Abstract
Gain and/or loss of function mediated by chimeric transcription factors generated by nonrandom translocations in leukemia is a key to understanding oncogenesis. E2A–hepatic leukemia factor (HLF), a chimeric basic region/leucine zipper (bZIP) transcription factor expressed in t(17;19)–positive leukemia cells, contributes to leukemogenesis through its potential to inhibit apoptosis. To identify physiologic counterparts of this chimera, we investigated the function of other bZIP factors that bind to the same DNA sequence recognized by E2A-HLF. Here, we show that thyrotroph embryonic factor (TEF), which shares a high level of sequence identity with HLF and recognizes the same DNA sequence, is expressed in a small fraction of each subset of hematolymphoid progenitors. When TEF was introduced into FL5.12 interleukin 3 (IL-3)–dependent cells, TEF protected the cells from apoptosis due to IL-3 deprivation. Unexpectedly, TEF also almost completely down-regulated expression of the common β (βc) chain of cytokine receptors. Consequently, TEF-expressing cells accumulated in G0/G1 phase without undergoing apoptosis. These findings suggest that TEF is one of the apoptotic regulators in hematopoietic progenitors and controls hematopoietic-cell proliferation by regulating the expression of the βc chain. In contrast, E2A-HLF promoted cell survival more efficiently than TEF but did not down-regulate βc chain expression, suggesting that E2A-HLF retains ideal properties for driving leukemic transformation.
Introduction
Since key systems that regulate cell survival have been conserved through evolution, relatively simple organisms such as Caenorhabditis elegans often provide insight into the complex mechanisms that control cell fate in mammals.1 The genetic approach demonstrated that 2 factors, cell death specification protein 1 (CES-1) and CES-2, regulate the programmed cell death of serotonergic neuro-secretory motor (NSM) neurons in C elegans in a cell type–specific fashion.2 Further studies revealed that CES-2 is a transcription factor that belongs to the basic region/leucine zipper (bZIP) superfamily (Figure 1).3 CES-2 negatively regulates CES-1, a member of the snail family of zinc finger transcription factors,4 and CES-1 appears to determine the fate of NSM sisters by negatively regulating the expression of egl1, a BH3-only proapoptotic member of the B cell lymphoma (Bcl-2) superfamily.5-7
Transcription factors that bind to the DNA sequence elements recognized by CES-2 might play important roles in cell type–specific cell death in mammals. The E2A–hepatic leukemia factor (HLF) fusion transcription factor is a good example. This chimera is derived from childhood acute pro-B-cell leukemia harboring a t(17;19) chromosomal translocation.8-10 It contains the transactivation domains of E2A and the bZIP DNA-binding and dimerization domain of HLF (Figure 1). E2A-HLF promotes the anchorage-independent growth of murine fibroblasts as a homodimer that depends on both the transactivation domain of E2A and the bZIP domain of HLF.11,12 E2A-HLF induces the expression of annexin II,13 annexin V, and sushi-repeat protein upregulated in leukemia (SRPUL),14 which have been postulated to play paraneoplastic roles in coagulopathies and bone invasion, as well as groucho-related genes, and E2A-HLF also suppresses Runt-related 1 (RUNX1).15 In addition, we and others established transgenic mice expressing E2AHLF that develop T-lineage lymphoid malignancies.16,17 Of note, we demonstrated that E2A-HLF binds avidly to the sequence recognized by CES-2 and protects cells from apoptosis due to growth factor deprivation without promoting cell proliferation.18-20 The close homology between the bZIP domains of HLF and CES-2 suggests that E2A-HLF subverts a cell-death pathway through a mechanism similar to that used by CES-2 in the worm. Indeed, we have identified SLUG, a zinc finger transcription factor closely related to CES-1, as one of the downstream targets of E2A-HLF in human leukemias associated with the 17;19 translocation and found that SLUG has antiapoptotic potential.21,22
These findings suggest that a cell-death pathway in mammalian hematopoietic cells is normally regulated by bZIP factors that exhibit a DNA-binding specificity similar to that of CES-2 or E2A-HLF. HLF is obviously a good candidate for the regulator of apoptosis in hematopoiesis; however, HLF is not expressed in hematopoietic progenitors and, moreover, enforced expression of HLF in cytokine-dependent cells failed to inhibit apoptosis.18,19 We then identified a bZIP factor, E4 promoter binding protein (E4BP4)/nuclear factor regulated by IL-3 (NFIL3) (Figure 1), as a physiologic counterpart of E2A-HLF. E4BP4 has nearly identical DNA-binding specificity to CES-2 and its expression is tightly regulated by interleukin (IL-3) in IL-3–dependent cell lines such as FL5.12 and Baf-3. Moreover, enforced expression of E4BP4 delays apoptosis of IL-3–deprived cells.23,24 These data suggested that subversion of the roles of antiapoptotic bZIP factors normally regulated by cytokines is 1 critical aspect of leukemogenesis induced by E2A-HLF.
Other bZIP factors may also play important roles in the regulation of hematopoietic cell survival. HLF is a member of the proline- and acidic amino acid–rich (PAR) bZIP family, which also includes thyrotroph embryonic factor (TEF)25 and albumin promoter D-box–binding protein (DBP)26 (Figure 1). TEF was originally cloned as a factor expressed in the developing anterior pituitary gland that could trans-activate the TSHβ promoter,25 whereas DBP was cloned as a liver-enriched transcriptional activator that binds to the D element of the albumin promoter.26 It has been well documented that the PAR proteins as well as E4BP4 are implicated in circadian control.27-33 PAR proteins may also be involved in the regulation of cell death because all 3 PAR proteins exhibit nearly identical DNA-binding specificity to that of E4BP4 and CES-2.25,26,28,29,34-37 Here we demonstrate that TEF promotes the survival of FL5.12 cells due to IL-3 starvation. Unexpectedly, we also found that TEF potently represses the expression of the β subunits of the IL-3 receptor, blocking proliferation and leading to arrest of the cell cycle in the G0/G1 phase, even in the presence of IL-3.
Materials and methods
Constructs of eukaryotic expression vectors
Expression plasmids containing the cDNAs of wild-type TEF (pMT-TEF), a mutated TEF (pMT-TEF basic mutant [BX]), and E2AHLF (pMT–E2A-HLF) were constructed with the pMT-CB6+ eukaryotic expression vector (a gift from F. Rauscher III, Wistar Institute, Philadelphia, PA), which contains the inserted cDNA under the control of a sheep metallothionein promoter as well as the neomycin resistance gene driven by the simian virus 40 early promoter. The cDNA encoding a basic mutant of TEF (TEF/BX) that contained substitutions for 6 of the amino acid residues in the basic region critical for DNA binding (TRRKKNNVAAK mutated to TSPKSYNVPPK; single-letter code) was made by polymerase chain reaction (PCR) mutagenesis.
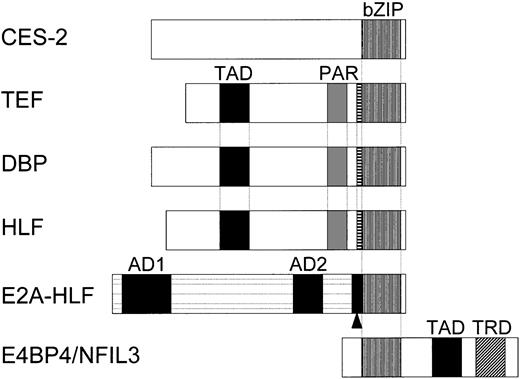
Schematic representation of the CES-2, TEF, DBP, HLF, E2A-HLF, and E4BP4/NFIL3 proteins. The CES-2 protein in C elegans contains a basic region/leucine zipper (bZIP) domain in the carboxyl-terminal region and acts as a transrepressor. TEF, DBP, and HLF contain a proline- and acidic amino acid–rich (PAR) domain as well as a bZIP domain in the carboxyl-terminal region, which shows high level of sequence identity to that of CES-2. TEF, DBP, and HLF each also contain a trans-activation domain (TAD) and can act as transcriptional activators. The E2A-HLF fusion protein, which is expressed in t(17;19)–positive leukemia cells, retains the 2 transactivation domains in the amino terminal of E2A (AD1 and AD2) but not its basic region/helix-loop-helix domain, which is replaced by the bZIP domain of HLF. E2A-HLF has a joining region (▪ with arrowhead) generated at the breakpoint but not the basic region extension (BRE; indicated with ▤) nor the PAR domain of HLF, which contribute to sequence-specific DNA binding. E4BP4/NFIL3 contains a bZIP domain in its amino-terminal region and has nearly identical DNA-binding specificity as that of CES-2, TEF, DBP, HLF, and E2A-HLF. E4BP4 acts as either a transactivator or transrepressor depending on complexes formed by its TAD or transrepression domain (TRD) in its carboxyl-terminal region.
Schematic representation of the CES-2, TEF, DBP, HLF, E2A-HLF, and E4BP4/NFIL3 proteins. The CES-2 protein in C elegans contains a basic region/leucine zipper (bZIP) domain in the carboxyl-terminal region and acts as a transrepressor. TEF, DBP, and HLF contain a proline- and acidic amino acid–rich (PAR) domain as well as a bZIP domain in the carboxyl-terminal region, which shows high level of sequence identity to that of CES-2. TEF, DBP, and HLF each also contain a trans-activation domain (TAD) and can act as transcriptional activators. The E2A-HLF fusion protein, which is expressed in t(17;19)–positive leukemia cells, retains the 2 transactivation domains in the amino terminal of E2A (AD1 and AD2) but not its basic region/helix-loop-helix domain, which is replaced by the bZIP domain of HLF. E2A-HLF has a joining region (▪ with arrowhead) generated at the breakpoint but not the basic region extension (BRE; indicated with ▤) nor the PAR domain of HLF, which contribute to sequence-specific DNA binding. E4BP4/NFIL3 contains a bZIP domain in its amino-terminal region and has nearly identical DNA-binding specificity as that of CES-2, TEF, DBP, HLF, and E2A-HLF. E4BP4 acts as either a transactivator or transrepressor depending on complexes formed by its TAD or transrepression domain (TRD) in its carboxyl-terminal region.
Cell culture and cell survival assay
FL5.12 cells, which are murine IL-3–dependent cells, were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum and 0.5% 10T1/2 cell-conditioned medium as a source of IL-3. Transfectants were generated by electroporation using 2 × 107 cells and 80 μg of DNA with a gene pulser (Bio-Rad, Hercules, CA) set at 300 mV and 960 μF. Cells were then cultured in 24-well dishes and selected in the presence of the neomycin analog G418 (0.6 mg/mL) for 2 weeks. The induction of protein expression with Zn in G418-resistant cells was confirmed by immunoblot analysis. Cell growth and cell survival experiments were performed using 3 independent pools of cells, and the results of representative data of multiple experiments are shown. For cell survival assays, cells growing exponentially in IL-3–containing medium were precultured for 16 hours in the presence or absence of 100 μM ZnSO4. Cells were washed with IL-3–free medium twice and were adjusted to 5 × 105 cells per milliliter. Viable cell counts were determined by trypan blue dye exclusion.
Immunoblot analysis
Cells were solubilized in Nonidet P-40 lysis buffer (150 mM NaCl, 1.0% Nonidet P-40, 50 mM Tris [tris(hydroxymethyl)aminomethane; pH 8.0]), and total cellular proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. After wet electrotransfer onto polyvinylidene difluoride membranes, proteins were detected with the following primary antibodies: anti-HLF c-terminal (C),35 anti-p44/42 mitogen-activated protein kinase (MAPK; New England BioLabs, Beverly, MA), anti-p70S6K (New England BioLabs), anti-Akt (New England BioLabs), antiphospho–signal transducers and activators of transcription 5 (anti–phospho-STAT5; Tyr694; New England BioLabs), anti–phospho-p44/42MAPK (Thr202/Tyr204; New England BioLabs), anti–phospho-p70S6K (Thr421/Ser424; New England BioLabs), and anti–phospho-Akt (Ser473; New England BioLabs) rabbit serum, or anti–β-tubulin (Pharmingen, San Diego, CA), antiphosphotyrosine (Upstate Biotechnology, Lake Placid, NY), and anti-STAT5 (Transduction Laboratories, Lexington, KY) mouse monoclonal antibodies. Then, the blots were stained by horseradish peroxidase–conjugated anti–rabbit or anti–mouse immunoglobulin G (IgG) and IgM secondary antibodies (MBL, Nagoya, Japan), respectively, and subjected to enhanced chemiluminescence detection (Amersham Life Science, Arlington Heights, IL).
Flow cytometric analysis
Cells (2 × 105) were washed with phosphate-buffered saline (PBS) and incubated with rat monoclonal antibodies specific for the mouse IL-3 receptor α chain (5B11)38 or the mouse IL-3 receptor β chain (AIC2A; βIL3; 9D3)39 on ice for 30 minutes. Cells were washed with PBS and stained with a fluorescein isothiocyanate (FITC)–conjugated goat anti–rat IgG (Jackson Immuno Research Laboratories, West Grove, PA). For detection of the common β chain of the IL-3 receptor (AIC2B; βC), cells were incubated with an anti–mouse βC chain hamster monoclonal antibody (MBL), followed by incubation with an FITC-conjugated goat anti–hamster IgG secondary antibody (Jackson Immuno Research Laboratories). Cells were analyzed by flow cytometry (FACS Calibar; Becton Dickinson, San Jose, CA).
Northern blot analysis
Total cellular RNA was isolated by the guanidinium-cesium chloride method. RNA samples (approximately 20 μg per lane) were separated by electrophoresis in 1% agarose gel containing 2.2 M formaldehyde, transferred to nylon membranes, and hybridized with the appropriate probes according to the standard procedure. For Northern blot analysis of multiple human hematolymphoid tissues, the blot was purchased from Clontech (Palo Alto, CA). Unique cDNA probes in the 3′ untranslated region of the PAR genes,40 a 1.3-kilobase (kb) XhoI fragment of mouse IL-3 receptor α-chain cDNA, and a 0.8-kb XhoI/BamHI fragment of mouse IL-3 receptor βIL3 chain cDNA were used as probes.
Analysis of gene expression in subsets of hematopoietic stem and progenitor cells
Cells (5 × 103) with surface marker expression patterns typical of hematopoietic stem cells (HSCs) and various types of myeloid and lymphoid progenitors were sorted from mouse bone marrow by fluorescence-activated cell sorting (FACS), as previously described.41 Total RNA was prepared with TRIZOL reagent and subjected to cDNA synthesis with the SuperScript first-strand cDNA synthesis system (GIBCO, Carlsbad, CA). PCR was performed for 40 cycles of 94°C for 30 seconds, 55°C for 45 seconds, and 72°C for 45seconds with the following primers: 5′-ACCATCTTCCTCTACTGCCATCTTTCAG and 5′-GTACTTGGTCTCGTACTTGGACACGATG for first-round PCR; 5′-GTGATCTGGTTCTCCTTCAG for nested PCR amplification of Tef; and 5′-CACAGGACTAGAACACCTGC and 5′-GCTGGTGAAAAGGACCTCT for PCR amplification of Hprt.
Electrophoretic mobility shift assay
Binding reactions in electrophoretic mobility shift assay (EMSA) were performed with a 32P–end-labeled DNA oligonucleotide probe (2 × 104 counts per minute [cpm]) in 10 μL of binding buffer (12% glycerol, 12 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.9], 4 mM Tris [pH 7.9], 133 mM KCl, 300 mg of bovine serum albumin per milliliter) and 5 μL of nuclear proteins extracted from FL5.12 cells by standard procedures, as previously described.35,42 The oligonucleotide probes used were wild-type HLF consensus sequence (CS) (5′-GCTACATATTACGTAATAAGCGTT-3′); wild-type β-casein for STAT5 (5′-AGATTTCTAGGAATTCAATCC-3′); and mutant β-casein (5′-AGATTTAGTTTAATTCAATCC-3′). As a carrier DNA, 1.5 μg of sheared calf thymus DNA or poly-d(I)d(C) was added to the reaction mixture testing for binding to HLF-CS or β-casein probe, respectively. The entire mixture was incubated at 30°C for 15 minutes. In the competition inhibition experiments, an approximately 100-fold molar excess of the unlabeled oligonucleotide was added to the reaction mixture. One microliter of polyvalent HLF(C) antiserum or preimmune rabbit serum was added to the nuclear lysates and incubated at 4°C for 30 minutes prior to the DNA-binding reaction. Nondenaturing polyacrylamide gels containing 4% acrylamide and 2.5% glycerol were prerun at 4°C in a high–ionic-strength Tris-glycine buffer for 30 minutes, loaded with the samples containing protein–DNA complexes, run at 35 mA for approximately 90 minutes, dried under vacuum, and analyzed by autoradiography.
Results
Expression of TEF in hematolymphoid tissues
The 3 members of the PAR family of proteins are differentially expressed in tissues and organs.8,10,40 In contrast to HLF, which shows tissue-specific expression, TEF and DBP are expressed ubiquitously, although their expression in normal hematopoietic and lymphoid tissues has not been clarified. First, we performed Northern blot analysis of human hematolymphoid tissues using a specific cDNA probe. As shown in Figure 2A, TEF was expressed in the spleen, lymph node, thymus, and fetal liver but was nearly undetectable in peripheral blood leukocytes and bone marrow. DBP was widely expressed in hematolymphoid tissues, whereas HLF was expressed in the fetal liver alone. Second, subsets of mouse hematolymphoid progenitors were sorted from the bone marrow and were subjected to reverse transcriptase–PCR (RT-PCR) for Tef (Figure 2B).22 Although Tef was detectable in unpurified bone marrow mononuclear cells, the RT-PCR product was virtually undetectable in each subset of progenitors (Figure 2B top panel). Upon performing nested PCR using each PCR product as a template, Tef was detected in each subset (Figure 2B middle panel). These results suggested that Tef is expressed in hematolymphoid progenitors but its expression is low or restricted to a small fraction of each subset of progenitors.
Expression of TEF during hematopoiesis. (A) Northern blot analysis of human hematolymphoid tissues. The blot was hybridized with human TEF, DBP, HLF, and β-actin cDNA probes. Lane 1, spleen; lane 2, lymph node; lane 3, thymus; lane 4, peripheral blood leukocyte; lane 5, bone marrow (BM); and lane 6, fetal liver. The mobility of 285 and 185 rRNA was indicated. (B) Patterns of Tef expression in murine myeloid and lymphoid progenitors in different stages of development. cDNA was synthesized from total RNA extracted from 5000 cells of each hematopoietic progenitor subset that had been sorted from bone marrows by FACS (lanes 1-7) as well as unpurified bone marrow mononuclear cells (lane 8). The cDNA was subjected to PCR with primers specific for the murine Tef (top panel) and Hprt (bottom panel) genes. The PCR product of amplification of Tef was subjected to nested PCR (middle panel). HSC indicates hematopoietic stem cells; CMP, common myeloid progenitor; GMP, granulocyte-monocyte progenitor; MEP, megakaryocyte-erythrocyte progenitor; and CLP, common lymphoid progenitor.
Expression of TEF during hematopoiesis. (A) Northern blot analysis of human hematolymphoid tissues. The blot was hybridized with human TEF, DBP, HLF, and β-actin cDNA probes. Lane 1, spleen; lane 2, lymph node; lane 3, thymus; lane 4, peripheral blood leukocyte; lane 5, bone marrow (BM); and lane 6, fetal liver. The mobility of 285 and 185 rRNA was indicated. (B) Patterns of Tef expression in murine myeloid and lymphoid progenitors in different stages of development. cDNA was synthesized from total RNA extracted from 5000 cells of each hematopoietic progenitor subset that had been sorted from bone marrows by FACS (lanes 1-7) as well as unpurified bone marrow mononuclear cells (lane 8). The cDNA was subjected to PCR with primers specific for the murine Tef (top panel) and Hprt (bottom panel) genes. The PCR product of amplification of Tef was subjected to nested PCR (middle panel). HSC indicates hematopoietic stem cells; CMP, common myeloid progenitor; GMP, granulocyte-monocyte progenitor; MEP, megakaryocyte-erythrocyte progenitor; and CLP, common lymphoid progenitor.
Establishment of FL5.12 cells conditionally expressing TEF
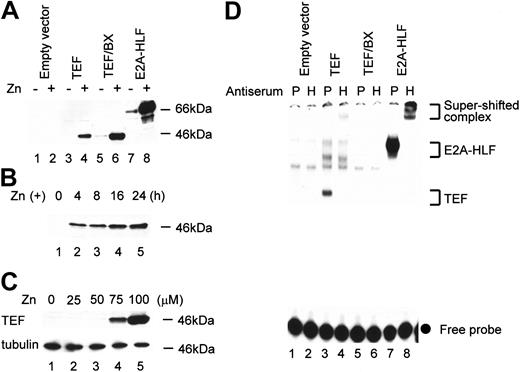
To analyze the function of TEF in hematopoietic cells, we established FL5.12 cells that inducibly expressed TEF by the addition of Zn using an expression vector (pMT-CB6+) under the control of a metallothionein promoter. Immunoblot analysis using the anti-HLF(C) antibody that effectively recognizes HLF, DBP, and TEF proteins34 revealed the induction of TEF protein in cells transfected with pMT-TEF when cultured in medium containing 100 μM Zn (Figure 3A lane 4). No endogenous or leaky expression of TEF protein was detected (Figure 3A lanes 1-3). Time course analysis revealed that protein expression of TEF was detectable within 4 hours after the addition of Zn (Figure 3B lane 2) and that it reached a plateau after 16 hours (Figure 3B lanes 4-5). The expression level of TEF was related to the concentration of Zn. Upon the addition of 75 μM Zn, the TEF expression level was approximately 20% of that induced by 100 μM Zn (Figure 3C lanes 4-5), whereas no significant induction was obtained by the addition of less than 50 μM Zn (Figure 3C lanes 2-3). We also established FL5.12 cells that conditionally expressed TEF/BX (Figure 3A lanes 5-6), which is a mutant form of TEF (see “Constructions of eukaryotic expression vectors”), or E2A-HLF (Figure 3A lanes 7 and 8). EMSA using the HLF-CS probe that contains the consensus binding sequence of the PAR proteins17 detected specific protein-DNA complexes in nuclear extracts from cells expressing either TEF (Figure 3D lanes 3-4) or E2A-HLF (Figure 3D lanes 7-8) but not in nuclear extracts from cells expressing TEF/BX (Figure 3D lanes 5-6). The lack of DNA-binding ability of TEF/BX was not due to aberrant subcellular localization because immunofluorescence studies showed that TEF/BX was localized in the nucleus (data not shown).
Establishment of FL5.12 cells conditionally expressing TEF, TEF/BX, or E2A-HLF. (A) Immunoblot analysis of FL5.12 cells using HLF(C) antiserum. Representative pools of FL5.12 cells transfected with the empty vector (lanes 1-2), pMT-TEF (lanes 3-4), pMT-TEF/BX (lanes 5-6), or pMT–E2A-HLF (lanes 7-8) were cultured in the presence (even lanes) or absence (odd lanes) of 100 μM ZnSO4 for 16 hours. (B) Time course analysis of TEF expression in pMT-TEF–transfected FL5.12 cells. FL5.12 cells transfected with pMT-TEF were cultured in IL-3–containing medium in the presence of 100 μM Zn for the indicated periods of time and subjected to immunoblot analysis using the HLF(C) antiserum. (C) TEF protein expression in pMT-TEF–transfected FL5.12 cells cultured in the presence of different concentrations of Zn. FL5.12 cells transfected with pMT-TEF were cultured in the presence of Zn at the indicated concentrations for 16 hours and subjected to immunoblot analysis using the HLF(C) antiserum. The expression of tubulin was also analyzed as control. (D) Antibody-perturbed electrophoretic mobility shift analysis (EMSA) with the HLF-CS probe. FL5.12 cells transfected with the empty vector (lanes 1-2), pMT-TEF (lanes 3-4), pMT-TEF/BX (lanes 5-6), or pMT–E2A-HLF (lanes 7-8) were cultured with 100μM of Zn for 16 hours. Nuclear extracts from these cells were incubated with either preimmune (P; odd lanes) or anti-HLF(C) (H; even lanes) antiserum. Brackets show the mobility of DNA-protein complexes containing the indicated proteins and the • indicates unbound, labeled oligonucleotide probes.
Establishment of FL5.12 cells conditionally expressing TEF, TEF/BX, or E2A-HLF. (A) Immunoblot analysis of FL5.12 cells using HLF(C) antiserum. Representative pools of FL5.12 cells transfected with the empty vector (lanes 1-2), pMT-TEF (lanes 3-4), pMT-TEF/BX (lanes 5-6), or pMT–E2A-HLF (lanes 7-8) were cultured in the presence (even lanes) or absence (odd lanes) of 100 μM ZnSO4 for 16 hours. (B) Time course analysis of TEF expression in pMT-TEF–transfected FL5.12 cells. FL5.12 cells transfected with pMT-TEF were cultured in IL-3–containing medium in the presence of 100 μM Zn for the indicated periods of time and subjected to immunoblot analysis using the HLF(C) antiserum. (C) TEF protein expression in pMT-TEF–transfected FL5.12 cells cultured in the presence of different concentrations of Zn. FL5.12 cells transfected with pMT-TEF were cultured in the presence of Zn at the indicated concentrations for 16 hours and subjected to immunoblot analysis using the HLF(C) antiserum. The expression of tubulin was also analyzed as control. (D) Antibody-perturbed electrophoretic mobility shift analysis (EMSA) with the HLF-CS probe. FL5.12 cells transfected with the empty vector (lanes 1-2), pMT-TEF (lanes 3-4), pMT-TEF/BX (lanes 5-6), or pMT–E2A-HLF (lanes 7-8) were cultured with 100μM of Zn for 16 hours. Nuclear extracts from these cells were incubated with either preimmune (P; odd lanes) or anti-HLF(C) (H; even lanes) antiserum. Brackets show the mobility of DNA-protein complexes containing the indicated proteins and the • indicates unbound, labeled oligonucleotide probes.
TEF but not E2A-HLF induced G0/G1 arrest
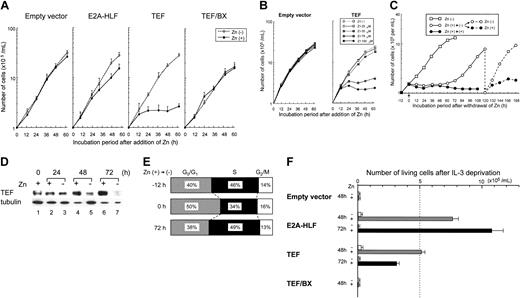
When TEF expression was induced by the addition of Zn in proliferating pMT-TEF–transfected FL5.12 cells cultured in IL-3–containing medium, cell growth was gradually disturbed and finally arrested within 36 hours (Figure 4A). In the dye exclusion assay, the viability of TEF-transfected cells was over 90% at 60 hours after the addition of Zn (data not shown). Growth arrest was observed when the Zn concentration was higher than 75 μM (Figure 4B). Cells transfected with pMT-TEF/BX or the empty vector proliferated exponentially in culture medium containing 100 μM Zn (Figure 4A), indicating that the growth inhibition depends on the sequence-specific DNA-binding activity of TEF and not on the cytotoxic effects of Zn. Cells transfected with pMT–E2A-HLF grew more slowly than cells transfected with the empty vector but did not undergo growth arrest (Figure 4A). Growth arrest observed in pMT-TEF–transfected FL5.12 cells was reversible after the withdrawal of Zn (Figure 4C). Cells preincubated in IL-3–containing medium in the presence of 100 μM Zn for 12 hours were washed with medium and subsequently cultured in IL-3–containing medium either in the presence or absence of 100 μM Zn. The expression of TEF was gradually decreased and became undetectable 72 hours after withdrawal of Zn (Figure 4D lanes 3, 5, 7), whereas TEF levels were maintained in the cells that were continued in culture in the presence of Zn (Figure 4D lanes 2, 4, 6). Interestingly, the growth arrest of cells cultured in the absence of Zn was reversed and cells began cycling approximately 72 hours after withdrawal of Zn, whereas the growth arrest and cell viability were maintained in the cells cultured in the presence of Zn (Figure 4C). Moreover, the growth of cells rescued by the withdrawal of Zn could be arrested again upon the subsequent readdition of Zn (Figure 4C).
To determine the cause of growth arrest, cell cycle analysis was performed using flow cytometry (Table 1). Upon the addition of Zn, TEF-expressing cells in the G0/G1 phase gradually accumulated and nearly 80% of the cells were in the resting phase after 48 hours. On the other hand, it was unlikely that cell death contributed to this growth arrest because the population in the sub-G0/G1 phase always comprised less than 10% of TEF-expressing cells (data not shown). In addition, the G0/G1-arrested pMT-TEF–transfected FL5.12 cells were capable of re-entering the cell cycle when Zn was withdrawn from the culture medium (Figure 4E). By contrast, in cells transfected with the empty vector, regardless of the presence or absence of Zn, more than 50% of the cells were in the S phase whereas nearly 40% of the cells were in the G0/G1 phase. Consistent with the slow growth rate, the number of E2A-HLF–expressing cells in the S phase slightly decreased after the addition of Zn.
Cell-cycle analysis of FL5.12 cells in the presence of IL-3
. | Empty vector, % . | . | . | TEF, % . | . | . | E2A-HLF, % . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zn+, h* . | G0/G1 . | S . | G2/M . | G0/G1 . | S . | G2/M . | G0/G1 . | S . | G2/M . | ||||||
| 0 | 37 | 56 | 7 | 32 | 54 | 14 | 30 | 63 | 7 | ||||||
| 8 | ND | ND | ND | 39 | 43 | 17 | ND | ND | ND | ||||||
| 16 | ND | ND | ND | 54 | 27 | 18 | ND | ND | ND | ||||||
| 24 | 41 | 52 | 7 | 61 | 19 | 20 | 47 | 40 | 13 | ||||||
| 48 | 39 | 53 | 8 | 79 | 9 | 12 | 35 | 50 | 14 | ||||||
. | Empty vector, % . | . | . | TEF, % . | . | . | E2A-HLF, % . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zn+, h* . | G0/G1 . | S . | G2/M . | G0/G1 . | S . | G2/M . | G0/G1 . | S . | G2/M . | ||||||
| 0 | 37 | 56 | 7 | 32 | 54 | 14 | 30 | 63 | 7 | ||||||
| 8 | ND | ND | ND | 39 | 43 | 17 | ND | ND | ND | ||||||
| 16 | ND | ND | ND | 54 | 27 | 18 | ND | ND | ND | ||||||
| 24 | 41 | 52 | 7 | 61 | 19 | 20 | 47 | 40 | 13 | ||||||
| 48 | 39 | 53 | 8 | 79 | 9 | 12 | 35 | 50 | 14 | ||||||
Data of representative pools of each transfectant are indicated.
ND indicates not determined.
Hours of incubation with Zn
TEF and E2A-HLF promoted cell survival
Next, we analyzed cell viability in the absence of IL-3 (Figure 4F). Cells were precultured in the presence or absence of Zn in IL-3–containing medium for 16 hours, washed with IL-3–free medium twice, and cultured in IL-3–free medium with or without Zn. Cells expressing E2A-HLF survived more than 72 hours in IL-3–free medium, as previously reported.18,19 TEF also promoted cell survival in IL-3–free medium for 48 hours, but the number of viable cells gradually decreased after 72 hours even though TEF expression was maintained (data not shown). Cells expressing TEF/BX rapidly underwent apoptosis in a manner similar to that of the control cells transfected with the empty vector. These data indicated that TEF protects IL-3–dependent FL5.12 cells from apoptosis in the absence of the cytokine and that this activity depends on its DNA-binding activity.
TEF inactivated IL-3 signaling pathways
To clarify the mechanism of G0/G1 arrest induced by TEF, we analyzed the tyrosine phosphorylation of cellular proteins. TEF-transfected cells were preincubated in IL-3–containing medium with Zn for 18 hours, then incubated in IL-3–deficient medium with Zn for 12 hours, and then restimulated by IL-3. As previously reported by others,43-45 rapid tyrosine phosphorylation of a wide variety of proteins was observed in cells transfected with the empty vector (Figure 5A lanes 1-4) but not in cells expressing TEF (Figure 5A lanes 5-8), suggesting that TEF inactivated signals from the IL-3 receptor. To verify the observation made in Figure 5A more precisely, we analyzed the STAT5 pathway, MAPK pathway, and phosphatidylinositol 3-kinase (PI3K) pathway by monitoring the phosphorylation status of not only STAT546 and p44/42 MAPK47 but also p70S6K48 and Akt,49 which are downstream proteins in the PI3K pathway47 (Figure 5B). Each FL5.12 transfectant was cultured in IL-3–free medium in the presence or absence of Zn for 12 hours following 18 hours' preincubation in IL-3–containing medium with or without Zn, respectively; subsequently exposed to IL-3 for 5 minutes; and processed for immunoblot analysis. In the absence of Zn, IL-3 stimulation of cells transfected with pMT-TEF (Figure 5B lanes 3-4) as well as cells transfected with pMT-TEF/BX (Figure 5B lanes 5-6), pMT–E2A-HLF (Figure 5B lanes 7-8), or the empty vector (Figure 5B lanes 1-2) rapidly induced the phosphorylation of STAT5. In the presence of Zn, phosphorylation of STAT5 was completely abrogated in cells transfected with the TEF expression vector (Figure 5B lanes 11-12), whereas it was rapidly induced in the other transfectants (Figure 5B lanes 9-10 and 13-16). The same results were observed with regard to the phosphorylation of p44/42 MAPK, p70S6K, and Akt. These results indicated that TEF, acting through its DNA-binding activity, blocked IL-3–signaling pathways whereas E2A-HLF did not. This was supported by the results of EMSA (Figure 5C), which demonstrated rapid induction of protein–DNA complexes containing STAT5 following restoration of IL-342 in nuclear extracts from cells transfected with the empty vector (Figure 5C lanes 1-6) but not in cells transfected with pMT-TEF (Figure 5C lanes 7-10).
Growth arrest and cell survival of FL5.12 cells transfected with the TEF expression vector. (A) Growth curves of FL5.12 cells transfected with the empty vector, pMT-TEF, pMT-TEF/BX, or pMT–E2A-HLF. Cells were adjusted to 1 × 105 cells per mL and cultured in IL-3–containing medium in the presence (•) or absence (○) of 100 μM Zn. Representative data of multiple experiments using 3 independent pools are shown. Bars indicate standard error. (B) Growth of FL5.12 cells transfected with the empty vector or pMT-TEF in IL-3–containing medium in the presence of 0, 25, 50, 75, or 100 μM of Zn. (C) Growth curves of pMT-TEF–transfected FL5.12 cells after withdrawal of Zn. □ indicates the growth of cells cultured in IL-3–containing medium in the absence of Zn. Cells were preincubated in the presence of 100 μM Zn for 12 hours, washed with medium, adjusted to 1.5 × 105 cells per mL, and then cultured in IL-3–containing medium either in the presence (•) or absence (○) of 100 μM Zn. One hundred twenty hours after withdrawal of Zn, cells cultured in the absence of Zn were adjusted to 1 × 105 cells per mL and cultured in the presence (•- - -•) or absence (○- - -○) of 100 μM Zn. The means of triplicate samples are indicated. (D) Time course analysis of TEF expression in pMT-TEF–transfected FL5.12 cells after withdrawal of Zn. Cells were preincubated in the presence of 100 μM Zn for 12 hours, washed with medium (lane 1), then cultured in IL-3–containing medium in the presence (lanes 2, 4, and 6) or absence (lanes 3, 5, and 7) of 100 μM Zn for the indicated periods of time and subjected to immunoblot analysis using the HLF(C) antiserum. The expression of tubulin was also analyzed as control. (E) Cell cycle analysis of pMT-TEF–transfected FL5.12 cells. Cells cultured in IL-3–containing medium (top; - 12 hours) were preincubated with 100 μM of Zn for 12 hours (middle; 0 hour), washed with medium, and subsequently cultured in IL-3–containing medium in the absence of Zn for 72 hours (bottom; 72 hours). ▨ indicates G0/G1; ▪, S; and □, G2/M. (F) Number of viable cells after IL-3 deprivation of FL5.12 cells. FL5.12 cells transfected with the empty vector, pMT-TEF, pMT-TEF/BX, or pMT–E2A-HLF were precultured in IL-3–containing medium in the presence or absence of 100 μM of Zn for 16 hours. Cells were then washed with IL-3–free medium, adjusted to 5 × 105 cells per mL, and cultured without IL-3 either in the presence (and ▪) or absence (□) of Zn, respectively. Upon deprivation of IL-3 for 48 or 72 hours, the numbers of viable cells as determined by trypan-blue dye exclusion are indicated. The mean data of 3 independent experiments are shown. Bars indicate standard error.
Growth arrest and cell survival of FL5.12 cells transfected with the TEF expression vector. (A) Growth curves of FL5.12 cells transfected with the empty vector, pMT-TEF, pMT-TEF/BX, or pMT–E2A-HLF. Cells were adjusted to 1 × 105 cells per mL and cultured in IL-3–containing medium in the presence (•) or absence (○) of 100 μM Zn. Representative data of multiple experiments using 3 independent pools are shown. Bars indicate standard error. (B) Growth of FL5.12 cells transfected with the empty vector or pMT-TEF in IL-3–containing medium in the presence of 0, 25, 50, 75, or 100 μM of Zn. (C) Growth curves of pMT-TEF–transfected FL5.12 cells after withdrawal of Zn. □ indicates the growth of cells cultured in IL-3–containing medium in the absence of Zn. Cells were preincubated in the presence of 100 μM Zn for 12 hours, washed with medium, adjusted to 1.5 × 105 cells per mL, and then cultured in IL-3–containing medium either in the presence (•) or absence (○) of 100 μM Zn. One hundred twenty hours after withdrawal of Zn, cells cultured in the absence of Zn were adjusted to 1 × 105 cells per mL and cultured in the presence (•- - -•) or absence (○- - -○) of 100 μM Zn. The means of triplicate samples are indicated. (D) Time course analysis of TEF expression in pMT-TEF–transfected FL5.12 cells after withdrawal of Zn. Cells were preincubated in the presence of 100 μM Zn for 12 hours, washed with medium (lane 1), then cultured in IL-3–containing medium in the presence (lanes 2, 4, and 6) or absence (lanes 3, 5, and 7) of 100 μM Zn for the indicated periods of time and subjected to immunoblot analysis using the HLF(C) antiserum. The expression of tubulin was also analyzed as control. (E) Cell cycle analysis of pMT-TEF–transfected FL5.12 cells. Cells cultured in IL-3–containing medium (top; - 12 hours) were preincubated with 100 μM of Zn for 12 hours (middle; 0 hour), washed with medium, and subsequently cultured in IL-3–containing medium in the absence of Zn for 72 hours (bottom; 72 hours). ▨ indicates G0/G1; ▪, S; and □, G2/M. (F) Number of viable cells after IL-3 deprivation of FL5.12 cells. FL5.12 cells transfected with the empty vector, pMT-TEF, pMT-TEF/BX, or pMT–E2A-HLF were precultured in IL-3–containing medium in the presence or absence of 100 μM of Zn for 16 hours. Cells were then washed with IL-3–free medium, adjusted to 5 × 105 cells per mL, and cultured without IL-3 either in the presence (and ▪) or absence (□) of Zn, respectively. Upon deprivation of IL-3 for 48 or 72 hours, the numbers of viable cells as determined by trypan-blue dye exclusion are indicated. The mean data of 3 independent experiments are shown. Bars indicate standard error.
TEF induces down-regulation of the IL-3 receptor β chains
The inactivation of IL-3 signaling observed in Figure 5 could be explained by down-regulation of the IL-3 receptor by TEF. In mice, the high-affinity IL-3 receptor is a heterodimer of the IL-3–specific α chain and either of the βIL3 chain or the βC chain, the latter of which is shared with the IL-5 and granulocyte-macrophage colony-stimulating factor (GM-CSF) receptors.50 Northern blot analysis was performed using an α-chain cDNA probe and a βIL3 chain cDNA probe; the latter recognizes both the βIL3 and βC transcripts because of their extremely high levels of sequence identity (Figure 6A).51 Upon the addition of Zn to TEF-transfected cells, the mRNA level of the β chains rapidly decreased and was almost undetectable within 16 hours. In contrast, the addition of Zn to cells transfected with either the empty vector or pMT-TEF did not change the expression level of α-chain mRNA.
Phosphorylation of cellular proteins by IL-3 restoration. FL5.12 cells transfected with the empty vector, pMT-TEF, pMT-TEF/BX, or pMT–E2A-HLF were cultured in IL-3–containing medium in the presence or absence of Zn for 16 hours, and then the cells were transferred to IL-3–deficient medium in the presence or absence of Zn for 12 hours. Thereafter, cells were restimulated with IL-3 for the indicated periods of time. (A) Immunoblot analysis of FL5.12 cells that had been transfected with the empty vector (lanes 1-4) or pMT-TEF (lanes 5-8) and then cultured in IL-3–containing medium in the presence of Zn using an antiphosphotyrosine monoclonal antibody. (B) Immunoblot analysis for the phosphorylated (p) and nonphosphorylated forms of STAT5, p44/42 MAPK, p70S6K, and Akt in transfectants that had been restored with IL-3 for 5 minutes in the absence (lanes 1-8) or the presence (lanes 9-16) of Zn. The blots were probed with antiphospho-STAT5, antiphospho-p44/42 MAPK, antiphospho-p70 S6K, and antiphospho-Akt as well as anti-STAT5, anti-p44/42 MAPK, anti-p70S6K, and anti-Akt antibodies. (C) Activation of STAT5 DNA-binding by IL-3 restoration. EMSA was performed using nuclear lysates from FL5.12 cells that had been transfected with either the empty vector (lanes 1-6) or pMT-TEF (lanes 7-10) with a β-casein sequence as a probe. In the competition inhibition, an approximately 100-fold molar excess of the unlabeled β-casein sequence oligonucleotide (C; lane 5) or β-casein sequence oligonucleotide with mismatches (M; lane 6) was added in the reaction mixture. A bracket indicates the mobility of specific DNA-protein complexes and • indicates unbound, labeled oligonucleotide probes.
Phosphorylation of cellular proteins by IL-3 restoration. FL5.12 cells transfected with the empty vector, pMT-TEF, pMT-TEF/BX, or pMT–E2A-HLF were cultured in IL-3–containing medium in the presence or absence of Zn for 16 hours, and then the cells were transferred to IL-3–deficient medium in the presence or absence of Zn for 12 hours. Thereafter, cells were restimulated with IL-3 for the indicated periods of time. (A) Immunoblot analysis of FL5.12 cells that had been transfected with the empty vector (lanes 1-4) or pMT-TEF (lanes 5-8) and then cultured in IL-3–containing medium in the presence of Zn using an antiphosphotyrosine monoclonal antibody. (B) Immunoblot analysis for the phosphorylated (p) and nonphosphorylated forms of STAT5, p44/42 MAPK, p70S6K, and Akt in transfectants that had been restored with IL-3 for 5 minutes in the absence (lanes 1-8) or the presence (lanes 9-16) of Zn. The blots were probed with antiphospho-STAT5, antiphospho-p44/42 MAPK, antiphospho-p70 S6K, and antiphospho-Akt as well as anti-STAT5, anti-p44/42 MAPK, anti-p70S6K, and anti-Akt antibodies. (C) Activation of STAT5 DNA-binding by IL-3 restoration. EMSA was performed using nuclear lysates from FL5.12 cells that had been transfected with either the empty vector (lanes 1-6) or pMT-TEF (lanes 7-10) with a β-casein sequence as a probe. In the competition inhibition, an approximately 100-fold molar excess of the unlabeled β-casein sequence oligonucleotide (C; lane 5) or β-casein sequence oligonucleotide with mismatches (M; lane 6) was added in the reaction mixture. A bracket indicates the mobility of specific DNA-protein complexes and • indicates unbound, labeled oligonucleotide probes.
Next, we analyzed the cell surface expression of the βIL3, βC, and α chains of the IL-3 receptor by flow cytometry using specific antibodies for each chain (Figure 6B). Consistent with the results of Northern blot analysis, within 24 hours after the addition of Zn, nearly complete down-regulation of the expression of both the β IL3 and βC chains was observed in cells transfected with pMT-TEF but not in cells transfected with the empty vector, pMT–E2A-HLF, or pMT-TEF/BX. These data indicated that TEF down-regulates expression of the β chains in a DNA-binding–dependent fashion. Moreover, we found up-regulation of the expression of the α chain in TEF-expressing cells but not in cells transfected with the empty vector, pMT-TEF/BX, or pMT-E2A-HLF. Since Northern blot analysis revealed that the level of mRNA expression of the α chain did not change (Figure 6A), up-regulation of the cell surface α-chain expression by TEF was due to posttranscriptional mechanism(s), probably inhibition of internalization caused by down-regulation of the expression of β chains.52 These observations indicated that TEF disrupts formation of the IL-3 receptor by down-regulating the cell surface expression of the β chains, thus rendering FL5.12 cells insensitive to IL-3.
Northern blot analysis and flow cytometric analysis of FL5.12 cells. (A) Northern blot analysis for the IL-3 receptor. Total RNA was extracted from FL5.12 cells that had been transfected with the empty vector (lanes 1-4) or pMT-TEF (lanes 5-8) and cultured in the presence of IL-3 with Zn for the indicated periods of time. The blot was hybridized with mouse cDNA probes specific for IL-3 receptor β chains and α chain. The 28S rRNA visualized with ethidium bromide staining is shown in the bottom panel. (B) Flow cytometric analysis for surface expression of the IL-3 receptor. FL5.12 cells transfected with the empty vector, pMT–E2A-HLF, pMT-TEF, or pMT-TEF/BX were cultured in IL-3–containing medium in the presence or absence of Zn for 24 hours. Cells were analyzed with the specific antibodies for mouse βIL3, βC, or α chains. Dotted or solid lines indicate the histograms of control staining, and filled curves indicate those of specific antibodies.
Northern blot analysis and flow cytometric analysis of FL5.12 cells. (A) Northern blot analysis for the IL-3 receptor. Total RNA was extracted from FL5.12 cells that had been transfected with the empty vector (lanes 1-4) or pMT-TEF (lanes 5-8) and cultured in the presence of IL-3 with Zn for the indicated periods of time. The blot was hybridized with mouse cDNA probes specific for IL-3 receptor β chains and α chain. The 28S rRNA visualized with ethidium bromide staining is shown in the bottom panel. (B) Flow cytometric analysis for surface expression of the IL-3 receptor. FL5.12 cells transfected with the empty vector, pMT–E2A-HLF, pMT-TEF, or pMT-TEF/BX were cultured in IL-3–containing medium in the presence or absence of Zn for 24 hours. Cells were analyzed with the specific antibodies for mouse βIL3, βC, or α chains. Dotted or solid lines indicate the histograms of control staining, and filled curves indicate those of specific antibodies.
Hypothetical roles of TEF and E2A-HLF. TEF down-regulates the expression of the common β (βC) chain and inactivates IL-3 signaling pathways, which are critical for both cell proliferation and cell survival. In addition, TEF promotes cell survival in the absence of survival signalings downstream of IL-3. Consequently, cells in the G0/G1 phase accumulated without undergoing apoptosis. In contrast, E2A-HLF blocks apoptosis without regulating the expression of cytokine receptors.
Hypothetical roles of TEF and E2A-HLF. TEF down-regulates the expression of the common β (βC) chain and inactivates IL-3 signaling pathways, which are critical for both cell proliferation and cell survival. In addition, TEF promotes cell survival in the absence of survival signalings downstream of IL-3. Consequently, cells in the G0/G1 phase accumulated without undergoing apoptosis. In contrast, E2A-HLF blocks apoptosis without regulating the expression of cytokine receptors.
Discussion
In this study, using a Zn-inducible system, TEF protein expression was induced in pMT-TEF–transfected FL5.12 cells within 4 hours after the addition of Zn (Figure 3B). The mRNA expression levels of the β subunits of the IL-3 receptor were reduced and became barely detectable within 16 hours (Figure 6A), and surface expression of the β chains was almost completely down-regulated within 24 hours (Figure 6B). Subsequently, the IL-3 signaling pathways were lost (Figure 5) and cells underwent cell cycle arrest in G0/G1 phase within 36 hours (Figure 4A; Table 1). However, the cells did not undergo apoptosis (Figure 4A) because TEF protected FL5.12 cells from apoptosis induced by IL-3 deprivation (Figure 4F). Thus, TEF induced FL5.12 cells to remain in a resting state, either in the presence or absence of IL-3 (Figure 7). These unique functions of TEF depended on its DNA-binding potential because a TEF mutant with amino acid substitutions in its basic region (TEF-BX) completely lost these activities (Figures 4, 5, 6). However, there is no potential binding site for TEF in the promoter regions of the 2 β-chain genes in the mouse51,53 and TEF is known as a transcriptional activator.29,37 Thus, TEF likely down-regulates transcription of the β-chain genes indirectly.
The βC chain is common to the receptors of a series of major cytokines including IL-5 and GM-CSF as well as IL-3 and is expressed by the majority of hematopoietic progenitors (H.I. and A.K., unpublished observation, May 2004; Militi et al54 ), which expand in response to multiple growth factors during normal hematopoiesis. Therefore, it is reasonable that the Tef expression level in each hematolymphoid progenitor was low (Figure 2B). TEF expression may be restricted to a small fraction of progenitors that are in the resting phase and do not express the βC chain. TEF might play an important role in preventing the exhaustion of hematopoietic progenitors by rendering a portion of the progenitors to be insensitive to growth factors. Establishment of Tef-deficient mice and extensive analysis of their hematopoiesis would elucidate the biologic significance of this unique transcription factor that regulates both cell survival and expression of cytokine receptors in hematopoietic progenitors.
This study also provides insights into functional differences between oncogenic chimeras and their normal counterparts. Similar to TEF, the E2A-HLF chimera inhibited apoptosis caused by IL-3 deprivation (Figure 4F; Inaba et al,18 Inukai et al,19 Altura et al20 ). However, unlike TEF, E2A-HLF did not down-regulate the expression of β subunits of the IL-3 receptor. As a result, cells expressing E2A-HLF proliferated in the presence of IL-3 (Figure 4A) and survived in the absence of IL-3 (Figure 4F). These findings suggest that the chimeric transcription factor retains the ability to drive leukemic transformation with maximum efficiency by disrupting multiple transcriptional networks. Since TEF and HLF share a high degree of sequence identity in their bZIP domains, the difference in the ability to regulate the levels of growth factor receptors between TEF and E2A-HLF is most likely caused by differences in their trans-activation domains (Figure 1). Alternatively, subtle changes in their DNA-binding specificity might contribute to the loss of activity of E2A-HLF to regulate cytokine receptors. The DNA-binding specificity of the PAR bZIP transcription factors is reported to be influenced by the sequences flanking the basic region, such as the basic region extension (BRE), fork region, and PAR domains.36 Because the BRE and PAR domains of HLF are not included in the E2A-HLF chimera (Figure 1), the DNA-binding potential of E2A-HLF to suboptimal sites is substantially impaired.34
The antiapoptotic potential of TEF that depends on its DNA-binding activity supports a model that the consensus binding sequence recognized by the PAR factors as well as E2A-HLF, E4BP4, and CES-2 could serve as a regulatory switch for programmed cell death.18-20,23,24 However, the detailed downstream pathways in mammalian systems have not yet been elucidated. In C elegans, CES-2 appears to down-regulate CES-1 expression.5,6 Although we previously identified the SLUG gene, a mammalian homologue of CES-1, as one of the downstream responders of E2A-HLF in human leukemia cells with t(17;19) translocation,23 neither TEF nor E2A-HLF was able to induce its expression in FL5.12 cells (data not shown). Thus, there must be other critical downstream pathway(s) for the antiapoptotic potential of TEF and E2A-HLF. Identification of downstream pathway(s) of TEF for its antiapoptotic potential and βIL3 and βC chain gene regulation will be critical for clarifying transcriptional regulation driving normal hematopoiesis as well as leukemogenesis by E2A-HLF.
Prepublished online as Blood First Edition Paper, January 21, 2005; DOI 10.1182/blood-2004-08-2976.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are indebted to Dr F. Rauscher III for providing the pMT-CB6+ Zn inducible vector. This research was supported by grants-in-aid from the Ministry of Education, Science and Culture of Japan.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal