Abstract
Interleukin-12 receptor β1 (IL12RB1) is expressed on a variety of immune cells, including T and natural killer (NK) cells and macrophages, and is involved in innate and adaptive immune responses. Levels of IL12RB1 mRNA are dynamically regulated by various cytokines, including interferon-γ (IFN-γ) and IL-15. To reveal the regulatory mechanisms governing IL12RB1 gene expression, we analyzed the transcriptional regulatory region of the mouse IL12RB1 gene. Promoter analyses in a mouse macrophage cell line, RAW264.7, revealed that the 2508-bp region upstream of the transcriptional start site is sufficient for the full transcriptional activation of the IL12RB1 gene by IFN-γ or IL-15. Analyses of the deletion mutants revealed critical roles of IRE/ISRE and ETS/PU.1 elements, to which IRF3 and PU.1, respectively, bound. Notably, chromatin immunoprecipitation (ChIP) assays revealed IL-15 rapidly induced histone H3 acetylation at the IL12RB1 promoter. Consistently, IL-15, as a histone deacetylase inhibitor, synergistically enhanced IL12RB1 gene expression and promoter activation by IFN-γ through increased protein binding to ETS/PU.1 and IRE/ISRE sites. Additionally, IL12RB1 promoter activation by IFN-γ was enhanced by the coexpression of a coactivator protein, CBP. Thus, IL-15 induces chromatin remodeling of the IL12RB1 gene promoter, increasing IL12RB1 mRNA expression in synergy with IFN-γ through the recruitment of PU.1 and IRF3.
Introduction
Interleukin-12 (IL-12) is a cytokine predominantly produced by antigen-presenting cells (APCs), such as dendritic cells (DCs) and macrophages, in response to various infectious agents, and it is essential for the development of efficient innate and T-helper 1 (TH1)–-type adaptive immunity. The distinctive role of IL-12 is to induce interferon-γ (IFN-γ) production by natural killer (NK) and T cells in response to a variety of inflammatory stimuli.1 IFN-γ in turn activates macrophages to express numerous genes associated with inflammatory responses, including IFN-γ production itself.1,2 Several recent studies, however, revealed that IL-12 also stimulates DCs and macrophages in autocrine and paracrine manners to produce abundant amounts of IFN-γ.3-5
IL-12 receptor (IL-12R) is composed of 2 subunits, β1 and β2,6,7 each of which belongs to the cytokine receptor superfamily and is highly homologous to gp130, granulocyte–colony-stimulating factor receptor (G-CSF-R), and leukemia inhibitory factor–receptor (LIF-R).8 Although IL12RB1 and IL-12Rβ2 share a similar structure, their binding affinities are distinct. In humans, IL12RB1 and IL-12Rβ2 separately bind IL-12 with low affinity, but together they bind IL-12 with high affinity.9 In mice, IL-12Rβ2 binds IL-12 at very low affinity, whereas IL12RB1 binds with low and high affinities.10 Genetic deficiency of IL12RB1 causes severe infection by low-pathogen bacteria, such as bacilli Calmette-Guerin (BCG), in humans and in mice, most likely because of insufficient IFN-γ production,7,11,12 indicating that IL12RB1 is essential for the host defense mechanisms. It has recently been reported that IL12RB1 is also a component of the receptor complex for another IFN-γ–inducing cytokine, IL-23.13
IL12RB1 expression is under dynamic regulation in response to various stimuli. In vivo, IL12RB1 mRNA levels in macrophages are increased by IL-12 as a consequence of IFN-γ expression.14 Consequently, direct stimulation of macrophages by IFN-γ up-regulated IL12RB1 mRNA.15 In peripheral blood mononuclear cells (PBMCs), IL12RB1 gene expression is induced by phytohemagglutinin (PHA), anti-CD3/CD28 monoclonal antibody (mAb), IL-2, IL-7, and IL-15 stimulation.16
IL-15 receptor is composed of the specific IL-15Rα and 2 receptor chains, IL-2Rβ and IL-2Rγc, which are shared by IL-2. Although IL-2Rα is predominantly expressed on activated T cells, IL-15Rα has been identified in multiple cell types, including APCs.17 Notably, the lipopolysaccharide (LPS)–induced IFN-γ response in severe combined immune deficient (SCID) mice was significantly reduced by pretreatment with a neutralizing anti–IL-15 antibody, indicating IL-15 participates in IFN-γ induction in a T cell–independent fashion.18 Moreover, splenic DCs and peritoneal macrophages from IL-15-/- mice produced significantly less IFN-γ than their control counterparts in response to IL-12, suggesting IL-12 controls IFN-γ production by APCs in an IL-15–dependent manner.19 This appeared to be caused by the essential role of IL-15 in IL12RB1 expression in APCs, because IL12RB1 mRNA expression is significantly reduced in APCs from IL-15, γc-/-, RAG-2-/-, and IL-2Rβ-/-RAG-2-/- mice.19 These lines of evidence indicated that IL-15–mediated IL12RB1 expression on APCs is important in the generation of TH1-type immune responses.
Thus, IL12RB1 mRNA expression in APCs is regulated by IFN-γ and IL-15, but the molecular mechanisms governing IL12Rβ1 transcription have not been extensively studied. In this study, we isolated mouse IL12RB1 genomic clones and investigated the transcriptional regulation in a mouse macrophage cell line. We here show that IL-15 and IFN-γ synergistically activated IL12RB1 gene expression through the cooperation of 2 transcription factors, PU.1 and IRF3. IL12RB1 gene transcription is also associated with histone acetylation and chromatin remodeling of the gene, which are efficiently induced by IL-15.
Materials and methods
Antibodies and reagents
Recombinant mouse IFN-γ and IL-15 were purchased from Peprotech (Seattle, WA). Anti–phospho-histone H3 (Ser 10), anti–acetyl-histone H3, and anti–acetyl-histone H4 antibodies were purchased from Upstate Biotechnology (Lake Placid, NY). Antihemagglutinin (HA), anti-p38, anti-JNK1, anti-PU.1, and anti-IRF3 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Phosphospecific anti-p38, anti-JNK, and antiphosphoserine antibodies were purchased from New England Biolabs (Beverly, MA). Lipopolysaccharide (LPS) from Escherichia coli serotype B5 055 and trichostatin A (TSA) were obtained from Sigma Chemical (St Louis, MO). Specific mitogen-activated protein kinase (MAPK) inhibitors SB203580 and SP600125 were purchased from Calbiochem (San Diego, CA).
Isolation and characterization of the mouse IL12RB1 genomic clones
The mouse IL12RB1 cDNA containing the 5′ portion of the coding region was prepared by reverse transcription–polymerase chain reaction (RT-PCR) using 2 primers, (5′-CTGCAGAAGTTCCTGGGGCCT and 5′-CCCGGGCAGGTGCTCAGCTG), from the total RNA of RAW264.7 cells, and was used for screening the mouse genomic phage library in Lambda Fix II (Stratagene, La Jolla, CA), as previously described.20
Primer extension analysis
Total RNA was prepared from IFN-γ–stimulated RAW264.7 cells. Contaminating DNA was removed by treatment with 10 μg/mL DNase I (Sigma) for 2 hours at 16°C. Remaining RNA was extracted twice with phenol/chloroform and was subjected to poly (A)+ RNA purification using the mRNA purification kit (Amersham Biosciences, Piscataway, NJ). An oligonucleotide, 5′-CCCATCATGTCCATGAGGAGCCGAGTCGTG, complementary to the 5′ untranslated region of the IL12RB1 cDNA (GenBank/EMBL accession number NM 008353) was 5′ end-labeled with 32 P. Annealing of the labeled primer to poly (A)+ RNA was performed in 11 μL of 2 × first-strand buffer (Invitrogen, Carlsbad, CA) by heating the mixture of 2 pmol labeled primer and 10 μg poly (A)+ RNA at 48°C for 40 minutes and by cooling to room temperature for 20 minutes. Reverse transcription was run on a 6% denaturing polyacrylamide urea gel, as previously described.21
Generation of the mouse IL12RB1 promoter constructs
A series of sense oligonucleotides—AGATCTGTGTTCCTGCAGAG (pGL3-3875), GGAATGTTAACCTGCGAGTG (pGL3-2508), ACTCCCTTGATTGGGCCACC (pGL3-1803), TAGGCAGGCCCCCCAGCCTG (pGL3-1483), CAGGACACTTGACCCCTGGG (pGL3-849), AGGGCCATATGGTATCTGTG (NF-κB del), TTGGTTTCCCAAAGCCGCCT (ETS/PU.1 del), CATGCAGCCTTTGCTCTGTG (IRE/ISRE del)—and an antisense oligonucleotide—CCAGGAACTTCTGCAGGGCTG—were used to generate a series of 5′ deletion DNA fragments. The PCR products were digested with KpnI/MluI (pGL3-3875) or XhoI/MluI (all the others) and was subcloned into the pGL3-basic vector (Promega). To generate the IRE/ISRE-mutated construct, 2 PCR products from 2 pairs of primers (GGAATGTTAACCTGCGAGTG and GCAGATGGGTTTTCAGACAG; CATGCAGCCTTTGCTCTCGT and CCAGGAACTTCTGCAGGGCTG) were amplified, digested with EcoRI, ligated, and cloned into the pGL3-basic vector. The mammalian expression plasmid pcDNA3-CBP-HA was a kind gift from Dr R Eckner (Institute for Molecular Biology, Zurich, Switzerland).22
Transfection and luciferase assays
A mouse macrophage cell line, RAW264.7, was obtained from RIKEN cell bank (Tsukuba, Japan). RAW264.7 stable transfectants were generated as previously described.23 For luciferase assays, cells were plated onto 35-mm plate at 1 × 105 cells/plate. Twenty-four hours after plating, cells were supplied with Dulbecco modified Eagle medium (DMEM) plus 1% fetal calf serum (FCS) and were left untreated or were treated with 10 ng/mL IFN-γ, 10 ng/mL IL-15, 1 μg/mL LPS, 100 nM TSA, or their combination for 8 hours. Luciferase activity in the cell lysates was measured using a single luciferase assay system (Promega) according to the manufacturer's instructions. All luciferase assays shown in the current study were performed in triplicate, and the average of 3 independent experiments was shown.
Northern blot analysis
Northern blotting was performed as previously described.23 For the probes, cDNA fragments containing the full coding regions of mouse IL12RB1, IRF1, IRF2, IRF3, and IRF7 were prepared by RT-PCR from total RNA of IFN-γ–treated RAW264.7 cells and were 32 P-labeled with the Prime-a-Gene labeling kit (Promega).
EMSA
Nuclear extracts were prepared from RAW264.7 cells as previously described.24,25 Primers used for electrophoretic mobility shift assay (EMSA) were 5′-TGGACTTCCC for NF-κB, 5′-GGTAGAGGAAGTCCAG for ETS/PU.1, 5′-AAAAAACTCAAACTG for IRE/ISRE, 5′-TTATTGATAACTGG for GATA1, 5′-TGTTCCTTGAAAACT for STAT/STAT5, and 5′-CACACGTGT for E-Box/USF. Approximately 1 × 105 cpm of an oligonucleotide, radiolabeled by γ32 P-ATP with T4 polynucleotide kinase, 2 μg nuclear extracts, and 1 μg poly (dI·dC), was added to the binding buffer (20 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid]–KOH [pH 7.9], 60 mM KCl, 1 mM dithiothreitol [DTT], 1 mM EDTA [ethylenediaminetetraacetic acid], and 5% glycerol)26 and was incubated for 30 minutes at 4°C. Specific IRF blockers used for the competition assays were C13 5′-AAGTGAAAGTGAAAGTGA for IRF1/IRF2,27 PRDIII 5′-GAAAACTGAAAGGG for IRF3, and 5′ GAAAGTGAACGC for IRF7.28 The mutated primer for IRF3/IRF7, 5′-GAAAGTCTTCGC,28 was used as a negative control. Competition assays were performed by incubating nuclear extracts with a 5- or a 50-fold excess of unlabeled oligonucleotide before the addition of the 32 P-labeled probe. For supershift assays, 2 μg anti-PU.1 or anti-IRF3 antibody was preincubated with nuclear extracts for 1 hour. Reaction mixtures were run through a 6% nondenaturing polyacrylamide gel at 4°C in TBE buffer.
Immunoblotting and immunoprecipitation
Total cellular lysate preparation, Western blotting, and immunoprecipitation were performed as previously described.23
Chromatin immunoprecipitation assay
Acetyl-histone chromatin immunoprecipitation (ChIP) assay was performed according to the manufacturer's instruction (Upstate Biotechnology). After IFN-γ, IL-15, or LPS stimulation, chromatin cross-linking, immunoprecipitation, and PCR amplification were performed essentially as previously described.21 The IL12RB1 promoter-specific primers were: sense, CATATGGTATCTGTGTGTTT; antisense, ACGAGAGCAAAGGCTGCATG. The locations of these primers are indicated in Figure 2C. PCR amplifies the genomic region containing ETS/PU.1 and IRE/ISRE consensus sites. PCR conditions were as follows: 94°C for 3 minutes; 20 to 25 cycles at 94°C for 30 seconds and at 66°C for 2 minutes; 72°C for 7 minutes, followed by 3 cycles with the γ32 P-ATP–labeled antisense primer. PCR products were analyzed by electrophoresis on a 6% denaturing polyacrylamide gel. For each reaction, 1% of the cross-link–released chromatin was saved and used as an input control.
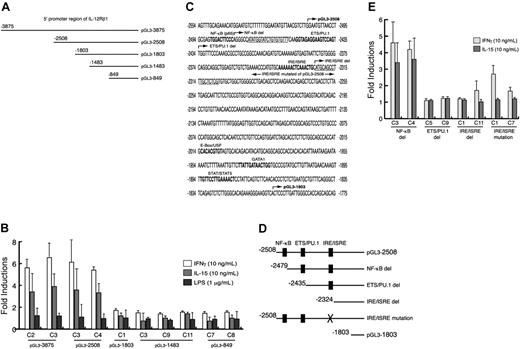
Functional elements required for IL12RB1 transcriptional activation. (A) Schematic presentation of the IL12RB1 promoter deletion constructs. A series of 5′ promoter deletion constructs were cloned into pGL3-basic vector and were stably integrated into RAW264.7 cells. The number of each construct corresponds to its 5′ end. (B) RAW264.7 cells stably integrated with IL12RB1 deletion constructs were cultured without serum at 1 × 105 cells/plate for 24 hours. Cells were left untreated or were treated with one of the following: 10 ng/mL IFN-γ, 10 ng/mL IL-15, or 1 μg/mL LPS for 8 hours before lysate preparation for luciferase assays. Luciferase activity from 2 independent clones of each deletion construct was examined. Basal luciferase activity of untreated cells was generally similar among all the examined clones. Results are expressed as fold induction over untreated controls (mean ± SD). (C) The nucleotide sequence of the region required for the IL12RB1 promoter activation by IFN-γ and IL-15, potential binding sites for transcription factors, are indicated by bold capital letters. Arrows identify the 5′ end of deletion constructs, and nucleotides used as primers for ChIP assays were underlined. (D) Schematic presentation of the second series of IL12RB1 promoter deletion mutant. Black boxes represent binding elements for indicated transcription factors. (E) RAW264.7 cells stably integrated with a second series of promoter deletion mutant (as in panel D) were cultured and plated as described in panel B. Cells were left untreated or were treated with 10 ng/mL IFN-γ or 10 ng/mL IL-15 for 8 hours before lysate preparation and luciferase assays. Basal luciferase activity of untreated cells was generally similar among the examined clones. Fold inductions by IFN-γ or IL-15 are expressed as described in panel B.
Functional elements required for IL12RB1 transcriptional activation. (A) Schematic presentation of the IL12RB1 promoter deletion constructs. A series of 5′ promoter deletion constructs were cloned into pGL3-basic vector and were stably integrated into RAW264.7 cells. The number of each construct corresponds to its 5′ end. (B) RAW264.7 cells stably integrated with IL12RB1 deletion constructs were cultured without serum at 1 × 105 cells/plate for 24 hours. Cells were left untreated or were treated with one of the following: 10 ng/mL IFN-γ, 10 ng/mL IL-15, or 1 μg/mL LPS for 8 hours before lysate preparation for luciferase assays. Luciferase activity from 2 independent clones of each deletion construct was examined. Basal luciferase activity of untreated cells was generally similar among all the examined clones. Results are expressed as fold induction over untreated controls (mean ± SD). (C) The nucleotide sequence of the region required for the IL12RB1 promoter activation by IFN-γ and IL-15, potential binding sites for transcription factors, are indicated by bold capital letters. Arrows identify the 5′ end of deletion constructs, and nucleotides used as primers for ChIP assays were underlined. (D) Schematic presentation of the second series of IL12RB1 promoter deletion mutant. Black boxes represent binding elements for indicated transcription factors. (E) RAW264.7 cells stably integrated with a second series of promoter deletion mutant (as in panel D) were cultured and plated as described in panel B. Cells were left untreated or were treated with 10 ng/mL IFN-γ or 10 ng/mL IL-15 for 8 hours before lysate preparation and luciferase assays. Basal luciferase activity of untreated cells was generally similar among the examined clones. Fold inductions by IFN-γ or IL-15 are expressed as described in panel B.
Results
IL12RB1 gene expression in RAW264.7 cells
As a model for APCs expressing IL12RB1, we analyzed a commonly used mouse macrophage cell line, RAW264.7. To reveal the expression pattern of IL12RB1 mRNA, cells were left untreated or were stimulated with LPS, IL-15, or IFN-γ for 4 hours, and total RNAs were subjected to Northern blot analyses. As shown in Figure 1A, a small amount of IL12RB1 mRNA was constitutively expressed in untreated RAW264.7 cells. Stimulation of the cells with IFN-γ or IL-15 (10 ng/mL each) significantly increased IL12RB1 gene expression, whereas no remarkable change was detected after LPS stimulation even at concentrations as high as 1 μg/mL.
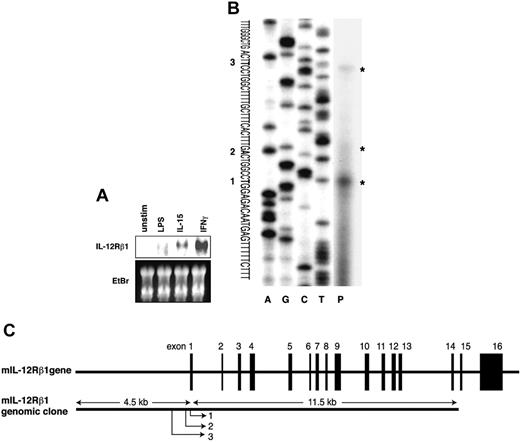
Expression and molecular cloning of the mouse IL12RB1 gene. (A) IL-15 and IFN-γ, but not LPS, induce IL12RB1 gene expression in a mouse macrophage cell line. RAW264.7 cells were left untreated (unstim) or treated for 4 hours with 1 μg/mL LPS, 10 ng/mL IL-15, or 10 ng/mL IFN-γ. Total RNA (20 μg each) was prepared for the Northern blot analysis using a 32 P-labeled IL12RB1 cDNA probe. Gene expression of IL12RB1 and a picture of the ethidium bromide (EtBr)–stained gel are shown. (B) Transcription start sites of the IL12RB1 gene were mapped by primer extension analysis. The transcribed product (P) was run along with sequencing ladder from the same primer on a 6% polyacrylamide/urea gel. Asterisks indicate the transcribed products; 3 transcriptional start sites are shown, and the proximal major site was designed as +1. (C) Schematic presentation of the mouse IL12RB1 gene. Black boxes represent exons. Positions of 3 transcriptional start sites are indicated.
Expression and molecular cloning of the mouse IL12RB1 gene. (A) IL-15 and IFN-γ, but not LPS, induce IL12RB1 gene expression in a mouse macrophage cell line. RAW264.7 cells were left untreated (unstim) or treated for 4 hours with 1 μg/mL LPS, 10 ng/mL IL-15, or 10 ng/mL IFN-γ. Total RNA (20 μg each) was prepared for the Northern blot analysis using a 32 P-labeled IL12RB1 cDNA probe. Gene expression of IL12RB1 and a picture of the ethidium bromide (EtBr)–stained gel are shown. (B) Transcription start sites of the IL12RB1 gene were mapped by primer extension analysis. The transcribed product (P) was run along with sequencing ladder from the same primer on a 6% polyacrylamide/urea gel. Asterisks indicate the transcribed products; 3 transcriptional start sites are shown, and the proximal major site was designed as +1. (C) Schematic presentation of the mouse IL12RB1 gene. Black boxes represent exons. Positions of 3 transcriptional start sites are indicated.
Isolation of mouse IL12RB1 genomic DNA containing 5′ promoter region
To identify the transcriptional regulatory region of the mouse IL12RB1 gene, we screened a mouse genomic library with an IL12RB1 cDNA probe. A 15-kb fragment of the IL12RB1 genomic DNA, which appeared to include approximately 4.5 kb of the putative 5′ flanking region, was isolated (Figure 1C). Primer extension analysis was then carried out to map the transcriptional start site. A synthetic oligonucleotide complementary to the 5′ end region of the reported IL12RB1 cDNA (GenBank/EMBL accession number NM 008353) was hybridized to poly (A)+ RNA from IFN-γ–stimulated RAW264.7 cells and was extended by reverse transcription. As shown in Figure 1B, the analysis revealed a major transcriptional start site (indicated as 1) and 2 minor sites (indicated as 2 and 3). The proximal major transcriptional start site (1) was located at 87 bp upstream from the 5′ end of the previously described cDNA sequence (GenBank/EMBL accession number NM 008353), and the nucleotide of this site (T) was designed as +1 throughout this report. The second and third minor transcriptional start sites were 7 and 30 bp upstream of the first major site, respectively. The schematic presentation of the mouse IL12RB1 gene structure is shown in Figure 1C.
Identification of the regulatory elements involved in IL12RB1 gene transcription
The nucleotide sequence upstream of the transcriptional start sites was searched for the potential binding sites for transcription factors using Genome Exploring and Modeling Software.29 Neither TATA nor CAAT boxes were present in the 5′ upstream region of each of the 3 transcriptional start sites. A series of 5′ deletion luciferase constructs (Figure 2A) was initially generated to locate the region required for IL12RB1 gene transcription. The generated plasmids were stably integrated into RAW264.7 cells, and several independent clones for each construct were isolated for stimulation with IFN-γ, IL-15, or LPS. Among the reporter constructs, the highest fold induction by IFN-γ or IL-15 was obtained from the longest pGL3-3875 construct, but no significant decrease was detected with the pGL3-2508 construct, indicating the 2508-bp region is sufficient for the full transcriptional activation of the IL12RB1 gene by these 2 cytokines. Further deletion to -1803 (pGL3-1803) completely abrogated promoter responsiveness to IFN-γ or IL-15, suggesting the region between -2508 and -1803 is essential for transcriptional induction. In contrast, neither of these constructs showed inducible promoter activity in response to LPS (Figure 2B).
Nucleotide analysis of the -2508 to approximately the -1803 region revealed binding sites for NF-κB, ETS/PU.1, IRE/ISRE, E-Box/USF, GATA1, and STAT/STAT5 (Figure 2C). We thus generated the second series of promoter deletion constructs, which specifically deleted NF-κB, ETS/PU.1, or IRE/ISRE (Figure 2D). As shown in Figure 2E, IL12RB1 promoter activation by IFN-γ or IL-15 was not affected by the removal of the NF-κB consensus element. In contrast, deletion of ETS/PU.1 (ETS/PU.1 del) abolished IL12RB1 promoter activation by IFN-γ or IL-15. To further examine the involvement of the IRE/ISRE binding site, the IRE/ISRE sequence of pGL3-2508 was independently mutated (IRE/ISRE mutation). The IRE/ISRE mutation significantly inhibited, but did not abolish, promoter activation by IFN-γ, whereas it completely abrogated the responsiveness to IL-15. Taken together, ETS/PU.1 and IRE/ISRE, but not NF-κB consensus elements, seemed essentially involved in IL12RB1 transcriptional activation by IFN-γ and IL-15.
Protein binding to the ETS/PU.1 and IRE/ISRE consensus elements in the IL12RB1 promoter
We next examined the protein-DNA binding responsible for IL12RB1 gene activation using EMSA. Nuclear extracts prepared from untreated, IFN-γ–, IL-15–, or LPS-treated RAW264.7 cells were incubated with 32 P-labeled double-strand oligonucleotides corresponding to the NF-κB, ETS/PU.1, IRE/ISRE, E-Box/USF, GATA1, or STAT/STAT5 consensus binding site of the IL12RB1 promoter (Figure 3A-B). EMSA revealed that stimulation with IFN-γ or IL-15 potently induced protein binding to ETS/PU.1 and IRE/ISRE consensus elements but not to NF-κB, E-Box/USF, GATA1, or STAT/STAT5 elements. The protein binding complexes of ETS/PU.1 and IRE/ISRE were specific because they were abrogated by the addition of excess unlabeled specific oligonucleotides.
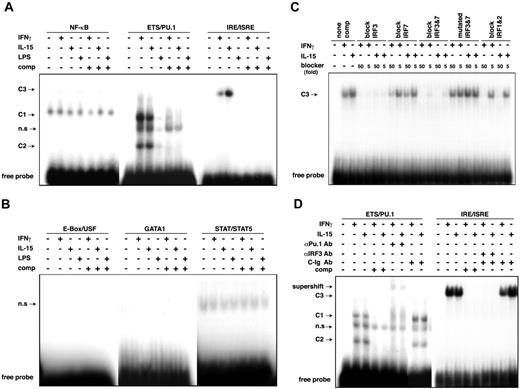
Specific protein binding of ETS/PU.1 and IRE/ISRE consensus sequences of IL12RB1 promoter. (A-B) RAW264.7 cells were left untreated or were treated with IFN-γ, IL-15, or LPS for 45 minutes, and the nuclear extracts were prepared for EMSA. Two micrograms nuclear extract was incubated with 32 P-labeled oligonucleotides complementary to the indicated consensus elements. DNA-protein complexes were separated on a 6% polyacrylamide gel. C1 and C2 indicate protein complex binding to ETS/PU.1; C3, protein complex binding to IRE/ISRE consensus sequence; n.s, nonspecific complex; comp, EMSA binding reaction containing 20-fold excess of the specific cold competitor. (C) Identification of IRF binding to the IRE/ISRE site of the IL12RB1 promoter. Cold oligonucleotides used in the competition assays are indicated. None comp indicates EMSA binding reaction that contained no competitor. (D) Supershift assay using anti-PU.1 antibody (αPU.1 Ab), anti-IRF3 antibody (αIRF3 Ab), or control isotype antibody (C-Ig Ab). EMSA was carried out as in panel A. For supershift experiments, nuclear extracts were preincubated with 2 μg indicated antibody for 1 hour before 32 P-labeled probe was added. Modified DNA-protein complexes are indicated.
Specific protein binding of ETS/PU.1 and IRE/ISRE consensus sequences of IL12RB1 promoter. (A-B) RAW264.7 cells were left untreated or were treated with IFN-γ, IL-15, or LPS for 45 minutes, and the nuclear extracts were prepared for EMSA. Two micrograms nuclear extract was incubated with 32 P-labeled oligonucleotides complementary to the indicated consensus elements. DNA-protein complexes were separated on a 6% polyacrylamide gel. C1 and C2 indicate protein complex binding to ETS/PU.1; C3, protein complex binding to IRE/ISRE consensus sequence; n.s, nonspecific complex; comp, EMSA binding reaction containing 20-fold excess of the specific cold competitor. (C) Identification of IRF binding to the IRE/ISRE site of the IL12RB1 promoter. Cold oligonucleotides used in the competition assays are indicated. None comp indicates EMSA binding reaction that contained no competitor. (D) Supershift assay using anti-PU.1 antibody (αPU.1 Ab), anti-IRF3 antibody (αIRF3 Ab), or control isotype antibody (C-Ig Ab). EMSA was carried out as in panel A. For supershift experiments, nuclear extracts were preincubated with 2 μg indicated antibody for 1 hour before 32 P-labeled probe was added. Modified DNA-protein complexes are indicated.
To further identify protein binding to the IRE/ISRE consensus element, we used 5- or 50-fold excesses of unlabeled, synthetic, double-strand oligonucleotides that specifically bind to interferon regulatory factor 1 and 2 (IRF1/2), IRF3, IRF7, and IRF3/7 transcription factors or to a mutated form of IRF3/7 consensus sequence to block the IRE/ISRE–protein complex formation.28 As shown in Figure 3C, protein binding to the IRE/ISRE consensus element of the IL12RB1 gene was completely abrogated by both high and low molar excesses of the unlabeled blocker specific for IRF3. The protein complex was also efficiently decreased by unlabeled IRF3/7 blocker. In contrast, only high molar excesses of unlabeled IRF7 and IRF1/2 blockers could block the protein binding, and the protein complex could not be blocked by the mutated form of IRF3/7 blocker. We then addressed whether an antibody against IRF3 could modify the DNA–protein complex binding to the IRE/ISRE element of the IL12RB1 gene. As shown in Figure 3D, the inducible DNA–protein complex formation was effectively inhibited by the anti-IRF3 antibody, indicating that this DNA–protein complex contained IRF3 at least as a component.
Next, to identify the protein binding to the ETS/PU.1 consensus element of the IL12RB1 gene, we applied an anti-PU.1 antibody to the EMSA. As shown in Figure 3D, DNA–protein complexes containing the ETS/PU.1 consensus element were effectively supershifted by the anti-PU.1 antibody, whereas the antibody did not modify the DNA–protein binding of IRE/ISRE sequence (data not shown). Thus, the binding of IRF3 and PU.1 appeared mutually independent. The identities of multiple DNA–protein complexes containing PU.1 are not certain. Although PU.1 can form complexes with various proteins, including IRFs and CBP, we failed to observe supershift or inhibition of the complexes with anti-IRF3 (Figure 3D) or anti-CBP (data not shown). It is possible that proteins other than IRF3 and CBP are involved in the complexes.
IL-15 and IFN-γ synergistically activated IL12RB1 gene transcription
Our data indicated that IL-15 and IFN-γ induced IL12RB1 promoter activation through ETS/PU.1 and IRE/ISRE elements (Figures 2, 3). To investigate the possible synergism between these 2 cytokines, we stimulated RAW264.7 cells with IFN-γ in combination with IL-15 and found that the combination showed a significantly synergistic effect (Figure 4A).30 Interestingly, TSA, a specific inhibitor of histone deacetylases, also showed synergistic IL12RB1 mRNA induction in combination with IFN-γ. In contrast, the synergy was less significant for TSA plus IL-15 (Figure 4A).
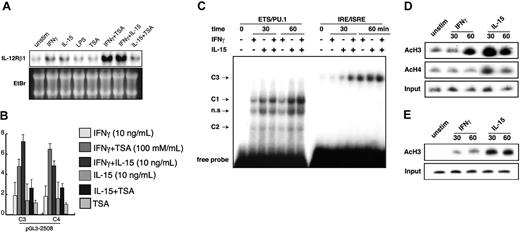
IL-15, synergistically with IFN-γ, activates IL12RB1 gene transcription. (A) RAW264.7 cells were left untreated (unstim) or were stimulated with either of the following: 1 μg/mL LPS, 10 ng/mL IL-15, 10 ng/mL IFN-γ, or combination IL-15 and IFN-γ for 4 hours. In some experiments, cells were pretreated with 100 mM TSA before stimulation. Total RNA was prepared for Northern blot analysis. Gene expression of IL12RB1 and a picture of the ethidium bromide (EtBr)–stained gel are shown. (B) RAW264.7 cells stably integrated with pGL3-2508 luciferase construct were prepared as described for Figure 2B. Cells were left untreated or were treated with 10 ng/mL IFN-γ, 10 ng/mL IL-15, combination IFN-γ and IL-15 for 6 hours with or without the addition of 100 mM TSA before lysate preparation and luciferase assays. Basal luciferase activity of untreated cells was generally similar among all the examined clones. Fold inductions by IFN-γ or IL-15 are expressed as described for Figure 2B. (C) IL-15 and IFN-γ synergistically enhanced protein binding to IRE/ISRE. RAW264.7 cells were left untreated or were treated with IFN-γ, IL-15, or combination IFN-γ and IL-15 for 30 or 60 minutes, and nuclear extracts were prepared for EMSA. EMSA was carried out as described in Figure 3A; inducible protein binding is shown. (D) Acetylation of histone H3 at the IL12RB1 gene is induced by IFN-γ and IL-15. RAW264.7 cells were left untreated (unstim) or were treated with 10 ng/mL IFN-γ or 10 ng/mL IL-15 for the indicated time. After treatment, chromatin was extracted and immunoprecipitated with anti–acetyl-histone H3 or anti–acetyl-histone H4. PCR analyses of DNA products from immunoprecipitation were carried out as described in “Materials and methods.” (E) Mouse peritoneal macrophages were prepared as previously described,30 and the ChIP assays were carried out as described for Figure 4D. Acetylation levels of histone H3 at the IL12RB1 gene are shown.
IL-15, synergistically with IFN-γ, activates IL12RB1 gene transcription. (A) RAW264.7 cells were left untreated (unstim) or were stimulated with either of the following: 1 μg/mL LPS, 10 ng/mL IL-15, 10 ng/mL IFN-γ, or combination IL-15 and IFN-γ for 4 hours. In some experiments, cells were pretreated with 100 mM TSA before stimulation. Total RNA was prepared for Northern blot analysis. Gene expression of IL12RB1 and a picture of the ethidium bromide (EtBr)–stained gel are shown. (B) RAW264.7 cells stably integrated with pGL3-2508 luciferase construct were prepared as described for Figure 2B. Cells were left untreated or were treated with 10 ng/mL IFN-γ, 10 ng/mL IL-15, combination IFN-γ and IL-15 for 6 hours with or without the addition of 100 mM TSA before lysate preparation and luciferase assays. Basal luciferase activity of untreated cells was generally similar among all the examined clones. Fold inductions by IFN-γ or IL-15 are expressed as described for Figure 2B. (C) IL-15 and IFN-γ synergistically enhanced protein binding to IRE/ISRE. RAW264.7 cells were left untreated or were treated with IFN-γ, IL-15, or combination IFN-γ and IL-15 for 30 or 60 minutes, and nuclear extracts were prepared for EMSA. EMSA was carried out as described in Figure 3A; inducible protein binding is shown. (D) Acetylation of histone H3 at the IL12RB1 gene is induced by IFN-γ and IL-15. RAW264.7 cells were left untreated (unstim) or were treated with 10 ng/mL IFN-γ or 10 ng/mL IL-15 for the indicated time. After treatment, chromatin was extracted and immunoprecipitated with anti–acetyl-histone H3 or anti–acetyl-histone H4. PCR analyses of DNA products from immunoprecipitation were carried out as described in “Materials and methods.” (E) Mouse peritoneal macrophages were prepared as previously described,30 and the ChIP assays were carried out as described for Figure 4D. Acetylation levels of histone H3 at the IL12RB1 gene are shown.
To analyze the synergistic effects on IL12RB1 promoter–driven luciferase activity, we used RAW264.7 cells stably integrated with the pGL3-2508 luciferase construct. The fold inductions by IFN-γ/IL-15 cotreatment were significantly higher than those by stimulation with each cytokine (Figure 4B). A similar synergistic effect was observed for costimulation with IFN-γ and TSA. In contrast, no synergistic effect was observed between TSA and IL-15.
We further analyzed the effects of combination IFN-γ and IL-15 on the kinetics of protein binding to the IL12RB1 promoter. As shown in Figure 4C, protein binding to ETS/PU.1 was similarly induced by IFN-γ and IL-15 stimulation. No synergistic increase of protein binding was detected when cells were stimulated with a combination of IL-15 and IFN-γ. In contrast, significant synergy was observed for the content of protein binding to the IRE/ISRE consensus element. Specifically, a synergistic effect on protein binding to IRE/ISRE was more evident at 30 minutes of stimulation than at 60 minutes.
Because IL-15 and TSA similarly activated IL12RB1 gene transcription in synergy with IFN-γ, we next investigate possible histone modification by IL-15 in the regulation of IL12RB1 gene. We applied the ChIP assay using IL12RB1 promoter-specific primers. As shown in Figure 4D-E, stimulation of RAW264.7 cells or mouse peritoneal macrophages with IL-15 rapidly induced the acetylation of histone H3 at the IL12RB1 promoter, whereas acetylation by IFN-γ stimulation was slower and less significant. On the other hand, only IL-15 induced the acetylation of histone H4.
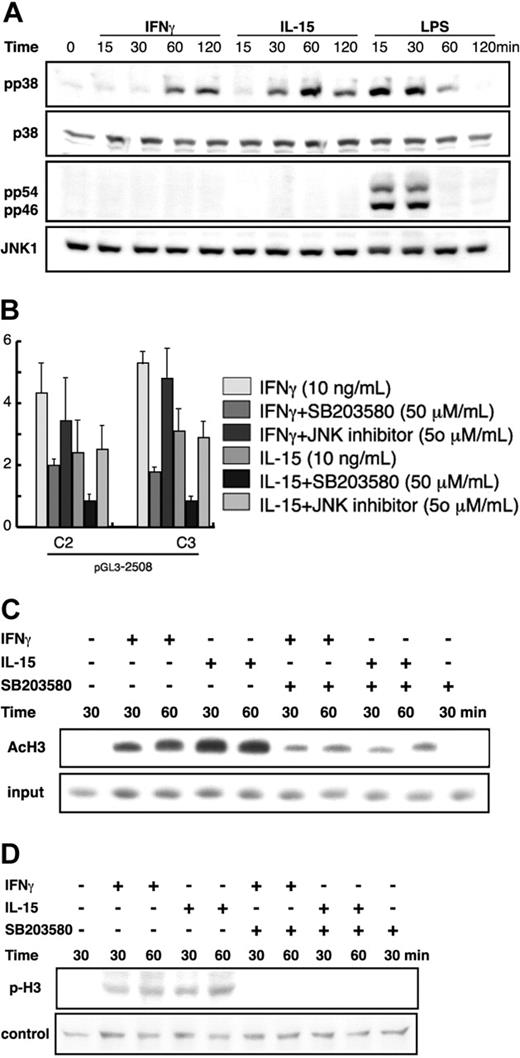
Activation of p38 MAPK is necessary for the transcriptional induction of IL12RB1
Previous studies have indicated p38 MAPK in association with the phospho-acetylation of histone H3 and the transcription of a subset of stimulus-induced cytokine and chemokine genes.31 To investigate whether p38 MAPK activation is involved in IL12RB1 gene induction in macrophages, we first examined p38 MAPK activation in RAW264.7 cells after IFN-γ and IL-15 stimulation. LPS stimulation was also carried out as a positive control. As shown in Figure 5A,32 treatment of RAW264.7 cells with IL-15 or LPS induced rapid phosphorylation of p38 MAPK within 30 minutes. In contrast, p38 MAPK phosphorylation by IFN-γ was slower, requiring 60 minutes for detectable phosphorylation. We also investigated JNK phosphorylation and found that, unlike LPS, neither IFN-γ nor IL-15 could induce detectable phosphorylation of JNK.
p38 MAPK is involved in the transcriptional regulation of IL12RB1. (A) RAW264.7 cells were treated with 10 ng/mL IFN-γ, 10 ng/mL IL-15, or 1 μg/mL LPS for the indicated time before total cellular lysate preparation. The phosphorylation of p38 (pp38) or JNK (pp54 and pp46) was detected by Western blot analysis using polyclonal antibody (pAb) specific for the phosphorylated p38 or mAb specific for the phosphorylated JNK. As a control, 10% of each lysate was used to detect p38 or JNK using anti-p38 or anti-JNK1 pAb, respectively. (B) RAW264.7 cells stably integrated with pGL3-2508 luciferase construct were prepared as described for Figure 2B. Cells were left untreated or were pretreated with 50 μM SB203580 or 50μM SP600125 for 30 minutes before stimulation with 10 ng/mL IFN-γ or 10 ng/mL IL-15 for an additional 8 hours. Lysates were prepared for luciferase assay. Basal luciferase activity of untreated cells was generally similar among all the examined clones, and the fold inductions are expressed as described for Figure 2B. (C) RAW264.7 cells were pretreated with SB203580 for 30 minutes before stimulation with 10 ng/mL IFN-γ or 10 ng/mL IL-15 for the indicated time. The acetylation of histone H3 was analyzed as described for Figure 4E. (D) RAW264.7 cells were treated as described in panel C. Nuclear lysates were prepared as previously described.32 The phosphorylation of histone H3 (p-H3) was detected by Western blot analyses using an antibody specific for the phosphorylated histone H3. As a control, 10% of each lysate was used to measure protein levels by Coomassie staining.
p38 MAPK is involved in the transcriptional regulation of IL12RB1. (A) RAW264.7 cells were treated with 10 ng/mL IFN-γ, 10 ng/mL IL-15, or 1 μg/mL LPS for the indicated time before total cellular lysate preparation. The phosphorylation of p38 (pp38) or JNK (pp54 and pp46) was detected by Western blot analysis using polyclonal antibody (pAb) specific for the phosphorylated p38 or mAb specific for the phosphorylated JNK. As a control, 10% of each lysate was used to detect p38 or JNK using anti-p38 or anti-JNK1 pAb, respectively. (B) RAW264.7 cells stably integrated with pGL3-2508 luciferase construct were prepared as described for Figure 2B. Cells were left untreated or were pretreated with 50 μM SB203580 or 50μM SP600125 for 30 minutes before stimulation with 10 ng/mL IFN-γ or 10 ng/mL IL-15 for an additional 8 hours. Lysates were prepared for luciferase assay. Basal luciferase activity of untreated cells was generally similar among all the examined clones, and the fold inductions are expressed as described for Figure 2B. (C) RAW264.7 cells were pretreated with SB203580 for 30 minutes before stimulation with 10 ng/mL IFN-γ or 10 ng/mL IL-15 for the indicated time. The acetylation of histone H3 was analyzed as described for Figure 4E. (D) RAW264.7 cells were treated as described in panel C. Nuclear lysates were prepared as previously described.32 The phosphorylation of histone H3 (p-H3) was detected by Western blot analyses using an antibody specific for the phosphorylated histone H3. As a control, 10% of each lysate was used to measure protein levels by Coomassie staining.
Pretreatment of RAW264.7 cells with 50 μM SB203580, a specific p38 MAPK inhibitor, effectively abrogated p38 phosphorylation by IFN-γ and IL-15 (data not shown). As shown in Figure 5B, pretreatment with SB203580 significantly inhibited IL12RB1 promoter-driven luciferase activity by IFN-γ and IL-15. In contrast, a JNK-specific inhibitor, SP600125, showed no inhibitory effect. Consistently, IL12RB1 mRNA increase by IFN-γ and IL-15 in RAW264.7 cells was effectively inhibited by SB203580 but not by SP600125 (data not shown). We also examined the acetylation of histone H3 in the presence and absence of SB203580 by ChIP. As shown in Figure 5C, pretreatment of RAW264.7 with the p38 MAPK inhibitor significantly reduced the acetylation level of histone H3 at the IL12RB1 promoter. In agreement with these data, SB203580 pretreatment completely abrogated phosphorylation of histone H3 (Figure 5D).
Roles of CBP and IRFs in IL12RB1 transcription
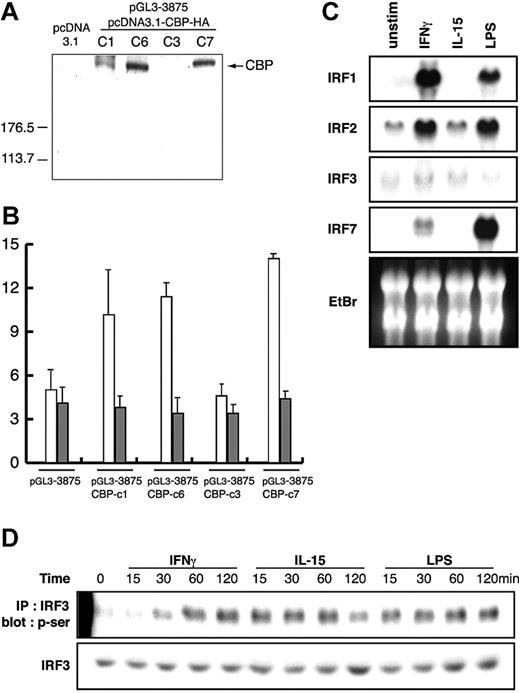
We were interested in determining the possible involvement of cyclic adenosine monophosphate (cAMP)–responsive elementbinding protein (CREB)–binding protein (CBP), which is an endogenous histone acetyltransferase (HAT), in IL12RB1 gene activation. For this purpose, RAW264.7 cells were stably integrated with pGL3-3875 and a CBP expression plasmid. Several G418-resistant clones were obtained. As confirmed by Western blotting, the HA-tagged CBP expression levels varied among these clones (Figure 6A). Using these stably integrated clones, we found that overexpression of CBP strongly enhanced IL12RB1 promoterdriven luciferase activity after IFN-γ treatment, whereas CBP expression had no effect on the promoter activation by IL-15 (Figure 6B). In addition, the inducible effect of CBP on IFN-γ–driven promoter activation correlated with its expression levels among the clones (Figure 6A-B).
Effects of CBP expression on IL12RB1 promoter activity and IRF3 phosphorylation. (A) RAW264.7 cells were stably integrated with pGL3-3875 of IL12RB1 luciferase construct, together with CBP-HA expression plasmid. Total cellular lysates were prepared from stably integrated clones, and the expression levels of CBP were analyzed by Western blotting using anti-HA mAb. (B) Overexpression of a coactivator protein CBP increases IFN-γ–mediated IL12RB1 transcription. The stably integrated clones shown in panel A were cultured and plated as described for Figure 2B. Cells were left untreated or were treated with either of the following: 10 ng/mL IFN-γ (□), 10 ng/mL IL-15 (▦), or 1 μg/mL LPS (not shown) for 8 hours before lysate preparation and luciferase assays. Basal luciferase activity of untreated cells was generally similar among all the examined clones. Fold inductions are shown. (C) Neither IFN-γ nor IL-15 induces gene expression of IRF3. RAW264.7 cells were left untreated (unstim) or were treated for 4 hours with 1 μg/mL LPS, 10 ng/mL IL-15, or 10 ng/mL IFN-γ. Total RNA was prepared for Northern blot analysis using IRF1, IRF2, IRF3, and IRF7 cDNA probes. Gene expression of the indicated gene and a picture of ethidium bromide (EtBr) are shown. (D) RAW264.7 cells were treated as described for Figure 5A. Total cellular lysates were prepared and immunoprecipitated with anti-IRF3 pAb. The phosphorylation of IRF3 was detected by Western blot analysis using antiphosphoserine mAb. As a control, 10% of each lysate was used to detect IRF3 by anti-IRF3 pAb.
Effects of CBP expression on IL12RB1 promoter activity and IRF3 phosphorylation. (A) RAW264.7 cells were stably integrated with pGL3-3875 of IL12RB1 luciferase construct, together with CBP-HA expression plasmid. Total cellular lysates were prepared from stably integrated clones, and the expression levels of CBP were analyzed by Western blotting using anti-HA mAb. (B) Overexpression of a coactivator protein CBP increases IFN-γ–mediated IL12RB1 transcription. The stably integrated clones shown in panel A were cultured and plated as described for Figure 2B. Cells were left untreated or were treated with either of the following: 10 ng/mL IFN-γ (□), 10 ng/mL IL-15 (▦), or 1 μg/mL LPS (not shown) for 8 hours before lysate preparation and luciferase assays. Basal luciferase activity of untreated cells was generally similar among all the examined clones. Fold inductions are shown. (C) Neither IFN-γ nor IL-15 induces gene expression of IRF3. RAW264.7 cells were left untreated (unstim) or were treated for 4 hours with 1 μg/mL LPS, 10 ng/mL IL-15, or 10 ng/mL IFN-γ. Total RNA was prepared for Northern blot analysis using IRF1, IRF2, IRF3, and IRF7 cDNA probes. Gene expression of the indicated gene and a picture of ethidium bromide (EtBr) are shown. (D) RAW264.7 cells were treated as described for Figure 5A. Total cellular lysates were prepared and immunoprecipitated with anti-IRF3 pAb. The phosphorylation of IRF3 was detected by Western blot analysis using antiphosphoserine mAb. As a control, 10% of each lysate was used to detect IRF3 by anti-IRF3 pAb.
Although our EMSA data have indicated that IRF3 is responsible for IL12RB1 gene activation (Figure 3B-C), mechanisms regarding how IRF3 regulates IL12RB1 gene transcription remain to be elucidated. We examined mRNA levels of IRFs in RAW264.7 cells after IFN-γ or IL-15 stimulation. Given that LPS stimulation is known to up-regulate gene expression of several IRF family members, LPS was used as a positive control. Northern blot analysis showed that gene expression of IRF1, IRF2, and IRF7 was induced by IFN-γ, but not with IL-15 stimulation (Figure 6C). Neither IL-15 nor IFN-γ could induce the IRF3 mRNA level, suggesting that IRF3 regulates IL12RB1 gene activation through posttranscriptional mechanisms. The studies on viral infection–responsive genes indicated that serine/threonine phosphorylation of IRF3 is associated with its nuclear translocation, DNA binding, and holocomplex formation with CBP.33-35 We investigated the serine-phosphorylation levels of IRF3 after IFN-γ and IL-15 stimulation of RAW264.7 cells. As shown in Figure 6D, IL-15 rapidly induced IRF3 phosphorylation, whereas IFN-γ induced the phosphorylation with a much longer time course.
Discussion
IRFs are a family of transcription factors first identified as the regulators of IFNA and IFNB gene expression.36,37 Among the 9 IRF members, IRF1, IRF3, and IRF-9 function as transcriptional activators, and the others can function as repressors or as activators/repressors.38 Although all IRF proteins recognize the core IRE/ISRE consensus sequence, each IRF has a clear sequence preference, enabling some oligonucleotides to be used as specific competitors for individual IRF.28 In our EMSA competition assays, an oligonucleotide specific for IRF3 completely abrogated protein binding to the IRE/ISRE element of the IL12RB1 gene. Involvement of IRF3 was also confirmed by the inhibition of the DNA–protein binding with a specific anti-IRF3 antibody (Figure 3C). On the other hand, competitive oligonucleotides for IRF1/2 and IRF7 partially blocked the complex formation (Figure 3C). These IRFs have DNA binding properties similar to those of IRF3, suggesting other IRF(s) might share the IRE/ISRE binding to the IL12RB1 promoter with IRF3. It is also possible that IRF7 forms a heterodimer complex with IRF3 on the IL12RB1 promoter, a phenomenon reported for the IFNB and certain IFNA family genes.39 However, mRNA for IRF1, IRF2, and IRF7 was induced significantly by IFN-γ but not IL-15 in RAW264.7 cells (Figure 6C), and modification of the DNA–protein binding by anti-IRF3 antibody was greater than 90% (data not shown), indicating that IRF3 is at least contained in most DNA-protein complexes.
In quiescent cells, IRF3 resides in the cytoplasm as an inactive form. IRF3 is activated by virus infection, LPS, and inflammatory cytokines through phosphorylation at multiple serine/threonine residues. Phosphorylation allows conformational changes and promotes the translocation of IRF3 from cytoplasm into the nucleus.40 Although mere nuclear translocation does not directly confer its transcriptional activity, increased DNA-binding affinity of the phosphorylated IRF3 results in transcriptional activation. In our cell system, IL-15 and IFN-γ did not increase IRF3 gene expression, but it did increase the IRF3 phosphorylation level (Figure 6D).
PU.1 appears to be another important transcription factor regulating IL12RB1 transcription (Figure 3D). PU.1 is a member of the ETS family, which is specifically expressed in B cells, neutrophils, mast cells, and macrophages.41,42 Like other ETS proteins, PU.1 contains a conserved ETS domain that confers its sequence-specific DNA binding to the 5′-C/AGGAA/T core sequence.41 PU.1 is a transcription factor involved in the regulation of a myriad of genes in the cell types listed here.43 Interestingly, as in the IL-12β1R gene, PU.1 plays a pivotal role in up-regulating IL-12 p40 gene expression in macrophages.44 Thus, our data indicated that activation of PU.1 increased IL-12–mediated signals in macrophages by up-regulating IL-12 p40 and IL12RB1 expression. It is unclear how the PU.1 pathway is activated by IFN-γ and IL-15. PU.1 activation through the formation of heterocomplexes with proteins such as IRFs and CBP has previously been reported. However, we could not detect heterocomplexes of PU.1 with IRF3 or CBP in RAW264.7 cells stimulated with IFN-γ or IL-15 (Figure 3D; data not shown). It is possible that these cytokines induce the association of PU.1 with the IL12RB1 gene promoter region through chromatin remodeling.
Our data indicated that IL-15 acted in concert with IFN-γ in the up-regulation of IL12RB1 mRNA and promoter activity in RAW264.7 cells (Figure 4A-C). This finding is compatible with previous reports on the role of IL-15. For example, decreased IL12RB1 expression and diminished IL-12 responsiveness are evident in IRF-1-/- mice,15 in which IL-15 expression is severely damaged.45 Moreover, splenic DCs and peritoneal macrophages from IL-15-/- mice produced significantly less IFN-γ than their control counterparts in response to IL-12, suggesting IL-15 directly affects APCs for IL-12 responsiveness.19 Furthermore, IL12RB1 mRNA expression is significantly reduced in APCs from IL-15-/-, γc-/-RAG-2-/-, and IL-2Rβ-/-RAG-2-/- mice,19 indicating IL-15 is responsible for IL12RB1 expression in APCs in a T cell–independent manner. These lines of evidence, together with our current finding, strongly indicated that IL-15 is pivotal in the generation of TH1-type immune responses by directly inducing IL12RB1 expression in APCs.
Combination IL-15 and IFN-γ treatment induced significant protein binding to the IRE/ISRE consensus sequence more quickly than treatment with IL-15 or IFN-γ alone (Figure 4C). The combination did not have any synergistic effects on PU.1 binding (Figure 4C). These data indicated that IL-15 cooperates with IFN-γ for IL12RB1 promoter activation by increasing protein binding to the IRE/ISRE element. Notably, as for the synergy with IFN-γ, IL-15 was similar to TSA (Figure 4A-C). The combination of IL-15 and TSA, on the other hand, showed no synergic effect. Because TSA is a well-known inhibitor of histone deacetylases, we sought to investigate the possibility that IL-15 may induce histone acetylation and chromatin remodeling of the IL12RB1 gene. ChIP assays showed that IFN-γ and IL-15 stimulation potently induced the acetylation of histone H3 at the IL12RB1 promoter. However, IFN-γ–induced acetylation was weaker and significantly slower than that by IL-15 (Figure 4D). These data indicated that the synergistic effect of IL-15 may be mediated by the rapid remodeling of the IL12RB1 gene promoter region into a more accessible structure for IRFs.
Histone H3 acetylation is closely related to its phosphorylation, and a recent report indicated that p38 MAPK is responsible for phospho-acetylation of histone H3 on a subset of stimulus-induced cytokine and chemokine genes.31 Our previous report also revealed that p38 MAPK is involved in phospho-acetylation and chromatin remodeling at mkp-m, a MAPK phosphatase gene.21 Our current data indicated that IL12RB1 appears to be another example of the genes regulated by p38 MAPK because pretreatment with SB203580, a specific inhibitor of p38 MAPK, significantly inhibited promoter responsiveness to IFN-γ and IL-15 (Figure 5B). Consistently, SB203580 effectively inhibited acetylation and phosphorylation of histone H3 induced by IFN-γ and IL-15 (Figure 5C-D). SB203580 inhibited IL12RB1 promoter activation by IL-15 more efficiently (almost completely abrogated) than that by IFN-γ, which seems consistent with the finding that IL-15 more potently induced histone H3 acetylation at the IL12RB1 gene (Figures 4D, 5D).
Conversely, however, activated p38 MAPK alone is not sufficient for IL12RB1 mRNA induction, given that LPS, an efficient activator of p38 MAPK (Figure 5A), only marginally induced IL12RB1 mRNA expression (Figure 1A). Interestingly, LPS-mediated p38 MAPK phosphorylation is temporary; it lasts only for 30 minutes (Figure 5A). Thus, it is possible that sustained activation of p38 MAPK may be required for the efficient transcriptional induction of IL12RB1. Additionally, IL-15 induced p38 MAPK phosphorylation more quickly than IFN-γ, which is consistent with the different time course of histone H3 acetylation by these 2 cytokines (Figure 5A). On the other hand, neither IFN-γ nor IL-15 induced JNK phosphorylation in RAW264.7 cells (Figure 5A), indicating that JNK activation is not necessary in the induction of IL12RB1 gene expression. Consistently, SP600125, a specific JNK inhibitor, failed to inhibit IL12RB1 promoter activation by IFN-γ or IL-15 (Figure 5B).
A transcriptional coactivator protein, CBP, is involved in the transcriptional regulation of various genes by providing a scaffold for transcription factors and modifying chromatin structure through its HAT activity.46-48 We demonstrated that the overexpression of CBP protein strongly enhanced IFN-γ–mediated IL12RB1 promoter activities. In contrast, IL-15–driven IL12RB1 promoter was not affected by CBP expression. Because IL-15 induces histone H3 acetylation in the IL12RB1 gene more potently than IFN-γ, it seems possible that IL-15 stimulation alone induced sufficient chromatin remodeling so that a further increase of HAT activity by CBP had no significant effects. The interaction between CBP and other transcription factors, including PU.1 and IRF3, is known to be involved in the transcriptional regulation of various genes.49 Notably, the phosphorylation of IRF3 promoted increased affinity for CBP, resulting in transcriptional activation.
In conclusion, we showed that IL-15 and IFN-γ stimulation of macrophages induce PU.1 and phosphorylated IRF3 binding to ETS/PU.1 and IRE/ISRE consensus motifs of the IL12RB1 promoter, respectively. IL-15 not only induces IL12RB1 mRNA by itself, it synergizes with IFN-γ to induce higher levels of IL12RB1 gene transcription. Understanding the roles of IL-15 and IFN-γ in the activation of IL12RB1 may promote our knowledge of host defense mechanisms against microbial infection.
Prepublished online as Blood First Edition Paper, September 2, 2004; DOI 10.1182/blood-2004-03-0842.
Supported in part by grants from the Ministry of Education, Science and Culture of the Japanese Government, the Yakult Bioscience Foundation, the Ohyama Health Foundation, and the Japan Society for the Promotion of Science.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Ms Itano and Ms Nishikawa for their technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal