Abstract
Both nuclear factor erythroid 2 45 kDa subunit (p45) and BTB and CNC homolog 1 (Bach) transcription factors can form dimers with one of the small Maf proteins, and these heterodimers bind to the musculoaponeurotic fibrosarcoma oncogene (Maf) recognition element (MARE). MARE is known to act as a critical cis-regulatory element of erythroid and megakaryocytic genes. Although detailed analyses of p45-null mutant mice and small maf compound mutant mice revealed that these factors are both critical for platelet production, the functional contributions of Bach1 and the relationship or redundancy between Bach1 and p45 in megakaryocytes remain to be clarified. To address these issues, we generated transgenic lines of mice bearing human BACH1 cDNA under the control of the GATA-1 locus hematopoietic regulatory domain. The transgenic mouse lines showed significant thrombocytopenia associated with impaired maturation of the megakaryocytes, and they developed myelofibrosis. The megakaryocytes in the transgenic mice exhibited reduced proplatelet formation, and the modal ploidy class of megakaryocytes was 2N, indicating the impairment of endomitosis. Transcription of the p45 target genes was down-regulated and we indeed found that BACH1 binds to the thromboxane synthase gene, one of the target genes for p45 in megakaryocytes. These findings thus provide evidence that BACH1 acts as a transcriptional repressor in the regulation of MARE-dependent genes in megakaryocytes.
Introduction
The musculoaponeurotic fibrosarcoma oncogene (Maf) recognition element (MARE) acts as a critical cis-regulatory element for erythroid and megakaryocytic genes, including various globin and heme biosynthetic enzyme genes.1-5 A MARE-specific binding activity was first identified as nuclear factor–erythroid 2 (NF-E2), a heterodimeric protein complex whose subunits consist of nuclear factor erythroid 2 45 kDa subunit (p45) and p18.6,7 p18 is one of the small Maf transcription factors (MafK, MafG, and MafF),7 whereas the p45 subunit is most closely related to the Drosophila nuclear regulatory protein Cap′n′collar (CNC).6,8 Both CNC and small Maf proteins possess a basic region-leucine zipper (bZip) structure, which mediates the DNA-binding and dimerization activities. The MARE can be bound by various sets of transcription factor heterodimers consisting of Maf and CNC family members, including p45, Nrf1, Nrf2, Nrf3, Bach1, and Bach2.9-13
Among the CNC family members, Bach1 and Bach2 function as transcription repressors, whereas the others are transcription activators.12-16 MAREs play critical roles in the enhancer function of the β-globin locus control region (LCR).1 Biochemical and chromatin immunoprecipitation (ChIP) studies have shown that the Bach1/small Maf heterodimer binds to MAREs within the β-globin LCR in murine erythroid leukemia (MEL) cells prior to terminal differentiation.17-19 Following dimethylsulfoxide (DMSO)–induced terminal differentiation of MEL cells, NF-E2 binds to MAREs to activate transcription.18,19 These results suggest that the exchange of p45 for Bach1 as a MafK dimerization partner is a key step in the switch from the repressed to the active state in erythroid lineage cells. This shift is associated with changes in the interaction of MafK with corepressors and coactivators.18 However, studies of CNC, Bach, and small Maf mutant mice have not been able to demonstrate the importance of these factors in globin gene expression. Thus, further studies are necessary.
The functions of CNC and small Maf proteins in megakaryocytes have been well documented by studies of mouse genetics. p45 is expressed in megakaryocytic as well as in erythroid cells.6,20 Homozygous disruption of the mouse p45 gene results in defective megakaryopoiesis and profound thrombocytopenia, leading to postnatal death, whereas erythroid abnormalities are only mild in these mice.21 When cultured in vivo, p45–/– megakaryocytes never develop proplatelets,22 which are considered to be the immediate precursors of blood platelets.23 Recent studies have shown that p45 is essential for the expression of a subset of specific genes in the late stages of megakaryopoiesis. To date, several genes that act downstream of p45 in megakaryocytes have been identified.24-27 ChIP studies have demonstrated recruitment of p45 to the MAREs within the promoters of the p45 targets in a cell line with a megakaryocytic lineage.28 Small maf mutant mice, as well as mafG and mafG::mafK compound mice also showed mild and severe thrombocytopenia, respectively, demonstrating that p45 and small Maf proteins function cooperatively in vivo.29,30 On the other hand, we previously reported that BACH1 is expressed in megakaryocytes as well as in erythrocytes.31 However, no abnormality of platelet counts was observed in bach1-null mutant mice (K.I., unpublished results, 2004). Therefore, the functions of Bach1 and the functional relationships between Bach1 and p45 in these cells remain unclear.
To investigate these relationships, we established transgenic mouse lines bearing the human BACH1 cDNA under control of the GATA-1 gene hematopoietic regulatory domain (G1-HRD), which restricts transgene expression to erythroid and megakaryocytic cells.32 Using this model system, we can evaluate the specific functions of the gene in the 2 lineages of cells. We show here that overexpression of BACH1 in vivo resulted in a significant reduction in the number of platelets in peripheral blood, due to a maturation arrest of megakaryocytes. Thus, BACH1 transgenic mice phenocopy p45-null mutant mice. This suggests that MARE-dependent genes are positively regulated by p45 and negatively regulated by Bach1 in megakaryocytes.
Materials and methods
Generation of transgenic mice
A cDNA fragment containing human BACH1 was cloned into an expression cassette downstream of the IE3.9int GATA-1 promoter. The construct was injected into fertilized BDF-1 eggs using standard procedures as described before.32 The BACH1 transgenic mice were identified by polymerase chain reaction (PCR) methods using primer pairs 5′-TGTCTCACAACCCTTTCTGTCC-3′ and 5′-GAGCTCTGCGTCAGT CTGGCCTAC-3′ to amplify the transgene.
Cell culture
UT-7/EPO33 and UT-7/TPO34 cell lines were maintained in Iscove modified Dulbecco medium (IMDM; Invitrogen Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Invitrogen Life Technologies) and 1 U/mL human recombinant erythropoietin (EPO; Kirin, Tokyo, Japan) or 10 ng/mL human recombinant thrombopoietin (TPO; Kirin), respectively.
ChIP assay
Chromatin fixation and purification procedures were as described previously.17 In brief, 3 × 107 UT-7/EPO or UT-7/TPO cells were fixed by adding formaldehyde to 1% final concentration for 7 minutes at room temperature. Cells were then sonicated to prepare chromatin suspensions containing DNA that was 500 to 1000 base pair (bp) in length. Immunoprecipitations were carried out as described previously.19 For the ChIP assay, an anti-Bach (A1-5) antibody was used.17 Normal mouse immunoglobulin G (IgG) (Santa Cruz Biotechnology, Santa Cruz, CA) was used for negative control experiments. PCRs were carried out by using Ex Taq DNA polymerase (Takara Bio, Otsu, Japan). PCR products were electrophoresed using a Bioanalyzer 2100 and the DNA 1000 LabChip kit (Agilent Technologies, Palo Alto, CA). Primers (5′ to 3′) were as follows: nfe2-1, ATTTTCCACCCCTCCGTCAACC and GTGATCTCAGCTCGTTGCAACCTC; nfe2-3 s, CAGAGTACAAGTCCGTGGTTAC and CAGGCAACCAAGCAAACATCAC; exon 13, ATTCAAAGAAAACCCTAAGTGTGG and AGAACCTCTTATCCTTGCATGC.
Construction of plasmid vectors
A 535-bp fragment corresponding to the 5′-flanking region of the TXAS promoter was amplified by PCR using the following set of primers (sense: 5′-CTACTCGAGTTGGTGGGATGGTTGTCC-3′ and antisense: 5′-GGTAAGCTTCCAGGTGAGACAGGAGGC-3′) and 0.1 μg gDNA extracted from the human myeloid leukemia cell line HEL.24 The PCR products were then subcloned into the Picagene basic vector (Toyo Ink, Tokyo, Japan). The human BACH1 expression vector was prepared by sucbloning the human BACH1 full-length cDNA clone no. 6335 into the mammalian expression vector pcDNA3.1 (Invitrogen Life Technologies). The Bach1-MafK fusion expression plasmids (B1K) were described before.36
Reporter gene assays
For transient transfection assays, quail fibroblast QT6 cells were maintained in Dulbecco modified Eagle medium (DMEM; Nissui, Tokyo, Japan) supplemented with 10% FBS and seeded in 12-well dishes 24 hours before transfection.35 Following replacement of the cell culture medium with Opti-MEM (Invitrogen-Gibco, Carlsbad, CA), cells were transfected in quadruplicate with 200 ng reporter plasmid and 100, 200, or 400 ng of the BACH1 expression plasmid or the B1K. For each transfection, 100 ng pENL, which encodes β-galactosidase, was included as an internal control.
The megakaryoblastic cell line CMK11-5 was maintained in RPMI 1640 medium supplemented with 10% FBS. Cells (3 × 106) were transiently transfected with 2 μg luciferase reporter vector driven by the 5′ upstream region of the TXAS gene, 1 μg pENL β-galactosidase expression vector, and 1 or 2 μg BACH1 or B1K expression plasmid. Electroporation was carried out using Nucleofector (Amaxa, Köln, Germany) according to the supplier's methods. The transfection reagent T and the program T-20 (Amaxa) were used. The cells were harvested 24 hours after transfection, and cell extracts were assayed using the Picagene luciferase assay system (Toyo Ink), following the supplier's protocol. The detection of β-galactosidase was performed using a chemiluminescent reporter-gene assay system according to the manufacturer's recommendation (Tropix, Bedford, MA).
Hematologic analysis
Caudal artery blood was collected from anesthetized mice, and the peripheral blood white blood cell count, hematocrit, hemoglobin concentration, and platelet count were determined using an automatic blood cell analyzer, MEK-6358 (Nihon Koden, Tokyo, Japan).
Histologic analysis
Samples of the femur, spleen, and liver were removed from wild-type littermates and transgenic mice after they were humanely killed. Samples were fixed in 10% buffered formaldehyde solution, embedded in paraffin, and stained with hematoxylin and eosin (HE) and silver impregnation. All stained samples were observed with LCPlanFI 20 ×/0.40 or 40 ×/0.60 objective lenses, and images were captured with a DP70 digital camera system (Olympus, Tokyo, Japan).
Ultrastructural analysis
Bone marrow samples were freshly obtained from transgenic mice and their wild-type littermates and were fixed in 2.5% glutaraldehyde and buffered in 0.1 M sodium cacodylate for 2 hours. Following postfixation with 0.1% osmium tetroxide for 1 hour, the specimens were dehydrated using an ascending series of ethanol solutions and embedded in Epon 812. Ultrathin sections stained with uranyl acetate and lead citrate were observed using a TEM-2000EX electron microscope (JOEL, Tokyo, Japan).
Western blot analysis
Whole tissue extract was prepared from spleens and livers of transgenic mice and their wild-type littermates according to methods described previously.17 Western blot analysis was performed according to standard protocols.35 Tissue extracts were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to Hybond-P membranes (Amersham Bioscience, Tokyo, Japan), and incubated with a 1:2000 dilution of anti-Bach1 rabbit antisera (A1-5)17 for 1 hour at room temperature. Horseradish peroxidase (HRP)–conjugated donkey antirabbit IgG (Amersham Bioscience) was used as a second antibody. HRP activity was detected using the enhanced chemiluminescence (ECL) system (Amersham Bioscience).
Northern blot analysis
Total cellular RNA was isolated by the guanidine thiocyanate-phenol chloroform method.37 Northern blot analysis was performed as described previously.35 Blots containing 10 μg RNA in each sample were hybridized with 32P-labeled human BACH1, murine p45, murine GATA-1, or chicken glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probes.
RT-PCR
Total RNA was prepared from cell lines and megakaryocytes using the Isogen kit according to the manufacturer's recommendations (Nippon Gene, Tokyo, Japan). Total RNA (1 μg) was reverse transcribed using a reverse transcription-PCR (RT-PCR) kit (Clontech, Palo Alto, CA). cDNAs were amplified by the GeneAmp PCR System (Applied Biosystems, Foster, CA) for 25 to 38 cycles using the following conditions: 1 minute at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C. The PCR primers were as follows: human BACH1, AAGGCTGTTCAAGCAATTTGG and CTGATTCAAGATTCTGTATACAGTCAA; human p45, TAGAGATGTACCCAGTGGAGT and TTAAAGTCATCTACCGGCAAG; human GATA-1, CAGGACAGGCCACTACCTATGCAA and CTCTACTACAAGCTACACCAGGTG; human GAPDH, TGAAGGTCGGAGTCAACGGATTTGGT and CATGTGGGCCATGAGGTCCACCAC; human and mouse TXAS, CCTCTGTGTGGGTACTATCTT and TGGGACCATTATGGATGGAAATG24 ; mouse GATA-1, ACTCGTCATACCACTAAGGT and AGTGTCTGTAGGCCTCAGCT38 ; mouse p45, TCAGCAGAACAGGAACAGGT and GCTTTGACACTGGTATAGCT38 ; mouse hypoxanthineguanine phosphoribosyl transferase (HPRT), GCTGGTGAAAAGGACCTCT and CACAGGACTAGAACACCGC38 ; mouse β1-tubulin coding sequence, ATCAGGGAGGAGTACCCGGATCGGA and CCCGGAATATACAAGCCACAGTCAG25 ; β1-tubulin 3′ untranslated region, GCATGATGCTGGATTCTCAAGTCCTGG and CGGTGTTTCTCCGTCCACAGCAAAGT25 ; mouse β5-tubulin, CGCCACGGCCGGTACCTCACAGTT and TCCGAAATCCTCTTCCTCTTCCGC25 ; mouse 3β-hydroxysteroid dehydrogenase (Hsd3b), AGGGCATCTCTGTTGTCATCCAand GGTTGGCTATGGAGAATTTGCC.26
Proplatelet formation assay
Bone marrow cells were collected from femurs and tibias of transgenic mice line 701 and its wild-type littermates. To enrich for megakaryocytes, bone marrow cells were centrifuged in a 50% Percoll/CATCH solution. The intermediate layer was recovered and placed on top of a bovine serum albumin (BSA) density gradient. Cells were collected from the bottom and resuspended in IMDM supplemented with Nutridoma-SP (Boehringer Mannheim, Mannheim, Germany). These enriched megakaryocytes were cultured in 5% CO2 at 37°C for 36 hours. Aliquots of the cells were mounted on slides by cytospin and stained with Giemsa. Acetylcholinesterase activity was detected before counting the megakaryocytes for proplatelet formation (PPF) assay.39
Culture and purification of primary megakaryocytes
Whole livers were recovered from mouse fetuses between embryonic days 13 and 15, and single-cell suspensions were prepared by successive passage through 22- and 25-gauge needles. Fetal liver cells were maintained in DMEM supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 50 U/mL penicillin, 50 μg/mL streptomycin, 0.1 mM nonessential amino acids, and 0.1 μg/mL recombinant human TPO (Kirin) for 5 or 8 days as described previously.40 To enrich for megakaryocytes, we performed the same purification method used for the PPF assay. The purified cell fraction contained more than 90% large acetylcholinesterase-positive cells.
Measurement of megakaryocyte DNA content
DNA ploidy of megakaryocytes in unfractionated bone marrow was determined as previously described.41 Bone marrows were harvested from the femurs of transgenic mice and their wild-type littermates. To label the megakaryocyte fraction, bone marrow cells were incubated with fluorescein isothiocyanate (FITC)–labeled anti–mouse CD41 monoclonal antibody (BD PharMingen, San Jose, CA) and DNA was stained with propidium iodide in hypotonic citrate buffer. Using FACScan flow cytometry (Becton Dickinson, San Jose, CA), the DNA content of CD41+ cells was measured in 2 × 106 nuclear bone marrow cells. For the control study, the isotype of FITC-labeled antibody (BD PharMingen) was used.
Results
Establishment of BACH1 transgenic mouse lines
To study the functional contribution of BACH1 to the differentiation and maintenance of erythroid and megakaryocytic lineages in vivo, we generated transgenic lines of mice expressing human BACH1 cDNA under the control of G1-HRD. Three independent transgenic lines of mice were established, and the expression level of the transgene was monitored by Northern blot analysis. Whereas all lines of transgenic mice expressed the transgenic BACH1 mRNA in the spleen, 2 lines (705 and 706) exhibited higher-level expression than did the remaining line (701; Figure 1A). We surmise that this may be due to the integrated position effect.
Transgenic expression of BACH1 in vivo. (A) Northern blot analysis of BACH1 mRNA using total RNA extracted from the spleens of the wild-type and transgenic mouse littermates shows transgene expression in transgenic mouse lines 701, 705, and 706. (B) Northern blot analysis of BACH1, GATA-1, and p45 mRNAs using total RNA derived from various tissues in the transgenic (Tg) and wild-type littermates. Note that BACH1 mRNA was detected only in the spleens of the transgenic mice, whereas the levels of GATA-1 and p45 expression in the BACH1 transgenic mice were consistent with those of their wild-type littermates. (C) Immunoblotting analysis of BACH1 using tissue extracts from the transgenic mice (705, 706) and their wild-type littermates. Extracts were prepared from the spleens, livers, and QT6 cells that were transfected with BACH1 expression vector. Tissue extracts (20 μg) and 2 μg QT6 protein were separated by 7% SDS-PAGE. After electrophoresis, samples were incubated with anti-Bach1 antisera (A1-5).17 Bound antibody was detected by a peroxidase-conjugated secondary antibody to rabbit immunoglobulin. After stripping off the A1-5 antibody, subsequent immunoblot analysis was performed using an anti–β-tubulin antibody as a control for the amount of protein loaded in each lane.
Transgenic expression of BACH1 in vivo. (A) Northern blot analysis of BACH1 mRNA using total RNA extracted from the spleens of the wild-type and transgenic mouse littermates shows transgene expression in transgenic mouse lines 701, 705, and 706. (B) Northern blot analysis of BACH1, GATA-1, and p45 mRNAs using total RNA derived from various tissues in the transgenic (Tg) and wild-type littermates. Note that BACH1 mRNA was detected only in the spleens of the transgenic mice, whereas the levels of GATA-1 and p45 expression in the BACH1 transgenic mice were consistent with those of their wild-type littermates. (C) Immunoblotting analysis of BACH1 using tissue extracts from the transgenic mice (705, 706) and their wild-type littermates. Extracts were prepared from the spleens, livers, and QT6 cells that were transfected with BACH1 expression vector. Tissue extracts (20 μg) and 2 μg QT6 protein were separated by 7% SDS-PAGE. After electrophoresis, samples were incubated with anti-Bach1 antisera (A1-5).17 Bound antibody was detected by a peroxidase-conjugated secondary antibody to rabbit immunoglobulin. After stripping off the A1-5 antibody, subsequent immunoblot analysis was performed using an anti–β-tubulin antibody as a control for the amount of protein loaded in each lane.
In accordance with our expectations, the expression profile of exogenous BACH1 mRNA was remarkably confined to the spleen (Figure 1B). To examine the expression of exogenous BACH1 protein, we carried out immunoblot analysis using antisera against Bach1.17 We found that BACH1 was abundantly expressed in the spleens of the transgenic mice (Figure 1C). In contrast, signals from endogenous Bach1 protein in the wild-type littermate mouse spleen were very faint. Either exogenous or endogenous Bach1 protein was not detectable in the liver. Taken together with the results of Northern blot analysis (Figure 1A-B), these results showed that exogenous BACH1 was expressed in the transgenic mice at a higher level compared with that of endogenous Bach1. We also examined the expression of GATA-1 and p45 mRNAs in the BACH1 transgenic mouse by Northern blot analysis. The expression levels of GATA-1 and p45 mRNAs in the BACH1 transgenic mice were similar to those of their wild-type littermates, suggesting that overexpression of Bach1 does not affect these factors, which are important in both erythroid and megakaryocytic cells (Figure 1B).
Thrombocytopenia in the BACH1 transgenic mice
To investigate whether or not transgenic expression of BACH1 disturbs normal hematopoiesis, we analyzed peripheral blood samples of the transgenic mice. Hematopoietic indices for the transgenic and wild-type littermates are shown in Figure 2. White blood cell counts and red cell indices of the transgenic mice were indistinguishable from those of wild-type mice (Figure 2A-C), and the morphology of the red cells, granulocytes, and lymphocytes in the peripheral blood was within the normal range in the transgenic mice (data not shown). In contrast, we found significant thrombocytopenia in the 2 transgenic mouse lines (705 and 706) that expressed BACH1 at a high level, whereas thrombocytopenia was mild in the line 701 mice that expressed a lower level of BACH1 (Figure 2D). These results thus demonstrate that the levels of transgene-derived BACH1 mRNA correlate well with the severity of thrombocytopenia. We next examined the change of platelet counts of the transgenic mice in the course of aging and found there was no significant difference in the platelet counts (Figure 2E).
Peripheral blood indices from transgenic mice and their wild-type littermates. (A) White blood cell (WBC) counts, (B) hemoglobin (Hb) levels, (C) red blood cell (RBC) counts, and (D) platelet (PLT) counts of the transgenic mice (•) and their wild-type littermates (○) in lines 701, 705, and 706 are shown. A satistically significant difference between wild-type and transgenic mice was found only in the platelet counts; *P < .01, **P < .001. Ages of the wild-type littermates and transgenic mice ranged from 1.30 to 50.40 weeks (n = 38; mean = 13.89; SD = 8.60) and 1.30 to 60.9 weeks (n = 62; mean = 15.65; SD = 10.47), respectively. (E) Platelet counts of the transgenic mice in 3 groups containing similar age ranges are shown. The ages of groups I, II, and III ranged from 6 to 12 weeks, 12 to 24 weeks, and 24 to 92 weeks, respectively (n = 126).
Peripheral blood indices from transgenic mice and their wild-type littermates. (A) White blood cell (WBC) counts, (B) hemoglobin (Hb) levels, (C) red blood cell (RBC) counts, and (D) platelet (PLT) counts of the transgenic mice (•) and their wild-type littermates (○) in lines 701, 705, and 706 are shown. A satistically significant difference between wild-type and transgenic mice was found only in the platelet counts; *P < .01, **P < .001. Ages of the wild-type littermates and transgenic mice ranged from 1.30 to 50.40 weeks (n = 38; mean = 13.89; SD = 8.60) and 1.30 to 60.9 weeks (n = 62; mean = 15.65; SD = 10.47), respectively. (E) Platelet counts of the transgenic mice in 3 groups containing similar age ranges are shown. The ages of groups I, II, and III ranged from 6 to 12 weeks, 12 to 24 weeks, and 24 to 92 weeks, respectively (n = 126).
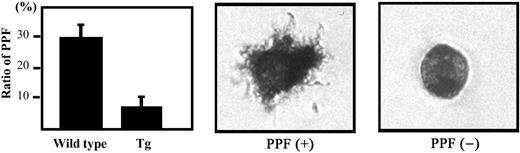
It has been suggested that PPF represents a terminal event in the course of platelet production.23 Indeed, PPF was decreased in the germline p45 and in small maf mutant mice that had thrombocytopenia.21,29,30 We therefore examined whether transgenic expression of BACH1 also affects PPF. Our results indicate that PPF was reduced about 30% in the BACH1 transgenic mice compared with their wild-type littermates (Figure 3). We also examined the frequency of megakaryocytic progenitors in the spleen by colony formation analysis, but we could not detect a significant difference in megakaryocyte colony-forming units (CFU-Megs) between the BACH1 transgenic mice and their wild-type littermates (data not shown). These results indicate that transgenic expression of BACH1 suppressed the terminal maturation of megakaryocytes.
PPF in BACH1 transgenic mice and their wild-type littermates. The megakaryocyte fractions (enriched by a BSA density gradient) were incubated in IMDM supplemented with 1 × Nutridoma-SP. After 20 hours of incubation, acetylcholine esterase activity was detected before counting the megakaryocytes to determine their PPF. The number of acetylcholine esterase-positive cells displaying the obvious filamentous cell projections was counted and the PPF ratio (number of megakaryocytes with PPF–total megakaryocytes) was calculated. (Left) The mean ± SD of PPF ratios from 3 independent experiments are shown. Typical proplatelet-producing (middle) and -nonproducing (right) megakaryocytes are shown. Cell images were obtained using an Olympus IX71 inverted microscope equipped with an LCPlan FI 40 ×/0.60 objective lens and an Olympus SC35 camera (Olympus).
PPF in BACH1 transgenic mice and their wild-type littermates. The megakaryocyte fractions (enriched by a BSA density gradient) were incubated in IMDM supplemented with 1 × Nutridoma-SP. After 20 hours of incubation, acetylcholine esterase activity was detected before counting the megakaryocytes to determine their PPF. The number of acetylcholine esterase-positive cells displaying the obvious filamentous cell projections was counted and the PPF ratio (number of megakaryocytes with PPF–total megakaryocytes) was calculated. (Left) The mean ± SD of PPF ratios from 3 independent experiments are shown. Typical proplatelet-producing (middle) and -nonproducing (right) megakaryocytes are shown. Cell images were obtained using an Olympus IX71 inverted microscope equipped with an LCPlan FI 40 ×/0.60 objective lens and an Olympus SC35 camera (Olympus).
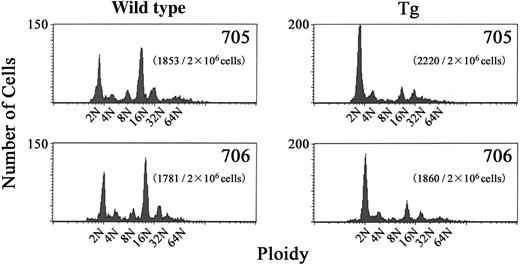
During the maturation process, megakaryocytes acquire a high DNA ploidy by endomitosis.42 To determine whether the process of megakaryocyte development was affected in BACH1 transgenic mice, we performed DNA ploidy analysis on their bone marrow cells. The results revealed that CD41+ megakaryocytes from the transgenic mice showed a significantly lower level of polyploidy than those derived from their wild-type littermates, whereas there were no significant differences in the number of bone marrow megakaryocytes between BACH1 transgenic mice and controls (data not shown). The modal ploidy class of megakaryocytes from transgenic mice was 2N, compared with 16N in their wild-type littermates (Figure 4), indicating that endomitosis was impaired in the transgenic mouse megakaryocytes.
DNA contents of CD41+ bone marrow cells in BACH1 transgenic mice and their wild-type littermates. Unfractionated bone marrow cells were labeled by FITC-conjugated anti-CD41 antibody and DNA content was stained with propidium iodide. Two-color flow cytometry was used to detect the DNA content in megakaryocytes. A typical histogram showing DNA content averaged over 3 experiments is shown. The transgenic lines and the number of CD41+ cells among 2 × 106 nucleated cells that were analyzed are indicated inside each graph.
DNA contents of CD41+ bone marrow cells in BACH1 transgenic mice and their wild-type littermates. Unfractionated bone marrow cells were labeled by FITC-conjugated anti-CD41 antibody and DNA content was stained with propidium iodide. Two-color flow cytometry was used to detect the DNA content in megakaryocytes. A typical histogram showing DNA content averaged over 3 experiments is shown. The transgenic lines and the number of CD41+ cells among 2 × 106 nucleated cells that were analyzed are indicated inside each graph.
Electron-microscopic analysis of megakaryocytes from BACH1 transgenic mice
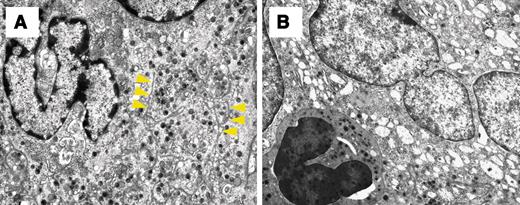
To further characterize the morphologic features of megakaryocytes in the transgenic mice, we performed electron-microscopic analysis. In contrast to megakaryocytes in wild-type mice, which exhibited well-formed platelet territories with the development of demarcation membranes, megakaryocytes from BACH1 transgenic mice showed serious impairment of the platelet field formation and lacked demarcation membranes. Megakaryocytes from the transgenic mice had a lower degree of nuclear complexity and different heterochromatin contents, compared to those from the wild-type littermates. Moreover, this ultrastructural analysis of megakaryocytes from the transgenic mice revealed the vacuolation of mitochondria and a deficit in the number of α granules (Figure 5B). Emperioporesis was also detected in many megakaryocytes from the transgenic mice (Figure 5B). Taken together, these results further support our contention that thrombocytopenia is caused primarily by a failure of megakaryocyte maturation and platelet development.
Ultrastructural examination of megakaryocytes derived from BACH1 transgenic mice. Panels show electron micrographs of megakaryocytes derived from the bone marrow of a wild-type littermate (A; original magnification × 4000) and a transgenic mouse (B; original magnification × 4000). Megakaryocytes derived from the transgenic mouse exhibited insufficient demarcation membrane formation and had a decreased number of cytoplasmic α granules. Yellow arrowheads indicate the demarcation membrane in megakaryocytes from a wild-type littermate.
Ultrastructural examination of megakaryocytes derived from BACH1 transgenic mice. Panels show electron micrographs of megakaryocytes derived from the bone marrow of a wild-type littermate (A; original magnification × 4000) and a transgenic mouse (B; original magnification × 4000). Megakaryocytes derived from the transgenic mouse exhibited insufficient demarcation membrane formation and had a decreased number of cytoplasmic α granules. Yellow arrowheads indicate the demarcation membrane in megakaryocytes from a wild-type littermate.
Histologic analysis of hematopoietic tissues in BACH1 transgenic mice
We next carried out histologic analyses of hematopoietic tissues from the BACH1 transgenic mice. An important observation was that a reduction of the femoral cavity was found on microscopic examination, and this was perhaps due to increased osteogenesis in the transgenic mice (Figure 6B). In addition, consistent with the electron microscopic analysis (see “Electron microscopic analysis of megakaryocytes from BACH1 transgenic mice”), we observed emperipolesis of granulocytes in megakaryocytes, specifically in the transgenic mice (Figure 6D).
Histologic examination of transgenic mouse bone marrow, spleen, and liver. The right column (A,C,E,G,I) and the left column (B,D,F,H,J) represent histologic examination of wild-type littermates and BACH1 transgenic mice, respectively. The photomicrographs in panels A-D show HE staining of bone marrow specimens. Yellow arrowhead in panel D indicates emperipolesis of granulocytes that was specifically observed in megakaryocytes from transgenic mice. (E-F) Silver impregnation staining of bone marrow. (G-H) Silver impregnation staining of spleen. Aggressive fibrosis was recognized in both tissues derived from transgenic mice. (I-J) HE staining of liver sections. Erythroid islets (white arrowhead in panel J) and megakaryocytes (white arrow in panel J) were observed in the sinusoids of the livers derived from transgenic mice. Representative staining sections were prepared using 4-month-old transgenic mice of line 705 and their corresponding wild-type littermates. Original magnifications × 200 (A-B,E-J) and × 400 (C-D).
Histologic examination of transgenic mouse bone marrow, spleen, and liver. The right column (A,C,E,G,I) and the left column (B,D,F,H,J) represent histologic examination of wild-type littermates and BACH1 transgenic mice, respectively. The photomicrographs in panels A-D show HE staining of bone marrow specimens. Yellow arrowhead in panel D indicates emperipolesis of granulocytes that was specifically observed in megakaryocytes from transgenic mice. (E-F) Silver impregnation staining of bone marrow. (G-H) Silver impregnation staining of spleen. Aggressive fibrosis was recognized in both tissues derived from transgenic mice. (I-J) HE staining of liver sections. Erythroid islets (white arrowhead in panel J) and megakaryocytes (white arrow in panel J) were observed in the sinusoids of the livers derived from transgenic mice. Representative staining sections were prepared using 4-month-old transgenic mice of line 705 and their corresponding wild-type littermates. Original magnifications × 200 (A-B,E-J) and × 400 (C-D).
It should also be noted that reticulin fibrosis in the bone marrow of the transgenic mice was markedly identifiable by silver impregnation (Figure 6F). Reticulin fibrosis was also detected in the spleen, but not in the liver, of the transgenic mice by silver impregnation. It was clearly demonstrated that reticulin fibers in the spleen reside in the marginal areas of megakaryocytes (Figure 6H). HE staining of spleen and liver sections revealed extramedullary hematopoiesis in the transgenic mice. Megakaryocytes and erythroid islets were detected in the sinusoid of the transgenic mouse livers (Figure 6J), whereas no hematopoietic foci were found in liver sections obtained from age-matched wild-type mice (Figure 6I).
Expression of p45 target genes in megakaryocytes of BACH1 transgenic mice
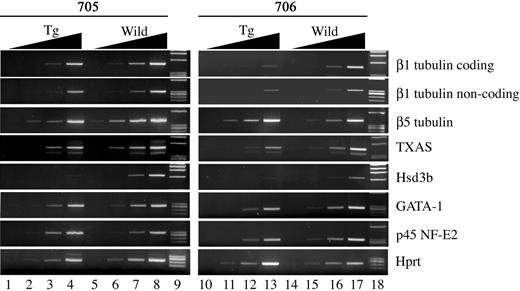
Because the phenotype of the BACH1 transgenic mice resembled that of the p45-deficient mice, it was thought that transgenic expression of BACH1 might repress the transcriptional activation mediated by p45. Therefore, to address this point, we examined the expression of the p45 NF-E2 target genes in the primary megakaryocytes from fetal liver cells of the BACH1 transgenic mice. RT-PCR analyses showed that expression of the hematopoietic-specific β1-tubulin, TXAS, and the 3β-hydroxy-steroid dehydrogenase (Hsd3b) genes was significantly down-regulated in the megakaryocytes from BACH1 transgenic mice (Figure 7). However, one of the β-tubulin isoforms, β5-tubulin, which was reported not to be affected in p45-deficient megakaryocytes,25 was expressed equally in BACH1 transgenic and control megakaryocytes. The expression of GATA-1 and p45 was marginally lower in BACH1 transgenic megakaryocytes than in control mice. These results thus support our contention that the expression of p45 target genes is specifically down-regulated in the BACH1 transgenic megakaryocytes.
Semiquantitative RT-PCR analysis of p45 target genes in megakaryocytes derived from BACH1 transgenic mice. Expression of β1-tubulin, β5-tubulin, TXAS, Hsd3b, GATA-1, p45, and Hprt was analyzed by RT-PCR using RNA extracted from cultured megakaryocytes derived from fetal liver cells of wild-type mice and their transgenic littermates. To ensure linearity of PCRs, 10-fold serial dilutions of template cDNA were used. Typical photographs from 3 independent experiments are shown.
Semiquantitative RT-PCR analysis of p45 target genes in megakaryocytes derived from BACH1 transgenic mice. Expression of β1-tubulin, β5-tubulin, TXAS, Hsd3b, GATA-1, p45, and Hprt was analyzed by RT-PCR using RNA extracted from cultured megakaryocytes derived from fetal liver cells of wild-type mice and their transgenic littermates. To ensure linearity of PCRs, 10-fold serial dilutions of template cDNA were used. Typical photographs from 3 independent experiments are shown.
In vivo binding of BACH1 to the TXAS gene enhancer
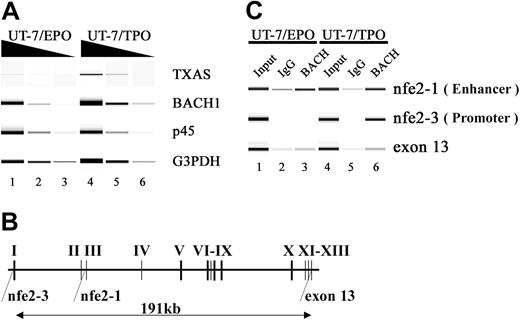
The TXAS gene is a well-known MARE-dependent gene containing functional MAREs in its promoter and in the second intron (Figure 8B). Therefore, to ask whether BACH1 actually binds to MARE in the megakaryocytic genes, we performed ChIP analysis with a BACH1-specific antibody. Because it is technically not feasible to use megakaryocytes from mouse bone marrow for this assay, we looked for a cultured megakaryocytic cell line. A megakaryocytic subline of UT-7 cells, UT-7/TPO,34 which has mature megakaryocytic features, seemed to be suitable for this analysis, along with the erythroid subline UT-7/EPO.33
Recognition of MAREs located in the TXAS gene by the BACH1 factor. (A) RT-PCR analysis of TXAS, BACH1, and p45 mRNA in UT-7/EPO and UT-7/TPO cells. The TXAS gene was expressed in UT-7/TPO, whereas the TXAS gene was detected only marginally in UT-7/EPO. Input RNA levels were normalized using G3PDH transcripts as controls. To ensure linearity of the PCRs, 10-fold serial dilutions of template cDNA were used. PCR products were electrophoresed using Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) and DNA1000 LabChip (Agilent Technologies) kits. (B) Schematic representation of the human TXAS gene. The regions amplified by the ChIP assay were shown to be nfe2-1, nfe2-3, and exon 13. The nfe2-1 and nfe2-3 sites contain the MARE, which has been implicated in TXAS gene expression. (C) ChIP analysis using an anti-Bach antibody (A1-5) in UT-7/EPO and UT-7/TPO. Gel images show PCR products corresponding to nfe2-1, nfe2-3, and exon 13, obtained using input (lanes 1 and 4) and precipitated chromatin as templates (lanes 2, 3, 5, and 6). The exon 13 template DNA was used as a control. Fragments containing MARE located in the second intron (nfe2-1) or in their proximal promoter (nfe2-3) were precipitated by an anti-Bach1 antibody in UT-7/EPO (lane 3) or in UT-7/TPO (lane 6). Normal mouse IgG was used as a control (lanes 2 and 5). These results represent 4 independent experiments.
Recognition of MAREs located in the TXAS gene by the BACH1 factor. (A) RT-PCR analysis of TXAS, BACH1, and p45 mRNA in UT-7/EPO and UT-7/TPO cells. The TXAS gene was expressed in UT-7/TPO, whereas the TXAS gene was detected only marginally in UT-7/EPO. Input RNA levels were normalized using G3PDH transcripts as controls. To ensure linearity of the PCRs, 10-fold serial dilutions of template cDNA were used. PCR products were electrophoresed using Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) and DNA1000 LabChip (Agilent Technologies) kits. (B) Schematic representation of the human TXAS gene. The regions amplified by the ChIP assay were shown to be nfe2-1, nfe2-3, and exon 13. The nfe2-1 and nfe2-3 sites contain the MARE, which has been implicated in TXAS gene expression. (C) ChIP analysis using an anti-Bach antibody (A1-5) in UT-7/EPO and UT-7/TPO. Gel images show PCR products corresponding to nfe2-1, nfe2-3, and exon 13, obtained using input (lanes 1 and 4) and precipitated chromatin as templates (lanes 2, 3, 5, and 6). The exon 13 template DNA was used as a control. Fragments containing MARE located in the second intron (nfe2-1) or in their proximal promoter (nfe2-3) were precipitated by an anti-Bach1 antibody in UT-7/EPO (lane 3) or in UT-7/TPO (lane 6). Normal mouse IgG was used as a control (lanes 2 and 5). These results represent 4 independent experiments.
We first confirmed that both BACH1 and TXAS genes are expressed in the UT-7/TPO subline (Figure 8A). A ChIP assay with UT-7/TPO cells demonstrated that BACH1 bound to the promoter and enhancer regions in vivo. In contrast, BACH1 was not enriched at the promoter region in UT-7/EPO cells, whereas it did bind to the enhancer (Figure 8C). To determine the specificity of these binding interactions, we looked for the exon 13 region in chromatin immunoprecipitates, which is approximately 190 kb downstream from the promoter. No significant enrichment of the exon 13 region was detected by ChIP analysis, confirming the specificity of this assay.
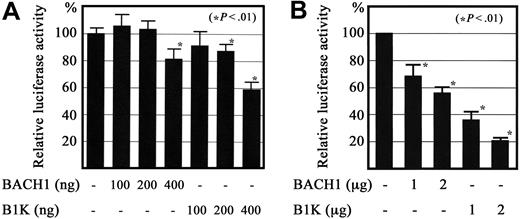
Previous reports showed that Bach1 can repress reporter gene expression when the reporter gene contains MARE in the promoter proximal region in transfect. We asked whether expression of the Bach1 protein repressed the reporter gene through the TXAS promoter. For this purpose, we prepared a reporter plasmid containing the TXAS promoter fused to the luciferase gene, and transfected the reporter with the expression vectors for BACH1 or Bach1-MafK fusion protein (B1K) into QT6 quail fibroblasts35 and into the megakaryoblast cell line CMK11-5. As expected, cotransfection with BACH1 or B1K expression plasmids repressed the reporter gene activity driven by the TXAS promoter in a dose-dependent manner (Figure 9). The transcriptional repression was more significant for B1K transfects than for the BACH1 transfects. Taken together, these results suggest that BACH1 binds to MARE in its regulatory region and acts as a repressor in megakaryocytes.
Regulation of TXAS gene promoter activity by BACH1. A luciferase reporter construct containing a 535-bp fragment that included the transcription initiation site for the TXAS gene was transiently cotransfected with BACH1 or the Bach1-MafK chimeric protein expression plasmid (B1K) into QT6 fibroblasts by Lipofectamin (A), as well as the megakaryoblast cell line CMK11-5, by Nucleofector (B). The respective relative luciferase activity (mean ± SD of 4 independent experiments in QT-6, and 3 in CMK11-5) is shown. Each activity was standardized by reference to the β-galactosidase activity from the cotransfected expression plasmid, which was driven by the elongation factor 1 promoter.
Regulation of TXAS gene promoter activity by BACH1. A luciferase reporter construct containing a 535-bp fragment that included the transcription initiation site for the TXAS gene was transiently cotransfected with BACH1 or the Bach1-MafK chimeric protein expression plasmid (B1K) into QT6 fibroblasts by Lipofectamin (A), as well as the megakaryoblast cell line CMK11-5, by Nucleofector (B). The respective relative luciferase activity (mean ± SD of 4 independent experiments in QT-6, and 3 in CMK11-5) is shown. Each activity was standardized by reference to the β-galactosidase activity from the cotransfected expression plasmid, which was driven by the elongation factor 1 promoter.
Discussion
We found in this study that transgenic expression of BACH1 in megakaryocytes in vivo results in thrombocytopenia. This is most likely due to maturation arrest of megakaryocytic cells and resulting dysfunctions in platelet formation. Transgenic BACH1 gives rise to these deleterious effects through heterodimerizing with one of the small Maf proteins. This heterodimer binds to MARE, and it has been well demonstrated that the small Maf-p45 heterodimer binds to MARE and activates gene expression.6,7 Thus, this study further supports our contention that cis-acting MARE acts as a critical regulatory element for megakaryocytic genes during megakaryocyte differentiation.29,30,39 Importantly, the phenotype of BACH1 transgenic mice resembles that of the p45-null mice; p45-null mice showed profound thrombocytopenia associated with increased production of abnormal megakaryocytes.21,22 Megakaryocytes lacking p45 show polyploidy, which is characterized by a large cytoplasm containing abundant but disorganized demarcation membrane and a reduced number of platelet-specific granules. Furthermore, megakaryocytes from p45 knockout mice and BACH1 transgenic mice both showed emperipolesis. These similarities in the megakaryocytic phenotype suggest the presence of common or overlapping mechanisms underlying the impaired megakaryopoiesis observed in these mutant mice. However, in contrast to p45-null mice, the number of megakaryocytes was not increased in BACH1 transgenic mice, and BACH1-overexpressing megakaryocytes did not complete endomitosis. These differences may indicate the disparity of the target genes of these transcription factors.
Transgenic mouse lines expressing small Maf proteins under the control of G1-HRD have been reported, and these lines of mice showed a reduction of PPF activity.39 Because small Maf proteins form unproductive homodimers by themselves, we surmise that transgenic small Maf proteins block small Maf-p45 heterodimer formation or binding. Similarly, it is plausible that BACH1 blocks the small Maf-p45 heterodimer function when BACH1 is overexpressed in a megakaryocytic lineage. Consistent with this notion, the expression of 3 p45 target genes was significantly repressed in the BACH1-expressing megakaryocytes. Although the physiologic function of BACH1 in megakaryocytes remains to be clarified, our results suggest that the dynamic exchange of BACH1-p45 heterodimer formation at MARE may also occur during differentiation of hematopoietic cells toward the megakaryocytic lineage. In this regard, it should be noted that a Bach1 gene-targeting experiment was recently carried out.43 The Bach1-null mutants are fertile and within a normal size range. Platelet counts in their peripheral blood are also within the normal range (K.I., unpublished results, 2004). To further clarify the roles of Bach1 in mice, therefore, it will be necessary to analyze thrombopoiesis in knockout mutant mice in which thrombocytopenia has been induced by irradiation or chemotherapy. We are now planning such a study.
In contrast to p45-null mutant mice, the BACH1 transgenic mice did not develop anemia. Recently, we demonstrated that heme inhibits the DNA-binding activity of the Bach1-MafK heterodimer by directly interacting with Bach1 through a dipeptide motif of cysteine and proline.44 Because erythroid cells synthesize large amounts of heme for hemoglobin, one plausible explanation for the absence of significant changes in the erythroid phenotype of the BACH1 transgenic mice is that there may be interference with Bach1 binding to its target sequence by heme that accumulates inside the erythroid cells.
One of the most striking phenotypes in the BACH1 transgenic mice is the development of myelofibrosis. A number of reports suggest that alterations in megakaryopoiesis, with accumulation of abnormal megakaryocytes may be essential for the development of myelofibrosis. GATA-1low mice, which are a genetically modified mouse line lacking upstream regions of the GATA-1 gene, show profound thrombocytopenia with dysregulated megakaryocyte proliferation and impaired cytoplasmic maturation of megakaryocytes.45-47 The spleens of GATA-1low mice showed an increase in the number of megakaryocytes as compared to littermate controls, and these megakaryocytes were smaller than wild-type megakaryocytes. They contained scant cytoplasm, condensed nuclei, and a disorganized demarcation membrane.
Recently, GATA-1low mice were reported to develop a disease resembling human idiopathic myelofibrosis (IM).48 IM is a myeloproliferative disorder of clonal origin that is characterized by fibrotic degeneration of the bone marrow and extramedullary hematopoiesis in the spleen and liver.49 IM cases also show the presence of teardrop poikilocytes, collagen fibers in the bone marrow and spleen, and hematopoietic foci in the liver. However, in the GATA-1low mice, early IM traits such as the presence of reticulin fibers in bone marrow and spleen were detected only after 10 months of age, and the disease progressed slowly to a frank IM after 15 months. In contrast, in the BACH1 transgenic mice the reticulin fibrosis could be detected as early as 5 weeks of age, but we did not observe progression to IM in these transgenic mice (data not shown). Although the numbers of megakaryocytes did not increase, megakaryocytes in the BACH1 transgenic mice showed abnormal morphology and function, suggesting that some alteration in regulatory pathways might occur in the BACH1-expressing megakaryocytes, and this may lead in turn to myelofibrosis.
In the GATA-1low mice, induced expression of several growth factors is implicated in the development of myelofibrosis. Growth factors, such as osteocalcin, transforming growth factor-β, platelet-derived growth factor, and endothelial growth factor were increased in the bone marrow of GATA-1low mice, but the expression of these cytokines within the GATA-1–deficient megakaryocytes was not affected.48 Further analysis of cytokine production in the BACH1 transgenic mice may help to reveal the mechanism of fibrosis.
The human BACH1 gene is localized to chromosome 21q22.150,51 within the Down syndrome–associated gene region.52 Down syndrome, which is caused by trisomy 21, is one of the most frequent human chromosomal abnormalities. Children with Down syndrome have an approximately 20-fold higher incidence of leukemia than unaffected children.53 The majority of leukemia cases associated with Down syndrome are acute megakaryocytic leukemia (AMKL).54 Recently, we and others have demonstrated that mutagenesis of the GATA1 gene is a very early event in the development of Down syndrome–related AMKL (DS-AMKL), during the process of multistep leukemogenesis. Acquired mutations of the GATA-1 gene have been detected in almost all cases in DS-AMKL and transient myeloproliferative disorder (TMD), a “preleukemia” that may be present in as many as 10% of newborn infants with Down syndrome.55-60 We envisage that mutations in GATA1 may not be the first event because the GATA1 gene is located on the X chromosome.
To further understand the mechanisms of a lineage-specific malignant conversion, it is very important to identify the gene or genes in chromosome 21 that cooperate with the GATA1 mutant during leukemogenesis. Although the BACH1 transgenic mice did not develop leukemia per se, the present data show that terminal differentiation of the megakaryocytes, as well as platelet production, were markedly impaired by the overexpression of BACH1. These results support our contention that BACH1 is one of the candidates for genes that cooperate with the mutated GATA1 at a very early stage of leukemogenesis in Down syndrome. In this regard, experiments are now under way to characterize a cross between the BACH1 transgenic mice described here and GATA1 gene knockdown mice rescued with an N-terminal domain-defective mutant of GATA1.61 It will be intriguing to further investigate the mechanisms of leukemogenesis in Down syndrome.
In summary, through the analysis of BACH1 transgenic mice, we have shown that overexpression of BACH1 in vivo resulted in significant thrombocytopenia, with down-regulation of the p45 target genes in megakaryocytic lineage cells, thus phenocopying the p45-null mutant mice. Together with the findings from ChIP assays showing that BACH1 bound to one of the p45 targets, that is, the TXAS gene, in vivo, the present results suggest that BACH1 plays an important role in transcriptional regulation through MARE in megakaryocytic lineage cells.
Prepublished online as Blood First Edition Paper, December 21, 2004; DOI 10.1182/blood-2004-07-2826.
Supported in part by Grants-in-Aid for Scientific Research, Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Technology, and grants from the Uehara Memorial Foundation, JST-ERATO, Naito Foundation, and Atherosclerosis Foundation. F.K. is a Japan Society for the Promotion of Science (JSPS) postdoctoral fellow.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Ms C. Tsushima for technical assistance.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal