietic progenitors, we purified Lin- Sca-1+ c-Kit+ bone marrow progenitors by a combination of magnetic bead depletion of lineage marker–positive cells and fluorescence-activated cell sorting (FACS). The Lin- Sca-1+ c-Kit+ fraction is highly enriched for HSCs.28 Lin- Sca-1+ c-Kit+ cells were cocultured with OP-9S/LXSN or OP-9S/LMJSN in the presence of IL-7 and Flt3L. IL-2 was added to half of the cocultures on day 10. Flow cytometry was performed on day 19 of cocultivation. Again, significantly more CD3-NK1.1+ NK cells developed in OP-9S/LMJSN cocultures (∼30%) than in the control cocultures (∼5%; Figure 2). The absolute numbers of CD3-NK1.1+ NK cells in the OP-9S/LMJSN cocultures were 7.5-fold higher than those in the negative control group. Virtually no CD4+ or CD8+ cells were detected (Figure 2, right panels), indicating that mJagged2 had no stimulatory effect on T-cell development.
Jagged2 induces the development of NK cells from Lin- Sca-1+ c-Kit+ HSCs. Lin- Sca-1+ c-Kit+ HSCs were cocultured with OP-9S/LXSN (negative control) or OP-9S/LMJSN (“J2”) in the presence of IL-7 and Flt3L for 19 days without or with IL-2 on days 10 to 19 and analyzed for the expression of NK1.1 versus CD3, CD19 versus B220, and CD8 versus CD4. The percent of CD3-NK1.1+ cells (left upper quadrant) is indicated.
Jagged2 induces the development of NK cells from Lin- Sca-1+ c-Kit+ HSCs. Lin- Sca-1+ c-Kit+ HSCs were cocultured with OP-9S/LXSN (negative control) or OP-9S/LMJSN (“J2”) in the presence of IL-7 and Flt3L for 19 days without or with IL-2 on days 10 to 19 and analyzed for the expression of NK1.1 versus CD3, CD19 versus B220, and CD8 versus CD4. The percent of CD3-NK1.1+ cells (left upper quadrant) is indicated.
Establishment and characterization of KIL
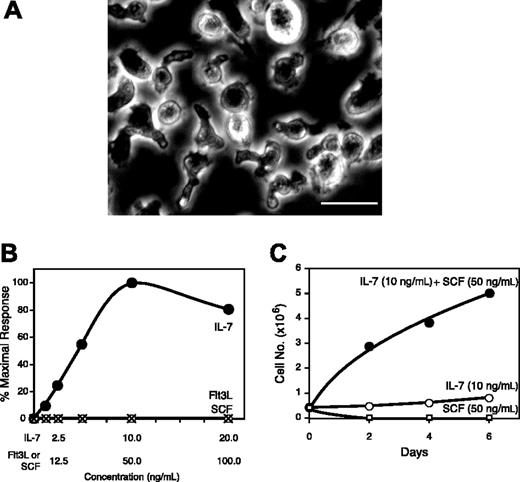
The CD3-NK1.1+ NK cells that developed in the cocultures of post-5-FU bone marrow MNCs and OP-9S/LMJSN exhibited extensive proliferative capacity. They continued to proliferate as long as fresh media, IL-7, and Flt3L were provided every 2 to 4 days. After 2 to 3 months, they became the predominant cell type. In contrast, the control OP-9S/LXSN cocultures remained dominated by CD19+B220+ or CD19-B220+ B-lymphoid precursors. Multiple spontaneously immortalized NK precursor cell lines emerged from the OP-9S/LMJSN (but not OP-9S/LXSN) cocultures after 3 months. The prototype is designated as KIL, for killer lymphocyte. KIL has been in continuous culture for 2 years. Under phase-contrast microscopy, KIL displayed the characteristic racket shape at 37°C (Figure 3A) but a spherical shape at room temperature (not shown), suggesting that maintaining the racket shape requires energy. The survival and proliferation of KIL depends on IL-7. In the absence of IL-7, KIL becomes apoptotic within 24 hours (Figure 3B). KIL cannot survive with SCF or Flt3L alone but SCF synergizes with IL-7 in stimulating the proliferation of KIL (Figure 3B-C). Flow cytometry demonstrates that KIL expresses CD45 (Ly5.1), CD25 (IL-2 receptor α chain), and NK1.1 (NKR P1C receptor), but not CD3, CD4, CD8, CD19 (Figure 4A), B220, TCRαβ, or TCRγδ (not shown). Less than 1% of the KILs express Mac-1 (Figure 4A). Like normal NK cells, the TCRαβ loci of KIL remain in the germ line configuration (Figure 4B-C). On Wright-Giemsa–stained cytospin preparations, KIL appears as large granular lymphocytes with fine azurophilic cytoplasmic granules.
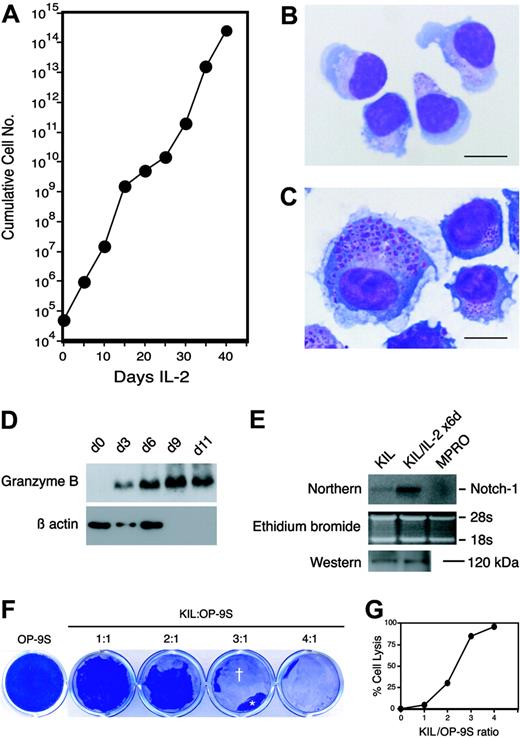
Morphology and growth factor responses of KIL. (A) Phase-contrast microscopy of KIL at 37°C. Most KIL cells display the characteristic racket shape. Bar = 30 μm. (B) Survival of KIL in response to single cytokines. KIL cells were washed with phosphate-buffered saline and exposed to various cytokines (⬡, IL-7; ○, SCF; ×, Flt3L) at the indicated concentrations. Viable cells were counted after 4 days and expressed as percent of the maximal response. Each point represents the mean of triplicates. (C) SCF synergized with IL-7 to stimulate the proliferation of KIL. Each culture was started with 5 × 105 washed KIL cells. Cell numbers were determined on days 2, 4, and 6. ⬡ indicates IL-7 + SCF; ○, IL-7 only; and □, SCF only. Each point represents the mean of triplicates.
Morphology and growth factor responses of KIL. (A) Phase-contrast microscopy of KIL at 37°C. Most KIL cells display the characteristic racket shape. Bar = 30 μm. (B) Survival of KIL in response to single cytokines. KIL cells were washed with phosphate-buffered saline and exposed to various cytokines (⬡, IL-7; ○, SCF; ×, Flt3L) at the indicated concentrations. Viable cells were counted after 4 days and expressed as percent of the maximal response. Each point represents the mean of triplicates. (C) SCF synergized with IL-7 to stimulate the proliferation of KIL. Each culture was started with 5 × 105 washed KIL cells. Cell numbers were determined on days 2, 4, and 6. ⬡ indicates IL-7 + SCF; ○, IL-7 only; and □, SCF only. Each point represents the mean of triplicates.
Phenotypic markers of the KIL cell line. (A) Flow cytometric analysis of the expression of Ly5.1 (CD45), CD3, NK1.1, CD25, CD4, CD8, CD19, and Mac-1. Open curve indicates isotype control. (B) Nested PCR analyses of D-J rearrangements of TCRβ loci. Genomic DNAs of W20 stromal fibroblasts (negative control),12 B6D2F1 spleen cells (positive control), and KIL were first amplified using external primer sets (Dβ1.1ext/Jβ1.7ext or Dβ2.1ext/Jβ2.7ext) and then re-amplified using internal primer sets (Dβ1.1int/Jβ1.7int or Dβ2.1int/Jβ2.7int). Aliquots of secondary PCR reactions were run on agarose gels and stained with ethidium bromide. DNA fragments resulting from amplification of the germline loci are indicated. (C) The same blots in panel B were probed with P32-labeled Dβ1.1-Jβ1.7 or Dβ2.1-Jβ2.7 probes. All DNAs amplified from the germ-line and rearranged D-J loci hybridize specifically to the respective probe.
Phenotypic markers of the KIL cell line. (A) Flow cytometric analysis of the expression of Ly5.1 (CD45), CD3, NK1.1, CD25, CD4, CD8, CD19, and Mac-1. Open curve indicates isotype control. (B) Nested PCR analyses of D-J rearrangements of TCRβ loci. Genomic DNAs of W20 stromal fibroblasts (negative control),12 B6D2F1 spleen cells (positive control), and KIL were first amplified using external primer sets (Dβ1.1ext/Jβ1.7ext or Dβ2.1ext/Jβ2.7ext) and then re-amplified using internal primer sets (Dβ1.1int/Jβ1.7int or Dβ2.1int/Jβ2.7int). Aliquots of secondary PCR reactions were run on agarose gels and stained with ethidium bromide. DNA fragments resulting from amplification of the germline loci are indicated. (C) The same blots in panel B were probed with P32-labeled Dβ1.1-Jβ1.7 or Dβ2.1-Jβ2.7 probes. All DNAs amplified from the germ-line and rearranged D-J loci hybridize specifically to the respective probe.
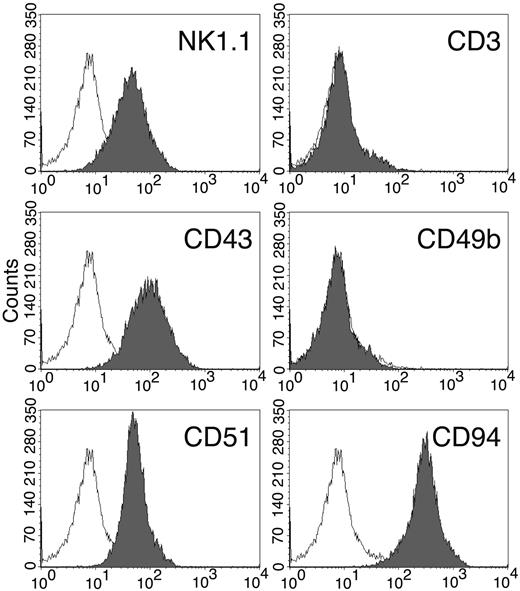
Five clonal lines of KIL, designated KIL C.1-C.5, were obtained by limiting dilution. KIL C.2 was chosen as the prototype because it had a shorter doubling time. KIL C.2 is very similar to the uncloned KIL in every respect examined, including morphology, growth factor responsiveness, cell surface markers, and cytotoxicity. In addition to all surface markers shown in Figure 4A, both KIL C.2 and the parental KIL also express CD43, CD51, and CD94 (Figure 5) and NKG2D (100%; not shown).
Cell surface markers of the KIL C.2 clone. In addition to those shown here, KIL C.2 displays all the phenotypic markers of the parental KIL as shown in Figure 4A. Open curve indicates isotype control.
Cell surface markers of the KIL C.2 clone. In addition to those shown here, KIL C.2 displays all the phenotypic markers of the parental KIL as shown in Figure 4A. Open curve indicates isotype control.
IL-2 induces proliferation and differentiation of KIL
When stimulated by IL-2 alone, KIL (or KIL C.2) proliferated extensively over a period of 5 to 6 weeks, during which the cell number increased by approximately 1010-fold (Figure 6A). Thereafter, proliferation ceased and all terminally differentiated NK cells disintegrated. The culture then became extinct. Serial phase-contrast microscopy revealed that KIL (or KIL C.2) became larger and very adherent to the tissue culture dishes after 4 to 6 days of stimulation with IL-2. Wright-Giemsa staining of KIL (or KIL C.2) that had been stimulated with IL-2 for 6 days showed that they contained numerous large azurophilic granules, cytoplasmic vacuoles, and ruffled cell membranes (Figure 6B-C). Western analysis showed that granzyme B was up-regulated by at least 10-fold in KIL stimulated with IL-2 for 6 to 9 days (Figure 6D). Both KIL and KIL C.2 expressed perforin I by Western analysis (not shown) and Notch-1 by Northern and Western analyses with or without IL-2 stimulation (Figure 6E).
IL-2 induces further proliferation and differentiation of KIL. (A) Growth curve of KIL stimulated with human IL-2 (20 ng/mL) alone. The vertical axis is plotted on a logarithmic scale. Each point represents the mean of triplicates. (B) Wright-Giemsa–stained KIL. Note the fine azurophilic granules in the cytoplasm. Bar = 20 μm. (C) Wright-Giemsa–stained KIL stimulated with IL-2 (20 ng/mL) alone for 6 days. Note the larger cell size, ruffled cell membrane, and prominent azurophilic granules. Bar = 20 μm. (D) Western analysis of the expression of granzyme B by KIL stimulated with IL-2 alone for 0, 3, 6, 9, and 11 days. The 32-kDa granzyme B is indicated. Low-level granzyme B expression was detectable in the day 0 sample on the original film. The same blot was reprobed with an anti–β-actin antibody. (E) Northern and Western analyses of Notch-1 expression in KIL stimulated with IL-2 alone for 0 and 6 days and in the promyelocyte cell line, MPRO (negative control).31 The Northern blot (top) was hybridized to a cDNA probe encoding the intracellular domain of mNotch1. The ethidium bromide–stained gel is shown in the middle row. The bottom row is a Western blot probed with affinity-purified antibodies against the carboxy terminus of mNotch1. The 120-kDa fragment of mNotch1 is indicated. (F) Cytolytic activity of KIL. KIL cells were incubated with monolayers of OP-9S at 1:1 to 4:1 effector-to-target ratios in a 12-well plate in growth medium containing IL-7 and SCF. KIL lysed the OP-9S monolayers in 24 to 48 hours. The OP-9S monolayers were then fixed and stained with Coomassie blue. Lysed or denuded areas (†) of the OP-9S monolayers appear clear whereas the intact areas (*) appear blue. (G) The relationship between the degree of cell lysis and the effector-to-target ratio.
IL-2 induces further proliferation and differentiation of KIL. (A) Growth curve of KIL stimulated with human IL-2 (20 ng/mL) alone. The vertical axis is plotted on a logarithmic scale. Each point represents the mean of triplicates. (B) Wright-Giemsa–stained KIL. Note the fine azurophilic granules in the cytoplasm. Bar = 20 μm. (C) Wright-Giemsa–stained KIL stimulated with IL-2 (20 ng/mL) alone for 6 days. Note the larger cell size, ruffled cell membrane, and prominent azurophilic granules. Bar = 20 μm. (D) Western analysis of the expression of granzyme B by KIL stimulated with IL-2 alone for 0, 3, 6, 9, and 11 days. The 32-kDa granzyme B is indicated. Low-level granzyme B expression was detectable in the day 0 sample on the original film. The same blot was reprobed with an anti–β-actin antibody. (E) Northern and Western analyses of Notch-1 expression in KIL stimulated with IL-2 alone for 0 and 6 days and in the promyelocyte cell line, MPRO (negative control).31 The Northern blot (top) was hybridized to a cDNA probe encoding the intracellular domain of mNotch1. The ethidium bromide–stained gel is shown in the middle row. The bottom row is a Western blot probed with affinity-purified antibodies against the carboxy terminus of mNotch1. The 120-kDa fragment of mNotch1 is indicated. (F) Cytolytic activity of KIL. KIL cells were incubated with monolayers of OP-9S at 1:1 to 4:1 effector-to-target ratios in a 12-well plate in growth medium containing IL-7 and SCF. KIL lysed the OP-9S monolayers in 24 to 48 hours. The OP-9S monolayers were then fixed and stained with Coomassie blue. Lysed or denuded areas (†) of the OP-9S monolayers appear clear whereas the intact areas (*) appear blue. (G) The relationship between the degree of cell lysis and the effector-to-target ratio.
KIL exhibits cytolytic activities
We noticed that the emergence of CD3-NK1.1+ NK cells in the OP-9S/LMJSN cocultures was accompanied by destruction of the OP-9S cells. This temporal linkage suggested that the emerging NK cells were cytotoxic to the OP-9S cells. To examine the cytolytic activity of KIL against OP-9S, we added KIL (or KIL C.2) cells to monolayers of OP-9S fibroblasts at various ratios in growth medium containing IL-7 and SCF. Phase-contrast microscopy revealed the presence of microscopic cytolytic foci in the OP-9S monolayers within 12 hours of the addition of KIL (or KIL C.2). After 24 to 48 hours, large areas of the OP-9S monolayers were denuded (Figure 6F). The degree of cytolysis correlated with effector-to-target cell ratios (Figure 6G). Similar findings were made using NIH3T3 fibroblasts that had been sublethally irradiated (900 rad) to prevent rapid regrowth (not shown). Attempts at the more traditional chromium-51 release assay were complicated by the strong tendency of the activated KIL to adhere to the tissue-culture dishes and their ability to re-uptake chromium-51 released by the lysed target cells during the assay period. Our experience suggests that the cytotoxicity assay described here is more robust and more easily performed than the traditional chromium-51 release assay. It is also less hazardous as no radioisotopes are used. As an added bonus, the fibroblast-based cytotoxicity assay provides a permanent visual record. Regardless of the assay method, the results clearly demonstrate that KIL is cytotoxic.
Discussion
In this report, we provide experimental evidence that Jagged2 has a strong stimulatory effect on the development of NK cells in an environment that otherwise supports B-cell development selectively.20,21,32 Our finding lends further support to the notion that different DSL ligands may have nonredundant roles in lymphopoiesis.33 It also raises the possibility of ex vivo production of nascent NK cells for experimental and therapeutic purposes.
Based on the results of in vitro and in vivo studies, a model of NK cell development has been proposed34,35 in which HSCs give rise to common lymphoid progenitors (CLPs) or common myeloid/lymphoid progenitors (CMLPs), which upon further differentiation give rise to bipotent pT/NK progenitors. The bipotent pT/NK progenitors then give rise to either committed T progenitors (pT) or committed NK progenitors (pNK). While the concept of CLPs or CMLPs is still evolving,36,37 there is substantial evidence supporting the existence of the bipotent pT/NK progenitors.32,38 The differentiation of committed pNK progenitors to mature NK can be further divided into 5 stages based on cell surface markers and functionality.34,35 Stage I is characterized by the expression of CD122 (IL-2/IL-15Rβ common chain) and the absence of most markers of NK cells including NK1.1 (in C57BL/6 mice), CD94, NKG2D, and Ly49. The expression of NK1.1 and the NK receptors CD94/NKG2 and NKG2D marks the beginning of stage II. Stage III is distinguished by the expression of Ly49 (C-type lectin superfamily), c-kit (CD117), and additional markers. Stage IV is characterized by all markers present in stage III plus high levels of α2 integrin (CD49b) and low levels of cytotoxicity and interferon-γ (IFN-γ). Stage IV is also the stage when the developing NK cells undergo major population expansion. Stage V (mature NK) cells express high levels of the integrin Mac-1 α chain (CD11b), CD43 (leukosialin), cytotoxicity, and IFN-γ.
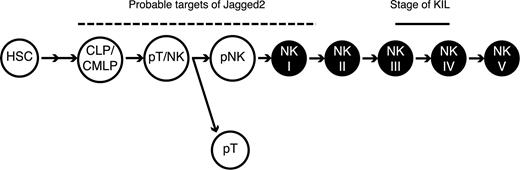
Since the NK cells that developed in the OP-9S/LMJSN cocultures were similar to the KIL cell line in all respects examined, the developmental stage of KIL provides useful clues to the developmental stage of NK cells that appeared in the OP-9S/LMJSN cocultures. Judging from the phenotypic profile of KIL (NK1.1+ CD3- TCRαβ- TCRγδ- CD4- CD8- CD19- CD25+ CD43+ CD45+ CD49b- CD51+ CD94+ NKG2D+ Mac-1-/low B220- c-kit+ perforin I+ granzyme B+ Notch-1+) and the capacity of KIL to undergo extensive proliferation and differentiation in response to IL-2 (Figure 6A), we believe that KIL is the equivalent of stage III/IV pNK. Taking into account the time it took for NK cells to develop in the OP-9S/LMJSN cocultures (∼2 weeks), the presumptive stage of KIL and the fact that Lin- Sca-1+ c-Kit+ HSCs also generate NK cells when cocultured with OP-9S/LMJSN (Figure 2), we believe that the target progenitors of Jagged2 may include stage I NK (CD122+ NK1.1-)/pNK, the bipotent pT/NK, CLPs (or CMLPs), and HSCs (Figure 7).
The potential targets of Jagged2 regulation during NK cell development. The schema depicts the differentiation of hematopoietic stem cell (HSC) to mature NK cell. CLP: common lymphoid progenitor36 ; CMLP: common myeloid/lymphoid progenitor36 ; pT/NK: bipotent progenitor for T and NK; pT: committed T progenitor; pNK: committed NK progenitor; NK I-V: stages of NK development as defined by Yokoyama et al34,35 ; short dash: the equivalent stage of KIL; dotted line: potential targets of mJagged2 regulation.
The potential targets of Jagged2 regulation during NK cell development. The schema depicts the differentiation of hematopoietic stem cell (HSC) to mature NK cell. CLP: common lymphoid progenitor36 ; CMLP: common myeloid/lymphoid progenitor36 ; pT/NK: bipotent progenitor for T and NK; pT: committed T progenitor; pNK: committed NK progenitor; NK I-V: stages of NK development as defined by Yokoyama et al34,35 ; short dash: the equivalent stage of KIL; dotted line: potential targets of mJagged2 regulation.
Northern and Western analyses revealed that KIL expressed significant levels of Notch-1 mRNA and protein with or without IL-2 stimulation (Figure 6E). This raises the possibility that Jagged2 may have simply amplified pre-existing NK1.1+ stage III/IV pNK in the OP-9S/LMJSN cocultures. This scenario is rather unlikely for several reasons. First, the kinetics of the appearance of CD3-NK1.1+ NK cells in the OP-9S/LMJSN cocultures (∼2 weeks) suggests the involvement of more primitive progenitors. Second, mJagged2 also stimulated the development of NK cells from CD3-NK1.1- (double-negative) post-5-FU bone marrow progenitors (not shown) and the highly purified Lin- Sca-1+ c-Kit+ HSCs (Figure 2), both of which were depleted of NK1.1+ progenitors. Third, sorted CD3-NK1.1+ post-5-FU bone marrow MNCs failed to proliferate under the same coculture conditions. Finally, neither sorted Lin- Sca-1+ c-Kit+ HSCs nor CD3-NK1.1- post-5-FU bone marrow progenitors proliferated in response to IL-2 alone (not shown).
We noticed that there was also low-level production of NK cells in the cocultures of Lin- Sca-1+ c-Kit+ HSCs and OP-9S/LXSN (negative control; Figure 2). Cocultures of Lin- Sca-1+ c-Kit+ HSCs and OP-9 cells also exhibited low-level NK production in the cited Dll-1 studies whether Dll-1 was present or not.20,21 Several possible mechanisms may account for the basal-level production of NK cells in these control cocultures, including expression of mJagged2 by OP-9 or OP-9S cells, autocrine or paracrine stimulation of HSCs by endogenous mJagged2, or stimulation by unidentified molecules expressed by OP-9 cells. It is also possible that the generation of committed NK progenitors is a strictly stochastic process requiring no environmental input. Although it was reported that OP-9 stromal fibroblasts expressed mJagged2 based on the results of reverse transcriptase (RT)–PCR,20 we found no detectable levels of mJagged2 mRNA or protein in OP-9S by Northern or Western analyses (Figure 1A). This does not rule out the possibility of very low-level expression. On the other hand, there is evidence that primitive hematopoietic progenitors including Lin- c-kit+ Rhodaminelow Hoechst33342low (ie, long-term repopulating), Lin- c-kit+ RhodamineHigh Hoechst33342low (ie, short-term repopulating), and Lin- c-kit+ (ie, unipotent and multipotent) hematopoietic progenitors express endogenous Jagged2.12 Thus, autocrine or paracrine stimulation of primitive hematopoietic progenitors by endogenous Jagged2 may account for the low-level NK production in the negative-control cocultures in these studies.
A major difference between T and NK cells is that while each T-cell clone expresses only one type of T-cell receptor, each NK cell clone expresses multiple NK receptors (activation, inhibitory, and costimulatory) on the same cell.39 In order to understand the intricate mechanisms controlling the activation or inhibition of NK cells, it may be necessary to know the entire NK receptor repertoires of individual NK clones. However, unlike T or B cells, NK cell clones are extremely difficult to establish.39,40 To our knowledge, the KIL cell line described in this report is perhaps the only murine NK cell line that has preserved most properties of its normal counterpart. As such, KIL provides a valuable model system for NK research.
Prepublished online as Blood First Edition Paper, January 13, 2005; DOI 10.1182/blood-2004-11-4237.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors would like to thank AnneMarie Yang and Jeanne Pierce for superb technical assistance; Namsom Hawk-Oh, Wayne Green, and Scott Perry for assistance with flow cytometry; and Wayne Yokoyama and Gerald Spangrude for monoclonal antibodies and helpful discussions. S.T. is a Research Scholar of the American Cancer Society.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal