Abstract
Fanconi anemia (FA) is characterized by congenital abnormalities, bone marrow failure, chromosome fragility, and cancer susceptibility. Eight FA-associated genes have been identified so far, the products of which function in the FA/BRCA pathway. A key event in the pathway is the monoubiquitination of the FANCD2 protein, which depends on a multiprotein FA core complex. In a number of patients, spontaneous genetic reversion can correct FA mutations, leading to somatic mosaicism. We analyzed the FA/BRCA pathway in 53 FA patients by FANCD2 immunoblots and chromosome breakage tests. Strikingly, FANCD2 monoubiquitination was detected in peripheral blood lymphocytes (PBLs) in 8 (15%) patients. FA reversion was further shown in these patients by comparison of primary fibro-blasts and PBLs. Reversion was associated with higher blood counts and clinical stability or improvement. Once constitutional FANCD2 patterns were determined, patients could be classified based on the level of FA/BRCA pathway disruption, as “FA core” (upstream inactivation; n = 47, 89%), FA-D2 (n = 4, 8%), and an unidentified downstream group (n = 2, 4%). FA-D2 and unidentified group patients were therefore relatively common, and they had more severe congenital phenotypes. These results show that specific analysis of the FA/BRCA pathway, combined with clinical and chromosome breakage data, allows a comprehensive characterization of FA patients.
Introduction
Fanconi anemia (FA) is an autosomal recessive condition characterized by congenital abnormalities, progressive bone marrow failure, chromosome fragility, and cancer susceptibility.1,2 Complementation experiments have defined at least 11 groups,3,4 and to date, 8 corresponding FA-associated genes have been identified (FANCA, FANCC, FANCD1/BRCA2, FANCD2, FANCE, FANCF, FANCG/XRCC9, and FANCL/PHF9).3,5-8 Products of these genes function in the common FA/BRCA pathway.9 The FANCA, -C, -E, -F, -G, and -L proteins are part of a nuclear multiprotein core complex, which triggers activating monoubiquitination of the FANCD2 protein during S phase and after exposure to DNA crosslinking agents.8,10 Monoubiquitinated FANCD2 colocalizes in nuclear foci with BRCA1, BRCA2 (identified as the FANCD1 protein), and NBS1.7,10-13 In addition, FANCD2 can be phosphorylated in an ATM/ATR-dependent manner in response to DNA damage, and it is involved in an S-phase checkpoint.14-16 Although cellular targets of the pathway are unknown, the FA/BRCA pathway is likely to be involved in an integrated cellular response to S-phase DNA damage, especially due to DNA interstrand crosslinks, in order to maintain genomic integrity.9
There is a remarkably high clinical variability among FA patients with regard to both the congenital abnormalities and the subsequent progressive development of bone marrow failure and neoplasia.1,2,17,18 The variability appears only partially linked to the different genotypes.17,19-22 The emergence of revertant cells, leading to hematopoietic mosaicism (the presence of genetically distinct hematopoietic populations), might also participate in the hematological variability of the disease over time in a number of FA patients. In revertant cells, acquired genetic events produce a functional FA gene that confers cellular resistance to genotoxic agents.23-26 It is likely that a growth advantage of such revertant cells favors the progressive replacement of defective cells in the bone marrow. Although the frequency and clinical consequences of somatic mosaicism are largely unknown, the presence of revertant cells can confuse FA diagnostic tests, giving ambiguous or even false-negative results.26-30
The large number of possible mutations in FA-associated genes, and the existence of as-yet-unknown genes, precludes specific molecular analysis as a first-line diagnosis of FA in unselected patients.3 Diagnosis of FA is therefore based on functional cellular tests, namely chromosome breakage tests and cell cycle tests, which take advantage of the hypersensitivity of FA cells to DNA crosslinking agents.31-33 The identification of the FANCD2 gene and further description of the unique FA/BRCA pathway have suggested new specific tests based on the detection of posttranslational modifications of the FANCD2 target protein.34 We here evaluated the FA/BRCA pathway by analysis of FANCD2 immunoblots in a prospective cohort of FA patients. We show that this analysis, combined with clinical data and chromosome breakage tests, allows a fine characterization of FA patients by detection of somatic mosaicism and classification of the disease according to the level of the FA/BRCA pathway disruption. Clinical correlations were addressed, considering both the revertant status and the level of FA/BRCA pathway disruption.
Patients, materials, and methods
Patients and cell lines
From February 2002 to February 2004, 53 FA patients were included in the study (allografted patients were excluded). All patients were seen at Saint-Louis Hospital (Paris, France) except 4 patients who were seen elsewhere in France by expert collaborators and whose samples were sent to our laboratory together with clinical data. Informed consents were obtained from the patients and/or their relatives. The study was approved by the review board of the Fédération d' Hématologie, Hoôpital Saint-Louis, Paris. Unique patient numbers were used to refer to FA patients and samples once the FA diagnosis was established: EGFA (European Group for Fanconi Anemia) followed by identification number and “-L” for lymphocytes, “-F” for fibroblasts, and “-E” for Epstein-Barr virus (EBV)-immortalized cell lines. The diagnosis of FA was based on clinical data, including personal and familial history, and physical examination, together with chromosome breakage tests. Main characteristics of the patients are given in Table 1. For all patients, the following information was available: sex, birthdate, date of diagnosis, date of entry in the study, description of the clinical phenotype, and blood cell counts. Short stature was defined by a height below 2 standard deviations (SDs, which corresponds to the 3rd percentile) at inclusion in the study, or before the use of androgens. Facial appearance was defined as characteristic or not after examination. Features taken into account were microcephaly, broad nasal base with micrognathia and triangular face, and eye abnormalities (microphtalmia and epicanthal folds). Skin abnormalities were defined by any combination of café-au-lait spots, hypopigmented spots or areas, and hyperpigmented areas. Renal and cardiac malformations were evaluated by echography, and limb abnormalities were searched for by examination and X-rays. The extent of the malformation syndrome was classified as limited (< 3 sites) or extensive (3 or more sites) according to the classification that we used in a previous study.35 Overall, there were 29 males and 24 females. Median age at diagnosis, and median age at entry in this study, were 7 years (range, birth to 36 years), and 10 years (range, 2 to 36 years), respectively. At last examination, 18 patients had short stature (range, -2SDto -5 SD); among the others, no patient had an height greater than normal median value, except for 2 patients treated with androgens. Facial appearance was characteristic in 43 patients, 42 had skin abnormalities, and limb abnormalities were detected in 28 patients. The malformation syndrome was extensive in 16 patients and limited in 37.
Clinical and biological data in 53 FA patients
. | . | . | . | . | Blood cell counts . | . | . | . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients . | Sex . | Extent of malformation . | Age at diagnosis, y . | Age at study, y . | WBC, × 109/L . | ANC, × 109/L . | Hb, g/L . | Ptl, × 109/L . | Bone marrow karyotype . | PBL breaks . | PBL FANCD2 . | |||
| FA-D2 | ||||||||||||||
| EGFA050 | M | E | 0.3 | 4 | 6.1 | 2.1 | 124 | 176 | N | + | 2 | |||
| EGFA014 | F | E | 3 | 6 | 4.3 | 1.2 | 126 | 26 | N | + | 0/2f | |||
| EGFA021 | M | E | 2.8 | 7 | 5.5 | 1.9 | 108 | 187 | N | + | 0/2f | |||
| EGFA066 | M | E | 6 | 7 | 5.2 | 2.4 | 109 | 310 | N | + | 0/2f | |||
| Unidentified downstream group | ||||||||||||||
| EGFA006 | F | E | 0.25 | 9 | 6.4 | 3.3 | 98 | 350 | N | — | 2 | |||
| EGFA053 | M | E | 0.3 | 10 | 4.0 | 1.2 | 118 | 83 | N | AMB | 2 | |||
| FA core | ||||||||||||||
| EGFA008 | M | L | 13 | 23 | 3.7 | 1.7 | 134 | 89 | N | — | 2 | |||
| EGFA012 | M | E | 0.1 | 6 | 5.2 | 2.1 | 127 | 104 | N | + | 2 | |||
| EGFA013 | M | L | 9 | 21 | 5.5 | 3.4 | 153 | 233 | N | + | 2 | |||
| EGFA039 | M | L | 10 | 10 | 7.2 | 4.0 | 139 | 98 | N | + | 2 | |||
| EGFA047 | M | L | 6 | 34 | 5.3 | 2.9 | 157 | 111 | N | — | 2 | |||
| EGFA001* | F | L | 6 | 13 | 3.8 | 1.1 | 131 | 117 | N | + | 1 | |||
| EGFA002* | F | L | 6 | 13 | 3.4 | 0.9 | 136 | 109 | N | + | 1 | |||
| EGFA003 | F | L | 11 | 11 | 2.3 | 1.0 | 92 | 10 | ABN | + | 1 | |||
| EGFA004 | F | L | 10 | 10 | 3.0 | 1.2 | 117 | 109 | N | + | 1 | |||
| EGFA005 | M | E | 10 | 34 | 1.4 | 0.6 | 82 | 35 | N | + | 1 | |||
| EGFA007 | F | E | 2.3 | 7 | 2.7 | 0.4 | 108 | 53 | N | + | 1 | |||
| EGFA010 | F | L | 5 | 10 | 3.5 | 0.9 | 112 | 51 | N | + | 1 | |||
| EGFA015 | M | L | 14 | 21 | 5.5 | 2.0 | 95 | 11 | ABN† | — | 1 | |||
| EGFA016 | M | L | 5 | 13 | 3.4 | 1.1 | 78 | 32 | N | + | 1 | |||
| EGFA017 | M | L | 5 | 27 | 3.4 | 1.7 | 136 | 197 | N | + | 1 | |||
| EGFA019 | F | L | 8 | 10 | 3.4 | 2.1 | 121 | 30 | N | + | 1 | |||
| EGFA023‡ | M | L | 7 | 16 | 3.5 | 1.6 | 121 | 26 | ABN | + | 1 | |||
| EGFA024‡ | F | L | 7 | 16 | 2.8 | 0.9 | 94 | 42 | N | + | 1 | |||
| EGFA025 | M | L | 5 | 5 | 4.3 | 0.9 | 47 | 15 | N | + | 1 | |||
| EGFA026 | M | L | 8 | 9 | 2.0 | 0.3 | 76 | 20 | N | + | 1 | |||
| EGFA028 | M | L | 4 | 6 | 1.6 | 0.3 | 62 | 63 | ABN§ | + | 1 | |||
| EGFA029‖ | F | L | 6 | 8 | 2.7 | 0.5 | 101 | 40 | N | + | 1 | |||
| EGFA030‖ | M | E | 1 | 3 | 7.6 | 1.8 | 118 | 118 | N | + | 1 | |||
| EGFA031 | M | E | 1.8 | 2 | 2.5 | 0.3 | 119 | 81 | ABN | + | 1 | |||
| EGFA032 | M | L | 9 | 9 | 2.6 | 1.1 | 99 | 45 | N | + | 1 | |||
| EGFA033 | F | L | 8 | 8 | 2.6 | 1.2 | 102 | 16 | N | + | 1 | |||
| EGFA034¶ | M | L | 5 | 10 | 3.0 | 0.8 | 96 | 45 | N | + | 1 | |||
| EGFA035¶ | F | L | 6 | 11 | 2.4 | 0.3 | 78 | 42 | N | + | 1 | |||
| EGFA036 | F | L | 27 | 32 | 4.3 | 0.6 | 112 | 97 | N | + | 1 | |||
| EGFA037 | M | L | 6 | 8 | 4.7 | 1.5 | 125 | 27 | N | + | 1 | |||
| EGFA040 | F | E | 5 | 5 | 3.4 | 1.6 | 95 | 46 | N | + | 1 | |||
| EGFA041 | M | L | 4 | 4 | 6.0 | 2.2 | 83 | 34 | N | + | 1 | |||
| EGFA042 | F | L | NA | 20 | 2.8 | 1.4 | 125 | 148 | N | + | 1 | |||
| EGFA044 | F | L | NA | 11 | 4.6 | 2.8 | 118 | 157 | N | + | 1 | |||
| EGFA045 | F | L | 15 | 35 | 2.9 | 1.7 | 111 | 103 | N | + | 1 | |||
| EGFA046 | M | E | 5 | 7 | 2.8 | 0.8 | 108 | 49 | N | + | 1 | |||
| EGFA048 | F | L | 6 | 10 | 3.6 | 0.8 | 104 | 48 | N | + | 1 | |||
| EGFA049 | M | L | 7 | 8 | 4.8 | 1.4 | 86 | 78 | N | + | 1 | |||
| EGFA051 | F | E | 6 | 8 | 3.5 | 0.9 | 46 | 39 | N | + | 1 | |||
| EGFA055 | F | L | 7 | 25 | 4.0 | 1.5 | 125 | 108 | N | + | 1 | |||
| EGFA057 | M | E | 7 | 7 | 3.2 | 0.8 | 115 | 31 | N | + | 1 | |||
| EGFA059 | M | L | 36 | 36 | 3.9 | 2.2 | 74 | 61 | N | + | 1 | |||
| EGFA060 | F | L | 4 | 4 | 2.6 | 0.4 | 90 | 58 | N | + | 1 | |||
| EGFA061 | M | E | 4 | 19 | 4.2 | 2.7 | 123 | 300 | ABN | + | 1 | |||
| EGFA062 | M | L | 10 | 18 | 3.2 | 1.0 | 102 | 55 | ABN | + | 1 | |||
| EGFA063 | F | L | 9 | 26 | 2.2 | 0.7 | 114 | 101 | N | + | 1 | |||
| EGFA067 | F | L | 9 | 20 | 5.8 | 3.5 | 119 | 55 | N | + | 1 | |||
. | . | . | . | . | Blood cell counts . | . | . | . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients . | Sex . | Extent of malformation . | Age at diagnosis, y . | Age at study, y . | WBC, × 109/L . | ANC, × 109/L . | Hb, g/L . | Ptl, × 109/L . | Bone marrow karyotype . | PBL breaks . | PBL FANCD2 . | |||
| FA-D2 | ||||||||||||||
| EGFA050 | M | E | 0.3 | 4 | 6.1 | 2.1 | 124 | 176 | N | + | 2 | |||
| EGFA014 | F | E | 3 | 6 | 4.3 | 1.2 | 126 | 26 | N | + | 0/2f | |||
| EGFA021 | M | E | 2.8 | 7 | 5.5 | 1.9 | 108 | 187 | N | + | 0/2f | |||
| EGFA066 | M | E | 6 | 7 | 5.2 | 2.4 | 109 | 310 | N | + | 0/2f | |||
| Unidentified downstream group | ||||||||||||||
| EGFA006 | F | E | 0.25 | 9 | 6.4 | 3.3 | 98 | 350 | N | — | 2 | |||
| EGFA053 | M | E | 0.3 | 10 | 4.0 | 1.2 | 118 | 83 | N | AMB | 2 | |||
| FA core | ||||||||||||||
| EGFA008 | M | L | 13 | 23 | 3.7 | 1.7 | 134 | 89 | N | — | 2 | |||
| EGFA012 | M | E | 0.1 | 6 | 5.2 | 2.1 | 127 | 104 | N | + | 2 | |||
| EGFA013 | M | L | 9 | 21 | 5.5 | 3.4 | 153 | 233 | N | + | 2 | |||
| EGFA039 | M | L | 10 | 10 | 7.2 | 4.0 | 139 | 98 | N | + | 2 | |||
| EGFA047 | M | L | 6 | 34 | 5.3 | 2.9 | 157 | 111 | N | — | 2 | |||
| EGFA001* | F | L | 6 | 13 | 3.8 | 1.1 | 131 | 117 | N | + | 1 | |||
| EGFA002* | F | L | 6 | 13 | 3.4 | 0.9 | 136 | 109 | N | + | 1 | |||
| EGFA003 | F | L | 11 | 11 | 2.3 | 1.0 | 92 | 10 | ABN | + | 1 | |||
| EGFA004 | F | L | 10 | 10 | 3.0 | 1.2 | 117 | 109 | N | + | 1 | |||
| EGFA005 | M | E | 10 | 34 | 1.4 | 0.6 | 82 | 35 | N | + | 1 | |||
| EGFA007 | F | E | 2.3 | 7 | 2.7 | 0.4 | 108 | 53 | N | + | 1 | |||
| EGFA010 | F | L | 5 | 10 | 3.5 | 0.9 | 112 | 51 | N | + | 1 | |||
| EGFA015 | M | L | 14 | 21 | 5.5 | 2.0 | 95 | 11 | ABN† | — | 1 | |||
| EGFA016 | M | L | 5 | 13 | 3.4 | 1.1 | 78 | 32 | N | + | 1 | |||
| EGFA017 | M | L | 5 | 27 | 3.4 | 1.7 | 136 | 197 | N | + | 1 | |||
| EGFA019 | F | L | 8 | 10 | 3.4 | 2.1 | 121 | 30 | N | + | 1 | |||
| EGFA023‡ | M | L | 7 | 16 | 3.5 | 1.6 | 121 | 26 | ABN | + | 1 | |||
| EGFA024‡ | F | L | 7 | 16 | 2.8 | 0.9 | 94 | 42 | N | + | 1 | |||
| EGFA025 | M | L | 5 | 5 | 4.3 | 0.9 | 47 | 15 | N | + | 1 | |||
| EGFA026 | M | L | 8 | 9 | 2.0 | 0.3 | 76 | 20 | N | + | 1 | |||
| EGFA028 | M | L | 4 | 6 | 1.6 | 0.3 | 62 | 63 | ABN§ | + | 1 | |||
| EGFA029‖ | F | L | 6 | 8 | 2.7 | 0.5 | 101 | 40 | N | + | 1 | |||
| EGFA030‖ | M | E | 1 | 3 | 7.6 | 1.8 | 118 | 118 | N | + | 1 | |||
| EGFA031 | M | E | 1.8 | 2 | 2.5 | 0.3 | 119 | 81 | ABN | + | 1 | |||
| EGFA032 | M | L | 9 | 9 | 2.6 | 1.1 | 99 | 45 | N | + | 1 | |||
| EGFA033 | F | L | 8 | 8 | 2.6 | 1.2 | 102 | 16 | N | + | 1 | |||
| EGFA034¶ | M | L | 5 | 10 | 3.0 | 0.8 | 96 | 45 | N | + | 1 | |||
| EGFA035¶ | F | L | 6 | 11 | 2.4 | 0.3 | 78 | 42 | N | + | 1 | |||
| EGFA036 | F | L | 27 | 32 | 4.3 | 0.6 | 112 | 97 | N | + | 1 | |||
| EGFA037 | M | L | 6 | 8 | 4.7 | 1.5 | 125 | 27 | N | + | 1 | |||
| EGFA040 | F | E | 5 | 5 | 3.4 | 1.6 | 95 | 46 | N | + | 1 | |||
| EGFA041 | M | L | 4 | 4 | 6.0 | 2.2 | 83 | 34 | N | + | 1 | |||
| EGFA042 | F | L | NA | 20 | 2.8 | 1.4 | 125 | 148 | N | + | 1 | |||
| EGFA044 | F | L | NA | 11 | 4.6 | 2.8 | 118 | 157 | N | + | 1 | |||
| EGFA045 | F | L | 15 | 35 | 2.9 | 1.7 | 111 | 103 | N | + | 1 | |||
| EGFA046 | M | E | 5 | 7 | 2.8 | 0.8 | 108 | 49 | N | + | 1 | |||
| EGFA048 | F | L | 6 | 10 | 3.6 | 0.8 | 104 | 48 | N | + | 1 | |||
| EGFA049 | M | L | 7 | 8 | 4.8 | 1.4 | 86 | 78 | N | + | 1 | |||
| EGFA051 | F | E | 6 | 8 | 3.5 | 0.9 | 46 | 39 | N | + | 1 | |||
| EGFA055 | F | L | 7 | 25 | 4.0 | 1.5 | 125 | 108 | N | + | 1 | |||
| EGFA057 | M | E | 7 | 7 | 3.2 | 0.8 | 115 | 31 | N | + | 1 | |||
| EGFA059 | M | L | 36 | 36 | 3.9 | 2.2 | 74 | 61 | N | + | 1 | |||
| EGFA060 | F | L | 4 | 4 | 2.6 | 0.4 | 90 | 58 | N | + | 1 | |||
| EGFA061 | M | E | 4 | 19 | 4.2 | 2.7 | 123 | 300 | ABN | + | 1 | |||
| EGFA062 | M | L | 10 | 18 | 3.2 | 1.0 | 102 | 55 | ABN | + | 1 | |||
| EGFA063 | F | L | 9 | 26 | 2.2 | 0.7 | 114 | 101 | N | + | 1 | |||
| EGFA067 | F | L | 9 | 20 | 5.8 | 3.5 | 119 | 55 | N | + | 1 | |||
Revertant patients are indicated in italics. Blood cell counts have been determined before any transfusion. WBC indicates white blood cells; ANC, absolute neutrophil count; Hb, hemoglobin; Plt, platelets; E, extensive malformation syndrome as defined in “Patients, materials, and methods”; L, limited; NA, information not available; N, no detected abnormality; ABN, clonal chromosomal abnormalities; +, positive; —, negative; AMB, ambiguous at the time of the study but formerly found positive at diagnosis; 2, detection of both the short and the long monoubiquitinated FANCD2 isoforms; 1, one single short isoform; and 0/2f, no FANCD2 bands or 2 faint bands in long exposure (Fig 3A).
Siblings
Siblings
Siblings
Siblings
Acute myeloid leukemia
Severe myelodysplasia
Control cell lines were the FANCD2-deficient PD20 and stably corrected PD20,6 and the NBS1-deficient EUFA1020-L and BRCA2-deficient EUFA2030-L (kindly provided by H. Joenje, Vrije Universiteit Medical Center, Amsterdam, Netherlands).
Sample preparations and immunoblots
Mononuclear peripheral blood cells were purified from fresh blood samples using Ficoll-Paque Plus (Amersham Biosciences, Saclay, France). 10 × 106 cells were cultured at a concentration of 106/mL in RPMI-FBS (RPMI 1640, 10% vol/vol fetal bovine serum [FBS]), and T-cells were stimulated with phytohemagglutin (PHA) for 72 hours without exposure to a DNA-damaging agent. For primary fibroblast culture, 4-mm-square skin biopsies were performed using local anesthesia with Emla (Astra, Rueil, France) and the Biopsy Punch (Stiefel, Rueil, France). Skin samples were thinly sliced, and the resulting fragments were adhered on plastic plates for 15 minutes then covered with modified Eagle medium (MEM) supplemented with antibiotic, HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), MEM nonessential amino acids (Invitrogen, Cergy Pontoise, France), and 20% fetal calf serum (FCS) (GIBCO-BRL Life Technologies, Cergy Pontoise, France), at 37°C with 5% CO2. Individual growing clones were picked up after 2 to 3 weeks and expanded. Cells were grown and tested without exposure to DNA damaging agent. Thirty micrograms of proteins from PHA-stimulated peripheral blood lymphocytes (PBLs), or 50 μg from growing nonconfluent fibroblasts, were loaded in a 80 mm × 80 mm preloaded 3.8% Nu-PAGE Novex Tris-Acetate gel (Invitrogen). Proteins were migrated for 85 minutes using a Xcell SureLock Mini-Cell tank (Invitrogen) then transferred onto nitrocellulose membrane using the Xcell II Blot module (Invitrogen). After blocking for 2 hours or overnight in 0.5% milk, a rabbit polyclonal anti-FANCD2 serum (A.D.D.'s laboratory) was used at 1:1000 for 1 hour. Detection was performed using horseradish peroxide (HRP)-linked secondary antibodies with the ECL PLUS kit (Amersham Biosciences). BRCA2 and NBS1 immunoblots were performed on PHA-stimulated lymphocytes and fibroblasts from patients EGFA008 and EGFA053, and from controls using BRCA2 Ab-2 antibodies (Oncogene Research Products, Darmstadt, Germany) and NBS1 antibodies (AbCam, Cambridge, United Kingdom).
Chromosome breakage tests
Chromosome breakage tests were done as previously described,32 independently from the FANCD2 tests. Fresh PBLs were stimulated by PHA for 24 hours and further incubated with and without DNA damaging agent for 48 hours. We used the nitrogen mustard mechlorethamine (Caryolysine; Synthelabo, Le Plessis Robinson, France) freshly diluted at a final concentration of 0.05 μg/mL.32 Examination of 100 mitoses (50 with and 50 without DNA damaging agents) allowed the scoring of chromatid breaks and chromosome breaks as one break, and chromosome rearrangements and radials as 2 breaks; chromatid gaps were not scored. Results were compared with healthy controls and positive controls run in parallel. In our laboratory, cytogenetic diagnosis of FA is based on the combined analysis of several criteria: (1) the mean number of breaks per metaphase in nitrogen mustard (NM)-exposed cells (a mean number greater than 2 suggests FA); (2) the ratio of this mean between NM-exposed cells and nonexposed cells (a ratio greater than 10:1 is required); (3) the number of NM-dependent breaks per aberrant mitosis (5 or more breaks in several cells strongly suggests FA); and (4) the percentage of mitoses with break(s) in NM-exposed cells (a minimum of 20% of aberrant mitoses is required). If all criteria are fulfilled, FA is diagnosed. Cases with significantly more breaks than in normal controls, but less than the Fanconi range, are considered “ambiguous”; however, this does not necessary rule out the diagnosis of FA, and additional confirmatory testing may be required. Somatic mosaicism is suspected when more than 20% of the metaphases in NM-exposed cells show no break. In primary fibroblasts, chromosome breakage tests were performed by culturing growing cells with dilutions of mechlorethamine using 2 exposure times (48 hours and 4 days). Positivity was determined by comparison with FA and non-FA control fibroblasts run in parallel (thresholds were lower than those used in PBLs, and the complete experiments in fibroblasts were run 3 times).28,36
Determination of complementation groups
In a subset of patients (FA core patients with reversion), subtyping was performed in the fibroblasts according to the FANCD2-based rapid FA subtyping assay.34 Briefly, stably expressing cell lines were established using the pMMP retroviral vectors containing the FANCA, FANCC, and FANCG cDNA, and empty vector.37 Growing fibroblasts from patients and control fibroblasts of known groups were transduced with each of the supernatant in parallel. After 48 hours, cell lysates were analyzed by FANCD2 immunoblot. In 12 nonrevertant FA core patients, subtyping was determined in fibroblasts, blood, or EBV cell lines (FA-A, n = 11; FA-G, n = 1).
Statistical analysis
All analyses were performed with PRISM (GraphPad, San Diego, CA) and were 2 tailed. Categorial variables were compared using the chi-square test. A P value less than .05 was considered to indicate statistical significance.
Results
Comparison of PBLs and fibroblasts using FA/BRCA tests detects somatic mosaicism
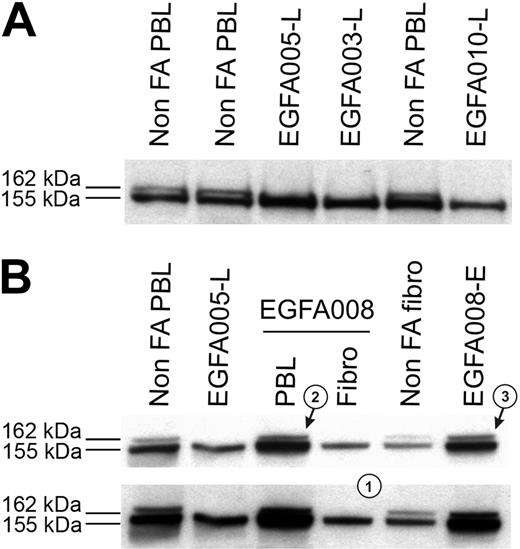
Clinical data, chromosome breakage tests, and FANCD2 immunoblots were analyzed in all the 53 FA patients, and results are summarized in Table 1. Most patients demonstrated abnormal FANCD2 patterns in PBLs, confirming the specific FA/BRCA pathway defect. Specifically, a single FANCD2 small isoform (FANCD2-S) but no large isoform (FANCD2-L) was detected in 42 patients (Figure 1A), whereas no FANCD2 bands were detected in 3 patients (D2 group; see classification of FA patients in the next subsection); in total, n = 45 patients out of 53. Consistently, positive chromosome breakage tests were found in these cases, except 1 patient who presented with acute myeloblastic leukemia (EGFA015; no more analysis could be done in this patient). Strikingly, 8 additional patients had normal FANCD2 patterns in PBLs (ie, the 2 FANCD2 isoforms), demonstrating an ability of PHA-stimulated lymphocytes to monoubiquitinate the FANCD2 protein. Four of these patients (EGFA012, -013, -039 and -050) exhibited increased chromosomal breakage in a subset of mitoses (20% to 56%), suggesting somatic mosaicism (Table 2). In the 4 remaining patients (EGFA006, -008, -047, and -053), chromosome breakage tests were negative or ambiguous (Table 2). Primary fibroblasts were grown from skin biopsies in all the 8 patients in order to obtain constitutional cells. Abnormal FANCD2 patterns were found in 6 out of the 8 fibroblast samples, demonstrating functional FA reversion in the PBLs (Figure 1B; Table 2). In the 2 remaining questionable patients (EGFA006 and -053), primary fibroblasts showed normal FANCD2 patterns but positive chromosome breakage tests, suggesting somatic mosaicism (reversion in unidentified group patients; see Table 2). Clinical records showed that these 2 patients had positive breakage tests in PBLs at diagnosis, 8 and 10 years before the present analysis, respectively, which is consistent with emergence of somatic mosaicism since this time. Altogether, the combined data from PBLs and fibroblasts using FANCD2 and chromosomal breakage tests allowed us to detect reversion in the PBLs of 8 (15%) FA patients out of 53. Dilution experiments of normal PBLs within FA PBLs showed that the threshold for detection of reversion by FANCD2 Western blot was approximately 15% of revertant cells by sample (data not shown).
FANCD2 immunoblot patterns in FA patients, including FA revertant. (A) FA core samples failed to monoubiquitinate FANCD2 and show only the lower unmodified short (FANCD2-S) isoform, whereas non-FA samples show both the FANCD2-S and the long monoubiquitinated (FANCD2-L) isoforms. FA patients are referred to by unique EGFA identification numbers. (B) Detection of FA reversion by the FANCD2 pattern analysis. Revertant patient EGFA008 samples show typical FA in primary fibroblasts pattern (a single FANCD2-L isoform, indicated as 1), whereas fresh PHA-stimulated lymphocytes (arrow 2) and the EBV-immortalized B-cell line (arrow 3) show both the FANCD2-S and FANCD2-L isoforms. Normal and nonrevertant EGFA005 are shown as controls. Short (top panel) and medium (bottom panel) exposures are shown.
FANCD2 immunoblot patterns in FA patients, including FA revertant. (A) FA core samples failed to monoubiquitinate FANCD2 and show only the lower unmodified short (FANCD2-S) isoform, whereas non-FA samples show both the FANCD2-S and the long monoubiquitinated (FANCD2-L) isoforms. FA patients are referred to by unique EGFA identification numbers. (B) Detection of FA reversion by the FANCD2 pattern analysis. Revertant patient EGFA008 samples show typical FA in primary fibroblasts pattern (a single FANCD2-L isoform, indicated as 1), whereas fresh PHA-stimulated lymphocytes (arrow 2) and the EBV-immortalized B-cell line (arrow 3) show both the FANCD2-S and FANCD2-L isoforms. Normal and nonrevertant EGFA005 are shown as controls. Short (top panel) and medium (bottom panel) exposures are shown.
Fanconi anemia patients with reversion (somatic mosaicism)
. | . | . | PBL breaks . | . | . | . | . | . | . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | 0 . | . | 1 . | . | ≥ 2 . | . | . | FANCD2 test . | . | . | . | . | ||||||
| Patient no. . | Age, y . | Group . | NM– . | NM+ . | NM– . | NM+ . | NM– . | NM+ . | Result . | PBL . | Fibro . | Breaks, fibro . | Follow-up, y . | Comments . | ||||||
| EGFA012 | 6 | A | 46 | 27 | 3 | 2 | 1 | 21* | POS | 2 | 1 | ND | 4 | FA reversion in a fraction of PBLs | ||||||
| Mild pancytopenia with recent improvement | ||||||||||||||||||||
| EGFA013 | 21 | A | 47 | 40 | 2 | 0 | 1 | 10* | POS | 2 | 1 | ND | 13 | FA reversion in a fraction of PBLs | ||||||
| Normal blood cell counts since diagnosis 13 years before ps | ||||||||||||||||||||
| Dysplasia in mouth | ||||||||||||||||||||
| EGFA039 | 10 | A | 44 | 22 | 5 | 6 | 1 | 22* | POS | 2 | 1 | ND | 1 | FA reversion in a fraction of PBLs | ||||||
| New diagnosis | ||||||||||||||||||||
| Thrombocytopenia; normal WBC and Hb | ||||||||||||||||||||
| EGFA050 | 4 | D2 | 49 | 37 | 1 | 4 | 0 | 9* | POS | 2 | 0 | ND | 4 | FA reversion in a fraction of PBLs | ||||||
| Normal blood cell counts since diagnosis | ||||||||||||||||||||
| EGFA008 | 23 | A | 50 | 46 | 0 | 4 | 0 | 0 | NEG | 2 | 1 | ND | 10 | FA reversion in PBLs (complete) | ||||||
| Mild pancytopenia since diagnosis | ||||||||||||||||||||
| Negative PBL breaks at diagnosis 10 years before ps | ||||||||||||||||||||
| EGFA006 | 8 | Unid | 49 | 48 | 1 | 2 | 0 | 0 | NEG | 2 | 2† | POS | 3 | FA reversion in PBLs (complete) | ||||||
| Persistent mild anemia since diagnosis | ||||||||||||||||||||
| Positive PBL breaks at diagnosis 8 years before ps | ||||||||||||||||||||
| EGFA047 | 34 | A | 49 | 48 | 1 | 1 | 0 | 1 | NEG | 2 | 1 | POS | 27 | FA reversion in PBLs (complete) | ||||||
| Anemia and severe thrombopenia at diagnosis; improvement and subnormal blood counts since 27 years with low-dose androgens (Figure 2) | ||||||||||||||||||||
| Dysplasia in mouth | ||||||||||||||||||||
| EGFA053 | 10 | Unid | 48 | 44 | 2 | 3 | 0 | 3* | AMB | 2 | 2† | POS | 6 | FA reversion in a fraction of PBLs | ||||||
| Mild pancytopenia since diagnosis | ||||||||||||||||||||
| Positive PBL breaks at 10 years before ps | ||||||||||||||||||||
. | . | . | PBL breaks . | . | . | . | . | . | . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | 0 . | . | 1 . | . | ≥ 2 . | . | . | FANCD2 test . | . | . | . | . | ||||||
| Patient no. . | Age, y . | Group . | NM– . | NM+ . | NM– . | NM+ . | NM– . | NM+ . | Result . | PBL . | Fibro . | Breaks, fibro . | Follow-up, y . | Comments . | ||||||
| EGFA012 | 6 | A | 46 | 27 | 3 | 2 | 1 | 21* | POS | 2 | 1 | ND | 4 | FA reversion in a fraction of PBLs | ||||||
| Mild pancytopenia with recent improvement | ||||||||||||||||||||
| EGFA013 | 21 | A | 47 | 40 | 2 | 0 | 1 | 10* | POS | 2 | 1 | ND | 13 | FA reversion in a fraction of PBLs | ||||||
| Normal blood cell counts since diagnosis 13 years before ps | ||||||||||||||||||||
| Dysplasia in mouth | ||||||||||||||||||||
| EGFA039 | 10 | A | 44 | 22 | 5 | 6 | 1 | 22* | POS | 2 | 1 | ND | 1 | FA reversion in a fraction of PBLs | ||||||
| New diagnosis | ||||||||||||||||||||
| Thrombocytopenia; normal WBC and Hb | ||||||||||||||||||||
| EGFA050 | 4 | D2 | 49 | 37 | 1 | 4 | 0 | 9* | POS | 2 | 0 | ND | 4 | FA reversion in a fraction of PBLs | ||||||
| Normal blood cell counts since diagnosis | ||||||||||||||||||||
| EGFA008 | 23 | A | 50 | 46 | 0 | 4 | 0 | 0 | NEG | 2 | 1 | ND | 10 | FA reversion in PBLs (complete) | ||||||
| Mild pancytopenia since diagnosis | ||||||||||||||||||||
| Negative PBL breaks at diagnosis 10 years before ps | ||||||||||||||||||||
| EGFA006 | 8 | Unid | 49 | 48 | 1 | 2 | 0 | 0 | NEG | 2 | 2† | POS | 3 | FA reversion in PBLs (complete) | ||||||
| Persistent mild anemia since diagnosis | ||||||||||||||||||||
| Positive PBL breaks at diagnosis 8 years before ps | ||||||||||||||||||||
| EGFA047 | 34 | A | 49 | 48 | 1 | 1 | 0 | 1 | NEG | 2 | 1 | POS | 27 | FA reversion in PBLs (complete) | ||||||
| Anemia and severe thrombopenia at diagnosis; improvement and subnormal blood counts since 27 years with low-dose androgens (Figure 2) | ||||||||||||||||||||
| Dysplasia in mouth | ||||||||||||||||||||
| EGFA053 | 10 | Unid | 48 | 44 | 2 | 3 | 0 | 3* | AMB | 2 | 2† | POS | 6 | FA reversion in a fraction of PBLs | ||||||
| Mild pancytopenia since diagnosis | ||||||||||||||||||||
| Positive PBL breaks at 10 years before ps | ||||||||||||||||||||
Patients are ordered in this table as presented in the text. Results shown in this table were obtained during the course of the present study (ps). Unid indicates unidentified downstream group(s). Chromosome breakage tests performed with (NM+) and without (NM–) exposure to nitrogen mustard; nos. of mitosis with 0, 1, and ≥ 2 breaks are indicated; conclusions of the chromosomal breakage tests are indicated as POS, positive, NEG, negative, AMB, ambiguous result. FANCD2 tests: 2, both the short and long FANCD2 isoforms were detected using FANCD2 immunoblot; 1, only the FANCD2-S isoform was detected; 0, no FANCD2 protein was detected; †, 2 and 4 independant primary fibroblasts clones were tested in patients EGFA006 and EGFA053, respectively. Breaks fibro : breakage tests in primary fibroblasts; nd, not tested. The association of a positive chromome breakage test which includes a population of resistant cells to the detection of FANCD2 monoubiquitination is consistent with FA reversion present in a fraction of the PBLs. Follow up, years of clinical follow up in our institution (it can be shorter than the time from the diagnosis as indicated in Table 1 since a number of patients were referred in our center years after diagnosis). Partial mosaicism was defined by the persistence of a number of mitosis with multiples breaks (≥ 5 breaks); complete mosaicism was defined by normal breakage tests in PBLs.
Including mitosis with ≥ 5 breaks by mitosis
Analysis of blood cell counts demonstrated that patients with FA reversion had significantly higher blood cell counts compared to nonrevertants (Table 3). Moreover, with a median follow up of 5 years (range, 1 to 27 years) of the 8 revertant patients in our institution, we did not observe any evolution to bone marrow aplasia or leukemia (Table 2; Figure 2). Two revertant patients, aged 21 and 34 years old, developed mouth dysplasias which are currently being treated and followed for detection of progression to squamous cell carcinoma. In nonrevertant FA patients, 31 out of 45 patients had aplasia (absolute neutrophil count [ANC] < 1 × 109/L, hemoglobin [Hb] < 80 g/L [< 8 g/dL], and/or Plt < 50 × 109/L, P < 10-3; Table 1), 1 patient had acute myeloblastic leukemia, and 1 had a severe myelodysplasia. This small number of leukemia and severe myelodysplasia cases during the time of the study precluded statistical analysis. No mouth dysplasia or other cancers were detected in the nonrevertant FA patients at the time of the study.
FA reversion is associated with higher blood cell counts
. | Revertant, median (range) . | Nonrevertant, median (range) . | P . |
|---|---|---|---|
| WBC, × 109/L | 5.4 (3.7-7.2) | 3.4 (1.4-7.6) | .00017 |
| ANC, × 109/L | 2.5 (1.2-4.0) | 1.1 (0.3-2.8) | .0001 |
| Hb, g/L | 131 (98-157) | 108 (46-136) | .001 |
| Platelets, × 109/L | 107.5 (83-350) | 51 (10-300) | .001 |
| MCV, fL | 96.1 (84-107) | 101.1 (82-125) | .2* |
. | Revertant, median (range) . | Nonrevertant, median (range) . | P . |
|---|---|---|---|
| WBC, × 109/L | 5.4 (3.7-7.2) | 3.4 (1.4-7.6) | .00017 |
| ANC, × 109/L | 2.5 (1.2-4.0) | 1.1 (0.3-2.8) | .0001 |
| Hb, g/L | 131 (98-157) | 108 (46-136) | .001 |
| Platelets, × 109/L | 107.5 (83-350) | 51 (10-300) | .001 |
| MCV, fL | 96.1 (84-107) | 101.1 (82-125) | .2* |
For revertant, n = 8; for nonrevertant, n = 45. MCV indicates mean corpuscular volume.
Nonsignificant
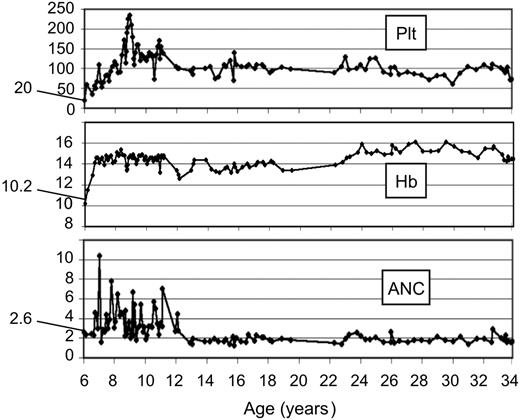
Blood cell curves in FA revertant patient EGFA047. Blood cell count curves from patient EGFA047 are shown over a 27-year follow-up in our institution. A total of 213 recorded blood cell counts were analyzed from age 6 years to 34 years (from diagnosis to present study). Starting values at diagnosis are indicated. Plt, platelets ×109/L; Hb, hemoglobin (g/dL); ANC, absolute neutrophil counts × 109/L.
Blood cell curves in FA revertant patient EGFA047. Blood cell count curves from patient EGFA047 are shown over a 27-year follow-up in our institution. A total of 213 recorded blood cell counts were analyzed from age 6 years to 34 years (from diagnosis to present study). Starting values at diagnosis are indicated. Plt, platelets ×109/L; Hb, hemoglobin (g/dL); ANC, absolute neutrophil counts × 109/L.
FANCD2 immunoblot analysis identifies the level of disruption of the FA/BRCA pathway and allows classification of FA patients
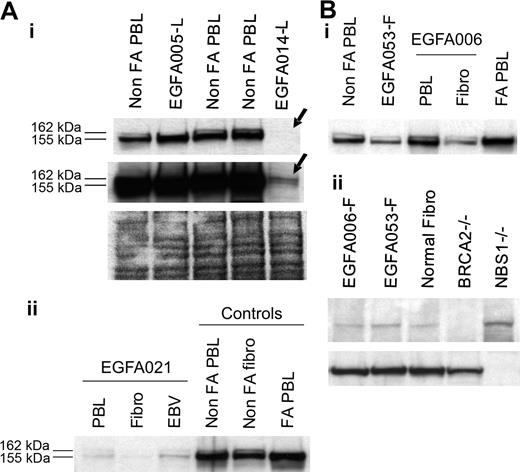
Considering PBL data in nonrevertant patients and fibroblast data in revertant patients, FANCD2 immunoblot patterns allowed determination of the level at which the FA/BRCA pathway is disrupted. In 47 (89%) out of 53 patients, including 5 FA revertant patients, one single, short, nonubiquitinated FANCD2 isoform was detected (Figure 1). These patients were therefore classified as “FA core,” corresponding to the upstream inactivation of the pathway. The 5 revertant FA core patients were shown to be complementation group A by retroviral transduction of their fibroblasts (Table 2). In 4 patients (EGFA014, -021, -050, -066), including 1 revertant (EGFA050), no FANCD2 bands were observed in several independent samples (Table 1; Figure 3A). When Western blots were overexposed, 2 barely detectable bands could be seen indicating faint residual levels of the protein, similar to the FA-D2 reference cell line PD20.6,10 These patients were therefore classified as “FA-D2.” In 2 additional patients (EGFA008, EGFA053), no FANCD2 abnormalities were detected by immunoblot in either the PBLs or fibroblasts (Figure 3B). Two distinct primary fibroblast clones from patient EGFA006, and 4 from patient EGFA053, were tested with similar results (data not shown). Together with their strong FA clinical features, including typical FA face and thumb abnormalities in both patients, short stature in patient EGFA053, positive chromosomal breakage testing in fibroblasts, and a history of a positive breakage test in PBLs at diagnosis, these 2 patients were provisionally classified as FA of unidentified group (Table 2). These findings are consistent with an inactivation of the FA/BRCA pathway downstream of FANCD2. Considering reports on D1/BRCA2 and NBS1 mutated phenotypes,7,11 we analyzed the PBLs and primary fibroblasts of these 2 patients by BRCA2 and NBS1 immunoblots but detected no alteration in level of protein expression (Figure 3B).
Analysis of FA-D2 and unidentified downstream group FA cells. (A) FANCD2 immunoblots detect the FA-D2 group. (i) No FANCD2 protein was observed at normal exposure of the immunoblot in sample EGFA014-L (top arrow). Long exposure revealed 2 barely detectable FANCD2 bands (bottom arrow), indicating residual levels of the FANCD2 protein, as previously shown in the reference D2 cell line PD20.6,10 Ponceau S staining is shown as loading control. Sequencing data further confirmed mutations in the FANCD2 gene in this patient (A.S., data not shown). (ii) FANCD2 immunoblot of PHA-stimulated primary fibroblasts and EBV-immortalized cells from FA-D2 patient EGFA021. (B, i) Unidentified group patients EGFA006 and EGFA053 have normal FANCD2 patterns in both their primary fibroblasts and lymphocytes (EGFA053-L; not shown). (ii) BRCA2 and NBS1 immunoblots detected no abnormalities in the fibroblasts of these 2 patients. EUFA243 and EUFA1020 EBV cell lines were used as positive controls for BRCA2 and NBS1 deficiency, as indicated.
Analysis of FA-D2 and unidentified downstream group FA cells. (A) FANCD2 immunoblots detect the FA-D2 group. (i) No FANCD2 protein was observed at normal exposure of the immunoblot in sample EGFA014-L (top arrow). Long exposure revealed 2 barely detectable FANCD2 bands (bottom arrow), indicating residual levels of the FANCD2 protein, as previously shown in the reference D2 cell line PD20.6,10 Ponceau S staining is shown as loading control. Sequencing data further confirmed mutations in the FANCD2 gene in this patient (A.S., data not shown). (ii) FANCD2 immunoblot of PHA-stimulated primary fibroblasts and EBV-immortalized cells from FA-D2 patient EGFA021. (B, i) Unidentified group patients EGFA006 and EGFA053 have normal FANCD2 patterns in both their primary fibroblasts and lymphocytes (EGFA053-L; not shown). (ii) BRCA2 and NBS1 immunoblots detected no abnormalities in the fibroblasts of these 2 patients. EUFA243 and EUFA1020 EBV cell lines were used as positive controls for BRCA2 and NBS1 deficiency, as indicated.
Clinical features of the FA-D2 (EGFA014, -021, -050, and -067) and unidentified group patients (EGFA006 and -053) were analyzed. All the 6 patients had extensive congenital abnormalities, compared to 10 out of 46 FA core patients (P < 10-3; Table 1). Furthermore, these patients were diagnosed early, at ages of 3 months, 4 months, 4 months, 2.8 years, 3 years, and 6 years. Three of the 6 patients were therefore diagnosed in infancy, and interestingly, these 3 patients developed reversion with somatic mosaicism, as did one of the 2 FA core patients also diagnosed during infancy (EGFA012; Table 2).
Discussion
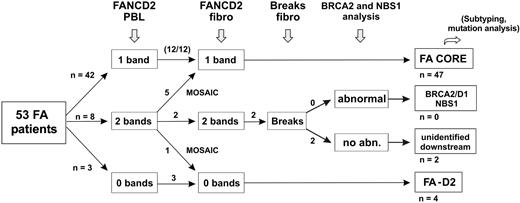
We prospectively analyzed a series of FA patients by clinical examination, FANCD2 immunoblot, and chromosome breakage tests. From this study, we propose an evaluation strategy for FA, as shown in Figure 4. This is based on the analysis of the FA/BRCA pathway in order to detect functional FA reversion in PBLs and to classify patients. First, FA patients' PBLs are tested by FANCD2 immunoblot. Most patients demonstrate one single nonubiquitinated FANCD2 isoform and are considered nonrevertant FA core patients. Absence of detection of the FANCD2 protein suggests FA-D2 group. Patients with FANCD2 monoubiquitination in PBLs are further investigated by FANCD2 immunoblot in fibroblasts. When fibroblasts demonstrate an FA pattern (FA core or FA-D2), FANCD2 monoubiquitination in PBLs indicates FA reversion. When FANCD2 in fibroblasts is monoubiquitinated to normal levels, chromosomal breakage tests in fibroblasts can be performed if necessary to confirm FA, and BRCA2 or NBS1 abnormalities should be investigated by immunoblotting and/or molecular analysis. Remaining patients with hypersensitivity to DNA crosslinks but in an unidentified complementation group are provisionally classified as “unidentified downstream group” patients. This evaluation strategy is rapid and cost-effective, and in most FA cases it allows confirmation of FA diagnosis, detection of potential FA reversion in PBLs, and classification of FA patients. It further facilitates determination of the complementation group and identification of the constitutional FA mutations by molecular analysis. Ultimately, reverse genetic events may be searched for by molecular analysis to demonstrate molecular reversion and somatic mosaicism.
Evaluation strategy for detection of reversion and classification of FA patients. The evaluation strategy proposed for FA patients in addition to PBL chromosome breakage test is shown. The number of patients at each step is indicated when this strategy is applied to the present series of 53 FA patients.
Evaluation strategy for detection of reversion and classification of FA patients. The evaluation strategy proposed for FA patients in addition to PBL chromosome breakage test is shown. The number of patients at each step is indicated when this strategy is applied to the present series of 53 FA patients.
We found that comparison of fibroblasts and PBLs using FANCD2 immunoblot allowed easy detection of functional reversion in the PBLs of 5 FA-core patients by positively and specifically showing the ability of the PBLs to monoubiquitinate FANCD2. In one FA-D2 patient, FA reversion was detected in PBLs by the normal level of FANCD2 expression compared with fibroblasts. In 2 patients from unidentified complementation groups, we had to analyze chromosome breaks in fibroblasts to confirm the constitutional FA phenotype and subsequently to detect functional reversion in PBLs by comparison. Altogether, FA reversion was found in the PBLs of 8 (15%) of 53 FA patients (Figure 4). We demonstrated that FA reversion as detected here was associated with significantly higher blood cell counts, as was previously suggested from case reports.23,26,27 Clinical records in the 8 revertant patients showed normal or mildly decreased blood cell counts and stability or improvement with a median follow-up of 5 years (range, 1 to 27 years) in our institute (Tables 2, 3; Figure 2). It is therefore likely that FA reversion can partially or completely overcome bone marrow failure in FA. Notably, a persistent mild thrombocytopenia was observed in several revertant patients (Tables 1, 3; Figure 2). It can be hypothesized that the pool of stem cells had been impaired before reversion,38 perhaps due to telomere erosion.39 Considering neoplasia predisposition, it was expected that patients with stable hematological correction could still have predisposition to cancer, including head and neck squamous carcinoma. Indeed, mouth dysplasia was diagnosed in 2 out of the 8 revertant patients, at the ages of 21 and 34 years. We did not observe leukemic transformation in the revertant patients, including those followed for a long term. Since the primary objective of the study was not to define prognosis factors, our data could not be used to analyze prediction of adverse evolution. However, our finding that, at the time of the study, revertant patients had significant higher blood cells counts and no aplasia, indicates that FA reversion might be associated with better prognosis. Long-term follow-up studies in large series of FA patients will be necessary to define the prognostic consequences of FA reversion over time, its variations, and interactions with marrow failure and neoplasic events, including leukemia. Notably, clonal karyotypic abnormalities were detected in 7 FA patients whose PBLs were found to be nonrevertant, including one with acute myeloid leukemia and one with severe myelodysplasia at the time of the analysis (Table 1). However, these chromosome abnormalities were detected in mitoses from unstimulated bone marrow but not in the PHA-stimulated PBLs, and therefore the FANCD2 status of the bone marrow clonal cells is elusive. Addressing the FA/BRCA pathway in these clones will be of interest, considering recent work associating leukemic progression to reversion.40 Leukemia may also develop from residual nonrevertant FA cells,25 and in non-FA patients an acquired FA defect might cause FA-like leukemia.41
Once constitutional cells were analyzed in all patients (PBLs in nonrevertant patients, and fibroblasts in revertant patients), the FANCD2 status allowed a classification based on the level of the FA/BRCA pathway inactivation (Figures 4, 5). Most patients (n = 47; 89%) were defined as “FA core,” consistent with the known high prevalence of the FA-A, and to a lesser extent, FA-G and FA-C groups.3,17,20 4 unrelated patients (8%) were classified as FA-D2, suggesting that this group might be more common than previously appreciated.4,17 Two of these patients were further analyzed by DNA sequencing, and FANCD2 mutations were detected, confirming D2 group (A.S., data not shown). In addition, 2 patients were provisionally considered as FA of unidentified downstream group (4%), considering their strong clinical FA features, normal FANCD2 monoubiquitination but chromosome breaks in fibroblasts, and a history of positive breakage tests in PBLs in infancy. No personal or familial cancer history was found in these patients now aged 9 and 10 years, and no BRCA2 or NBS1 protein abnormalities were detected. Although extensive DNA analysis must be done to definitely rule out BRCA2 gene mutation and discard D1 group, it is likely that these cases belong to additional downstream group(s), such as the FA-J group, which has been defined recently by complementation analysis.4 Notably, the 6 FA-D2 and unidentified downstream group patients had severe phenotype with extensive congenital abnormalities and early diagnosis (Table 1), consistent with pleiotropic functions of the FANCD2 and downstream proteins.9,14-16,42 Interestingly, 3 of these 6 patients had FA reversion, compared to 5 out of 47 FA core patients, and these 3 patients were diagnosed in infancy. A severe specific cellular defect might favor early reversion, due to a high level of genetic instability and apoptosis.42,43
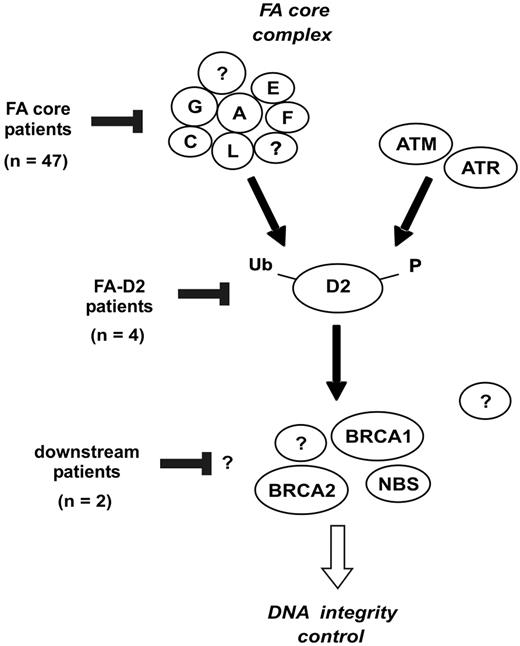
Classification of FA patients based on the location of the FA/BRCA pathway disruption. The FANCA, FANCC, FANCE, FANCF, FANCG, and FANCL proteins are required for monoubiquitination of FANCD2. During S phase, and following DNA damage, FANCD2 is monoubiquitinated and targeted to nuclear foci containing BRCA1, FANCD1/BRCA2, and NBS1.9 Based on the strategy shown in Figure 4, the presumptive location of FA/BRCA pathway disruption in patients of the present series is indicated.
Classification of FA patients based on the location of the FA/BRCA pathway disruption. The FANCA, FANCC, FANCE, FANCF, FANCG, and FANCL proteins are required for monoubiquitination of FANCD2. During S phase, and following DNA damage, FANCD2 is monoubiquitinated and targeted to nuclear foci containing BRCA1, FANCD1/BRCA2, and NBS1.9 Based on the strategy shown in Figure 4, the presumptive location of FA/BRCA pathway disruption in patients of the present series is indicated.
The present study also allowed evaluation of the FANCD2 immunoblot as a diagnostic test in FA patients. We found that FANCD2 Western blot is a rapid and specific FA test, as previously reported, and it can be used to screen broad patient populations.34 In rare FA cases, it appeared that the chromosome breakage test was more sensitive as a diagnosis test than the FANCD2 immunoblot, due to the ability to detect minor residual nonrevertant populations in PBLs (n = 4 cases of the present study), and because of unidentified downstream group (n = 2 cases; Table 2). Therefore, we believe that the chromosomal breakage test remains the standard to-date for initial FA diagnosis. On the other hand, it has been reported that increased chromosomal breakage may also be detected in other syndromes with FA-like features, such as Nijmegen breakage syndrome.11 A normal FANCD2 pattern in these patients would show that more investigations are required. In addition, in unassigned patients with negative or ambiguous FA tests in PBLs but a strong clinical suspicion of FA, fibroblast analysis can be essential to search for FA with reversion.30 To that purpose, we found it much more convenient to analyze primary fibroblasts in first line by FANCD2 immunoblot than by chromosome breakage test or MMC sensitivity. Finally, primary fibroblasts from allografted FA patients can also be analyzed by FANCD2 immunoblot to identify FA subtype (n = 3 allografted FA patients not included in this study, including one further FA-D2 patient; data not shown).
In conclusion, we found that the specific analysis of the FA/BRCA pathway by FANCD2 test gives helpful information in addition to the chromosome breakage data. Most important, it allows easy detection of FA reversion, and it pinpoints the level at which the FA/BRCA pathway is disrupted, including the FANCD2 protein itself. We propose that the FANCD2 analysis might be used as a research center approach, in combination with the chromosome breakage analysis, for a comprehensive characterization of FA and FA-like patients.
Prepublished online as Blood First Edition Paper, September 21, 2004; DOI 10.1182/blood-2004-05-1852.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the FA patients and families, and the French family support group AFMF (Association Française de la maladie de Fanconi) for their kind cooperation. We thank C. Poulet, and the nurses of the clinical units. We appreciate excellent technical assistance in the Cytogenetic Unit of Saint-Louis Hospital, and from R. Nancel and E. Savariau. We thank O. Boespflug-Tanguy (Clermond-Ferrand), O. Hermine (Paris), C. Picard (Paris), and Y. Reguerre (Saint-Denis, Réunion) for referring clinical data and sending samples, as well as P. S. Jouk (Grenoble) and C. Leonard (Bicêtre) for communication of archived data. We thank members of the Société d'Hématologie et Immunologie Pédiatrique (SHIP) for clinical collaboration. We are indepted to C. M. Green, D. Papadopoulo, A. Aurias, and Y. Beuzard for helpful discussions. The EGFA (European Group for Fanconi Anemia) is supported by the AFMF and the Association Francaise pour les Myopathies (AFM).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal