Abstract
To develop murine models of leukemogenesis, a series of transgenic mice expressing BCR-ABL in different hematopoietic cell subsets was generated. Here we describe targeted expression of P210 BCR-ABL in stem and progenitor cells of murine bone marrow using the tet-off system. The transactivator protein tTA was placed under the control of the murine stem cell leukemia (SCL) gene 3′ enhancer. Induction of BCR-ABL resulted in neutrophilia and leukocytosis, and the mice became moribund within 29 to 122 days. Autopsy of sick mice demonstrated splenomegaly, myeloid bone marrow hyperplasia, and extramedullary myeloid cell infiltration of multiple organs. BCR-ABL mRNA and protein were detectable in the affected organs. Fluorescence-activated cell sorter (FACS) analysis demonstrated a significant increase in mature and immature myeloid cells in bone marrow and spleen, together with increased bilineal B220+/Mac-1+ cells in the bone marrow. tTA mRNA was expressed in FACS-sorted hematopoietic stem cells expanded 26-fold after BCR-ABL induction. Thirty-one percent of the animals demonstrated a biphasic phenotype, consisting of neutrophilia and subsequent B-cell lymphoblastic disease, reminiscent of blast crisis. In summary, this mouse model recapitulates many characteristics of human chronic myeloid leukemia (CML) and may help elucidate basic leukemogenic mechanisms in CML stem cells during disease initiation and progression. (Blood. 2005;105:324-334)
Introduction
Chronic myeloid leukemia (CML) was the first human malignancy shown to be associated with a specific genetic lesion, the Philadelphia chromosome, an abnormality first described as a shortened chromosome 221 and subsequently identified as chromosomal translocation t(9;22),2 which fuses parts of the breakpoint cluster region (BCR) with the gene encoding the tyrosine kinase (ABL). The BCR-ABL protein is a constitutively active tyrosine kinase that is associated with different forms of leukemia. Animal models and clinical trials with the ABL kinase inhibitor imatinib have implicated BCR-ABL as the cause of the leukemia.3-5 The most common form, p210 BCR-ABL, is found in more than 90% of patients with CML, a malignant disorder of hematopoietic stem cells (HSCs).6 Previous studies have demonstrated that the phenotype of p210 BCR-ABL transgenic mice is dependent on the expression pattern and cell-type specificity of the regulatory elements used to drive expression of the oncogene.4,7 We developed a series of transgenic mice in which expression of the BCR-ABL fusion protein in different subsets of hematopoietic progenitor cells is adjustable by the administration of tetracycline.8,9 These models demonstrated that BCR-ABL alone is sufficient to induce leukemia and that its continued expression is required for maintenance of the leukemic phenotype. The regulation of the tetracycline-controlled transactivator (tTA) by the mammary tumor virus long terminal repeat (MMTV-LTR) resulted in the rapid and exclusive development of a pre-B-cell leukemia.8 This phenotype was consistent with high expression of MMTV-LTR in early B cells.8 We reasoned that promoter or enhancer elements derived from HSC-specific genes should result in a more accurate model of the development of CML. Previously, regulatory elements derived from the Ly-6E.1 gene10 and the H2K gene11 have been used to target adult HSCs. However, expression of the Ly-6E1 gene is dependent on the genetic background of the mice,12,13 and the H2K gene is expressed in most tissues,14 which makes these elements less desirable. We isolated and characterized regulatory elements of the human CD34 gene locus that are capable of expressing human CD34 in HSCs15 and subsequently modified the expression cassette to allow for inducible expression of a heterologous transgene in mice.16 These animals were bred to a line of mice that carries p210 BCR-ABL under the control of a tetracycline-responsive element (TRE). Double-transgenic mice consistently developed a myeloproliferative syndrome with preferential expansion of the megakaryocytic lineage causing thrombocytosis,9 a feature also seen in patients with CML. However, the expression pattern of tTA in these transgenic mice with highest expression in megakaryocyte-erythrocyte progenitor (MEP) cells was slightly different from that of human CD34 expressed from an unmodified human CD34 PAC clone.9
The SCL gene encodes a basic helix-loop-helix transcription factor expressed in hematopoietic cells, endothelial cells, the central nervous system, and the embryonic skeleton.17-19 Within the hematopoietic system, SCL is normally expressed in erythroid cells, mast cells, megakaryocytes, and multipotent progenitor cells.20-23 Studies with null mutants have shown that SCL is a critical regulator of hematopoiesis.24-27 Based on these results, it was predicted that normal SCL-expressing progenitor cells should be multipotent, and a knock-in strategy has indeed confirmed this hypothesis.28 Detailed analysis of enhancer elements within the murine SCL gene has identified a fragment within the 3′ region of the gene, which restricted expression of a heterologous LacZ transgene within adult murine bone marrow to hematopoietic stem and progenitor cells and megakaryocytes.29,30 We have modified this construct to allow inducible expression of transgenes by placing tTA expression under the control of this 3′ enhancer fragment. Here we describe that inducing BCR-ABL expression invariably resulted in neutrophilia, subsequent splenomegaly, and organ invasion by myeloid cells, thus recapitulating major features of CML in patients.
Materials and methods
Generation of the SCLtTA construct and of transgenic mice
The transgenic construct (Figure 1A), consisting of the SV40 minimal promoter, an intervening sequence (intron), the tTA gene, and the 5.5-kb BglII fragment from the murine 3′-SCL enhancer,29,31 was generated by inserting the rabbit β-globin intron 2 into the SfiI site in the SV40 minimal promoter contained in pGL2. The tTA sequence with polyadenylation (polyA) site was isolated by digestion of pUHD 15-132 with HindIII/EcoRI and then was ligated into the SalI-digested SV40/intron plasmid. This fragment was released by HindII-blunted/NheI digestion and was ligated into the multiple cloning site 5′ of the 5.5-kb murine SCL 3′-enhancer in pBluescript KS digested with BglII-blunted/XbaI (Figure 1A). The transgenic construct was released from the vector backbone by digestion with XhoI, and the fragment was purified using QiaEx II (Qiagen, Valencia, CA) following the manufacturer's instructions. The DNA was resuspended in microinjection buffer (1 mM Tris/HCl, pH 8.0, 0.1 mM EDTA [ethylenediaminetetraacetic acid]) and was injected into the pronucleus of BDF mice. Nine different founder animals were identified, from which 6 transgenic lines were subsequently established. To test for expression of a functional transactivator protein, SCLtTA mice were bred with Tet-O-cre transresponder mice.16 SCLtTA/BCR-ABL double-transgenic mice were generated by cross-breeding of male SCLtTA mice with female TRE-BCR-ABL mice (founder line 2)8 in the presence of tetracycline administered with the drinking water (0.5 g/L) to prevent expression of the transgene during embryogenesis.8
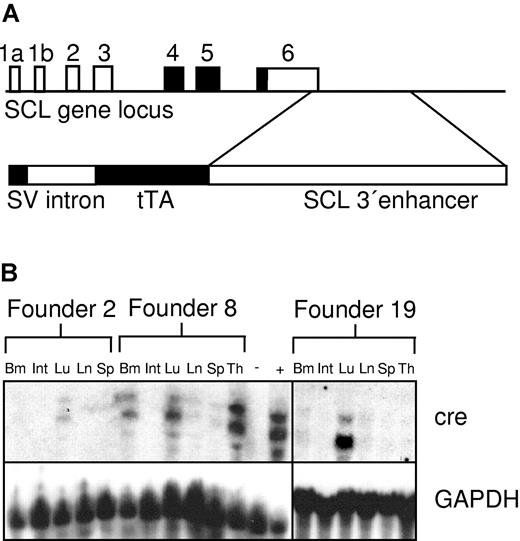
SCLtTA transactivator construct and expression of cre in SCLtTA x Tet-O-cre reporter mice. (A, upper portion) Structure of the murine SCL locus (coding exons shaded in black) indicating the position of the 3′ enhancer fragment contained in the SCLtTA construct. (A, lower portion) Transgenic construct consisting of the SV40 minimal promoter upstream of the rabbit β-globin intron 2, the tetracycline-dependent transactivator protein tTA, and a 5.5-kb BglII fragment of the enhancer of the murine SCL gene. (B) Cre mRNA expression under the control of the murine SCL enhancer in different tissues of the 3 independent founder lines (founders 2, 8, and 19), which demonstrated detectable cre expression after being crossed to the Tet-O-cre responder strain. Twenty micrograms per lane of RNA extracted from the following tissues were loaded as indicated: bone marrow (Bm), intestines (Int), lung (Lu), lymph node (Ln), spleen (Sp), and thymus (Th). RNA from the spleen of an MMTVtTA-Tet-O-cre double-transgenic mouse (+) and from the thymus of an SCLtTA single-transgenic mouse (-) served as positive and negative controls, respectively. The Northern blot for cre expression showed 2 bands, as described previously,16 using a 1.0-kb fragment of the Tet-O-cre transgenic construct34 as a probe, as described in “Materials and methods.” The Northern blot was stripped and reprobed using a GAPDH probe.
SCLtTA transactivator construct and expression of cre in SCLtTA x Tet-O-cre reporter mice. (A, upper portion) Structure of the murine SCL locus (coding exons shaded in black) indicating the position of the 3′ enhancer fragment contained in the SCLtTA construct. (A, lower portion) Transgenic construct consisting of the SV40 minimal promoter upstream of the rabbit β-globin intron 2, the tetracycline-dependent transactivator protein tTA, and a 5.5-kb BglII fragment of the enhancer of the murine SCL gene. (B) Cre mRNA expression under the control of the murine SCL enhancer in different tissues of the 3 independent founder lines (founders 2, 8, and 19), which demonstrated detectable cre expression after being crossed to the Tet-O-cre responder strain. Twenty micrograms per lane of RNA extracted from the following tissues were loaded as indicated: bone marrow (Bm), intestines (Int), lung (Lu), lymph node (Ln), spleen (Sp), and thymus (Th). RNA from the spleen of an MMTVtTA-Tet-O-cre double-transgenic mouse (+) and from the thymus of an SCLtTA single-transgenic mouse (-) served as positive and negative controls, respectively. The Northern blot for cre expression showed 2 bands, as described previously,16 using a 1.0-kb fragment of the Tet-O-cre transgenic construct34 as a probe, as described in “Materials and methods.” The Northern blot was stripped and reprobed using a GAPDH probe.
Genotyping
Transgenic mice were identified by Southern blot analysis (for the identification of founder animals) or by polymerase chain reaction (PCR) with tTA gene-specific primers. The upstream primer (5′-AGA AGA GAT GAG GTG TGA GGA T-3′) and the downstream primer (5′-GGA TCC ACAAGAATGAGCATGTA-3′) amplified a 405-base pair fragment within the tTA gene. SuperMix polymerase (Gibco BRL, Grand Island, NY) was used with amplification conditions of 35 cycles at 94°C for 40 seconds, 65°C for 30 seconds, and 72°C for 30 seconds. Southern blot analysis of EcoRI-restricted genomic DNA was performed using positively charged nylon membranes (Biotrans +; ICN, Irvine, CA), as described.8 Stable chromosomal incorporation of the transgene was detected with a 1.0-kb BamHI/HindII fragment of the tTA gene. Genotyping for the P210 BCR-ABL transgene was performed as described previously.8 Genotyping for the cre recombinase transgene was performed by PCR with primers 5′-ATGTTCAATTTACTGACCG-3′ and 5′-CGCCGCATAACCAGTGAAAC-3′. Amplification conditions consisted of 35 cycles of 30-second denaturation at 94°C, 30-second annealing at 51°C, and 30-second extension at 72°C, yielding an amplification product of 355 bp.
RNA isolation and Northern blot analysis
Total RNA for Northern blot analysis was isolated from murine tissue according to the CsCl method.33 Expression analysis was performed as described, and a 1.02-kb fragment of the tTA gene isolated from pUHD 15-132 and the joining region of p210 BCR-ABL were used as probes.8 Expression of the gene for cre recombinase was detected using a 1.0-kb probe generated by digestion of the Tet-O-cre transgenic construct16,34 with EcoRV/PvuII. Northern blots were stripped and reprobed with a GAPDH-specific probe.35
Western blotting
Western blotting was performed essentially as described.8 Briefly, cells in single suspension were washed with ice-cold phosphate-buffered saline (PBS) and were lysed in RIPA buffer or 10% trichloracetate. After pH adjustment, sample buffer was added immediately, and the samples were boiled for 10 minutes. Proteins were separated on a 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, transferred onto a nitrocellulose membrane (Bio-Rad, Hercules, CA), and probed with mouse anti-c-abl antibody (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA).
Analysis and monitoring of disease
Peripheral blood was collected from each retro-orbital plexus with a heparinized capillary tube, and total white blood cell and differential counts were performed once a week. Peripheral blood films were stained with Wright-Giemsa for differential analysis, and the number of white blood cells was determined with a hemocytometer after lysis of enucleated red blood cells with ACK lysis buffer (0.15 M NH4Cl, 1 mM KHCO3, 0.1 mM Na2-EDTA, pH 7.3).
Histology
Mice were killed by CO2 inhalation, and organs were removed and fixed in neutrally buffered 10% formalin at room temperature for 16 hours before they were embedded in paraffin and sectioned. All tissues were stained with hematoxylin and eosin for light microscopy. All tissues were stained with hematoxylin and eosin or Wright Giemsa as indicated. Light microscopy was performed with a Zeiss Axioplan microscope (Zeiss, Guttingen, Germany) using a 25 × Plan-Neofluar 0.80; 63 × Plan-Apochromat 1.4 oil; or 100 × Plan-Neofluar 1.30 oil lens. Images were captured using Adobe Photoshop version 5.5 (Adobe Systems, San Jose, CA) and Microsoft Powerpoint 2000 (Microsoft, Redmond, WA).
Flow cytometry analysis
Bone marrow cells were flushed from tibias and femurs with PBS. Enucleated red blood cells were lysed with ACK lysis buffer, and the remaining white blood cells were washed once with PBS. Lymph node and spleen tissue were squeezed through a 100-μm cell strainer (Becton Dickinson Labware, Franklin Lakes, NJ) with the plunger of a syringe. Cells were incubated with the appropriate antibodies. All analyses were performed on a dual-laser FACS (Becton Dickinson). The following antibodies were used: antimurine B220 (Caltag, San Francisco, CA) and murine CD34, CD41, Ter 119, Gr-1, Mac-1, c-kit, Sca-1, BP-1, CD43, and immunoglobulin M (IgM) (all from PharMingen (San Diego, CA).
Multicolor FACS sorting and analysis of HSCs and progenitor cells
For sorting of myeloid progenitors, red blood cell-lysed bone marrow cells were stained with tricolor-conjugated rat antibodies specific for the following lineage markers: CD3 (RM3406), CD4 (MCD 0406), CD8a (MCD0806), CD19 (RM7706), Ly-6G (Gr-1) (RM3006), and CD45R (B220) (RM2606) (all from Caltag, Burlingame, CA). Lin+ cells were removed with sheep antirat IgG-conjugated magnetic beads (Dynabeads M-450; Dynal A.S., Oslo, Norway), and the remaining cells were stained with rat antibodies anti-CD34-fluorescein isothiocyanate (FITC) (RAM34), anti-FcRγII/III-phycoerythrin (PE) (2.4G2), anti-CD117 (c-kit)-allophycocyanin (APC) (2B8), and anti-Sca-1 (Ly-6A/E)-biotin (E13-161.7) (all from PharMingen) and subsequently with antistreptavidin-APC/Cy7 (SA1014) (Caltag). Myeloid progenitors were sorted as Lin-Sca-1-c-Kit+CD34+FcγRII/IIIlo (CMP), Lin-Sca-1-c-Kit+CD34+FcγRII/IIIhi (GMP), or Lin-Sca-1-c-Kit+CD34-FcγRII/IIIlo (MEP), as described previously.36 For hematopoietic stem cells (HSCs) and common lymphocyte progenitors (CLPs), bone marrow cells were stained with lineage markers and with anti-Sca-1 (Ly-6A/E)-FITC), anti-CD117 (c-kit)-APC (2B8), and anti-interleukin-7 receptor α (IL-7Rα)-chain PE (A7R34; e-Bioscience, San Diego, CA). HSCs and CLPs were sorted as Lin-Sca-1hic-Kithi and IL-7Rα+Lin-Sca-1loc-Kitlo populations, respectively. Cells were sorted using a highly modified double laser (488 nm/350 nm Enterprise II + 647 nm Spectrum) high-speed FACS (Moflo-MLS; Cytomation, Fort Collins, CO). Cells were sorted into 500 μL buffer RLT with β-mercaptoethanol, and RNA was extracted using the RNAEasy Micro kit including the carrier RNA step and DNase on-column treatment according to the manufacturer's instructions (Qiagen).
RT-PCR analysis of isolated HSCs and progenitors
Reverse transcription (RT) was performed using SuperScript reverse transcriptase (Gibco BRL) with random primer hexamers according to the manufacturer's instructions. One microliter of each cDNA reaction was used for the detection of tTA gene expression. tTA was amplified with sense primer 5'-AAA AGC TGG ATC GAT CCT GAG AAC and antisense primer 5'-GGC GGC ATA CTA TCA GTA GTA GGT G. Murine GAPDH cDNA, which served as a positive control, was amplified using sense primer 5'-GGT GCT GAG TAT GTC GTG GAG TCT A and antisense primer 5'-CCT GGT TCA CCA CCT TCT TCT TGA TGT C. PCR conditions consisted of 35 cycles, with 30-second denaturation at 94°C, 30-second annealing at 55°C (tTA) or 58°C (GAPDH), and 90-second extension at 72°C.
CFU assay and megakaryocyte assay
Bone marrow was flushed with Iscove modified Dulbecco medium (IMDM) containing 5% fetal bovine serum (FBS) from femurs and tibias of double-transgenic and single-transgenic control animals. Spleen cells were isolated by squeezing tissue through a cell strainer. Enucleated red blood cells were removed with ACK cell lysis buffer, and colony assays were initiated according to the manufacturer's instructions in Methocult GF M3434 supplemented with growth factors, as provided by the manufacturer (Stem Cell Technologies, Vancouver, BC, Canada). Numbers of granulocyte colony-forming units (CFU-GMs), erythroid blast-forming units (BFU-Es), and granulocyte-macrophage-erythroid-megakaryocyte colony-forming units (CFU-GEMMs) were determined 8 days after plating.
Transplantation into NOD/SCID mice and real-time genomic PCR for engraftment assessment
Bone marrow cells were harvested from the femurs and tibias of double-transgenic and control mice. Cells were resuspended in PBS and injected (2 × 106 cells/0.3 mL) into the tail veins of sublethally irradiated (300 cGy) NOD/SCID mice. Mice were housed in microisolator cages under sterile conditions. Real-time genomic PCR was performed on an ABI Prism 7700 Sequence Detector (Applied Biosystems, Foster City, CA) using the following parameters: 40 cycles of 94°C (15 seconds) and 60°C (60 seconds). Primers and probe (FAM-labeled) were as follows: murine CCAAT enhancer-binding protein alpha (CEBPA) forward, 5′-AAA GCC AAG AAG TCG GTG GAC; CEBPA reverse, 5′-CTT TAT CTC GGC TCT TGC GC; CEBPA probe, 5′-CGA GTA CCG GGT ACG GCG; BCR-ABL forward, 5′-CTG GCC CAA CGA TGG CGA; BCR-ABL reverse, 5′-CAC TCA GAC CCT GAG GCT CAA; BCR-ABL probe, 5′-CCC TTC AGC GGC CAG TAG CAT CTG A.
Statistical analysis
Data are expressed as mean ± SD unless otherwise indicated and were compared using an unpaired Student t test, as described in “Results.” P less than .05 was considered significant.
Results
SCLtTA transactivator mice express within the bone marrow
To express tTA in blood stem and progenitor cells, we created a construct that would express tTA under control of the SCL 3′ stem cell enhancer (Figure 1A) and generated multiple lines of transgenic mice. Of 9 SCLtTA transgenic founder lines, 6 showed germ line transmission. To determine whether these founder lines expressed a functional transactivator protein and to compare the expression pattern, SCLtTA mice were crossbred with Tet-O-cre mice.16 Expression of the cre gene was used as an indicator. We detected expression of the cre reporter gene by Northern blot analysis in the bone marrow and lung of double-transgenic mice bred from 3 SCLtTA founder lines F2, F8, and F19 (Figure 1B). In addition, high expression was detected in the thymus in founder line 8. None of the other 3 founder lines showed any detectable cre RNA expression by Northern blot analysis (data not shown); therefore, these mice were not used for further studies. Animals derived from founder line 19 without detectable cre expression in the thymus were subsequently crossbred with TRE-BCR-ABL mice (founder line 2)8 to generate SCLtTA/BCR-ABL double-transgenic animals. Thirty-eight double-transgenic and 33 control animals (12 SCLtTA single transgene, 13 BCR-ABL single transgene, and 8 wild type) were induced. Fourteen mice were killed for the initial description of the disease phenotype (Table 1), and the other animals were used for specific studies such as HSC and CFU analysis and reversion experiments, as described below.
Clinical characteristics of induced human SCLtTA/BCR-ABL double-transgenic animals at time of death
Animal . | Clinical condition . | WBC/μL, neutrophils (%)* . | Hgb (g/dL) . | Spleen weight, mg . | Other organs involved, gross pathology/histology . | Time of analysis, d . |
|---|---|---|---|---|---|---|
| W98 | Moribund | 25 800 (33) | 10.5 | 410† | LN, skin, lung bleed, PB lymphoblasts | 29 |
| W107 | Moribund, tumor right cervival LN | 8 300 (32) | 12.5 | 240 | Axillary LN (923 mg), liver, lung bleed | 122 |
| W109 | Moribund | 3 900 (26) | ND | 310 | Liver, LN, lung bleed | 105 |
| W110 | Moribund | 29 200 (82) | 8.2 | 850 | Kidney, perinephric fluid, liver, skin, LN (all myeloid cells), brittle bones | 57 |
| W114 | Moribund | 18 500 (61) | 12 | 240† | LN, liver (myeloid cells), lung bleed, brittle bones | 46 |
| W118 | Leukocytosis | 15 500 (51) | 14 | 260 | Liver, pale BM | 43 |
| W119 | Leukocytosis, anemia | 70 000 (27) | 10.5 | 480 | LN, liver, pale BM, PB lymphoblasts (70%) | 34 |
| W184 | Moribund | > 30 000 (34) | ND | 130 | LN (mild), brittle bones | 67 |
| W185 | Neutrophilia, humanely killed | 9 200 (54) | ND | 300 | Pale BM | 42 |
| W194 | Found dead | 25 300 (ND) | ND | 440 | Kidney, liver | 66 |
| W202 | Neutrophilia, humanely killed | 9 500 (50) | ND | 270 | pale BM | 30 |
| W207 | Moribund, distended abdomen | 22 000 (53) | ND | 400 | Liver, kidney | 69 |
| W209 | Moribund | 19 600 (63) | ND | 130 | LN (mild) | 42 |
| W214 | Humanely killed | 6 800 (26) | ND | 120 | None | 7 |
Animal . | Clinical condition . | WBC/μL, neutrophils (%)* . | Hgb (g/dL) . | Spleen weight, mg . | Other organs involved, gross pathology/histology . | Time of analysis, d . |
|---|---|---|---|---|---|---|
| W98 | Moribund | 25 800 (33) | 10.5 | 410† | LN, skin, lung bleed, PB lymphoblasts | 29 |
| W107 | Moribund, tumor right cervival LN | 8 300 (32) | 12.5 | 240 | Axillary LN (923 mg), liver, lung bleed | 122 |
| W109 | Moribund | 3 900 (26) | ND | 310 | Liver, LN, lung bleed | 105 |
| W110 | Moribund | 29 200 (82) | 8.2 | 850 | Kidney, perinephric fluid, liver, skin, LN (all myeloid cells), brittle bones | 57 |
| W114 | Moribund | 18 500 (61) | 12 | 240† | LN, liver (myeloid cells), lung bleed, brittle bones | 46 |
| W118 | Leukocytosis | 15 500 (51) | 14 | 260 | Liver, pale BM | 43 |
| W119 | Leukocytosis, anemia | 70 000 (27) | 10.5 | 480 | LN, liver, pale BM, PB lymphoblasts (70%) | 34 |
| W184 | Moribund | > 30 000 (34) | ND | 130 | LN (mild), brittle bones | 67 |
| W185 | Neutrophilia, humanely killed | 9 200 (54) | ND | 300 | Pale BM | 42 |
| W194 | Found dead | 25 300 (ND) | ND | 440 | Kidney, liver | 66 |
| W202 | Neutrophilia, humanely killed | 9 500 (50) | ND | 270 | pale BM | 30 |
| W207 | Moribund, distended abdomen | 22 000 (53) | ND | 400 | Liver, kidney | 69 |
| W209 | Moribund | 19 600 (63) | ND | 130 | LN (mild) | 42 |
| W214 | Humanely killed | 6 800 (26) | ND | 120 | None | 7 |
Clinical condition refers to the condition of the animals at the time of analysis. Humanely killed indicates mice killed for analysis before the development of an obvious phenotype. All mice developed neutrophilia (defined as more than 20% neutrophils; mean neutrophil percentage of control animals was 10% ± 3% WBCs, and the neutrophil percentage in control animals did not exceed 20%) after induction of BCR-ABL. Platelet counts (normal range, 300-900/nL) were not significantly elevated. Normal spleen weight in single- transgenic animals was 80 to 90 mg (n = 3). Thirty-one percent of all animals demonstrated a biphasic phenotype, consisting of neutrophilia, subsequent development of lymphoid tumors, and acute lymphoblastic leukemia reminiscent of blast crisis. Hgb indicates hemoglobin (normal range, 12-16 g/dL); LN, lymphadenopathy; BM, bone marrow; PB, peripheral blood; ND, not determined at time of death.
Normal range of WBC counts was 4000 to 10,000/μL.
Myeloid cells.
Expression of BCR-ABL causes neutrophilia, leukocytosis, and organ invasion by myeloid cells
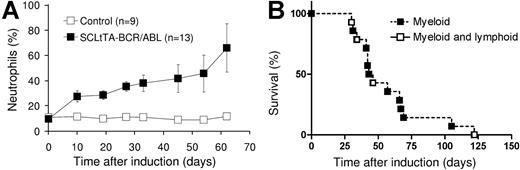
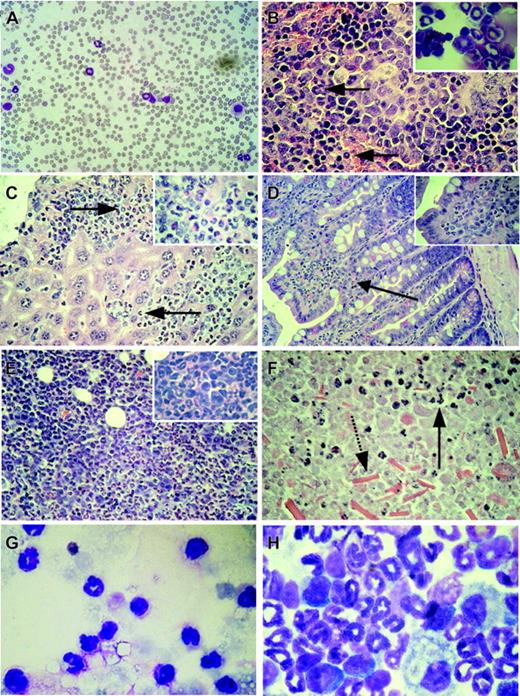
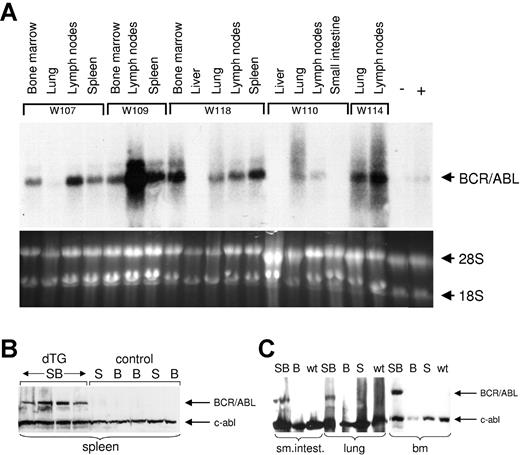
Induction of BCR-ABL expression by withdrawal of tetracycline from the drinking water led to significant 3-, 5-, and 2-fold increases in the percentages of neutrophils, absolute numbers of neutrophils, and absolute numbers of white blood cells (WBCs), respectively, in the peripheral blood of double-transgenic mice within 2 weeks of induction (Figures 2 and 3A). Anemia developed in some of the mice (Table 1). The WBC count and the percentage of neutrophils in the peripheral blood was increased at the time of death in 57% and 100% of the animals, respectively (Table 1). No increase in WBC counts was observed in single-transgenic (BCRABL only or SCLtTA only) animals or wild-type littermates housed in the same cages (Figure 2). In parallel, splenomegaly developed in all double-transgene-induced animals (Table 1). Histologic examination of the spleen performed at various time points after induction demonstrated expansion of the splenic red pulp by predominantly granulocytic myeloid cells (Figure 3B), and this was confirmed by flow cytometric analysis (Table 2). In addition, infiltration of myeloid cells was also seen in the liver (Figure 3C) (57% of the animals displayed hepatomegaly), lymph node, lamina propria of the gut (Figure 3D), and lung (occasionally resulting in focal pulmonary hemorrhages similar to those described in other BCR-ABL disease models37-39 ). In addition, skin chloromas were seen in 2 animals with fulminant disease (Table 1). The major cause of death in animals that had progressive disease before humane killing was related to myeloid cell infiltration of extramedullary organs, including the liver (n = 1), kidney (n = 3), and lung (n = 3), or to the development of lymphoblastic disease (n = 4; see Figure 2B and under “Evidence for progression to blast crisis”). Three animals developed fatal obstructive hydronephrosis 57, 66, and 69 days after the induction of BCR-ABL expression (Table 1). Autopsy revealed embedding of the kidney in a fluid-filled cyst, and histology revealed infiltration of the kidneys and ureters by myeloid cells, parenchymal necrosis, and crystal depositions (Figure 3E-F). Neutrophils were present in the fluid recovered from the cyst of 1 of the animals (Figure 3G). However, we cannot exclude the possibility of a contribution of BCR-ABL expression in nonhematopoietic cells, which, in conjunction with the myeloid infiltrate, could conceivably have resulted in additional tissue damage. Altogether, the survival time after induction of BCR-ABL expression ranged from 4 weeks to 10 weeks, with the exception of 1 mouse that survived for 17 weeks (Table 1; Figure 2B). BCR-ABL mRNA and protein expression was demonstrated in cells isolated from the affected organs (Figure 4A-B), including the lung of animals that showed disease involvement of this organ (hemorrhage and pleural effusion). Northern blotting failed to detect BCR-ABL mRNA in the liver (Figure 4A), but BCR-ABL mRNA was readily detected using RT-PCR (not shown).
Neutrophilia and survival of SCLtTA/BCR-ABL mice after induction of BCR-ABL. (A) The graph shows the percentages of neutrophils (shown as mean ± SE) among total white blood cells in SCLtTA/BCR-ABL double-transgenic mice (▪) or controls (□). Controls consisted of wild-type (n = 3) and BCR-ABL single-transgenic (n = 6) mice, which had similar values. The percentage of neutrophils was greater in SCLtTA/BCR-ABL mice than in controls (P < .05) for each time point up to day 45 (on day 54, the ranges were 20%-79% for SCLtTA/BCR-ABL and 5%-12% for control animals, and on day 62 the ranges were 47%-85% and 11%-14%, respectively). SE for the control mice was at each point less than 2.1 and, therefore does not show on the diagram. (B) Kaplan-Meier-style survival and morbidity curve of induced double-transgenic mice, depicting overall survival and disease phenotype (▪, myeloid; □, sequential myeloid and lymphoblastic) after induction of BCR-ABL by tetracycline removal.
Neutrophilia and survival of SCLtTA/BCR-ABL mice after induction of BCR-ABL. (A) The graph shows the percentages of neutrophils (shown as mean ± SE) among total white blood cells in SCLtTA/BCR-ABL double-transgenic mice (▪) or controls (□). Controls consisted of wild-type (n = 3) and BCR-ABL single-transgenic (n = 6) mice, which had similar values. The percentage of neutrophils was greater in SCLtTA/BCR-ABL mice than in controls (P < .05) for each time point up to day 45 (on day 54, the ranges were 20%-79% for SCLtTA/BCR-ABL and 5%-12% for control animals, and on day 62 the ranges were 47%-85% and 11%-14%, respectively). SE for the control mice was at each point less than 2.1 and, therefore does not show on the diagram. (B) Kaplan-Meier-style survival and morbidity curve of induced double-transgenic mice, depicting overall survival and disease phenotype (▪, myeloid; □, sequential myeloid and lymphoblastic) after induction of BCR-ABL by tetracycline removal.
Organ infiltration by myeloid cells after induction of BCR-ABL expression in double-transgenic animals. (A) Peripheral blood smear showing leukocytosis and neutrophilia in a diseased double-transgenic mouse. (B) Histology of the spleen demonstrating extramedullary myeloid hematopoiesis (arrows pointing to myeloid cells). Inset: Myeloid cells in the cytospin of the spleen from the same animal. (C) Histology of the liver. Perivascular infiltration of the liver by hematopoietic cells (arrows point to myeloid cells). Inset: Part of the same organ at higher magnification. (D) Histology of the small intestine showing infiltration of the lamina propria by myeloid cells (arrow, and magnified in the inset). (E) Histology of a kidney from mouse W207 showing disruption of the kidney architecture and massive invasion by hematopoietic cells. Inset: Cells at higher magnification. (F) Histology of the kidney of animal W194 demonstrating parenchymal tissue necrosis with some invading myeloid cells still present (arrow with solid line); note the presence of crystals in the lower part of the picture (arrow with broken line). (G) Fluid recovered from the perinephric cyst of the same animal containing high numbers of neutrophils. (H) Cytospin of the bone marrow showing predominance of myelopoiesis and suppression of the erythroid and lymphoid compartment in the bone marrow. Original magnification, × 60 (B-C) or × 100 (A;D, inset). Original magnifications, × 25 (D), × 60 (A-C, E-F), and × 100 (G-H, insets). Stains were Wright Giemsa for panels A, G, and H, and hematoxylin and eosin for panels B-F.
Organ infiltration by myeloid cells after induction of BCR-ABL expression in double-transgenic animals. (A) Peripheral blood smear showing leukocytosis and neutrophilia in a diseased double-transgenic mouse. (B) Histology of the spleen demonstrating extramedullary myeloid hematopoiesis (arrows pointing to myeloid cells). Inset: Myeloid cells in the cytospin of the spleen from the same animal. (C) Histology of the liver. Perivascular infiltration of the liver by hematopoietic cells (arrows point to myeloid cells). Inset: Part of the same organ at higher magnification. (D) Histology of the small intestine showing infiltration of the lamina propria by myeloid cells (arrow, and magnified in the inset). (E) Histology of a kidney from mouse W207 showing disruption of the kidney architecture and massive invasion by hematopoietic cells. Inset: Cells at higher magnification. (F) Histology of the kidney of animal W194 demonstrating parenchymal tissue necrosis with some invading myeloid cells still present (arrow with solid line); note the presence of crystals in the lower part of the picture (arrow with broken line). (G) Fluid recovered from the perinephric cyst of the same animal containing high numbers of neutrophils. (H) Cytospin of the bone marrow showing predominance of myelopoiesis and suppression of the erythroid and lymphoid compartment in the bone marrow. Original magnification, × 60 (B-C) or × 100 (A;D, inset). Original magnifications, × 25 (D), × 60 (A-C, E-F), and × 100 (G-H, insets). Stains were Wright Giemsa for panels A, G, and H, and hematoxylin and eosin for panels B-F.
Flow cytometry data
. | Bone marrow . | . | Spleen . | . | Lymph node . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Surface marker . | SCLtTA/BCR-ABL (n = 13) . | Controls (n = 11) . | SCLtTA/BCR-ABL (n = 13) . | Controls (n = 11) . | SCLtTA/BCR-ABL, % (n = 8) . | Controls, % (n = 3) . | |||
| Gr-1 | 62 ± 18† | 47 ± 11 | 21 ± 13† | 8 ± 2 | 0-45 | 11-14 | |||
| Gr-1loMac-1 +* | 23 ± 4† | 12 ± 2 | ND | ND | ND | ND | |||
| Gr-1 + Mac-1 +* | 52 ± 15† | 37 ± 8 | 17 ± 11† | 4 ± 2 | 0-25 | 2-7 | |||
| Mac-1 | 57 ± 16† | 42 ± 9 | 23 ± 11† | 13 ± 8 | 6-35 | 7-26 | |||
| Mac-1/B220 | 21 ± 10† | 8 ± 3 | 12 ± 6 | 9 ± 7 | 5-17 | 5-20 | |||
| Mac-1/Sca-1 | 12 ± 5† | 4 ± 2 | 10 ± 3 | 7 ± 5 | 1-15 | 6-43 | |||
| Mac-1/CD34 | 16 ± 6† | 6 ± 2 | 5 ± 3 | 2 ± 3 | 0-4 | 4-10 | |||
| B220 | 35 ± 13 | 30 ± 4 | 58 ± 13 | 54 ± 5 | 44-81 | 26-43 | |||
| Sca-1 | 20 ± 5† | 12 ± 5 | 48 ± 9† | 66 ± 10 | 10-72 | 50-83 | |||
| c-kit | 26 ± 9 | 22 ± 8 | 13 ± 8† | 4 ± 1 | 0-37 | 0-2 | |||
| BP-1/B220 | 15-43‡ | 8 ± 4 | 3-43‡ | 8 ± 4 | 0-91 | 1-4 | |||
| CD43/B220 | 10-37‡ | 8 ± 3 | 7-52‡ | 8 ± 3 | 15-77 | 6-25 | |||
| IgM/B220 | 13 ± 6 | 8 ± 2 | 17 ± 7† | 37 ± 4 | 7-31 | 9-26 | |||
| CD41 | 21 ± 10† | 11 ± 2 | 23 ± 7† | 14 ± 5 | 2-23 | 5-24 | |||
| Ter119 | 14 ± 7† | 22 ± 10 | 13 ± 9† | 5 ± 2 | 0-11 | 6-16 | |||
. | Bone marrow . | . | Spleen . | . | Lymph node . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Surface marker . | SCLtTA/BCR-ABL (n = 13) . | Controls (n = 11) . | SCLtTA/BCR-ABL (n = 13) . | Controls (n = 11) . | SCLtTA/BCR-ABL, % (n = 8) . | Controls, % (n = 3) . | |||
| Gr-1 | 62 ± 18† | 47 ± 11 | 21 ± 13† | 8 ± 2 | 0-45 | 11-14 | |||
| Gr-1loMac-1 +* | 23 ± 4† | 12 ± 2 | ND | ND | ND | ND | |||
| Gr-1 + Mac-1 +* | 52 ± 15† | 37 ± 8 | 17 ± 11† | 4 ± 2 | 0-25 | 2-7 | |||
| Mac-1 | 57 ± 16† | 42 ± 9 | 23 ± 11† | 13 ± 8 | 6-35 | 7-26 | |||
| Mac-1/B220 | 21 ± 10† | 8 ± 3 | 12 ± 6 | 9 ± 7 | 5-17 | 5-20 | |||
| Mac-1/Sca-1 | 12 ± 5† | 4 ± 2 | 10 ± 3 | 7 ± 5 | 1-15 | 6-43 | |||
| Mac-1/CD34 | 16 ± 6† | 6 ± 2 | 5 ± 3 | 2 ± 3 | 0-4 | 4-10 | |||
| B220 | 35 ± 13 | 30 ± 4 | 58 ± 13 | 54 ± 5 | 44-81 | 26-43 | |||
| Sca-1 | 20 ± 5† | 12 ± 5 | 48 ± 9† | 66 ± 10 | 10-72 | 50-83 | |||
| c-kit | 26 ± 9 | 22 ± 8 | 13 ± 8† | 4 ± 1 | 0-37 | 0-2 | |||
| BP-1/B220 | 15-43‡ | 8 ± 4 | 3-43‡ | 8 ± 4 | 0-91 | 1-4 | |||
| CD43/B220 | 10-37‡ | 8 ± 3 | 7-52‡ | 8 ± 3 | 15-77 | 6-25 | |||
| IgM/B220 | 13 ± 6 | 8 ± 2 | 17 ± 7† | 37 ± 4 | 7-31 | 9-26 | |||
| CD41 | 21 ± 10† | 11 ± 2 | 23 ± 7† | 14 ± 5 | 2-23 | 5-24 | |||
| Ter119 | 14 ± 7† | 22 ± 10 | 13 ± 9† | 5 ± 2 | 0-11 | 6-16 | |||
Bone marrow and spleen values are mean ± SD. ND indicates not determined.
Gr-1loMac-1 + cells were calculated as the fraction of the cells in gate R1 among the combined fractions of cells in R1 and R2, as shown in Figure 5B. Gr-1 + Mac-1 + cells represent the total number of Gr-1-and Mac-1-costaining cells.
P < .05 compared with controls (unpaired t test). Controls represent BCR-ABL (n = 8) or SCLtTA (n = 1) single-transgenic or wild-type (n = 2) mice.
Percentage range in mice with lymphoid phenotype.
BCR-ABL RNA and protein are expressed in hematopoietic tissues of diseased mice. (A) Northern blot analysis of RNA isolated from organs, as indicated above the blot from 5 different SCLtTA/BCR-ABL mice (W107, W109, W118, W110, W114, as described in Table 1). RNA from the bone marrow of a SCLtTA single-transgenic (-) mouse and from the affected meninges of an MMTV tTA-BCR-ABL8 double-transgenic (+) mouse served as negative and positive controls, respectively. The lower panel shows 18S and 28S RNA as loading controls. (B) Western blot showing BCR-ABL and c-abl protein expression in the spleens of 4 SCLtTA/BCR-ABL and 5 control mice. SB, B, and S designate SCLtTA/BCR-ABL double-transgenic (dTG), BCR-ABL single-transgenic, and SCLtTA single-transgenic mice, respectively. (C) Western blot of cells from the small intestine (sm.intest.), lung, and bone marrow (bm) of SCLtTA/BCR-ABL double-transgenic (SB), BCR-ABL single-transgenic (B), SCLtTA single-transgenic (S), and wild-type (wt) mice.
BCR-ABL RNA and protein are expressed in hematopoietic tissues of diseased mice. (A) Northern blot analysis of RNA isolated from organs, as indicated above the blot from 5 different SCLtTA/BCR-ABL mice (W107, W109, W118, W110, W114, as described in Table 1). RNA from the bone marrow of a SCLtTA single-transgenic (-) mouse and from the affected meninges of an MMTV tTA-BCR-ABL8 double-transgenic (+) mouse served as negative and positive controls, respectively. The lower panel shows 18S and 28S RNA as loading controls. (B) Western blot showing BCR-ABL and c-abl protein expression in the spleens of 4 SCLtTA/BCR-ABL and 5 control mice. SB, B, and S designate SCLtTA/BCR-ABL double-transgenic (dTG), BCR-ABL single-transgenic, and SCLtTA single-transgenic mice, respectively. (C) Western blot of cells from the small intestine (sm.intest.), lung, and bone marrow (bm) of SCLtTA/BCR-ABL double-transgenic (SB), BCR-ABL single-transgenic (B), SCLtTA single-transgenic (S), and wild-type (wt) mice.
Expansion of the myeloid compartment in the bone marrow
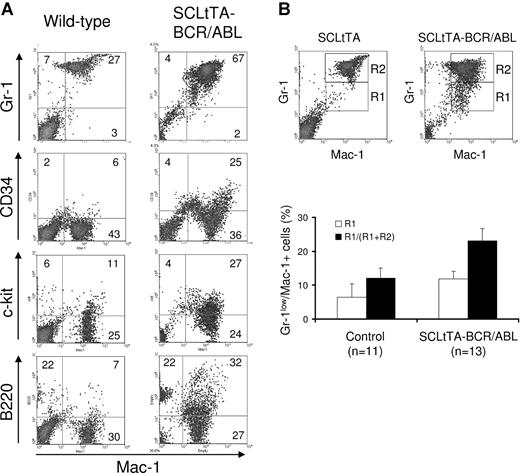
Flow cytometry of bone marrow cells demonstrated significantly increased numbers of cells staining positively for the myeloid markers Gr-1 and Mac-1, comprising up to 97% of the cells in the bone marrow (Figure 5A; Table 2), and this increase was confirmed by cytospin preparations of the bone marrow of diseased animals (Figure 3H). Histologic examination and immunohistochemical analysis for myeloperoxidase expression demonstrated hypercellularity and an increased ratio of myeloid and erythroid cells dominated by granulocytic forms (not shown). In addition, a highly significant increase in the number of immature myeloid cells was found, as characterized by Gr-1lo/Mac-1+ staining (P < .0001) (Figure 5B).40 Consistent with expression of BCR-ABL in progenitor cells, we observed an increase of cells that stained positively for Sca-1 and Mac-1 (P = .002) and for CD34 and Mac-1 (P = .001) (Figure 5A; Table 2). Cytospin analysis showed a predominance of myeloid cells (more than 90%) but no significant increase in myeloid blasts. Furthermore, an increased number of cells staining positively for markers of the lymphoid and the myeloid lineage (B220 and Mac-1) was found in the bone marrow of diseased mice (P < .001). Because the SCL gene is also expressed in megakaryocytes, we performed staining for CD41.41 Consistent with this expression pattern, we found higher numbers of CD41+ cells in the bone marrow (P = .007) and spleen (P = .001) of diseased mice compared with control mice. However, the number of platelets in the peripheral blood was not affected.
Bone marrow of SCLtTA/BCR-ABL mice has an expanded immature myeloid cell population. (A) Representative FACS profile of bone marrow cells from an induced double-transgenic (W202) and a wild-type animal. Cells were costained with PE-labeled Mac-1 and FITC-conjugated Gr-1, CD34, c-kit, or B220 antibodies, as shown. (B) (upper panel) Staining of the myeloid markers Gr-1 and Mac-1 in a single-transgenic SCLtTA mouse and a double-transgenic SCLtTA/BCR-ABL mouse (W185) after the induction of BCR-ABL following the withdrawal of tetracycline. R2 represents Gr-1+/Mac-1+ cells (neutrophils), and R1 represents Gr-1lo/Mac-1+ cells (more immature myeloid cells). (lower panel) Percentage of Gr-1lo/Mac-1+ cells as absolute values (R1) or as the fraction of these cells among all Gr-1+/Mac-1+- staining cells (R1/R1+R2). Both percentages proved to be significantly higher in double-transgenic mice compared with BCR-ABL (n = 8), SCLtTA (n = 1) single-transgenic, or wild-type animals (n = 2), which represented “control” animals shown in the figure. Values are shown as mean ± SD.
Bone marrow of SCLtTA/BCR-ABL mice has an expanded immature myeloid cell population. (A) Representative FACS profile of bone marrow cells from an induced double-transgenic (W202) and a wild-type animal. Cells were costained with PE-labeled Mac-1 and FITC-conjugated Gr-1, CD34, c-kit, or B220 antibodies, as shown. (B) (upper panel) Staining of the myeloid markers Gr-1 and Mac-1 in a single-transgenic SCLtTA mouse and a double-transgenic SCLtTA/BCR-ABL mouse (W185) after the induction of BCR-ABL following the withdrawal of tetracycline. R2 represents Gr-1+/Mac-1+ cells (neutrophils), and R1 represents Gr-1lo/Mac-1+ cells (more immature myeloid cells). (lower panel) Percentage of Gr-1lo/Mac-1+ cells as absolute values (R1) or as the fraction of these cells among all Gr-1+/Mac-1+- staining cells (R1/R1+R2). Both percentages proved to be significantly higher in double-transgenic mice compared with BCR-ABL (n = 8), SCLtTA (n = 1) single-transgenic, or wild-type animals (n = 2), which represented “control” animals shown in the figure. Values are shown as mean ± SD.
BCR-ABL increases the number of progenitor cells in the spleen
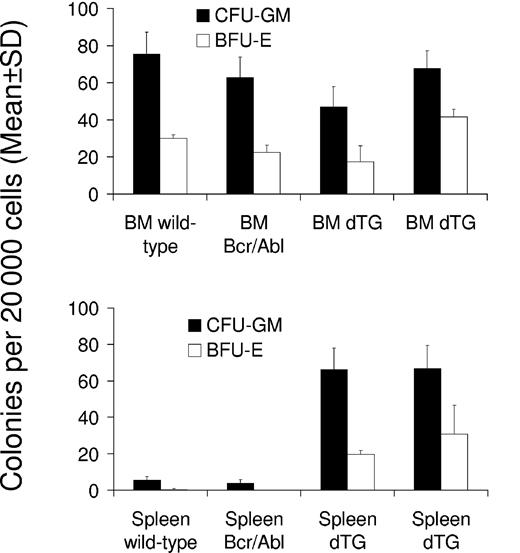
To test whether BCR-ABL expression increased the number of progenitor cells in the bone marrow and spleen, we performed CFU assays with cells isolated from these organs on days 30 and 42 after the induction of 2 double-transgenic mice (W202 and W185, respectively) and compared findings with those of control mice. No significant effect was seen on the number of CFU-GMs and BFU-Es within the bone marrow of induced mice (Figure 6). However, both types of colonies (CFU-GMs and BFU-Es) were significantly increased in the spleens of SCLtTA/BCR-ABL mice (P < .05 vs control mice) (Figure 6). The numbers of CFU-GEMMs were not significantly different in bone marrow or spleen of double-transgenic and control animals (data not shown).
SCLtTA/BCR-ABL mice have increased clonogenic progenitor cells in the spleen but not in the bone marrow. Numbers of CFU-GM (▪) and BFU-E (□) (mean ± SD) were assessed per 20 000 cells extracted from the bone marrow (BM) (upper panel) or spleen (lower panel) on days 42 and 30 after induction of 2 induced double-transgenic mice (dTG) (W185 and W202, respectively) or 2 control mice (wild-type or BCR-ABL single-transgenic, as indicated). Differences of CFU-GM and BFU-E between SCLtTA/BCR-ABL and control mice were statistically significant (P < .05).
SCLtTA/BCR-ABL mice have increased clonogenic progenitor cells in the spleen but not in the bone marrow. Numbers of CFU-GM (▪) and BFU-E (□) (mean ± SD) were assessed per 20 000 cells extracted from the bone marrow (BM) (upper panel) or spleen (lower panel) on days 42 and 30 after induction of 2 induced double-transgenic mice (dTG) (W185 and W202, respectively) or 2 control mice (wild-type or BCR-ABL single-transgenic, as indicated). Differences of CFU-GM and BFU-E between SCLtTA/BCR-ABL and control mice were statistically significant (P < .05).
Expansion of the hematopoietic stem cell compartment in the bone marrow
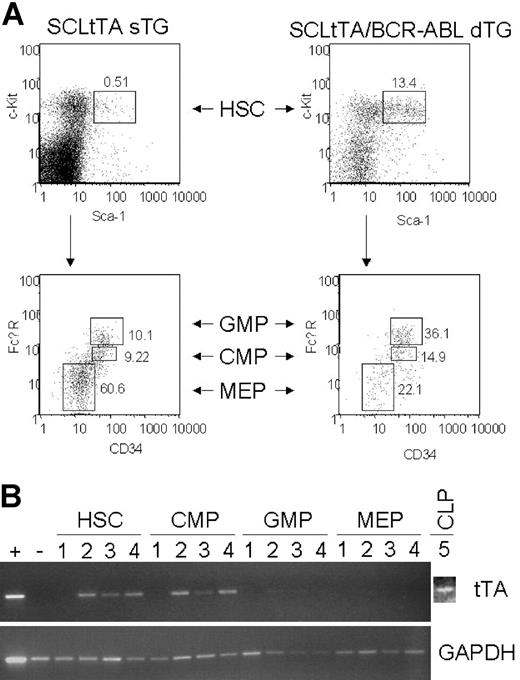
Multicolor-FACS sorting of pooled bone marrow-derived lineage-negative cells from 6 induced double-transgenic and 6 control (3 BCR-ABL, 2 SCLtTA, 1 wild-type) mice on day 12 after induction demonstrated a 7-fold increase of the percentage of HSCs (data not shown), and this compartment was further expanded (26-fold) after 21 days in a second experiment (Figure 7A). The percentage of granulocyte-macrophage progenitors (GMPs) was increased 3-fold at both time points. Conversely, the percentage of MEP was decreased 3-fold at both time points. Common myeloid progenitors (CMPs) were decreased 2-fold after 12 days and increased 1.5-fold after 21 days (Figure 7A). tTA mRNA expression was readily detected by RT-PCR in HSCs, CMPs, and CLPs but not in GMPs or MEPs (Figure 7B). However, Southern blotting of the PCR products revealed low-level tTA expression in GMPs and MEPs (not shown). GAPDH mRNA served as a positive control and was expressed in all progenitor populations (Figure 7B).
Expansion of hematopoietic stem and progenitor cells in double-transgenic animals. (A) Bone marrow-derived hematopoietic stem cells (HSC), common myeloid progenitors (CMP), granulocyte-macrophage progenitors (GMP), and megakaryocyte-erythrocyte progenitors (MEP) were analyzed after lineage depletion using multicolor FACS, as described in “Materials and methods.” Left panel and right panel show a representative analysis for an SCLtTA single-transgenic and an SCLtTA/BCR-ABL double-transgenic mouse, respectively; both mice were killed after 21 days of tetracycline removal. (upper panel) Percentage of Lin-c-kit+Sca-1+ HSC. (lower panel) Percentage of Lin-c-kit+Sca-1- GMP, CMP, and MEP populations in the bone marrow. (B) tTA mRNA expression in multicolor FACS-sorted hematopoietic stem cells (HSC), common myeloid progenitors (CMP), common lymphoid progenitors (CLP), granulocyte-macrophage progenitors (GMP), and megakaryocyte-erythrocyte progenitors (MEP). DNase-treated mRNA was reverse-transcribed into cDNA, as detailed in “Materials and methods,” and was subjected to 35 cycles of RT-PCR using primers specific for tTA or murine GAPDH. Sample labels indicate the following: 1, Tet-O-cre mouse; 2, SCLtTA single-transgenic mouse; 3 to 5, SCLtTA/BCR-ABL double-transgenic mice. The lymph node of an SCLtTA/BCR-ABL double-transgenic mouse (W107) served as a positive control, and the spleen of an MMTV/BCR-ABL double-transgenic mouse (W202) was used as a negative control.
Expansion of hematopoietic stem and progenitor cells in double-transgenic animals. (A) Bone marrow-derived hematopoietic stem cells (HSC), common myeloid progenitors (CMP), granulocyte-macrophage progenitors (GMP), and megakaryocyte-erythrocyte progenitors (MEP) were analyzed after lineage depletion using multicolor FACS, as described in “Materials and methods.” Left panel and right panel show a representative analysis for an SCLtTA single-transgenic and an SCLtTA/BCR-ABL double-transgenic mouse, respectively; both mice were killed after 21 days of tetracycline removal. (upper panel) Percentage of Lin-c-kit+Sca-1+ HSC. (lower panel) Percentage of Lin-c-kit+Sca-1- GMP, CMP, and MEP populations in the bone marrow. (B) tTA mRNA expression in multicolor FACS-sorted hematopoietic stem cells (HSC), common myeloid progenitors (CMP), common lymphoid progenitors (CLP), granulocyte-macrophage progenitors (GMP), and megakaryocyte-erythrocyte progenitors (MEP). DNase-treated mRNA was reverse-transcribed into cDNA, as detailed in “Materials and methods,” and was subjected to 35 cycles of RT-PCR using primers specific for tTA or murine GAPDH. Sample labels indicate the following: 1, Tet-O-cre mouse; 2, SCLtTA single-transgenic mouse; 3 to 5, SCLtTA/BCR-ABL double-transgenic mice. The lymph node of an SCLtTA/BCR-ABL double-transgenic mouse (W107) served as a positive control, and the spleen of an MMTV/BCR-ABL double-transgenic mouse (W202) was used as a negative control.
Transplantation into NOD/SCID mice
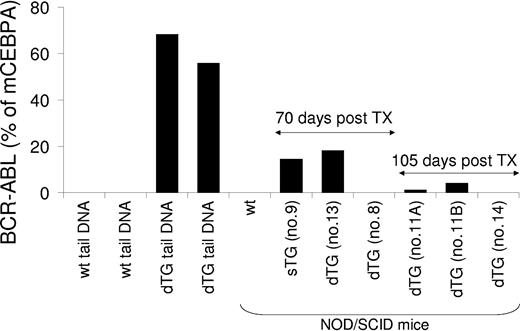
To assess whether myeloproliferative and lymphoid diseases could be treated through transplantation, bone marrow (2 × 106) and purified HSCs (up to 10 000 per transplant) from mice at different stages of disease were transplanted in multiple experiments into sublethally irradiated NOD/SCID mice. We chose NOD/SCID mice as recipients to circumvent possible transplant rejections because the SCLtTA/BCR-ABL mice are of mixed-strain background (BDF for SCLtTA, and FVBN for BCR-ABL). Recipient mice were kept under sterile conditions and were followed up for as long as 5 months. During this time, none of the recipients developed any signs of disease. However, the level of engraftment of BCR-ABL-positive donor cells in the bone marrow of the recipients decreased markedly during 105 days of observation, suggesting failure of long-term reconstitution under the present conditions using NOD/SCID mice (Figure 8). In support of this, recipient mice that had received purified HSCs or lymphoblastic cells showed no evidence of engraftment (Figure 8).
Assessment of engraftment of BCR-ABL-positive cells transplanted into NOD/SCID recipient mice. Twelve days after tetracycline removal from the drinking water, cells were isolated from the bone marrow of a single-transgenic (sTG) or a double-transgenic (dTG) BCR-ABL-positive donor mouse and were transplanted into sublethally (300 cGy) irradiated NOD/SCID mice (recipient 9 for sTG; recipients 11A, 11B, 13 for dTG). At this time point, the dTG donor mouse showed neutrophilia, anemia, splenomegaly, and granulocytic hyperplasia of the bone marrow and spleen. In separate experiments, bone marrow-derived FACS-sorted Lin-c-kit+Sca-1+ HSCs from pooled, diseased dTG donor mice were transplanted into a 300 cGy-irradiated NOD/SCID mouse (recipient 8), and lymphoblastic cells from an affected lymph node of a dTG mouse were transplanted into a nonirradiated NOD/SCID mouse (recipient 14). As indicated, 70 or 105 days after transplantation, genomic DNA was isolated from the bone marrow of the recipients. BCR-ABL genomic DNA was amplified by real-time PCR and expressed as the percentage of the amount of amplified murine CCAAT enhancer binding protein alpha (mCEBPA) genomic DNA to quantify the engraftment of BCR-ABL-positive donor cells. Genomic DNA from the bone marrow of a wild-type NOD-SCID mouse that did not undergo transplantation and from the tails of 2 wild-type (wt) and 2 SCLtTA/BCR-ABL (dTG) mice served as negative and positive controls. None of the recipient mice have shown any signs of disease within the 5-month observation period.
Assessment of engraftment of BCR-ABL-positive cells transplanted into NOD/SCID recipient mice. Twelve days after tetracycline removal from the drinking water, cells were isolated from the bone marrow of a single-transgenic (sTG) or a double-transgenic (dTG) BCR-ABL-positive donor mouse and were transplanted into sublethally (300 cGy) irradiated NOD/SCID mice (recipient 9 for sTG; recipients 11A, 11B, 13 for dTG). At this time point, the dTG donor mouse showed neutrophilia, anemia, splenomegaly, and granulocytic hyperplasia of the bone marrow and spleen. In separate experiments, bone marrow-derived FACS-sorted Lin-c-kit+Sca-1+ HSCs from pooled, diseased dTG donor mice were transplanted into a 300 cGy-irradiated NOD/SCID mouse (recipient 8), and lymphoblastic cells from an affected lymph node of a dTG mouse were transplanted into a nonirradiated NOD/SCID mouse (recipient 14). As indicated, 70 or 105 days after transplantation, genomic DNA was isolated from the bone marrow of the recipients. BCR-ABL genomic DNA was amplified by real-time PCR and expressed as the percentage of the amount of amplified murine CCAAT enhancer binding protein alpha (mCEBPA) genomic DNA to quantify the engraftment of BCR-ABL-positive donor cells. Genomic DNA from the bone marrow of a wild-type NOD-SCID mouse that did not undergo transplantation and from the tails of 2 wild-type (wt) and 2 SCLtTA/BCR-ABL (dTG) mice served as negative and positive controls. None of the recipient mice have shown any signs of disease within the 5-month observation period.
Evidence for progression to blast crisis
Although all mice developed neutrophilia and the predominant phenotype was an expansion of the myeloid cell lineage, 31% of the mice progressed to a syndrome similar to lymphoid blast crisis. This secondary disorder was characterized by lymph node enlargement and the appearance of lymphoblasts in the peripheral blood, bone marrow, and spleen (Table 1; Figure 2B). In all cases, lymphoid cells stained uniformly positive for the markers BP-1 and B220 (Figure 9B). Interestingly, the lymphoblasts were CD43+ in only 50% of these mice, and TdT of variable intensity was detected by immunohistochemistry within the tumor cell population (data not shown), suggesting arrest at different maturation stages (pro-B- or pre-B-cell stages, respectively).42 This is in contrast to the consistent pre-B-cell disease that developed in all animals when expression was driven by the long terminal repeat of the MMTV.8
Myeloid and lymphoblastic disease phenotypes are reversible. (A) Six SCLtTA/BCR-ABL double-transgenic mice (▪) and 2 BCR-ABL single-transgenic mice (controls, □) were repeatedly induced (tetracycline removed from the drinking water) and reverted (tetracycline readministered). Shown is the absolute neutrophil count (ANC) in the peripheral blood over the course of 129 days after the first removal of tetracycline. (striped box) Presence or absence of tetracycline (Tet). Where no error bars are visible, SD was less than 1. (B) Reversibility of the lymphoblastic disease in an SCLtTA/BLCR-ABL double-transgenic mouse. This mouse had developed cervical lymphoma on day 28 of induction of BCR-ABL by tetracycline removal from the drinking water, and FACS analysis revealed that 38% of the cells in the peripheral blood were BP-1+B220lo B-lymphoblastic tumor cells (left panel). Twelve days after tetracycline was readministered, there was no clinical evidence of lymphoma, and FACS analysis confirmed that the BP-1+B220lo cells had disappeared from the peripheral blood (right panel).
Myeloid and lymphoblastic disease phenotypes are reversible. (A) Six SCLtTA/BCR-ABL double-transgenic mice (▪) and 2 BCR-ABL single-transgenic mice (controls, □) were repeatedly induced (tetracycline removed from the drinking water) and reverted (tetracycline readministered). Shown is the absolute neutrophil count (ANC) in the peripheral blood over the course of 129 days after the first removal of tetracycline. (striped box) Presence or absence of tetracycline (Tet). Where no error bars are visible, SD was less than 1. (B) Reversibility of the lymphoblastic disease in an SCLtTA/BLCR-ABL double-transgenic mouse. This mouse had developed cervical lymphoma on day 28 of induction of BCR-ABL by tetracycline removal from the drinking water, and FACS analysis revealed that 38% of the cells in the peripheral blood were BP-1+B220lo B-lymphoblastic tumor cells (left panel). Twelve days after tetracycline was readministered, there was no clinical evidence of lymphoma, and FACS analysis confirmed that the BP-1+B220lo cells had disappeared from the peripheral blood (right panel).
Reversion of phenotype in the absence of BCR-ABL expression
We found that continued expression of BCR-ABL remained essential for the maintenance of the myeloid and lymphoid phenotypes. Readministration of tetracycline at various time points after induction led to stable improvement of the overall clinical condition, disappearance of palpable splenomegaly and lymphomas, and reversion of the neutrophilia and leukocytosis to normal values as early as 4 days after reapplication of tetracycline (Figure 9A). Similarly, BP-1 and B220 double-positive lymphoblasts disappeared, as demonstrated by FACS analysis of the peripheral blood (Figure 9B). Subsequent tetracycline withdrawal led to reinduction of neutrophilia and leukocytosis, and this was again reversible by readministration of tetracycline (Figure 9A). Lymphoblastic disease reappeared on the reinduction of BCR-ABL (data not shown). Among 16 mice that experienced reversion, only 1 animal had progressive disease after the readministration of tetracycline. In the absence of tetracycline, this mouse had developed a large tumor that protruded from the right side of the groin and that did not regress after the reapplication of tetracycline, and the animal was killed in moribund condition on day 42 after induction. Autopsy showed large amounts of ascites and a tumor that had completely infiltrated the retroperitoneum and was fixed to the abdominal wall. A portion of the tumor weighing 923 mg was removed, and histologic analysis revealed a lymphoblastic lymphoma with extensive areas of necrosis. Lymphoblasts expressed BP-1 and B220 by flow cytometry and TdT by immunohistochemistry (not shown). Tumor cells were also found in the ascitic fluid, bone marrow, and spleen. Western blotting demonstrated that the tumor cells had remained BCR-ABL positive in the presence of tetracycline. This was the only animal that had progressive disease after the readministration of tetracycline. In all other animals, the expression of BCR-ABL was tightly regulated, and none of them developed any signs of myeloproliferation or overt disease while on tetracycline.
Discussion
In this report, we describe the generation of a tet-off inducible CML murine model. After the induction of BCR-ABL, these mice developed chronic neutrophilia and leukocytosis (Figure 2), splenomegaly, extramedullary hematopoiesis, and invasion of nonhematopoietic organs by myeloid cells (Figure 3). These features closely resemble the natural course of human CML without treatment. Of note, the development of hydronephrosis and kidney invasion by myeloid cells occurred in animals with an exclusively myeloid but not a lymphoid phenotype and within a closely defined timeframe after induction (days 57-69), which suggests a similar rate of tumor progression in these animals (Figure 3). The described model could be useful for studying basic leukemogenic mechanisms in CML stem cells. In particular, it may be suitable for studying in vivo the initial events immediately after BCR-ABL expression, such as dysfunctions of DNA break repair, as described in a similar inducible model of myc-induced T-cell lymphomas and acute myeloid leukemia (AML),43 as well as the alterations leading to blastic transformation, including translational inhibition of critical transcription factors such as CCAAT enhancer-binding protein alpha44 and additional cytogenetic abnormalities.
One important discrepancy between our model and human CML is the difference in WBC counts. Although patients with CML typically have average WBC counts of 50 000/μL or higher, the WBC counts of the mice remained between 20 000 and 40 000/μL. Strain characteristics may provide a partial explanation because mice used in this study physiologically have a lower percentage of neutrophils and a higher percentage of lymphocytes in the peripheral blood compared with humans. On the other hand, the absolute neutrophil count (ANC) in the peripheral blood was increased 5-fold (Figure 9A), and this may be comparable to increases of ANCs in patients with CML.
Although the BCR-ABL-induced myeloid or lymphoid disease clearly resulted in an ultimately fatal organ dysfunction in many of these mice, we cannot exclude additional effects of BCR-ABL in nonhematopoietic cells in some mice. In mice with lung hemorrhages, the dense infiltrates of myeloid or lymphoblastic cells are likely causally related. No damage to the lung parenchyma was found in other mice that had low levels of infiltration by hematopoietic cells. Because the SCL 3′ enhancer may direct expression of BCR-ABL to endothelial cells, we also searched for signs of damage to the endothelium. However, we could not detect any histologically discernible endothelial alterations in the sections from bone marrow, spleen, lung, or small intestine of induced double-transgenic mice.
In addition to the SCLtTA founder line 19 described herein, the effect of BCR-ABL was also analyzed in a few animals from SCLtTA founder lines 2 and 8. Although there was evidence of a myeloid cell expansion in bone marrow and spleen, however, these animals rapidly developed severe malabsorption associated with enlargement, distention, and hypervascularization of the small bowel, which precluded analysis of the progression of the myeloid disorder (data not shown). Microscopic examination revealed hyperproliferation of intestinal epithelial cells and infiltration by neutrophils. This phenotype is similar to the disease described in a TEL/ABL mouse model, which was characterized by the simultaneous presence of a myeloproliferative syndrome and enlargement, distention, and hypervascularization of the small intestines.45
To assess whether the myeloid and lymphoid diseases were autonomous to the bone marrow of the induced double-transgenic mice or whether expression of BCR-ABL in nonhematopoietic tissues contributed to the phenotype, we used total bone marrow and purified HSCs from double-transgenic mice with either myeloid or both myeloid and lymphoid disease and transplanted these cells into NOD/SCID recipients. NOD/SCID mice were chosen because the strain background of the donor mice was heterogeneous. During 5 months of follow-up, we were unable to detect signs of disease in the recipients. However, when we assessed the level of engraftment by quantitative genomic PCR, we found that the donor cells failed to show significant engraftment and were unable to provide long-term reconstitution of the recipients (Figure 8). In particular, mice that underwent transplantation of purified HSCs at a high dose from double-transgenic mice did not show any engraftment. These results may indicate an alteration of the engraftment properties of double-transgenic HSCs, as has been shown for human CML cells transplanted into NOD/SCID mice,46 or they may indicate residual recipient immunity against the transplanted cells. Thus, it is at present not possible to determine the extent of contribution of extrahematopoietic BCR-ABL expression, and we are backcrossing the mice into a congenic background to address these issues. It will be interesting in future studies to transplant uninduced double-transgenic HSCs into recipients and subsequently to examine the effects of BCR-ABL induction.
An alternative explanation for the lack of signs of disease in the NOD/SCID recipient mice may be that the latency of developing disease has not yet been reached. Long latency times with preceding normal hematopoiesis have been described for other myeloproliferative diseases, especially when donor cells were derived from early stages of disease, as was the case in our studies.37,47 A third possibility is that these diseases, like other experimental myeloproliferative syndromes,48,49 are not transplantable.
We have generated 3 models of inducible BCR-ABL-mediated disease by targeting different hematopoietic subpopulations. Phenotypes of these mice range from pre-B-cell acute lymphoblastic leukemia,8 a myeloproliferative syndrome resembling essential thrombocythemia,9 to the herein-described CML-like disease. These models allow comparative investigations of the molecular mechanisms of BCR-ABL in different target cell populations, efficacy testing of new kinase inhibitors in vivo for different facets of the same disease, and inducible expression of BCR-ABL mutants to study their susceptibility to these drugs. Interestingly, the mean latency of disease lethality among the 3 models was different. Although MMTVtTA/BCR-ABL animals died within 4 weeks of induction of BCR-ABL, the survival span of the SCLtTA/BCR-ABL mice ranged from 4 weeks to 17 weeks for animals that were not humanely killed prematurely, and CD34tTA/BCR-ABL mice lived for 2 to more than 15 months (mean lifespan, more than 7 months). One possible explanation for these different latencies could be that the BCR-ABL expression level differs in the tumor cells, as was shown for the mouse models using MMTV and CD34 regulatory regions.9
Human CML is viewed as a stem cell disorder, based on findings that the BCR-ABL translocation is detectable in various hematopoietic lineages.6 In our SCLtTA/BCR-ABL model, the percentage of Lin-c-kit+Sca-1+ HSCs was increased 26-fold after 3 weeks of induction, consistent with the transformation of HSCs by BCR-ABL. In addition, we found evidence for an expanded pool of myeloid progenitor cells. The percentages of GMP and immature myeloid Gr-1loMac-1+ cells40 in the bone marrow were significantly increased, and the number of colony-forming cells was elevated at least 10-fold in the spleen of double-transgenic animals. Although the percentages of HSCs and GMP were increased in the bone marrow, those for CMPs were largely unchanged and those for MEPs were decreased. Expression of tTA mRNA was readily detected in HSCs and CMP but showed only low-level expression in GMP and MEP. This is consistent with the observation that expression levels of transgenes under the control of the SCL 3′ enhancer decrease with maturation of the target cell (B.G., unpublished results, June 2004). In addition, differences in cell cycle, maturation kinetics, and apoptosis might account for a differential expansion of GMP and CMP. We have repeatedly attempted to quantify BCR-ABL mRNA levels in sorted HSCs and progenitors using real-time PCR but though GAPDH expression was easily detected, BCR-ABL mRNA was below the detection limit (data not shown). A similar approach using cre expression in SCLtTA/Tet-O-cre indicator mice also showed very low expression of cre in these cells. These results are in keeping with published reports showing that BCR-ABL expression was very low in progenitor populations (CMP, GMP) of hMRP8bcl2/BCR-ABL transgenic mice and could only be detected by nested PCR.50 Moreover, disease penetrance and oncogene expression may be inversely correlated, as has been shown for promyelocytic leukemia-retinoic acid receptor α (PML-RARα).51
Cells that express markers for the myeloid (Mac-1) and the lymphoid (B220) lineages were found in the bone marrow of diseased mice, representative of a biphenotypic or a mixed-lineage derivation and consistent with the transformation of a stem cell or an early progenitor cell. This expression pattern is the hallmark feature of mixed-lineage leukemia (MLL)-associated leukemias52 and was recapitulated in a recently described model of MLL-GAS7 leukemia in mice.53 Previously, the generation of a population coexpressing B220 and Mac-1 in vitro from murine bone marrow cells that were either transformed with retroviruses carrying different oncogenes, such as abl,54 or were stimulated with Flt3 ligand55 has been described. Interestingly, when SCLtTA/BCRABL mice were compared with control mice, the percentage of B220+/Mac-1+ cells was significantly increased only in the bone marrow but not in the spleen, suggesting BCR-ABL-mediated transformation of these cells specifically in the bone marrow.
A hallmark of untreated human CML is the development of accelerated-phase CML and eventually myeloid or lymphoid blast crisis after 3 to 5 years of chronic-phase CML. One animal (W110) clearly showed rapid progression of the neutrophilia but died of massive organ invasion before the onset of blast crisis. Thirty-one percent of the SCLtTA/BCR-ABL mice showed evidence of blast crisis with exclusively BP-1+ lymphoblasts. However, in contrast to the uniformly CD43- pre-B-cell leukemia of MMTVtTA/BCRABL animals,8 lymphoblasts in this model were arrested at different stages of maturation (pro-B and pre-B). In addition, the lymphoid phenotype developed rapidly and overshadowed the preexisting CML. These findings may indicate the presence of a second genetic event in these lymphoblasts. Cytogenetic analysis was not performed, but additional cytogenetic changes have been described in 60% to 80% of patients with blast crisis.56 Alternatively, BCR-ABL might have transformed HSCs, CMPs, and CLPs independently. We have shown that the tTA gene was expressed in CLP, and it is tempting to speculate that BCR-ABL may confer self-renewal capacities to CLP cells and turn them into leukemic stem cells, as has been described for CMPs.50 It should be noted that the expression of BCR-ABL in B-lymphoid cells was surprising because SCL is not physiologically expressed in this population.36 However, given that BCR-ABL mRNA and protein were expressed in lymphoblasts isolated from several mice with lymphoid disease and given that tTA was expressed in CLPs, we hypothesize that the SCL 3′ enhancer directs expression to HSCs, CMPs, and CLPs and that a silencer element, which is present in the endogenous SCL gene locus and down-regulates SCL expression in lymphoid cells, may be missing from our transgenic construct. It may be argued that the initial phase of neutrophilia was reactive as a response to BCR-ABL-expressing cells. However, though 100% of the mice showed myeloid cell expansion, only 31% of the mice demonstrated a lymphoblastic phenotype, suggesting that myeloid progenitor cells were involved in the development of the disease.
In summary, our murine model of inducible SCLtTA/BCR-ABL expression recapitulates many of the features of human CML and may, therefore, be helpful in studying basic mechanisms and new therapeutic strategies of this disease.
Prepublished online as Blood First Edition Paper, August 26, 2004; DOI 10.1182/blood-2003-12-4369.
Supported by National Institutes of Health grants DF/HCC P30CA6516 (J.L.K.) and DK48660 (D.G.T.). C.S.H. is a fellow of the Jose Carreras Leukemia Foundation International (fellowship JC 2000). S.K. is a fellow of the Deutsche Forschungsgemeinschaft (research fellowship KO2155/1-1). B.G. is a fellow of the Leukaemia Research Fund.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Maris Fenyus for animal care, Joel Lawitts and the Beth Israel Deaconess Medical Center Trangenic Facility for production of transgenic mice, Maris Handley for cell sorter operation, and Roderick Bronson, Rick Van Etten, Frank Rosenbauer, and members of the Tenen laboratory for expert advice.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal