Abstract
Preclinical studies with the histone deacetylase (HDAC) inhibitor depsipeptide (FK228) in chronic lymphocytic leukemia (CLL) and acute myeloid leukemia (AML) have demonstrated that it effectively induces apoptosis at concentrations at which HDAC inhibition occurs. We initiated a minimum effective pharmacologic dose study of depsipeptide, targeting an in vivo dose at which acetylation of histone proteins H3 and H4 increased by 100% or more in vitro. Ten patients with CLL and 10 patients with AML were treated with 13 mg/m2 depsipeptide intravenously days 1, 8, and 15 of therapy. Neither life-threatening toxicities nor cardiac toxicities were noted, although the majority of patients experienced progressive fatigue, nausea, and other constitutional symptoms that prevented repeated dosing. Several patients had evidence of antitumor activity following treatment, but no partial or complete responses were noted by National Cancer Institute criteria. HDAC inhibition and histone acetylation increases of at least 100% were noted, as well as increases in p21 promoter H4 acetylation, p21 protein, and 1D10 antigen expression. We conclude that depsipeptide effectively inhibits HDAC in vivo in patients with CLL and AML, but its use in the current schedule of administration is limited by progressive constitutional symptoms. Future studies with depsipeptide should examine alternative administration schedules.
Introduction
Acute myeloid leukemia (AML) and chronic lymphocytic leukemia (CLL) are 2 of the most common types of leukemia diagnosed in adults.1 Most of the patients diagnosed with AML present with bone marrow failure and require immediate treatment. Of the patients receiving cytotoxic therapy as induction treatment, 50% to 75% will attain a complete response (CR).2 Among young patients with de novo AML, however, only approximately 30% will attain a long-term disease-free survival even with subsequent treatment intensification.3 The chance for cure with conventional therapies is even smaller in high-risk patients with AML such as the elderly,4 patients with high-risk cytogenetics,5 or patients with treatmentrelated AML.6 In contrast, patients with CLL do not always require immediate therapy. However, once symptomatic, the median survival of these patients ranges from 18 months to 6 years, depending on the clinical stage.7 While the introduction of fludarabine8-12 and more recently rituximab13-16 has improved the overall success of CLL treatment, therapy is still palliative with no curative potential. Identification of new therapies for both CLL and AML therefore remains a high priority.
One promising new therapeutic strategy is aimed at reversing tumor-related alterations in chromatin structure and concomitant transcriptional silencing. Epigenetic silencing or down-modulation of important tumor suppressor and differentiation genes, such as occurs by changes in histone modifications, can lead to neoplastic transformation and disease progression. Importantly, epigenetic silencing involving both aberrant methylation17-21 and histone deacetylation22-25 has been demonstrated to be active in the pathogenesis of AML and CLL.
Loss of gene function also occurs in neoplastic diseases through mutations or deletions, but these changes are not easily targeted by pharmacologic intervention, as the gene sequence itself is permanently altered. In contrast, epigenetic silencing, as occurs through deacetylation of histone proteins or methylation of DNA, is a reversible process. Preclinical data suggest that combining hypomethylating agents and histone deacetylase (HDAC)–inhibiting agents produces synergistic gene reexpression and apoptosis in vitro against a variety of different malignancies, including AML.26-29 Thus, development of effective agents that alone or in combination can target in vivo the machinery mediating epigenetic gene silencing is crucial. This development ideally will include detailed assessment of target validation to identify the minimum effective pharmacologic dose (MEPD) that inhibits the target enzyme or results in re-expression of an epigenetically silenced gene.
Depsipeptide is one of the HDAC inhibitors currently under clinical development. Initial preclinical studies demonstrated that depsipeptide causes down-regulation of c-myc and morphologic normalization of Ras-transformed tumor cells.30 More recent investigations demonstrated that this agent also effectively inhibits HDAC in human tumor cell lines27,31-33 and inhibits predominately type I HDACs.1-3,8,34,35 Subsequently, work done by our group demonstrated that depsipeptide promotes apoptosis in both primary CLL and AML tumor cells in vitro at a concentration corresponding to that at which H3 and H4 acetylation and HDAC inhibition occurs.36-38 These studies provide a strong rationale for pursuit of clinical trials with depsipeptide in AML and CLL. Importantly, they also provide justification for targeting an in vivo dose that corresponds to the concentration at which HDAC inhibition, histone acetylation, and apoptosis occurs in primary CLL and AML cells in vitro.
At present, 2 phase 1 studies using depsipeptide have been performed in patients with refractory solid-tumor malignancies.39,40 In these trials, dose-limiting toxicities were constitutional symptoms and thrombocytopenia. Clinical activity was observed in a small subset of patients with T-cell lymphoma enrolled in one of these studies, and this activity has been confirmed in an ongoing phase 2 clinical trial.41 While these trials established the safety of depsipeptide in solid tumors, they did not include patients with AML or CLL who have impaired marrow reserve and predisposition for infection. Furthermore, neither focused on identification of the MEPD required to produce the expected biologic effect (ie, inhibition of HDAC in tumor cells with subsequent increase in histone acetylation). Identifying both the safety and MEPD of depsipeptide in AML and CLL will facilitate combination strategies with other epigenetically targeted agents such as the hypo-methylating agent decitabine. Herein, we describe results from a detailed MEPD trial with depsipeptide in AML and CLL.
Patients, materials, and methods
Patients and inclusion criteria
Approval for this clinical trial and blood collection for laboratory studies was obtained from The Ohio State University Institutional Review Board. Each patient provided informed consent for their participation according to the Declaration of Helsinki. This MEPD study included 2 cohorts of patients. Cohort 1 included patients with CLL and small lymphoma as defined by the World Health Organization (WHO) classification.42 These patients were required to have received at least 1 prior therapy that included fludarabine, cladribine, or pentostatin. Patients with a contraindication to receiving purine analog therapy were allowed to enroll. Cohort 2 included patients with primary, refractory, or relapsed AML (≤1 year) who were not candidates for allogeneic or autologous stem cell transplantation. In addition, previously untreated patients with poor-risk leukemia, defined as having any of the following: age 65 years or older, poor risk cytogenetics as defined by Cancer and Leukemia Group B (CALGB),5 or poor candidates for aggressive chemotherapy, could enroll on this study without receipt of prior chemotherapy for AML. Patients in cohort 2 were also required to have a normal leukocyte count (equal to or below 10 × 109/L) or a stable count (equal to or below 40 × 109/L for greater than 1 week). Patients were allowed to receive hydroxyurea prior to starting therapy and during the first week of therapy if clinically indicated to maintain the leukocyte count equal to or below 40 × 109/L.
Other eligibility criteria for patients in both cohorts included age older than 18 years, Eastern Cooperative Oncology Group (ECOG) performance status less than or equal to 2, total bilirubin less than or equal to 18.6 μmol/L (1.5 mg/dL), creatinine less than 176.8 μmol/L (<2.0 mg/dL), and alanine aminotransferase (ALT) and aspartate aminotransferase (AST) less than or equal to 3 times the upper limit of normal. Given the potential for cardiac toxicity with depsipeptide, patients were required to have a cardiac ejection of at least 50% and no history of a myocardial infarction or unstable angina within the past 6 months. In addition, patients were required to have had no recent major surgery, radiotherapy, or chemotherapy (except hydroxyurea) within 28 days of treatment, and they could not have an active infection requiring oral or intravenous antibiotics. Patients with active autoimmune processes and HIV infection were also ineligible.
Treatment and response assessment
All patients underwent a screening assessment (history, physical examination, bone marrow aspirate and biopsy, cardiac ejection fraction, and electrocardiogram) within 10 days of beginning treatment. Depsipeptide (13 mg/m2) was administered intravenously through a central line over 4 hours on days 1, 8, and 15 of every 4-week cycle. All patients received granisetron hydrochloride 2 mg (or equivalent) for antiemetic prophylaxis. Other antiemetic treatment was administered based on patient symptoms. Patients could continue therapy for up to 1 year, provided they had a complete response, partial response, or continuing hematologic improvement. All patients were treated for a minimum of 2 months in the absence of progression of disease or unacceptable toxicity. Extensive cardiac monitoring, including serial electrocardiograms, multigated acquisition (MUGA) scans, troponin levels (before and after therapy) and noninvasive monitoring, were performed throughout treatment. Response was assessed using the previously reported criteria for CLL43 and AML.44
Dose escalation schedule
As the primary goal of this study was to define the MEPD of depsipeptide, the study design was intended to treat a total of 10 patients in each cohort at the dose of 13 mg/m2. The desired target change in histone acetylation considered to be a positive biologic effect was an increase in total H3 or H4 acetylation of at least 100% relative to the pretreatment sample. In our in vitro studies, this level of H3 and H4 acetylation was consistently observed in CLL cells at the tumor cell LC50 (lethal concentration to 50%) of depsipeptide (0.038 μM).37 Upon completion of the first cohort of patients, if clinical and biologic effects (disease improvement, partial response, complete response) were observed in 5 or more patients, a dose reduction of 25% would be given to the next cohort of patients. If clinical or biologic effects were not observed following treatment in at least 5 patients at 13 mg/m2 depsipeptide, the treatment dose would be increased by 25% (eg, to 16 mg/m2).
Dose-limiting toxicities were defined as nonhematologic toxicity of grade 3 or greater, or in some cases grade 2 (eg, irreversible renal, chronic pulmonary, neurologic, cardiac, or local toxicities). An exception to this was fatigue, unless it was grade 3 persisting more than 5 days or grade 4. The National Cancer Institute (NCI) Common Toxicity Criteria, version 3.0 (NCI, Bethesda, MD), were used to quantify toxicity. Hematologic dose-limiting toxicities for the CLL cohort included grade 4 cytopenias if the patient's baseline platelet count was above 75 × 109/L and neutrophil count above 2 × 109/L before initiation of depsipeptide infusion. Patients with neutrophil or platelet counts below these values were not considered evaluable for hematologic dose-limiting toxicity unless they failed to recover to 75% of baseline (in the absence of disease progression) by day 35, which would then be considered a dose-limiting toxicity. In cohort 2, hematologic dose-limiting toxicity was defined as failure to recover to baseline hematologic parameters by day 35 in the absence of residual leukemia.
Patients experiencing a dose-limiting toxicity were allowed to remain on study provided the toxicity reversed completely. These patients had a 25% dose reduction (9 mg/m2 depsipeptide) with subsequent treatments. Patients requiring more than 1 dose reduction were taken off study.
Histone protein acetylation and immunoblot studies
Peripheral blood mononuclear cells were collected before and at 4 and 24 hours following depsipeptide administration for patients with CLL. In cases where CLL cells were recovered in sufficient numbers and purity to perform this procedure at each time point, histone proteins and whole cellular isolates were isolated as previously described by our group.37 Total protein in each sample was quantified by the bicinchoninic acid (BCA) method (Pierce, Rockford, IL). Lysates or extracted histone proteins were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Protein equivalence on the gels was verified by SYPRO Red staining (Molecular Probes, Eugene, OR) followed by subsequent analysis via fluorescence (Storm 860 Imaging System/ImageQuant; Molecular Dynamics, Sunnyvale, CA). Histone modifications were determined via immunoblotting with antibodies specific for acetylated H3 or H4 (Upstate, Lake Placid, NY). Antibodies to p21 were from Santa Cruz Biotechnology (Santa Cruz, CA), and p21 signals were normalized to the housekeeping protein glyceraldehyde 3-phosphodehydrogenase (Chemicon, Temecula CA). Horseradish peroxidase–conjugated secondary antibodies were obtained from Amersham Biosciences (Piscataway, NJ). Immunoblots were developed as described previously37 and quantified using the Storm 860 Digital Imaging System and ImageQuant software.
Histone deacetylase assay
Assays were performed as described previously,37 using cell lysates containing 50 μg protein derived from patient samples taken before treatment and at 4 and 24 hours following depsipeptide infusion. HDAC activity in 3 serial CLL patient samples derived from different time points in the absence of any therapeutic intervention varies by ± 6.4%. To provide a positive and negative control, lysates from HeLa cells incubated with and without 250 mM sodium butyrate were included in all experiments. All samples were assayed in duplicate, and results were averaged.
Immunocytochemistry studies of acetylation on H3 and H4 histone proteins
Slides were prepared by depositing 1 × 105 cells isolated from CLL or AML patient samples onto glass slides using a Cytospin 3 centrifuge (Thermo Shandon, Pittsburgh, PA). Slides were Wright-Giemsa stained or fixed for 1 minute at room temperature in a solution of 95% ethanol and 5% glacial acetic acid, followed by washes in phosphate-buffered saline (PBS). Cells were then permeabilized for 10 minutes at room temperature with 0.2% Triton X-100 and then blocked in 10% normal donkey serum (NDS) in PBS for 1 hour at room temperature. Slides were incubated overnight at 4°C with anti-acetylated H3 (Upstate Biotechnology, Lake Placid, NY) diluted 1:150, or with anti-acetylated H4 (Upstate Biotechnology) diluted 1:100 in 2% NDS in PBS. After washing 3 times in PBS, the slides were stained with donkey anti–rabbit carbocyanine 3 (Cy3)–conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1.5 hours and then mounted with Cytoseal 60 (Electron Microscopy Science, Fort Washington, PA). In parallel control experiments, it was observed that omission of either primary antibody eliminated staining. Slides were analyzed using an Olympus BX51 microscope equipped with an Olympus PM30 camera (Olympus, Melville, NY), using an ocular lens of 10 ×/2.3 NA, and an objective lens of 40 ×/0.75 NA. The image was captured using analySIS FIVE software (Olympus).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed using the Chromatin Immunoprecipitation Assay Kit (Upstate Biotechnology) according to the manufacturer's recommended protocol with minor modifications: 1to2 × 106 cells were used for each assay. Formaldehyde crosslinking was performed for 20 minutes at room temperature. Cells were harvested in SDS lysis buffer with protease inhibitors (Protease Inhibitor Cocktail Set III; CalBioChem, La Jolla, CA) and left on ice for 15 minutes. The chromatin was broken down to an average size of 0.2 to approximately 1 kb through sonication (Sonic Dismembrator; Fisher Scientific, Pittsburgh PA). The input and immunoprecipitated chromatins were incubated at 65°C for 6 hours to reverse the formaldehyde crosslinking and were digested by proteinase K (RNA grade; 20μg/mL; Invitrogen, Carlsbad, CA) for 6 hours at 50°C to remove the proteins. Polymerase chain reaction (PCR) was performed using GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA) with primers specific for p21 gene promoter: forward, 5′-GACCGGCTGGCCTGCTGGAACTCG-3′, and reverse, 5′-CGGGCACGCTTGGCTCGGCTCTG-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter: forward, 5′-AAAAGCGGGGAGAAAGTAGG, and reverse, 5′-CTAGCCTCCCGGGTTTCTCT-3′. The cycle number and the amount of template were varied to ensure that results were within the linear range of the PCR. The PCR products were resolved on 2.0% agarose gel containing ethidium bromide and analyzed under UV. Densitometry was performed using ImageQuant 5.0 (Molecular Dynamics). Bands were quantified and corrected for the slight variation in GAPDH amplified by PCR in a linear range.
1D10 antigen analysis
Surface expression of the HLA subtype 1D10 antigen was assessed on CLL patient samples before treatment and at 4 and 24 hours after treatment. Peripheral blood mononuclear cells were isolated, washed in PBS, and stained with anti-1D10 antibody conjugated to phycoerythrin (PE). Cells were stained simultaneously with anti-CD19 conjugated to fluorescein isothiocyanate (FITC) and were run on an EPICS-XL flow cytometer (Beckman Coulter, Miami FL). Samples were gated to analyze only CD19+ cells, and expression of 1D10 was calculated as PE intensity of 1D10-stained cells relative to the same sample incubated with a PE-labeled isotype control.
Pharmacokinetic studies
Plasma samples were obtained at the following time points: before treatment; 30, 60, 120, and 180 minutes after the start of infusion; and 15 and 30 minutes, and 1, 3, 6, and 12 hours after the end of infusion. Urine samples were collected during treatment and for 24 hours after completion of therapy. Depsipeptide concentrations were measured using the liquid chromatography/mass spectrometry (LC/MS) assay previously described.35,45 This method provides a limit of quantification of 0.1 ng/mL. The analyses were performed using both compartmental and noncompartmental methods (WinNonlin, Version 3.0; Pharsight, Mountain View, CA),46 and the results were compared with those published previously.39
Results
Patient characteristics
A total of 20 (10 CLL and 10 AML) patients were enrolled to this study between September 27, 2001, and September 19, 2002. The pretreatment features for patients in each disease cohort are summarized in Tables 1 and 2. The ECOG performance status of all patients prior to enrollment was ECOG 0 (4 patients, 20%), ECOG 1 (15 patients, 75%), and ECOG 2 (1 patient, 5%).
Clinical features of patients with CLL
. | Data . |
|---|---|
| No. of patients | 10 |
| Median age, y (range) | 55 (46-68) |
| No. of men (%) | 6 (60) |
| Rai stage at treatment, no. | |
| Intermediate risk (I/II) | 2 |
| High risk (III/IV) | 8 |
| Median WBC, × 109/L (range) | 42.3 (1.8-305) |
| Median Hgb, g/dL (range) | 11.3 (9.2-13.9) |
| Median platelets, × 109/L (range) | 84 (22-232) |
| Median ECOG performance status (range) | 1 (0-1) |
| Median no. of prior therapies (range) | 3 (1-7) |
| No. refractory to fludarabine | 10 |
| No. refractory to last therapy | 6 |
| Interphase cytogenetics, no.* | |
| Del(17)(p13.1) | 3 |
| Del(11)(q22.3) | 3* |
| Trisomy 12 | 0 |
| Del(13)(q14) | 2 |
| No abnormalities detected | 2 |
. | Data . |
|---|---|
| No. of patients | 10 |
| Median age, y (range) | 55 (46-68) |
| No. of men (%) | 6 (60) |
| Rai stage at treatment, no. | |
| Intermediate risk (I/II) | 2 |
| High risk (III/IV) | 8 |
| Median WBC, × 109/L (range) | 42.3 (1.8-305) |
| Median Hgb, g/dL (range) | 11.3 (9.2-13.9) |
| Median platelets, × 109/L (range) | 84 (22-232) |
| Median ECOG performance status (range) | 1 (0-1) |
| Median no. of prior therapies (range) | 3 (1-7) |
| No. refractory to fludarabine | 10 |
| No. refractory to last therapy | 6 |
| Interphase cytogenetics, no.* | |
| Del(17)(p13.1) | 3 |
| Del(11)(q22.3) | 3* |
| Trisomy 12 | 0 |
| Del(13)(q14) | 2 |
| No abnormalities detected | 2 |
Hgb indicates hemoglobin.
Dohner prioritization schema used to categorize patients with multiple abnormalities [(17)(p13.1)> del(11)(q22.3)> +12 > del(13)(q14)].
Clinical features of patients with AML
. | Data . |
|---|---|
| No. of patients | 10 |
| Median age, y (range) | 63 (46-81) |
| No. of men (%) | 6 (60) |
| AML features, no. | |
| De novo | 8 |
| Arising from myelodysplasia | 2 |
| Arising following treatment | 0 |
| Status at time of treatment, no. | |
| Initial therapy | 2 |
| Primary refractory disease | 2 |
| Relapsed disease, less than 12 mo | 4 |
| Relapsed disease, more than 12 mo | 2 |
| Relapse after autologous transplantation | 2 |
| Relapse after allogeneic transplantation | 1 |
| Median WBC, × 109/L (range) | 2.2 (0.8-16.6) |
| Median Hgb, g/dL, (range) | 9.9 (8.3-12.7) |
| Median platelets, × 109/L (range) | 61 (10-187) |
| Median ECOG performance status (range) | 1 (0-2) |
| On hydroxyurea prior to treatment, no. | 2 |
| Active problem requiring hospitalization, no. | 1 |
| CALGB cytogenetic classification, no. | |
| Low risk | 0 |
| Intermediate risk | 8 |
| High risk | 2 |
. | Data . |
|---|---|
| No. of patients | 10 |
| Median age, y (range) | 63 (46-81) |
| No. of men (%) | 6 (60) |
| AML features, no. | |
| De novo | 8 |
| Arising from myelodysplasia | 2 |
| Arising following treatment | 0 |
| Status at time of treatment, no. | |
| Initial therapy | 2 |
| Primary refractory disease | 2 |
| Relapsed disease, less than 12 mo | 4 |
| Relapsed disease, more than 12 mo | 2 |
| Relapse after autologous transplantation | 2 |
| Relapse after allogeneic transplantation | 1 |
| Median WBC, × 109/L (range) | 2.2 (0.8-16.6) |
| Median Hgb, g/dL, (range) | 9.9 (8.3-12.7) |
| Median platelets, × 109/L (range) | 61 (10-187) |
| Median ECOG performance status (range) | 1 (0-2) |
| On hydroxyurea prior to treatment, no. | 2 |
| Active problem requiring hospitalization, no. | 1 |
| CALGB cytogenetic classification, no. | |
| Low risk | 0 |
| Intermediate risk | 8 |
| High risk | 2 |
Treatment outcome of patients with CLL
The toxicities observed in the CLL cohort are summarized in Table 3. Of these 10 patients,1 did not complete 1 cycle of therapy, 1 completed 1 cycle of therapy, 5 received 2 cycles, 1 received 3 cycles, and 2 received 4 treatments with depsipeptide. Two patients required dose reductions, 1 for fatigue and 1 for hyponatremia. The single patient who did not complete cycle 1 of therapy presented with fludarabine-refractory CLL with a del(17p13.1). This patient experienced tumor lysis syndrome (the absolute lymphocyte count before therapy, 303 × 109/L [303 000 cells/μL] decreased to 125 × 109/L [125 000 cells/μL] on day 6 after the first dose of depsipeptide accompanied by hyperkalemia, hyperphosphatemia, hypocalcemia, hyperuricemia, and renal insufficiency) that required hospitalization. Despite complete resolution of this event, the patient elected not to receive further therapy. The uniform reason for discontinuing treatment in the remaining 9 patients was acute nausea or, more commonly, progressively worsening chronic nausea, anorexia, and fatigue that made patients unwilling to receive further therapy. Of the patients who began therapy with elevated leukocyte counts (n = 7), all demonstrated improvement in peripheral leukocyte counts (average decrease 58%; range, 45%-76%), and 1 patient experienced a 46% reduction in lymphadenopathy. However, no patients met the NCI criteria43 for partial response during depsipeptide treatment. Therefore, due to the progressive onset of constitutional symptoms and modest evidence of clinical activity, further pursuit of this schedule of administration of depsipeptide in CLL was abandoned.
Toxicities observed in patients with CLL treated with depsipeptide
Toxicity . | Grade 1, no. . | Grade 2, no. . | Grade 3, no. . | Grade 4, no. . |
|---|---|---|---|---|
| Anorexia | 6 | 2 | 1 | 0 |
| Fatigue | 3 | 7 | 0 | 0 |
| Nausea | 3 | 6 | 0 | 0 |
| Vomiting | 1 | 5 | 0 | 1 |
| Myalgias | 5 | 2 | 0 | 0 |
| Headache | 4 | 1 | 0 | 0 |
| Diarrhea | 4 | 2 | 0 | 0 |
| Insomnia | 2 | 0 | 0 | 0 |
| Mood alteration | 0 | 2 | 0 | 0 |
| Taste loss | 4 | 0 | 0 | 0 |
| Dyspepsia | 1 | 1 | 0 | 0 |
| Stomatitis | 1 | 4 | 0 | 0 |
| Abdominal pain | 2 | 0 | 0 | 0 |
| Dyspnea | 0 | 3 | 0 | 0 |
| Cough | 0 | 2 | 0 | 0 |
| Diaphoresis | 3 | 1 | 0 | 0 |
| Rash | 0 | 1 | 0 | 0 |
| Edema | 2 | 1 | 0 | 0 |
| Chest pain | 1 | 0 | 0 | 0 |
| Hyponatremia | 0 | 0 | 2 | 0 |
| Dizziness | 1 | 1 | 0 | 0 |
| Ataxia | 0 | 1 | 0 | 0 |
| Infection | 1 | 1 | 1 | 0 |
| Tumor lysis | 0 | 0 | 0 | 0 |
| Hyperglycemia | 0 | 0 | 0 | 0 |
| Constipation | 5 | 1 | 0 | 0 |
| Arthralgia | 1 | 1 | 0 | 0 |
| Catheter-related infection | 1 | 0 | 0 | 0 |
| Sinus tachycardia | 1 | 0 | 0 | 0 |
| Epistaxis | 1 | 0 | 0 | 0 |
| Neutropenia | 2 | 1 | 1 | 6 |
| Anemia | 6 | 2 | 0 | 0 |
| Thrombocytopenia | 0 | 2 | 6 | 2 |
Toxicity . | Grade 1, no. . | Grade 2, no. . | Grade 3, no. . | Grade 4, no. . |
|---|---|---|---|---|
| Anorexia | 6 | 2 | 1 | 0 |
| Fatigue | 3 | 7 | 0 | 0 |
| Nausea | 3 | 6 | 0 | 0 |
| Vomiting | 1 | 5 | 0 | 1 |
| Myalgias | 5 | 2 | 0 | 0 |
| Headache | 4 | 1 | 0 | 0 |
| Diarrhea | 4 | 2 | 0 | 0 |
| Insomnia | 2 | 0 | 0 | 0 |
| Mood alteration | 0 | 2 | 0 | 0 |
| Taste loss | 4 | 0 | 0 | 0 |
| Dyspepsia | 1 | 1 | 0 | 0 |
| Stomatitis | 1 | 4 | 0 | 0 |
| Abdominal pain | 2 | 0 | 0 | 0 |
| Dyspnea | 0 | 3 | 0 | 0 |
| Cough | 0 | 2 | 0 | 0 |
| Diaphoresis | 3 | 1 | 0 | 0 |
| Rash | 0 | 1 | 0 | 0 |
| Edema | 2 | 1 | 0 | 0 |
| Chest pain | 1 | 0 | 0 | 0 |
| Hyponatremia | 0 | 0 | 2 | 0 |
| Dizziness | 1 | 1 | 0 | 0 |
| Ataxia | 0 | 1 | 0 | 0 |
| Infection | 1 | 1 | 1 | 0 |
| Tumor lysis | 0 | 0 | 0 | 0 |
| Hyperglycemia | 0 | 0 | 0 | 0 |
| Constipation | 5 | 1 | 0 | 0 |
| Arthralgia | 1 | 1 | 0 | 0 |
| Catheter-related infection | 1 | 0 | 0 | 0 |
| Sinus tachycardia | 1 | 0 | 0 | 0 |
| Epistaxis | 1 | 0 | 0 | 0 |
| Neutropenia | 2 | 1 | 1 | 6 |
| Anemia | 6 | 2 | 0 | 0 |
| Thrombocytopenia | 0 | 2 | 6 | 2 |
Treatment outcome of patients with AML
The toxicities observed in the AML cohort are summarized in Table 4. Of these 10 patients, 3 did not complete 1 cycle of therapy, 4 completed 1 cycle of therapy, and 3 received 2 cycles. Reasons for not completing the first cycle include (1) tumor lysis syndrome with hyperkalemia, hyperphosphatemia, hypocalcemia, hyperuricemia, and acute renal failure (1 patient); (2) development of persistent diarrhea with electrolyte disturbances (1 patient); and (3) grade 3 fatigue (1 patient). Of the 4 patients who completed 1 cycle of therapy, 3 had evidence of progressive disease and 1 had stable disease but was only able to complete 2 of the 3 treatments of cycle 2 because of increasing fatigue, nausea, and emesis. The remaining 3 patients all completed 2 cycles of therapy without evidence of response. All of these patients experienced nausea, emesis, and progressively worsening fatigue with continued depsipeptide treatment. While some patients experienced transient declines in blood or bone marrow blast counts and 1 developed tumor lysis syndrome with a decrease in peripheral blast count from 12.8 × 103/μL to less than 1 × 103/μL, no complete or partial responses were observed by NCI criteria.44 Therefore, due to both early and late constitutional symptoms and minimal evidence of clinical activity, further pursuit of this schedule of administration of depsipeptide in AML was abandoned.
Toxicity observed in patients with AML treated with depsipeptide
Toxicity . | Grade 1, no. . | Grade 2, no. . | Grade 3, no. . | Grade 4, no. . |
|---|---|---|---|---|
| Anorexia | 5 | 4 | 1 | NA |
| Fatigue | 4 | 3 | 2 | NA |
| Nausea | 2 | 5 | 1 | NA |
| Vomiting | 4 | 4 | NA | NA |
| Myalgias | 4 | 1 | 1 | NA |
| Headache | 2 | 1 | NA | NA |
| Diarrhea | 3 | 1 | 1 | NA |
| Headache | 2 | 1 | NA | NA |
| Petechiae | 1 | NA | NA | NA |
| Taste loss | 3 | NA | NA | NA |
| Stomatitis | 1 | NA | NA | NA |
| Abdominal pain | NA | NA | 1 | NA |
| Dyspnea | 1 | NA | NA | NA |
| Cough | 2 | NA | NA | NA |
| Chest Pain | 1 | NA | NA | NA |
| Dizziness | 2 | 1 | NA | NA |
| Ataxia | NA | 1 | NA | NA |
| Infection | NA | NA | NA | 1 |
| Epistaxis | 2 | NA | NA | NA |
| Constipation | 2 | NA | NA | NA |
| Ileus | NA | NA | 1 | NA |
| Pruritus | NA | 1 | NA | NA |
| Dehydration | NA | NA | 2 | NA |
| Tumor lysis syndrome | NA | NA | 1 | NA |
| Hypoxia | NA | NA | 1 | NA |
| Hypotension | 3 | 2 | NA | NA |
| Fever | 1 | NA | 2 | NA |
| Catheter-related infection | NA | 1 | NA | NA |
| Arthralgia | 2 | NA | NA | NA |
| Sinus tachycardia | 1 | NA | NA | NA |
| Rigors/chills | 1 | NA | NA | NA |
| Weight loss | 1 | NA | NA | NA |
Toxicity . | Grade 1, no. . | Grade 2, no. . | Grade 3, no. . | Grade 4, no. . |
|---|---|---|---|---|
| Anorexia | 5 | 4 | 1 | NA |
| Fatigue | 4 | 3 | 2 | NA |
| Nausea | 2 | 5 | 1 | NA |
| Vomiting | 4 | 4 | NA | NA |
| Myalgias | 4 | 1 | 1 | NA |
| Headache | 2 | 1 | NA | NA |
| Diarrhea | 3 | 1 | 1 | NA |
| Headache | 2 | 1 | NA | NA |
| Petechiae | 1 | NA | NA | NA |
| Taste loss | 3 | NA | NA | NA |
| Stomatitis | 1 | NA | NA | NA |
| Abdominal pain | NA | NA | 1 | NA |
| Dyspnea | 1 | NA | NA | NA |
| Cough | 2 | NA | NA | NA |
| Chest Pain | 1 | NA | NA | NA |
| Dizziness | 2 | 1 | NA | NA |
| Ataxia | NA | 1 | NA | NA |
| Infection | NA | NA | NA | 1 |
| Epistaxis | 2 | NA | NA | NA |
| Constipation | 2 | NA | NA | NA |
| Ileus | NA | NA | 1 | NA |
| Pruritus | NA | 1 | NA | NA |
| Dehydration | NA | NA | 2 | NA |
| Tumor lysis syndrome | NA | NA | 1 | NA |
| Hypoxia | NA | NA | 1 | NA |
| Hypotension | 3 | 2 | NA | NA |
| Fever | 1 | NA | 2 | NA |
| Catheter-related infection | NA | 1 | NA | NA |
| Arthralgia | 2 | NA | NA | NA |
| Sinus tachycardia | 1 | NA | NA | NA |
| Rigors/chills | 1 | NA | NA | NA |
| Weight loss | 1 | NA | NA | NA |
Cardiac effects of depsipeptide
Preclinical toxicology identified that cardiac toxicity may be dose limiting with depsipeptide. Outside of previously reported electrocardiogram (EKG) findings, including ST and T wave abnormalities,39 minimal cardiac effects were observed with depsipeptide treatment. One patient developed a transient left bundle branch block 22 hours after completion of the first dose of depsipeptide. This resolved spontaneously the following day and did not recur with subsequent depsipeptide treatments. One other patient developed an isolated elevated troponin level with the first dose of depsipeptide treatment that was transient and not associated with any other serologic, EKG, or echocardiographic evidence of myocardial ischemia or infarction. Serial cardiac ejection fractions as assessed by MUGA scans or echocardiogram demonstrated no diminishment in any patient following treatment with depsipeptide.
Pharmacokinetics
The pharmacokinetics of depsipeptide was studied in all 20 patients enrolled in this trial. All the plasma concentration time profiles fit a 2-compartment model with a 4-hour infusion input. Initially, pharmacokinetic parameters for the 2 patient populations (AML and CLL) were separately estimated but were found to be statistically equivalent. Therefore, these 2 patient populations were grouped together. The outcome of this analysis is shown in Table 5. The maximum clearance (Cmax) was 1.39 μM, t1/2, α was 0.25 hours, t1/2, β was 3.67 hours, and area under curve (AUC) 6.02 μM · hour. The parameter estimates all gave reasonable standard. Overall, the pharmacology observed in this study was similar to those reported in a previous study by Sandor et al,39 which used comparable doses of depsipeptide.
Relevant pharmacokinetic parameters of depsipeptide
Parameter . | Current study . | Sandor et al39 . |
|---|---|---|
| Dose, mg/m2 | 13 | 17.8 |
| AUC, μM · h | 6.02 ± 2.93 | 4.20 ± 2.48* |
| Cmax, μM | 1.39 ± 0.69 | 1.02 ± 0.56* |
| Cl, L/h/m2 | 4.81 ± 2.02 | 10.5 ± 6.4 |
| Vss, L/m2 | 3.29 ± 0.94 | 37.1 ± 47.8 |
| Uex, % dose | 1.18 ± 0.72 | Not given |
| T1/2, α, h (range) | 0.25 (0.1-0.6) | 0.42 ± 0.25 |
| T1/2, β, h (range) | 3.67 (2.3-6.5) | 8.1 ± 6.0 |
| MRT† | 0.82 ± 0.37 | NR |
Parameter . | Current study . | Sandor et al39 . |
|---|---|---|
| Dose, mg/m2 | 13 | 17.8 |
| AUC, μM · h | 6.02 ± 2.93 | 4.20 ± 2.48* |
| Cmax, μM | 1.39 ± 0.69 | 1.02 ± 0.56* |
| Cl, L/h/m2 | 4.81 ± 2.02 | 10.5 ± 6.4 |
| Vss, L/m2 | 3.29 ± 0.94 | 37.1 ± 47.8 |
| Uex, % dose | 1.18 ± 0.72 | Not given |
| T1/2, α, h (range) | 0.25 (0.1-0.6) | 0.42 ± 0.25 |
| T1/2, β, h (range) | 3.67 (2.3-6.5) | 8.1 ± 6.0 |
| MRT† | 0.82 ± 0.37 | NR |
Data are shown as mean ± SD. For the current study, n = 20; for Sandor et al39 , n = 13. AUC indicates area under the curve; Cmax, maximum clearance; Vss, volume at steady state; Uex, excreted in urine; T1/2, half-life; MRT, mean retention time; NR indicates not reported.
Recalculated from Sandor et al.39
Estimated from a noncompartment method.
Pharmacodynamic data
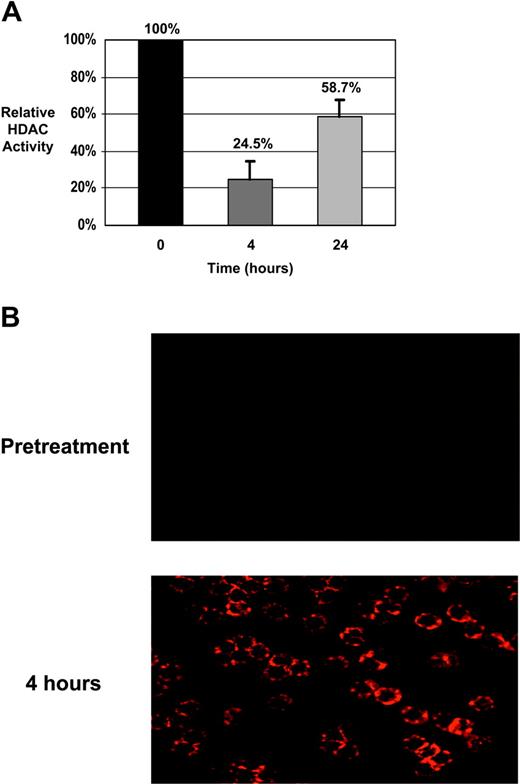
Patients with CLL. The extent of H3 and H4 histone acetylation was assessed in samples from 7 patients with CLL in which sufficient cell numbers were isolated. Histone proteins were extracted and separated on acrylamide gels. Total protein equivalence was verified using SYPRO Red fluorescent staining and acetylation of histones were detected by immunoblotting using antibodies specific for either acetylated H3 or H4. Amounts of acetylated histones were then calculated relative to the pretreatment sample for each case. Tables 6 and 7 show the results from these analyses. The MEPD, defined as an increase of at least 100% (ie, doubling) in histone acetylation relative to the pretreatment sample, was attained for H3 in all 7 patients at 4 hours and in 6 of 7 patients at 24 hours. At least a 100% increase in H4 acetylation was also observed in all 7 patients with CLL at 4 hours and 6 of 7 patients at 24 hours. Also shown in Table 6 are the effects of depsipeptide on HDAC enzymatic activity. In all patients tested, HDAC activity was notably reduced 4 hours after the initiation of treatment and partially but not completely recovered to pretreatment levels after 24 hours. In most cases, the levels of H3 and H4 acetylation were higher at the 4-hour time point than at 24 hours. This initial increase in histone acetylation at 4 hours, followed by a decline at 24 hours, corresponded to the pattern of HDAC enzymatic inhibition in 5 of 7 cases. When acetylation of specific lysine (K) residues was analyzed, increases were noted in H4 K5, H4 K12, H4 K8, and H3 K9 as previously observed in our in vitro work37 (not shown). Interestingly, increases in H3 K14 and H4 K16 acetylation were also detected that we had not previously seen. As analysis of changes in histone acetylation by immunoblotting does not distinguish between large increments in a small proportion of tumor cells versus equivalent increments among the majority of tumor cells, we also examined changes in H3 acetylation by immunocytochemistry. As shown in Figure 1A, these experiments revealed increase in H3 and H4 acetylation in virtually all the CLL cells examined. Finally, to verify that these changes in histone acetylation correspond to inhibition of HDAC, we performed serial assessment of HDAC activity on these same patients. HDAC activity declined to 24.6% of baseline immediately after treatment but rose again to 59% of baseline 24 hours after treatment, as shown in Figure 1B. Thus, temporally the pattern of HDAC inhibition corresponded well to the changes in histone hyperacetylation.
In vivo histone protein acetylation in CLL patient cells following depsipeptide treatment
. | Relative increase in H4 acetylation, % before treatment . | . | Relative increase in H3 acetylation, % before treatment . | . | Relative HDAC enzyme activity, % before treatment . | . | Maximum reduction in lymphocyte count, % . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient no. . | 4 h after treatment . | 24 h after treatment . | 4 h after treatment . | 24 h after treatment . | 4 h after treatment . | 24 h after treatment . | . | |||
| 1 | 260 | 170 | 240 | 800 | 26.0 | 52.5 | 45 | |||
| 2 | 630 | 440 | 460 | 370 | 19.4 | 64.1 | 76 | |||
| 3 | 200 | 230 | 240 | 410 | 30.6 | 45.3 | 61 | |||
| 4 | 390 | 770 | 510 | 330 | 16.2 | 86.8 | 59 | |||
| 5 | 800 | 270 | 440 | 260 | 24.2 | 58.3 | 50 | |||
| 6 | 340 | 240 | 260 | 100 | 37.5 | 66.2 | 50 | |||
| 7 | 300 | 200 | 630 | 390 | 36.2 | 80.2 | 62 | |||
. | Relative increase in H4 acetylation, % before treatment . | . | Relative increase in H3 acetylation, % before treatment . | . | Relative HDAC enzyme activity, % before treatment . | . | Maximum reduction in lymphocyte count, % . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient no. . | 4 h after treatment . | 24 h after treatment . | 4 h after treatment . | 24 h after treatment . | 4 h after treatment . | 24 h after treatment . | . | |||
| 1 | 260 | 170 | 240 | 800 | 26.0 | 52.5 | 45 | |||
| 2 | 630 | 440 | 460 | 370 | 19.4 | 64.1 | 76 | |||
| 3 | 200 | 230 | 240 | 410 | 30.6 | 45.3 | 61 | |||
| 4 | 390 | 770 | 510 | 330 | 16.2 | 86.8 | 59 | |||
| 5 | 800 | 270 | 440 | 260 | 24.2 | 58.3 | 50 | |||
| 6 | 340 | 240 | 260 | 100 | 37.5 | 66.2 | 50 | |||
| 7 | 300 | 200 | 630 | 390 | 36.2 | 80.2 | 62 | |||
Data are shown as percentage relative to the pretreatment sample in each case, set at 100%. Increases of H3 and H4 acetylation of at least 100% more than the pretreatment sample (ie, 200% before treatment or greater) are considered to meet the MEPD criteria.
In vivo histone protein acetylation in AML patient cells 4 hours after depsipeptide treatment
Patient no. . | Relative increase in H4 acetylation, % before treatment . | Relative increase in H3 acetylation, % before treatment . | Maximum reduction in bone marrow blast count . |
|---|---|---|---|
| 8 | 160 | 380 | NA |
| 9 | 200 | 340 | NA |
| 10 | 100 | 290 | NA |
| 12 | 340 | 550 | NA |
| 13 | 330 | 330 | NA |
Patient no. . | Relative increase in H4 acetylation, % before treatment . | Relative increase in H3 acetylation, % before treatment . | Maximum reduction in bone marrow blast count . |
|---|---|---|---|
| 8 | 160 | 380 | NA |
| 9 | 200 | 340 | NA |
| 10 | 100 | 290 | NA |
| 12 | 340 | 550 | NA |
| 13 | 330 | 330 | NA |
Data are shown as percentage relative to the pretreatment sample in each case, set at 100%. Increases of at least 100% over the pretreatment sample (ie, 200% or greater) are considered to meet the MEPD criteria. NA indicates no decrease in percentage of bone marrow blast at any time point evaluated.
Biologic effects of depsipeptide in CLL patient cells. (A) CLL patient cells were isolated prior to therapy with depsipeptide, as well as immediately before treatment and 4 and 24 hours after the start of infusion. Lysates were prepared and analyzed for specific HDAC activity. Activity is shown as a percentage relative to baseline (pretreatment). Significant HDAC inhibition is noted at completion of infusion (4 hours) and diminishes by 24 hours. (B) CLL patient cells isolated before therapy and at 4 hours following the start of the infusion were analyzed by fluorescent immunocytochemistry using an antibody to acetylated H3. Acetylation of H3 is minimal at baseline but is evident in the majority of the cells at the 4-hour time point.
Biologic effects of depsipeptide in CLL patient cells. (A) CLL patient cells were isolated prior to therapy with depsipeptide, as well as immediately before treatment and 4 and 24 hours after the start of infusion. Lysates were prepared and analyzed for specific HDAC activity. Activity is shown as a percentage relative to baseline (pretreatment). Significant HDAC inhibition is noted at completion of infusion (4 hours) and diminishes by 24 hours. (B) CLL patient cells isolated before therapy and at 4 hours following the start of the infusion were analyzed by fluorescent immunocytochemistry using an antibody to acetylated H3. Acetylation of H3 is minimal at baseline but is evident in the majority of the cells at the 4-hour time point.
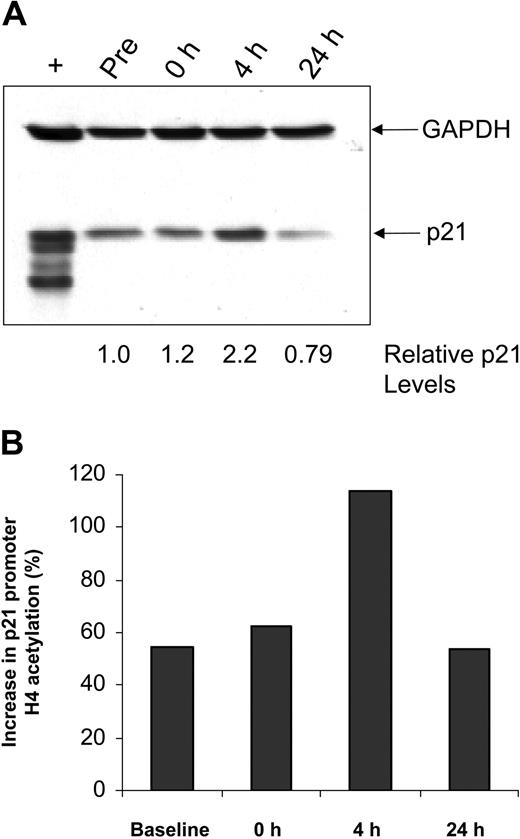
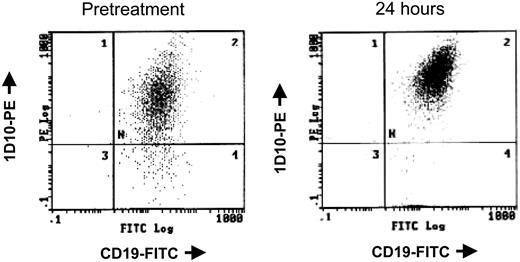
The cell cycle regulatory protein p21 closely controls cell cycle and is induced by treatment with HDAC inhibitors, including depsipeptide.47 In vitro studies with alternative HDAC inhibitors48,49 in cell lines have demonstrated that p21 induction occurs concurrent with p21 promoter region acetylation. Such up-regulation of p21 protein expression is also seen in a subset of patients with CLL examined, including the patient sample shown in Figure 2A. Chromatin immunoprecipitation was performed to ascertain whether this increase in p21 expression was a nonspecific response to DNA damage, as can occur with irradiation or fludarabine, or directly due to depsipeptide influence on the p21 promoter. Here it is demonstrated that, following treatment with depsipeptide, an increase in p21 protein expression (Figure 2A) occurs concurrently with H4 acetylation of the p21 promoter (Figure 2B). Direct or indirect modulation of other proteins, including HLA-DR50 and cellular Fas-associated death domain protein (FADD)–like interleukin-1β-converting enzyme (FLICE)–inhibitory protein (c-FLIP),37 has been previously noted in vitro with depsipeptide. Figure 3 demonstrates this also occurs in vivo following treatment with depsipeptide, where the 1D10 (whose target is the HLA-DR β-chain) antigen is increased following 24-hour treatment with depsipeptide. Similar to our in vitro work with HDAC inhibitors,37,51 we observed that the antiapoptotic protein c-FLIP is decreased in some patients following treatment with depsipeptide, but this decrease was not consistently seen (not shown).
Effects of depsipeptide on p21 in vivo. (A) Lysates from CLL patient cells obtained before and 4 or 24 hours after depsipeptide administration were analyzed by immunoblot using an anti-p21 antibody. Simultaneous incubation with anti-GAPDH antibody was performed to detect protein-loading differences. (B) Cells from these same time points were subjected to chromatin immunoprecipitation analysis using anti-acetylated H4 antibody. The graph shows the increase in p21 promoter acetylation relative to GAPDH.
Effects of depsipeptide on p21 in vivo. (A) Lysates from CLL patient cells obtained before and 4 or 24 hours after depsipeptide administration were analyzed by immunoblot using an anti-p21 antibody. Simultaneous incubation with anti-GAPDH antibody was performed to detect protein-loading differences. (B) Cells from these same time points were subjected to chromatin immunoprecipitation analysis using anti-acetylated H4 antibody. The graph shows the increase in p21 promoter acetylation relative to GAPDH.
Depsipeptide causes increased 1D10 antigen expression in vivo. Before and 24 hours following treatment with 13 mg/m2 depsipeptide of a patient with CLL, peripheral mononuclear cells were isolated and analyzed simultaneously for surface expression of CD19 and the HLA-DR antigen 1D10.
Depsipeptide causes increased 1D10 antigen expression in vivo. Before and 24 hours following treatment with 13 mg/m2 depsipeptide of a patient with CLL, peripheral mononuclear cells were isolated and analyzed simultaneously for surface expression of CD19 and the HLA-DR antigen 1D10.
Patients with AML. Due to the need to perform serial bone marrow biopsies to obtain sufficient cell counts, limited pharmacodynamic studies were performed on the patients with AML. Sufficient cell numbers were obtained from 5 of the 10 patients with AML for these experiments at 4 hours following the start of therapy. The MEPD (increase in acetylation of at least 100% over the pretreatment sample) for H3 was obtained in all 5 patients with AML at this time point, and the MEPD for H4 was attained in 3 of these 5 patients (Table 7). Similar patterns of lysine-specific acetylation were observed in the patients with AML as seen in the patients with CLL (not shown).
Discussion
In this report, we describe the results of the first disease-specific MEPD study of the HDAC inhibitor depsipeptide in patients with AML and CLL. This trial used a starting dose of depsipeptide of 13 mg/m2 over 4 hours administered on days 1, 8, and 15 every 28 days. In both groups of patients we demonstrated some antileukemia activity that was more evident in the patients with CLL. Indeed, all 7 of the patients with CLL with elevated leukocyte counts had a significant decrease during treatment (average decrease, 58%; range, 45%-76%). The decline in leukocyte count generally occurred within 1 to 2 treatment cycles and persisted to decrease in patients who continued therapy. The reductions in lymphocyte counts occurred over several days following treatment, often when histone acetylation had partially recovered to baseline levels. This suggests the presence of a secondary event such as re-expression of a previously silenced gene, versus depsipeptide-induced histone acetylation leading directly to death. Although no significant cardiac toxicity was observed in the patients treated, nausea, fatigue, anorexia, and other constitutional symptoms developed in the great majority of patients despite the use of aggressive pretreatment and later antiemetic treatment. This toxicity limited the ability to continue therapy and to administer continued sustained doses of depsipeptide even in the presence of continuing clinical benefit. These constitutional symptoms did not meet grade 3 by the NCI common toxicity criteria, but it must be emphasized that this grading system better differentiates acute as opposed to chronic persistent toxicities observed in this study. In neither group did the clinical benefit observed meet the NCI criteria for partial responses. Furthermore, in patients that were dose-reduced because of these constitutional symptoms, lack of continued improvement and even progression was noted. Therefore, pursuit of the day 1, 8, and 15 depsipeptide schedule of administration at the current or lower doses than used in this MEPD study as initially proposed in patients with acute or chronic leukemia is not recommended.
Nevertheless, from this trial we determined that the 13-mg/m2 dose of depsipeptide administered in a 4-hour infusion attained the MEPD biologic effect of increasing acetylation of H3 and H4 by at least 100% over pretreatment levels in the majority of patients, even though we were unable to obtain sufficient cells for analysis in every case. Furthermore, in all 7 patients with CLL tested, we demonstrated that HDAC activity was substantially reduced within 4 hours following the initiation of depsipeptide treatment. This enzymatic activity in each case had partially but not completely recovered at the 24-hour time point. This finding correlates well with our pharmacokinetic data, which demonstrated a depsipeptide half-life of approximately 3.7 hours in this patient group. The reduction of histone acetylation at 24 hours after therapy, relative to the 4-hour time point, corresponded in 5 of the 7 CLL cases to partial recovery of HDAC activity. This suggests that there may in some cases be factors other than HDAC enzymatic activity that regulate the time course of histone deacetylation. Finally, in a subset of patients we demonstrated that the in vivo induction of p21 protein by depsipeptide in part corresponds to increased acetylation of the promoter region of the p21 gene. Other effects of depsipeptide previously noted in vitro were also observed, including up-regulation of the HLA-DR 1D10 antigen on the surface of CLL cells from a subset of patients. This detectable increase in 1D10 expression presents intriguing possibilities for future studies investigating combination therapies with monoclonal antibodies, such as Hu1D10, directed at cell-surface antigens. In agreement with our earlier in vitro data,37 we also confirmed down-regulation of c-FLIP, an important protein in preventing activation of the extrinsic pathway of apoptosis in CLL cells. Application of this biologic pharmacodynamic data to a new schedule of depsipeptide that allows dose escalation or de-escalation as initially proposed but not performed in this trial will be pursued in a separate MEPD study. Our study supports the notion that pharmacodynamic data can be obtained in real time to allow interpretation of biologic endpoints as part of MEPD study designs using targeted therapies such as depsipeptide.
The pharmacokinetics of depsipeptide were similar to both previous solid-tumor trials39,40 with this agent. Interestingly, the total depsipeptide AUC inversely correlated with increased acetylation of H4, and depsipeptide clearance directly correlated with H4 acetylation increases. These findings support the hypothesis that depsipeptide as administered is actually a less-active HDAC inhibitor that is then metabolized in vivo to an intermediate with increased potency, as was suggested in 2 recent studies.34,35 Importantly, our work demonstrates that real time in vivo studies of target validation can be performed in patients with CLL and AML, and that the 13 mg/m2 dose may be sufficient to mediate the biologic effects of depsipeptide in vivo. These findings may enable the use of depsipeptide in single-agent studies with alternative schedules of administration, or in combination with other agents such as DNA hypomethylating agents or monoclonal antibodies.
The findings of significant nausea, fatigue, anorexia, and other constitutional symptoms with depsipeptide therapy has also been noted in the trial performed by Marshall et al,40 in which a day-1, -8, and -15 schedule of administration was also used. Of the 6 patients treated at a 13.2-mg/m2 dose, constitutional symptoms developed during the first (3 patients) or second (4 patients) course of therapy, with no patients proceeding past this point. Of the patients with CLL who received 3 months (n = 1) or 4 months (n = 2) of depsipeptide treatment, all discontinued therapy because of chronic increasing constitutional symptoms. The only other depsipeptide phase 1 trial reported to date used a day-1 and -5 schedule in which the recommended phase 2 dose was 17.8 mg/m2 administered over 4 hours. Of the 9 patients treated at this dose, the great majority experienced grade 1 or 2 fatigue, nausea, emesis, and hypocalcemia. Fatigue, nausea, emesis, and electrolyte disturbances (hypophosphatemia and hypocalcemia) were noted in all 8 patients treated at the higher dose level of 24.9 mg, and in some patients these effects were dose limiting. It is likely that these constitutional symptoms are not specific to depsipeptide, but rather are a clinical manifestation of inhibition of HDAC, as they have been observed with other HDAC inhibitors, including phenylbutyrate,52,53 CI-994,54 and high doses of valproic acid.55 Given the transient hypotension, low-grade fever, and constitutional symptoms observed in many patients receiving depsipeptide and other HDAC inhibitors, future studies of cytokine release (eg, tumor necrosis factor α [TNF-α] and interleukin 6 [IL-6], as is seen in a similar flavopiridol inflammatory syndrome) should be considered.56 Understanding the pathophysiology of HDAC inhibitor–induced constitutional symptoms should be an important focus of future investigation with these agents. Additionally, preclinical studies need to be performed to assure that therapies added to diminish the side effects of depsipeptide (eg, corticosteroids) do not negate the desired biologic effects of these agents.
Epigenetic silencing by altered promoter histone acetylation or methylation can occur in normal57 as well as malignant cells,17-25 including CLL and AML. Thus, histone deacetylase inhibitors that directly alter histone acetylation in both normal and malignant cells have the potential to reactivate similar genes and therefore lack specificity. Additionally, genes such as MDR1 that mediate drug resistance to a variety of natural products (including depsipeptide) also can be activated directly by depsipeptide.58 Despite this hypothetical concern, we have demonstrated that with 2 structurally different histone deacetylase inhibitors depsipeptide36,37 and MS27551 that selective cytotoxicity is observed relative to normal hematopoietic cells. Future studies examining induction of MDR1 and other potential genes that mediate resistance to apoptosis that are re-expressed by depsipeptide induced inhibition of histone acetylation should be included in clinical studies of this agent.
In conclusion, our data demonstrate that the HDAC inhibitor depsipeptide has some clinical activity in AML and CLL and in vivo consequences on histone protein acetylation, enzyme inhibition, and down-stream targets, similar to effects previously shown in vitro. However, non–life-threatening but chronic constitutional symptoms occur frequently with depsipeptide and mandate exploration of alternative schedules to allow repeated dosing.
Prepublished online as Blood First Edition Paper, October 5, 2004; DOI 10.1182/blood-2004-05-1693.
Supported by The Chronic Lymphocytic Leukemia (CLL) Research Consortium (P01 CA81534) (J.C.B., M.R.G.), National Cancer Institute (NCI) (R21 CA096323) (J.C.B.) and (K08 CA90469) (G.M.), The Sidney Kimmel Cancer Research Foundation (J.C.B.), The Leukemia and Lymphoma Society of America (J.C.B), National Health, Lung, and Blood Institute/National Institutes of Health (NHLBI/NIH) (K24-HLO 4208) (P.B.), The D. Warren Brown Foundation (J.C.B.), and The American Cancer Society (M.R.P.).
J.C.B. and G.M. contributed equally to this project.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal