Abstract
Type 1 diabetes is a systemic autoimmune disease that can be cured by transplantation of hematopoietic stem cells (HSCs) from disease-resistant donors. Nonobese diabetic (NOD) mice have a number of features that distinguish them as bone marrow transplant recipients that must be understood prior to the clinical application of chimerism to induce tolerance. In the present studies, we characterized NOD HSCs, comparing their engraftment characteristics to HSCs from disease-resistant strains. Strikingly, NOD HSCs are significantly enhanced in engraftment potential compared with HSCs from disease-resistant donors. Unlike HSCs from disease-resistant strains, they do not require graft-facilitating cells to engraft in allogeneic recipients. Additionally, they exhibit a competitive advantage when coadministered with increasing numbers of syngeneic HSCs, produce significantly more spleen colony-forming units (CFU-Ss) in vivo in allogeneic recipients, and more granulocyte macrophage–colony-forming units (CFU-GMs) in vitro compared with HSCs from disease-resistant controls. NOD HSCs also exhibit significantly enhanced chemotaxis to a stromal cell–derived factor 1 (SDF-1) gradient and adhere significantly better on primary stroma. This enhanced engraftment potential maps to the insulin-dependent diabetes locus 9 (Idd9) locus, and as such the tumor necrosis factor (TNF) receptor family as well as ski/sno genes may be involved in the mechanism underlying the autonomy of NOD HSCs. These findings may have important implications to understand the evolution of autoimmune disease and impact on potential strategies for cure.
Introduction
Nonobese diabetic (NOD) mice spontaneously develop autoimmune diabetes and share more than 20 genetic loci that confer diabetes susceptibility with humans with type I diabetes, including insulin-dependent diabetes locus 1 (Idd1) that maps to the major histocompatibility complex (MHC).1-4 NOD mice exhibit multiple hematopoietic defects including impaired myeloid differentiation and function, decreased antigen presenting cell function,5-8 lack of a hemolytic complement system, and impaired natural killer (NK) and NK/T-cell function.5,7-11 In both humans and NOD mice, bone marrow transplantation (BMT) induces disease prevention and reverses the underlying autoimmunity.12-15 BMT has therefore been suggested as a potential approach to treat type 1 diabetes.13,14,16 Ikehara et al14 were the first to show that diabetes progression is eliminated in NOD mice through BMT. Li et al12 subsequently demonstrated that mixed chimerism not only eliminates diabetes progression, but also reverses active autoimmunity. These studies indicate that the defect that leads to autoimmunity in NOD mice is closely linked to the hematopoietic stem cells (HSCs). It is therefore important to characterize the function of HSCs from NOD mice in developing novel therapeutic strategies to treat diabetes.
NOD mice have a number of features that distinguish them as recipients of BMT due to the underlying autoimmune process.12,15,17 They require significantly more cells and higher levels of conditioning to establish allogeneic engraftment compared with wild-type mice.12 In spite of these barriers, and in contrast with disease-resistant mouse strains, once allogeneic engraftment is achieved in NOD mice, the donor cells exhibit a competitive advantage over the endogenous NOD bone marrow cells (BMCs).12,17 We therefore postulated that the hematopoietic environment might somehow be dysregulated by the autoimmunity.
In the present study, we compared the function of NOD HSCs to HSCs from other strains using purified c-Kit+/Sca-1+/lin- (KSL) cells. Unlike KSL cells from disease-resistant mouse strains, purified KSL cells from NOD mice engraft more readily in allogeneic recipients. They exhibit a competitive advantage over KSL cells from donors syngeneic to the recipient in a dose-dependent fashion. This autonomy is not due to increased numbers of HSCs or progenitors, which are present at comparable levels in the KSL population to those of diabetes-resistant mice. As with engraftment in vivo, NOD KSL cells are more capable of forming day-12 splenic colonies representing multilineage progenitor cells in allogeneic recipients compared with B10.BR HSCs. Unlike normal BMCs, NOD BMCs are less dependent on MHC-matching between donor and recipient. Finally, NOD HSCs migrate significantly more efficiently to a stromal cell–derived factor 1 (SDF-1) gradient, exhibit enhanced adhesion on primary stroma, and form higher numbers of granulocyte macrophage–colony-forming units (CFU-GMs) in vitro. Notably, this autonomy maps to the insulin-dependent diabetes locus 9 (Idd9). Taken together, these data indicate that HSCs from NOD mice are more efficient and are capable of withstanding the immune assault of the allogeneic microenvironment and reconstitute ablated recipients independent of MHC disparity. These findings may have important implications in the design of HSC-based therapies to treat diabetes and other autoimmune diseases.
Materials and methods
Animals
Female 4- to 6-week-old NOD (H-2g7) mice (Taconic Laboratories; Germantown, NY) and B10.BR/SgSn (B10.BR: H-2k), C3H/HeJ (H-2k), C57BL/10Sn (B10; H-2b), B10.D2/NSn (B10.D2; H-2d), and nonobese resistant (NOR) (H-2g7) mice (Jackson Laboratory; Bar Harbor, ME) were used. NOD.B10 congenic breeding pairs were purchased from Taconic. Animals were housed in a barrier animal facility at the Institute for Cellular Therapeutics (Louisville, KY) and cared for according to National Institutes of Health animal care guidelines.
Antibodies
All monoclonal antibodies (mAbs) used in this study were purchased from BDPharmingen (San Diego, CA). KSL cell (c-Kit+/Sca-1+/Lin-) sorting experiments used the following mAbs: stem cell antigen-1 (Sca-1) phycoerythrin (PE) (E13-161.7); c-Kit allophycocyanin (2B8); and the lineage negative (Lin-) panel consisting of: CD8α fluorescein isothiocyanate (FITC) (53-6.7); CD11b FITC (M1/70); CD45R FITC (RA3-6B2); Gr-1 FITC (11-26c.2a); β-TCR FITC (H57-597); γδ-TCR FITC (GL3). Sorting for facilitating cells (FCs) used anti-β-TCR FITC, γδ-TCR FITC and CD8α PE. For the assessment of chimerism, H-2Kd FITC (SF1-1.1), H-2Kk PE (AF3-12.1), and H-2Kb PE (AF6-88.5) mAbs were used as previously described.18
Cell sorting
KSL cells and FCs were sorted from bone marrow as previously described.18 A single cell suspension of BMCs was diluted to a concentration of 100 × 106 cells/mL in sterile cell sorting media (CSM) containing 1 × Hanks balanced salt solution without phenol red (GIBCO, Grand Island, NY), 2% heat-inactivated fetal bovine serum (FBS; GIBCO), 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) buffer (GIBCO), and 30 μg/mL gentamicin (GIBCO). BMCs were incubated with saturating concentrations of directly labeled mAbs for 30 minutes on ice, washed twice in CSM, and resuspended to 2.5 × 106 cells/mL in CSM and sorted using the FACSVantage (Becton Dickinson, Mountain View, CA).
Chimera preparation
Recipient mice were conditioned with 950 cGy total body irradiation (TBI) from a Cesium source (Nordion, Ontario, ON, Canada) and reconstituted with 10 000 KSL cells with or without 30 000 FCs by lateral tail vein injection at least 6 hours after irradiation. The FCs and HSCs were mixed prior to injection.
Cobblestone area–forming cell (CAFC) assay
Confluent monolayers of the stromal cell line FBMD-1 were established in 96-well tissue-culture plates coated with 0.1% gelatin (Sigma). Purified KSL cells were then seeded on the monolayers at 2-fold serial dilutions in Iscove modified Dulbecco medium (IMDM; GIBCO) supplemented with 20% horse serum (GIBCO), 10-4 M 2-ME (Sigma), 10-5 M hydrocortisone (Sigma), 100 U/mL penicillin, and 100 μg/mL streptomycin (GIBCO) between 500 and 31 cells/well (33°C, 5% CO2). Wells were screened for cobblestone area formation using reverse microscopy at days 14, 21, 28, and 35. Frequencies of CAFCs were determined for each time point using the L-Calc Limiting Dilution Analysis software, version 1.1 (Stem Cell Technologies, Vancouver, BC, Canada) where the frequency equals 1 divided by the initial number of cells that yield 37% negative wells.
Spleen colony-forming unit (CFU-S)
BMCs were resuspended to 2 × 105 cells/mL in media 199 (GIBCO) containing 50 μg/mL gentamycin and 0.5 mL (1 × 105 cells total) administered intravenously to conditioned (950 cGy TBI) NOD or B10.BR mice. After 12 days, the mice were euthanized and the spleens fixed in Bouin Fixative (Polysciences). The macroscopic colonies formed on the surface of the spleens from the recipient animals were counted at least 24 hours later.
Migration to SDF-1
BMCs were resuspended in RPMI with 0.5% bovine serum albumin (BSA) and equilibrated for 10 minutes at 37°C.19 Prewarmed serum-free medium containing either no chemoattractant or SDF-1 (200 ng/mL) was added to the lower chambers of a Costar Transwell 24-well plate (Costar Corning, Cambridge, MA) with 6.5-mm diameter and 5 μm pore filter. Aliquots of the cell suspension (1 × 105 cells/100 μL) were loaded onto the upper chambers. The cultures were incubated (37°C, 95% humidity, 5% CO2) for 3 hours, after which the cells in the lower chamber were harvested. In some experiments, BMCs recovered from the lower chambers were plated in methylcellulose (Methocult; Stem Cell Technologies) cultures supplemented with artifical serum (prepared by M.Z.R.) and G-CSF. CFU-GM colonies were evaluated at 7 days.
Adhesion assay
Confluent monolayers of bone marrow stroma cells were established in 24-well plates and grown in IMDM supplemented with 12.5% horse serum and 12.5% FBS.20 Purified Sca-1+ cells isolated from NOD or NOR mice were seeded on the stroma cell monolayers for 1 or 6 hours. Cells were harvested by trypsin digestion, washed, and resuspended in methylcellulose supplemented with murine IL-3 and GM-CSF (Stem Cell Technologies). On day 7, the number of CFU-GM colonies was scored using an inverted microscope (Olympus America, Melville, NY).
Modified competitive reconstitution assay
Previous reconstitution assays have used genetic differences between the 2 competing donor cell types to track the engraftment of cells into recipients that were either syngeneic or congenic at MHC to both donor cell types.21-23 In the present assay, the number of NOD KSL cells was kept constant at 10 000 and increasing numbers of competing syngeneic HSC (B10.BR) were added, ranging from 1000 to 10 000. Inocula were transplanted into B10.BR recipients conditioned with 950 cGy.
Adhesion molecule analysis
NOD and NOR BMCs were purified by sorting as described. Sorted cells were stained with Sca1 PE, c-Kit APC, and the FITC lineage panel plus one of the following mAbs: CD106 (VCAM-1), CD102 (ICAM-2); CD62L (L-selectin); CD62P (P-selectin); CD11a (integrin α-chain); CD184 (CXCR4, 2B11 clone); CD49d (integrin β-chain); CD44 (Pgp-1); CD54 (ICAM-1), or the appropriate isotype control FITC (Pharmingen/BD Bio-Science). Cells were analyzed for 4 colors on FACSCalibur using the CellQuest program.
Statistical analysis
Survival comparisons were via Kaplan-Meier analysis using the SPSS 12.0 Windows statistical software package. P values shown were obtained through log-rank comparisons.
Results
NOD KSL cells have decreased Sca-1 expression
We first evaluated the KSL population in bone marrow of NOD mice. Sca-1+ cells from NOD comprise a similar percentage of the c-Kit+/Lin- cells compared with B10.BR HSCs. However, the mean fluorescence intensity (MFI) of Sca-1 is significantly decreased on NOD KSL cells (MFI = 206 vs 76, respectively; Figure 1A). NOD KSL cells are capable of engraftment as reflected by establishing long-term radioprotection in syngeneic recipients, with all blood cell lineages present, indicating that these are indeed functional HSCs (data not shown).
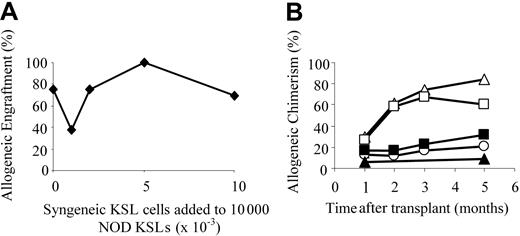
NOD HSCs have lower cell-surface expression of Sca-1 and exhibit autonomy in vivo. (A) Bone marrow cells from NOD and B10.BR mice were harvested and stained for KSL cells. Marker 1 (M1) indicates all Sca-1 positive-staining cells. MFI of the Sca-1 staining in the c-Kit+/Lin- population is indicated. Results are from one purification sort and are representative of at least 20 individual sorts per strain. (B) Engraftment B10.BR recipients conditioned with 950 cGy TBI and given 10 000 KSL cells from KSL (□), C57BL/10 (⬡), or B10.D2 (○) mice. B10.BR mice receiving no KSL cells were used as irradiation controls (dotted line). Survival B10.BR recipients of 10 000 NOD KSL cells and 30 000 NOD FCs are also shown (▪). (C) Survival of C57BL/10 recipients given NOD KSL cells alone (□) or in the presence of NOD FCs (▪). (D) Kinetics of percent donor chimerism in the peripheral blood lymphocytes (PBLs) in B10.BR recipients of NOD KSL cells alone (□) or in the presence of NOD FCs (▪). (E) B10.BR recipients were given decreasing numbers of NOD HSCs (dotted line indicates none; ○, 2000 cells; ▵, 5000;  , 7500; and □, 10 000) after 950 cGy TBI. Survival of the recipients is shown based on time after transplantation. P values versus 10 000 HSCs are less than .05 and all P values versus 0 HSCs are less than .05. n.a. indicates not applicable.
, 7500; and □, 10 000) after 950 cGy TBI. Survival of the recipients is shown based on time after transplantation. P values versus 10 000 HSCs are less than .05 and all P values versus 0 HSCs are less than .05. n.a. indicates not applicable.
NOD HSCs have lower cell-surface expression of Sca-1 and exhibit autonomy in vivo. (A) Bone marrow cells from NOD and B10.BR mice were harvested and stained for KSL cells. Marker 1 (M1) indicates all Sca-1 positive-staining cells. MFI of the Sca-1 staining in the c-Kit+/Lin- population is indicated. Results are from one purification sort and are representative of at least 20 individual sorts per strain. (B) Engraftment B10.BR recipients conditioned with 950 cGy TBI and given 10 000 KSL cells from KSL (□), C57BL/10 (⬡), or B10.D2 (○) mice. B10.BR mice receiving no KSL cells were used as irradiation controls (dotted line). Survival B10.BR recipients of 10 000 NOD KSL cells and 30 000 NOD FCs are also shown (▪). (C) Survival of C57BL/10 recipients given NOD KSL cells alone (□) or in the presence of NOD FCs (▪). (D) Kinetics of percent donor chimerism in the peripheral blood lymphocytes (PBLs) in B10.BR recipients of NOD KSL cells alone (□) or in the presence of NOD FCs (▪). (E) B10.BR recipients were given decreasing numbers of NOD HSCs (dotted line indicates none; ○, 2000 cells; ▵, 5000;  , 7500; and □, 10 000) after 950 cGy TBI. Survival of the recipients is shown based on time after transplantation. P values versus 10 000 HSCs are less than .05 and all P values versus 0 HSCs are less than .05. n.a. indicates not applicable.
, 7500; and □, 10 000) after 950 cGy TBI. Survival of the recipients is shown based on time after transplantation. P values versus 10 000 HSCs are less than .05 and all P values versus 0 HSCs are less than .05. n.a. indicates not applicable.
NOD HSCs exhibit enhanced engraftment in allogeneic recipients
Purified HSCs from normal mouse strains face a greater barrier to engraftment in allogeneic recipients compared with whole BMCs.24,25 We therefore assessed the efficiency of NOD HSC engraftment in allogeneic B10.BR recipients conditioned with 950 cGy TBI and that received a transplant of 10 000 NOD KSL cells. The genetics of each mouse strain used are listed in Table 1. Surprisingly, NOD KSL cells exhibited significantly enhanced engraftment potential compared with KSL cells from B10 or B10.D2 mouse strains (P < .001; Figure 1B). B10 and B10.D2 mice are disparate with B10.BR recipients across the entire MHC (Table 1). B10.D2 mice are congenic to NOD mice at H-2K but disparate at the other MHC loci (Table 1). Because we previously determined that matching at MHC H-2K between HSC donor and recipient enhances HSC engraftment,18 we also evaluated engraftment of NOD KSL cells in B10 recipients (Figure 1C). Enhanced engraftment of NOD HSCs was also observed in these MHC-disparate recipients.
Haplotype pedigree of mice used in transplant experiments
. | H-2 complex . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse strain . | K . | I-Aβ . | I-Aα . | I-Eβ . | I-Eα . | D . | |||||
| B10.BR | k | k | k | k | k | k | |||||
| C3H | k | k | k | k | k | k | |||||
| C57BL/10 | b | b | b | b | null | b | |||||
| B10.D2 | d | d | d | d | d | d | |||||
| NOD and NOD.B10 congenics | d | g7 | d | d | null | b | |||||
| NOR | d | g7 | d | d | null | b | |||||
. | H-2 complex . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse strain . | K . | I-Aβ . | I-Aα . | I-Eβ . | I-Eα . | D . | |||||
| B10.BR | k | k | k | k | k | k | |||||
| C3H | k | k | k | k | k | k | |||||
| C57BL/10 | b | b | b | b | null | b | |||||
| B10.D2 | d | d | d | d | d | d | |||||
| NOD and NOD.B10 congenics | d | g7 | d | d | null | b | |||||
| NOR | d | g7 | d | d | null | b | |||||
The addition of CD8+/TCR- graft FCs was not required for durable engraftment of NOD HSCs into either B10.BR or B10 recipients (Figure 1B-C), indicating that NOD HSCs engraft in allogeneic recipients without the need for accessory cells. Engraftment of the NOD KSL cells differed depending on recipient strain (Figure 1B-C). Survival of B10.BR (H-2k) recipients was significantly impaired when FCs were coadministered with KSL cells compared with KSL cells alone, whereas B10 recipients exhibited similar long-term survival. Notably, B10 mice are congenic to NOD mice at H-2D, whereas B10.BR mice are fully MHC disparate (Table 1). The percentage donor chimerism in the NOD KSL cell recipients was initially mixed (40%-50%), but increased with time after transplantation to nearly 100% donor by 5 months (Figure 1D).
To further characterize the enhanced engraftment ability of NOD KSL cells, B10.BR recipients received transplants of decreasing numbers of NOD KSL cells. Durability of engraftment was directly correlated with the dose of NOD KSL cells administered (R2 = 0.9: Figure 1E). Interestingly, the administration of less than 10 000 KSL cells did not result in durable engraftment (P < .05 compared with 10 000 HSCs; Figure 1E).
NOD KSL cells exhibit a competitive advantage in the presence of syngeneic KSL cells
The efficiency of engraftment of NOD KSL cells in allogeneic recipients was compared with that of syngeneic KSL cells using a modified competitive repopulation assay.21-23 B10.BR mice were conditioned and received transplants of 10 000 NOD KSL cells plus increasing numbers of B10.BR KSL cells until the syngeneic/allogeneic KSL cell ratio reached 1:1 (Figure 2). NOD KSL cells were capable of engrafting in allogeneic B10.BR recipients in the presence of increasing numbers of B10.BR KSL cells (Figure 2A). As the ratio of B10.BR KSL cells to NOD KSL cells increased, engraftment remained relatively consistent. Seventy-five percent (9 of 12) of the B10.BR recipients of 10 000 NOD KSL cells alone engrafted whereas 70% (9 of 13) of those given 10 000 B10.BR KSL cells plus 10 000 NOD KSL cells engrafted. Although the percent allogeneic chimerism was similar in all groups one month after transplantation, it increased more with time in the recipients of NOD KSL cells either alone or in the presence of only 1000 B10.BR KSL cells (Figure 2B). The average percent allogeneic chimerism in recipients of NOD KSL cells alone or with 1000 B10.BR KSL cells increased from 29.9% ± 25.3% or 27.1% ± 17.4% at one month after transplantation to 60.9% ± 29.1% or 58.7% ± 3.8%, respectively, 2 months after transplantation. For recipients of KSL cells at a ratio of 1:1 the percentage NOD chimerism remained stable at approximately 20%. These data illustrate that NOD KSL cells exhibit significantly enhanced engraftment efficiency even in the presence of equal numbers of competing KSL cells from the recipient strain.
NOD HSCs engraft in B10.BR recipients in the presence of equal numbers of B10.BR HSCs. The ability of NOD HSCs to engraft in allogeneic B10.BR recipients was assessed using a modified competitive reconstitution assay.21-23 (A) Percent engraftment of B10.BR mice given 10 000 NOD KSL cells in the presence of increasing numbers of B10.BR KSL cells. Groups shown are: no B10.BR KSL cells (n = 12); 1000 B10.BR KSL cells (n = 8); 2000 B10.BR KSL cells (n = 4); 5000 B10.BR KSL cells (n = 3); and 10 000 B10.BR KSL cells (n = 13). (B) The kinetics of allogeneic chimerism in the engrafting B10.BR mice are shown based on the number of B10.BR KSL cells coadministered with the 10 000 NOD KSL cells (▵ represents none; □, 1 × 103; ○, 2 × 103; ▴, 5 × 103; and ▪, 10 × 103).
NOD HSCs engraft in B10.BR recipients in the presence of equal numbers of B10.BR HSCs. The ability of NOD HSCs to engraft in allogeneic B10.BR recipients was assessed using a modified competitive reconstitution assay.21-23 (A) Percent engraftment of B10.BR mice given 10 000 NOD KSL cells in the presence of increasing numbers of B10.BR KSL cells. Groups shown are: no B10.BR KSL cells (n = 12); 1000 B10.BR KSL cells (n = 8); 2000 B10.BR KSL cells (n = 4); 5000 B10.BR KSL cells (n = 3); and 10 000 B10.BR KSL cells (n = 13). (B) The kinetics of allogeneic chimerism in the engrafting B10.BR mice are shown based on the number of B10.BR KSL cells coadministered with the 10 000 NOD KSL cells (▵ represents none; □, 1 × 103; ○, 2 × 103; ▴, 5 × 103; and ▪, 10 × 103).
The KSL population from NOD mice is not enriched in long-term repopulating cells
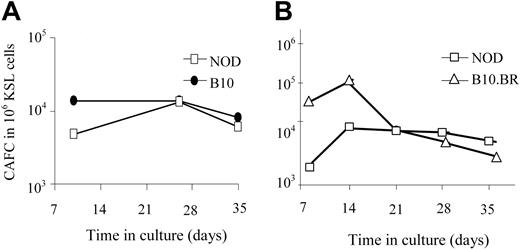
One explanation for the enhanced engraftment capability of NOD KSL cells could be an increase in their long-term repopulating ability compared with other mouse strains. The number of HSCs and hematopoietic progenitor cells in the NOD KSL cell population was therefore compared with B10.BR and B10 KSL cells using the CAFC assay. The CAFC assay uses limiting dilution analysis to quantify the frequency of hematopoietic progenitor cells. The time at which a given cell produces a cobblestone area correlates with the maturation state of the cell when it was placed in culture.26 Later-appearing cobblestone areas (days 28-35) correlate with the more primitive HSCs, whereas CAFC on days 14 to 21 enumerate the early hematopoietic progenitor cells (eHPCs), which are more committed than HSCs but retain self-renewing capacity and multilineage differentiation potential.27,28 When purified NOD KSL cells were compared with KSL cells from disease-resistant mouse strains, there was no significant difference in the frequency of CAFCs obtained (Figure 3), indicating no difference in the frequency of the more primitive subpopulations of HSCs. Therefore, the autonomous behavior exhibited by NOD HSCs is not due to an increase in the frequency of either the primitive hematopoietic cells or eHPCs in the populations transplanted into recipient mice.
The frequency of the primitive HSCs in the purified KSL populations does not differ in the NOD HSCs compared with HSCs from either B10 or B10.BR mice. CAFC assays were performed in parallel on KSL cells from NOD mice (□) and either C57BL/10 (A, ⬡) or B10.BR (B, ▵) mice. Results are from one representative experiment of 3 (A) and 5 (B) performed. Results are represented as frequency of CAFC/106 purified KSL cells.
The frequency of the primitive HSCs in the purified KSL populations does not differ in the NOD HSCs compared with HSCs from either B10 or B10.BR mice. CAFC assays were performed in parallel on KSL cells from NOD mice (□) and either C57BL/10 (A, ⬡) or B10.BR (B, ▵) mice. Results are from one representative experiment of 3 (A) and 5 (B) performed. Results are represented as frequency of CAFC/106 purified KSL cells.
Recently, Orschell-Traycoff et al29 demonstrated that long-term HSCs could be separated from short-term progenitor cells based on the expression of the adhesion molecules CD49d and CD49e. Long-term engrafting HSCs are CD49e+/CD49ddim, whereas short-term progenitor cells are CD49edim/CD49dbright. The average percent CD49e+/CD49ddim population in sorted KSL cells from B10, B10.BR, and NOD strains was similar (Table 2), confirming that there is no significant increase in the primitive HSC population.
Long-term repopulating cells as assessed by CD49e and CD49d adhesion marker expression
Mouse strain . | CD49e+/CD49ddim . | P* . |
|---|---|---|
| NOD | 67.90 ± 6.98 | NA |
| B10.BR | 77.73 ± 0.78 | > .05 (NS) |
| C57BL/10 | 74.95 ± 2.19 | > .05 (NS) |
Mouse strain . | CD49e+/CD49ddim . | P* . |
|---|---|---|
| NOD | 67.90 ± 6.98 | NA |
| B10.BR | 77.73 ± 0.78 | > .05 (NS) |
| C57BL/10 | 74.95 ± 2.19 | > .05 (NS) |
KSL cells from NOD, B10.BR, and B10 mice were analyzed for CD49e and CD49d expression. Results are percent of KSL population plus or minus standard deviation of 3 replicates.
NA indicates not applicable; NS, not significant.
t test versus NOD
BMCs from NOD mice have an increased ability to form day-12 CFU-S colonies in allogeneic recipients
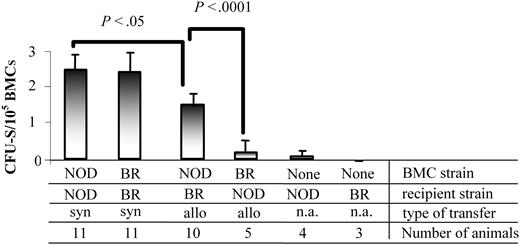
The CFU-S assay has long been used to characterize multilineage progenitor cells in vivo.30 The colonies observed on day 12 are indicative of HSCs or eHPCs, and their formation is dependent on MHC matching.31 We next determined the ability of NOD BMCs to produce CFU-S colonies in allogeneic (B10.BR) or syngeneic (NOD) recipients. As expected, when NOD BMCs were administered into syngeneic NOD recipients, significantly more day-12 CFU-Ss were obtained than when they were transplanted to allogeneic B10.BR recipients (P < .05; Figure 4). B10.BR recipients of syngeneic B10.BR BMCs had a similar number of CFU-Ss as the NOD syngeneic transfers. The number of CFU-Ss was significantly decreased when B10.BR BMCs were given to allogeneic NOD recipients (P < .0001). Although NOD BMCs produced fewer CFU-Ss in allogeneic recipients compared with syngeneic recipients, the number produced was significantly increased compared with B10.BR BMCs in allogeneic transplants (P < .0001). These data indicate that NOD hematopoietic progenitor cells are able to colonize allogeneic recipients at a significantly enhanced rate compared with other mouse strains, representing a significant competitive advantage and enhanced engraftment capability.
NOD BMCs form CFU-S colonies in both allogeneic and syngeneic recipients. BMCs from NOD or B10.BR mice were isolated and administered to either NOD or B10.BR (BR) recipient mice 6 hours after receiving 950 cGy TBI. Day-12 splenic colonies were enumerated. Data are the mean and standard deviation from 3 separate experiments. P values were determined using a 2-tailed Student t test with the assumption of equal variance. n.a. indicates not applicable.
NOD BMCs form CFU-S colonies in both allogeneic and syngeneic recipients. BMCs from NOD or B10.BR mice were isolated and administered to either NOD or B10.BR (BR) recipient mice 6 hours after receiving 950 cGy TBI. Day-12 splenic colonies were enumerated. Data are the mean and standard deviation from 3 separate experiments. P values were determined using a 2-tailed Student t test with the assumption of equal variance. n.a. indicates not applicable.
NOD KSL cells are more resistant to ionizing radiation than KSL cells from nonautoimmune mouse strains
Geiger et al32 demonstrated that normal HSC survival is linked to various DNA repair and cell cycle regulation genes on mouse chromosomes 2, 7, and 11. Strain-specific changes in the number of HSCs during aging were linked to these genetic loci. Interestingly, these markers and others are located in close proximity to several Idd genes.3,32,33 Because NOD mice are more resistant to ionizing radiation than other strains of mice,12 we evaluated radiation-sensitivity of NOD KSL cells.
Whole bone marrow from NOD, B10, and B10.BR mice was exposed to increasing amounts of ionizing radiation and cultured for 20 hours. Survival of the KSL cells was compared with those cultured without radiation exposure. KSL cells from NOD mice survive better than B10.BR BMCs between 650 cGy and 950 cGy (Figure 5A). No significant difference in BMC survival between NOD and B10 KSL cells was detected (data not shown).
NOD HSCs are more radioresistant. The ability of HSCs to survive after exposure to ionizing radiation was determined by exposing whole BMCs (A) or purified KSL cells (B) to the indicated doses of irradiation. (A) The percent KSL cells surviving after a 20-hour culture was determined by flow cytometry. ○ indicates NOD; ▪, B10.BR. Results are the average survival plus and minus the standard deviation of cells from 3 individual animals in one of 2 different experiments (*P < .05). (B) The ability of irradiated KSL cells to produce colonies in methylcellulose plus growth factors was determined. Data expressed as plus and minus the standard deviation (*P < .05). Results are the average of quadruplicate wells from one of 2 (NOR [□] and B10 [⬡]) or 3 (NOD [⋄] and B10.BR [▴]) experiments, expressed as percent of unirradiated KSL cells.
NOD HSCs are more radioresistant. The ability of HSCs to survive after exposure to ionizing radiation was determined by exposing whole BMCs (A) or purified KSL cells (B) to the indicated doses of irradiation. (A) The percent KSL cells surviving after a 20-hour culture was determined by flow cytometry. ○ indicates NOD; ▪, B10.BR. Results are the average survival plus and minus the standard deviation of cells from 3 individual animals in one of 2 different experiments (*P < .05). (B) The ability of irradiated KSL cells to produce colonies in methylcellulose plus growth factors was determined. Data expressed as plus and minus the standard deviation (*P < .05). Results are the average of quadruplicate wells from one of 2 (NOR [□] and B10 [⬡]) or 3 (NOD [⋄] and B10.BR [▴]) experiments, expressed as percent of unirradiated KSL cells.
To further analyze resistance to radiation, KSL cells were sorted from NOD, B10.BR, and B10 mice as well as from NOD congenic nonobese resistant (NOR) mice that share 88% of the NOD genome. The KSL cells were exposed to increasing amounts of ionizing radiation and their function was evaluated in CFC assays. NOD KSL cell populations formed more colonies in methylcellulose than the KSL cells from B10.BR or B10 mice at 200 cGy and more than B10 KSL cells when treated with 350 cGy (Figure 5B). NOR KSL cells formed colonies at a similar rate as NOD KSL cells, withstanding the effects of ionizing irradiation better than those obtained from other strains.
NOD BMCs exhibit enhanced migration in response to SDF-1
Because migration of HSCs is critical for engraftment, we assessed NOD BMCs in an in vitro migration assay. SDF-1 is the only chemokine known to mediate HSC migration.34 HSCs express CXCR4, the receptor for SDF-1.34 The response of NOD BMCs to an SDF-1 gradient was compared with that for diabetes-resistant NOR controls in vitro. Notably, chemotaxis of total BMCs was significantly enhanced (over 2-fold; P < .0001) for NOD BMCs in migration to an SDF-1 gradient, compared with NOR BMCs (Figure 6A). Moreover, when the migrated cells were subsequently placed in a clonogenic assay, there was a significant (P = .001) increase in the number of CFU-GM colonies observed in the cultures initiated by NOD mice SDF-1–migrating cells from NOD mice as compared with SDF-1–responsive cells from NOR animals (Figure 6B).
NOD HSCs exhibit enhanced migration to SDF-1 and adhesion to primary stroma. (A) Chemotaxis of NOR and NOD BMCs to SDF-1 gradient (200 ng/mL) compared with control without SDF-1. Data are expressed as percentage increase in migration compared with control (percentage migration without SDF-1 gradient). All experiments were repeated at least 2 times and were performed in quadruplicate (*P < .0001). (B) Number of CFU-GM colonies formed by BMMNCs from NOR or NOD mice after migration to SDF-1 (*P < .001). The number of colonies formed by cells migrating to SDF-1 gradient is shown as percent increase of colonies that were observed in cultures initiated with cells showing chemotaxis to control (no SDF-1) medium (arbitrary assumed to be 100%). (C) Adhesion assay: BM Sca-1+ cells from NOD (▪) or NOR mice (□) were allowed to adhere for 1 or 6 hours to primary BM stroma. Adherent cells were then placed in methylcellulose cultures and stimulated to grow CFU-GM colonies (mrIL-3 + mrGM-CSF) that were counted 7 days later. Data represent 2 experiments performed in triplicate (*P = .005). (D) Adhesion molecule expression on NOD versus NOR KSL cells. Whole BM cells from NOD and NOR mice were stained for KSL cells and either adhesion molecule using mAbs against CD106, CD54, CD102, CD44, or CD62L FITC-labeled or the appropriate isotype control Abs. Data show the overlay of the different adhesion molecules (black) and the isotype control (gray). One representative experiment of 2 performed.
NOD HSCs exhibit enhanced migration to SDF-1 and adhesion to primary stroma. (A) Chemotaxis of NOR and NOD BMCs to SDF-1 gradient (200 ng/mL) compared with control without SDF-1. Data are expressed as percentage increase in migration compared with control (percentage migration without SDF-1 gradient). All experiments were repeated at least 2 times and were performed in quadruplicate (*P < .0001). (B) Number of CFU-GM colonies formed by BMMNCs from NOR or NOD mice after migration to SDF-1 (*P < .001). The number of colonies formed by cells migrating to SDF-1 gradient is shown as percent increase of colonies that were observed in cultures initiated with cells showing chemotaxis to control (no SDF-1) medium (arbitrary assumed to be 100%). (C) Adhesion assay: BM Sca-1+ cells from NOD (▪) or NOR mice (□) were allowed to adhere for 1 or 6 hours to primary BM stroma. Adherent cells were then placed in methylcellulose cultures and stimulated to grow CFU-GM colonies (mrIL-3 + mrGM-CSF) that were counted 7 days later. Data represent 2 experiments performed in triplicate (*P = .005). (D) Adhesion molecule expression on NOD versus NOR KSL cells. Whole BM cells from NOD and NOR mice were stained for KSL cells and either adhesion molecule using mAbs against CD106, CD54, CD102, CD44, or CD62L FITC-labeled or the appropriate isotype control Abs. Data show the overlay of the different adhesion molecules (black) and the isotype control (gray). One representative experiment of 2 performed.
NOD HSCs exhibit enhanced adhesion in vitro
The ability of HSCs to adhere to stromal cell monolayers is a well-established in vitro correlate to homing.35-38 To evaluate the adhesion properties of NOD HSCs, adhesion to wild-type stroma was assessed. Sca-1+ BMCs were incubated for 1 or 6 hours on stroma and the adherent cells were assessed in secondary methylcellulose cultures for the number of CFU-GMs after 7 days. Notably, the number of CFU-GMs that adhered to stroma from NOD mice was significantly higher compared with NOR mice at both time points (P < .005; Figure 6C), correlating with the enhanced engraftment observed in vivo. Therefore, there was a significant difference in migration of NOD HSCs compared with NOR HSCs, as well as a significant difference in the fraction of clonogenic progenitors in NOD BMCs that were capable of adhering to stromal cell monolayers.
The increased ability of NOD HSCs to adhere to stroma was further confirmed by increased expression of adhesion molecules such as VCAM-1 (CD106; 26% for NOD versus 13% for NOR), ICAM-1 (CD54; 34% for NOD versus 27% for NOR), or ICAM-2 (CD102; 36.5% for NOD versus 30% for NOR) on NOD KSL cells (Figure 6D). There were no differences for the other adhesion molecules tested including CD44, CD62 (Figure 6D), CD62P, CD11a, or CD184 (data not shown). This increase in selected adhesion molecules suggests a role for these molecules in the increased adhesion ability of NOD HSCs.
NOD HSC autonomy maps to the Idd9 genetic locus
In order to define the genetic regions responsible for the increased autonomy of NOD HSCs, KSL cells were purified from NOD, NOR, and subcongenic NOD.B10 mice with defined degrees of genetic disparity (Table 3). NOR and NOD mice are MHC congenic (Table 1). The Idd congenic NOD.B10 strains are genetically identical to NOD mice with the exception that a single B10-derived locus is present (Table 3). When KSL cells from NOR mice were transplanted into C3H recipients, early engraftment (up to 120 days) resembled that for NOD HSCs (Student t test: P > .05). However, by 150 days only 30% of the recipients of a transplant of NOR KSL cells remained durably engrafted compared with 100% of those given NOD KSL cells (P = .014) (Figure 7A).
Idd loci in the NOD B10 mice and the effect of the B10 gene on autoimmunity
Locus . | Chromosome . | Effect of congenic on diabetes . | Candidate factors . | Reference . |
|---|---|---|---|---|
| Idd3 | 3 | Complete diabetes resistance when linked with Idd10 and Idd18 | IL2 | 39-41 |
| Idd10 | 3 | Complete diabetes resistance when linked with Idd3 and Idd18 | CD53, Kcna3 | 42 |
| Idd18 | 3 | Complete diabetes resistance when linked with Idd3 and Idd10 | CD53, Kcna3 | 43 |
| Idd3/10/18 | Reduces diabetes and insulitis incidence | IL2, CD53, Kena3 | 43 | |
| Idd5 | 1 | Reduces diabetes and insulitis incidence | FLIP, CD28, CTLA4, Nramp | 43, 44 |
| Idd9.1/9.2/9.3 | 4 | Near-complete protection from diabetes, all progress to insulitis | TNFR superfamily molecules | 2 |
| Idd9.1 | 4 | Near-complete protection from diabetes, all progress to insulitis | Ick, insulin-like protein 5, sno | 2 |
| Idd9.2* | 4 | Near-complete protection from diabetes, all progress to insulitis | TNFR-II, CD30, CD137, ski | 2 |
| Idd9.3 | 4 | Near-complete protection from diabetes, all progress to insulitis | CD137 CD134 | 2 |
Locus . | Chromosome . | Effect of congenic on diabetes . | Candidate factors . | Reference . |
|---|---|---|---|---|
| Idd3 | 3 | Complete diabetes resistance when linked with Idd10 and Idd18 | IL2 | 39-41 |
| Idd10 | 3 | Complete diabetes resistance when linked with Idd3 and Idd18 | CD53, Kcna3 | 42 |
| Idd18 | 3 | Complete diabetes resistance when linked with Idd3 and Idd10 | CD53, Kcna3 | 43 |
| Idd3/10/18 | Reduces diabetes and insulitis incidence | IL2, CD53, Kena3 | 43 | |
| Idd5 | 1 | Reduces diabetes and insulitis incidence | FLIP, CD28, CTLA4, Nramp | 43, 44 |
| Idd9.1/9.2/9.3 | 4 | Near-complete protection from diabetes, all progress to insulitis | TNFR superfamily molecules | 2 |
| Idd9.1 | 4 | Near-complete protection from diabetes, all progress to insulitis | Ick, insulin-like protein 5, sno | 2 |
| Idd9.2* | 4 | Near-complete protection from diabetes, all progress to insulitis | TNFR-II, CD30, CD137, ski | 2 |
| Idd9.3 | 4 | Near-complete protection from diabetes, all progress to insulitis | CD137 CD134 | 2 |
Table data obtained from Mouse Genome Informatics.45
9.2/9.3 congenic: delayed diabetes progression, decreased incidence of diabetes; there is no congenic with only 9.1 or 9.2 alone
Autonomy of NOD HSCs maps to Idd9 locus. Purified KSL cells were administered to allogeneic recipients and long-term survival was evaluated. (A) Survival of C3H recipients exposed to 950 cGy TBI and given 10 000 KSL cells from NOD (□) or NOR (▵) mice. Conditioned controls receiving no KSL cells are also shown (solid line). (B) Survival curves of C3H recipients given 10 000 KSL cells from NOD mice (□), NOD B10.Idd5 congenic mice (○), or NOD B10.Idd3, 10, and 18 congenic mice (▵). (C) Survival curves of B10.BR recipients given 10 000 KSL cells from NOD mice (□), NOD B10.Idd9.3 congenic mice (⋄), NOD Idd9.1, 9.2, and 9.3 congenic mice (○), or NOD Idd9.2, 9.3 congenic mice (▵). Results shown for each donor type combine the survival data from 2 to 5 separate transplant experiments. Survival of the different transplant recipient groups was compared via Kaplan-Meier survival curves using the SPSS 12.0 for Windows statistical software package. Log-rank comparisons were performed and the P values for each comparison are shown. n.d. indicates not determined; n.a., not applicable.
Autonomy of NOD HSCs maps to Idd9 locus. Purified KSL cells were administered to allogeneic recipients and long-term survival was evaluated. (A) Survival of C3H recipients exposed to 950 cGy TBI and given 10 000 KSL cells from NOD (□) or NOR (▵) mice. Conditioned controls receiving no KSL cells are also shown (solid line). (B) Survival curves of C3H recipients given 10 000 KSL cells from NOD mice (□), NOD B10.Idd5 congenic mice (○), or NOD B10.Idd3, 10, and 18 congenic mice (▵). (C) Survival curves of B10.BR recipients given 10 000 KSL cells from NOD mice (□), NOD B10.Idd9.3 congenic mice (⋄), NOD Idd9.1, 9.2, and 9.3 congenic mice (○), or NOD Idd9.2, 9.3 congenic mice (▵). Results shown for each donor type combine the survival data from 2 to 5 separate transplant experiments. Survival of the different transplant recipient groups was compared via Kaplan-Meier survival curves using the SPSS 12.0 for Windows statistical software package. Log-rank comparisons were performed and the P values for each comparison are shown. n.d. indicates not determined; n.a., not applicable.
HSCs purified from NOD mice congenic with nondiabetic B10 mice at specific Idd loci including Idd5, Idd9 (and Idd9 subcongenics), and the linked loci Idd3, 10, and 18 strain (Table 3) were also transplanted into H-2k (C3H, B10.BR) recipients. When KSL cells from NOD.B10 Idd5 or Idd3, 10, and 18 congenic mice were transplanted into C3H mice, engraftment was not significantly different from NOD KSL cells (Figure 7B), suggesting that those loci are not involved in the efficiency of NOD KSL engraftment. KSL cells from NOD mice congenic with B10 at the entire Idd9 locus or only the Idd9.3 portion of the locus exhibited an autonomous engraftment pattern similar to NOD KSL cells (Figure 7C). Strikingly, KSL grafts from NOD mice congenic with B10 at Idd9.2 and 9.3 lost this property and exhibited engraftment potential similar to normal KSL in allogeneic recipients. By 60 days after transplantation, the engraftment pattern of KSL cells from Idd9.2 and 9.3 donors differed significantly from that of NOD KSL cells (P < .03). This engraftment kinetic differed from that observed in recipients of NOR HSCs and suggests that the Idd9.2 and 9.3 loci are involved in the autonomous engraftment ability of NOD HSCs (Figure 7A). These data demonstrate multiple mechanisms are likely to work in concert to contribute to the autonomy of NOD HSCs and that the autonomy of NOD HSC engraftment is multigenic in nature. Taken together, these findings implicate the Idd9 locus, especially 9.1 and 9.2, as contributing to the autonomy of NOD HSCs.
Discussion
Multiple immune system defects have been defined in NOD mice and in humans with type 1 diabetes, including aberrant MHC class II molecules that confer susceptibility to diabetes development, abnormalities in myeloid APC and dendritic cells that correlate with diabetes progression, impaired generation of regulatory T cells, and impaired NK cell cytolytic function.5,7-11 We previously reported that NOD mice have a number of features related to engraftment that distinguish them from normal mice as stem cell recipients. They require more conditioning and higher numbers of allogeneic BMCs to establish durable engraftment,15,17 are highly sensitive to T-cell–depletion graft failure,17 and once engraftment does occur, the disease-resistant marrow exhibits a competitive advantage.17 An understanding of the mechanism underlying these differences may lead to a better understanding of how the autoimmune process develops and impacts on the development of therapeutic approaches to cure diabetes.
In the present studies, we evaluated the NOD HSC function using sorted KSL cells. We found that NOD KSL cells engraft readily in recipients with full MHC disparities. As few as 104 NOD KSL cells establish durable engraftment in 90% to 100% of allogeneic recipients, an engraftment potential that is significantly enhanced compared with KSL cells from normal allogeneic donors (P < .001). Moreover, the autonomy of NOD HSCs was observed in more than one strain combination. In contrast with normal mouse strains, the addition of graft FCs was not required and did not improve engraftment of NOD HSCs.24,25 Although outside the scope of this report, our ongoing studies confirm that NOD mice lack facilitating cells (S.T.I. and Y.H., unpublished data, Spring 2004). One could hypothesize that the autonomy of NOD HSCs reflects a compensatory mechanism due to this defect. This high efficiency of NOD HSC engraftment was further confirmed by their ability to generate day-12 CFU-Ss, reflective of multilineage progenitors in allogeneic recipients. A similar requirement for MHC restriction between HSCs and stromal cells in vivo was demonstrated by Hashimoto et al,46 where they demonstrated that transplanted HSCs preferentially colonized syngeneic bone grafts compared with allogeneic bone grafts. This preference could be eliminated if the bones were irradiated prior to transplantation.46 The ability of the totally allogeneic NOD KSL cells to efficiently compete with KSL cells that are syngeneic to the recipient indicates that the HSCs from NOD mice do not have this MHC restriction requirement. It was previously reported that transplantation of a mixture of NOD plus BALB/c bone marrow cells into NOD recipients resulted in complete NOD chimerism with failure of engraftment of the allogeneic BALB/c cells.47 However, because NOD mice are resistant to allogeneic bone marrow engraftment,12,17 it was unclear whether the failure of engraftment of the allogeneic marrow was due to autonomy of NOD BMCs or impaired microenvironment in the NOD recipient for allogeneic HSCs. This autonomy may be reflective of an abnormal bone marrow microenvironment, which could explain the decreased ability of HSCs from disease-resistant strains to engraft in NOD recipients.12,17 Kawamura et al48 previously showed that HSCs from autoimmune W/BFI mice similarly did not require MHC matching to generate CFU-Ss. Although durable engraftment was not studied in this report, it indicates that HSC autonomy in allogeneic recipients is not limited to one autoimmune model and may represent a feature common to some autoimmune diseases.
Previous studies have shown that megadoses of purified HSCs can engraft in allogeneic recipients, an effect hypothesized to occur via a veto mechanism.49 Although 104 purified KSL cells is a slightly larger dose than our laboratory routinely uses for other allogeneic KSL strain combinations,18 it is still 50- to 100-fold lower than the 5 × 105 to 1 × 106 cell dose that has been shown to be required for the megadose effect.50 It is therefore possible that the KSL cells from NOD mice themselves have an enhanced ability to act as veto cells in allogeneic microenvironments.
The autonomy of NOD HSCs is further reflected by the fact they significantly out-compete syngeneic HSCs in competitive engraftment assays. When mice were administered increasing numbers of syngeneic KSL cells with a fixed number of allogeneic NOD KSL cells in a modified competitive repopulation assay, stable and durable engraftment of NOD KSL cells occurred even when the NOD and syngeneic HSCs were given at a 1:1 ratio. When fewer syngeneic KSL cells were given in conjunction with the NOD KSL cells, the percentage of NOD-derived cells increased over time to approach 90% donor chimerism. When the frequency of progenitor cells in the B10.BR and NOD KSL populations was determined, similar numbers of long-term repopulating cells were found, indicating that cell ratios based on total KSL cells would be similar if expressed as CAFC ratios.
One possible mechanism for the increased autonomy of NOD HSCs is that the number of long-term or short-term repopulating cells could be increased compared with normal strains. The NOD KSL population did not contain an increased frequency of primitive HSCs (d35 CAFC) and there was a decrease in the committed progenitor population (d8 CAFC). These data were further confirmed by the fact that there was no difference in the CD49e+/CD49ddim long-term repopulating cell phenotype between the KSL populations obtained from NOD, B10.BR, or B10. Therefore, a difference in the composition of the NOD HSCs is an unlikely explanation for their autonomous engraftment ability in allogeneic recipients.
We found here that NOD KSL cells have a high resistance to irradiation, reflected by both cell survival in culture 20 hours after irradiation and the in vitro CFC assay, which further suggests even more resistant primitive HSCs. Taken together, these data demonstrate that NOD HSCs exhibit an enhanced ability to escape alloreactivity and to compete with normal HSCs. This can be correlated to the fact that it is difficult to establish allogeneic engraftment in NOD mice with whole bone marrow and even more difficult with T-cell–depleted marrow,12,47 and the fact that purified HSCs encounter an even greater barrier to engraftment in NOD recipients.15
The homing of HSCs after transplantation is critical to successful engraftment and is a complex process involving migration, diapedesis through the endothelium, and adhesion to stroma. The CXCR4/SDF-1 axis has been demonstrated to be central to HSC homing and migration.20 We found that NOD HSCs migrate significantly more efficiently to an SDF-1 gradient and exhibit enhanced adhesion on primary stroma compared with NOR controls. Moreover, there was up-regulation of specific adhesion molecules on NOD KSL cells, including ICAM-1, ICAM-2, and VCAM-1. Our present data therefore suggest an important role for the SDF-1/CXCR4 axis as well as an increase in adhesion molecules that would enhance adhesion and homing as a mechanism underlying NOD HSC autonomy.
In order to assess the genetic requirements behind the increased autonomy of NOD HSCs, we transplanted KSL cells from NOR and NOD.B10 congenic mice, which express diabetes-resistant genes from B10 mice at defined Idd loci, into allogeneic recipient mice (Table 3). Notably, we observed that the autonomy of the NOD HSCs is multigenic, in that autonomous engraftment potential segregated into 2 separate phases, early and late. Recipients given KSL cells from NOR mice, congenic to NOD in 88% of the genome, engrafted and survived past the initial short-term phase; however, survival decreased between 4 and 5 months after transplantation compared with recipients of NOD KSL cells (P = .03). The majority of the recipients of KSL cells from NOD congenic mice containing the B10 Idd9.2/9.3 locus exhibited less autonomy. As early as 2 months after transplantation they exhibited a survival pattern similar to recipients of allogenic KSL cells alone. Interestingly, when the entire B10 Idd9 locus (Idd9.1/9.2/9.3) or only the 9.3 locus was present in donors of the KSL cells, engraftment was similar to NOD KSL cells. Taken together, these data implicate the involvement of Idd9.2 and 9.3 loci in HSC/engraftment potential and a regulatory role of Idd9.1.
As shown in Table 3, the Idd9 locus encodes the genes for multiple TNF-receptor superfamily members, including TNFR-II, CD30, and CD137. These members are involved in T-cell costimulation. In a recent study, the treatment of CD34+ human progenitor cells with CD137 ligand resulted in the increased differentiation of cells to mature myeloid dendritic cells.51 Moreover, both CD30 and CD137 have been demonstrated to have roles in the onset of diabetes. The administration of an antagonistic mAb against the CD30 ligand abrogated the development of diabetes in NOD mice and in vitro T-cell proliferation to islet-specific antigens.52 Moreover, the transgenic expression of an agonistic CD137 mAb in NOD mice dramatically increased the onset and severity of diabetes in NOD mice.53 If these molecules exert influence over alloreactive T cells, they may indeed hold the key to explain the increased ability of NOD HSCs to survive in allogeneic recipients. Alternatively, since TNF-α is involved in survival and self-renewal of quiescent HSCs,54 a defect in the TNFRII gene may indeed cause the NOD HSCs to better resist the cell death induced by TNF-α, and therefore results in enhanced autonomy.54 Additionally, TNF-α has been shown to act on HSCs via hyaluronic acid, the ligand for CD44, to up-regulate adhesion of HSCs in the marrow.55
The pluripotent HSC itself is of great interest in understanding type 1 diabetes and other autoimmune diseases because it has the capacity to both transfer and reverse the autoimmune process. In the present studies, we have shown for the first time that NOD HSCs exhibit increased autonomy in vivo and in vitro as reflected by increased engraftment potential in allogeneic recipients and increased adhesion to primary stroma. Notably, this autonomy is associated with the Idd9.2 and 9.3 genetic loci. Taken together, these findings may be of clinical interest in the mechanism-driven design of strategies to apply HSC transplantation to treat autoimmune diseases such as diabetes as well as understanding the mechanism of diabetes progression.
Prepublished online as Blood First Edition Paper, November 2, 2004; DOI 10.1182/blood-2004-07-2757.
Supported in part by the National Institutes of Health (R01 AI45486 and NHLBI HL 63442), the Juvenile Diabetes Foundation, The Commonwealth of Kentucky Research Challenge Trust Fund, The Jewish Hospital Foundation, and the University of Louisville Hospital.
P.M.C. and F.R. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Dr Christina Kaufman for careful review of the manuscript and helpful comments, Carolyn DeLautre and Kim Nichols for manuscript preparation, the staff of the animal facility for outstanding animal care, and Dr Gary Van Zant (University of Kentucky) and Hartmut Geiger (University of Cincinnati) for supplying FBMD7 cell line and instruction on the CAFC assay.


![Figure 5. NOD HSCs are more radioresistant. The ability of HSCs to survive after exposure to ionizing radiation was determined by exposing whole BMCs (A) or purified KSL cells (B) to the indicated doses of irradiation. (A) The percent KSL cells surviving after a 20-hour culture was determined by flow cytometry. ○ indicates NOD; ▪, B10.BR. Results are the average survival plus and minus the standard deviation of cells from 3 individual animals in one of 2 different experiments (*P < .05). (B) The ability of irradiated KSL cells to produce colonies in methylcellulose plus growth factors was determined. Data expressed as plus and minus the standard deviation (*P < .05). Results are the average of quadruplicate wells from one of 2 (NOR [□] and B10 [⬡]) or 3 (NOD [⋄] and B10.BR [▴]) experiments, expressed as percent of unirradiated KSL cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/5/10.1182_blood-2004-07-2757/6/m_zh80050575140005.jpeg?Expires=1769086702&Signature=04YSdtv8gtd0WR5vXnKLrL2Nu2fWszqvPdlXmkun0wOZ0cq1DwFGJhGTDgjjegXO6nPRRUSKQxCut6lkBEqXpJ7ix9Bep7L5ORzfHwDeHG1w9FSe2SxB3t3ElgucwsIEFwBskXmmsIZ1EHU4YsVOu1OeDKUxB6QiUOURyDmVjHXDlObISfthqpCaFN3s3xEYI5AqvmEyeUMMh9Uph08waS7Hyp9A1y6BH2OVwlTmoRVeIsi-oH25xHviGTVDFQ~b2BPBPdp2jCf65qq4WnM6sAA1qjloYSsE1bJGIlszdcazyN159Ej8U69meKb743Nu9c9OJ1fxSSDiOcEOsZFCVA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal