Abstract
The role of natural killer (NK) cells in multiple myeloma is not fully understood. Here, NK susceptibility of myeloma cells derived from distinct disease stages was evaluated in relation to major histocompatibility complex (MHC) class I, MHC class I chain-related protein A (MICA), MHC class I chain-related protein B (MICB), and UL16 binding protein (ULBP) expression. MHC class I molecules were hardly detectable on bone marrow cells of early-stage myeloma, while late-stage pleural effusion-derived cell lines showed a strong MHC class I expression. Conversely, a high MICA level was found on bone marrow myeloma cells, while it was low or not measurable on pleural effusion myeloma cells. The reciprocal surface expression of these molecules on bone marrow- and pleural effusion-derived cell was confirmed at mRNA levels. While bone marrow-derived myeloma cells were readily recognized by NK cells, pleural effusion-derived lines were resistant. NK protection of pleural effusion cells was MHC class I dependent. Receptor blocking experiments demonstrated that natural cytotoxicity receptor (NCR) and NK receptor member D of the lectinlike receptor family (NKG2D) were the key NK activating receptors for bone marrow-derived myeloma cell recognition. In ex vivo experiments patient's autologous fresh NK cells recognized bone marrow-derived myeloma cells. Our data support the hypothesis that NK cell cytotoxicity could sculpture myeloma and represents an important immune effector mechanism in controlling its intramedullary stages. (Blood. 2005;105:251-258)
Introduction
Natural killer (NK) cells are cytotoxic and cytokine-producing lymphocytes, involved in the immune defense against viral infections and tumors.1 Their homeostasis is regulated by cytokines and membrane associate receptors able to inhibit or activate cellular programs.2 The inhibitory receptors are well characterized and described extensively in several reviews.3-5 Triggering of NK cells depends largely on NK receptor member D of the lectinlike receptor family (NKG2D) and natural cytotoxicity receptors (NCRs): NKp46, NKp44, NKp30.6,7 NCRs are involved in the recognition of several tumor cell lines, although their ligands remain elusive.6 NKG2D recognizes the MHC (major histocompatibility complex) class Ichain-related (MIC) protein A (MICA) and B (MICB); both are nonclassic class I molecules. The UL16-binding proteins (ULBP1-3 or RAET1 proteins; ULBP1-3 in this paper) are the second group of NKG2D ligands in humans. MICs are expressed during virus infection or cell transformation; ULBP expression in fresh tumor cells is essentially unknown; only long-term cultured in vitro cell lines have been looked at so far.8-10
Cytotoxic T lymphocytes (CTLs) and interferons (IFNs) have a key role in tumor progression and tumor “immune-editing process.”11 MHC class I molecule loss is a frequent event in tumor progression and could prevent CTL recognition. However, theoretically, NK cells could recognize MHC class I-defective tumors, according with the “missing self hypothesis.”12
So far, only in mouse models NK cells were demonstrated to destroy in vivo lymphoma and melanoma tumors with reduced MHC class I expression and/or with high levels of activating target structures.13-15 Even though almost 30 years ago human NK cells were discovered for their in vitro antitumor cytotoxicity, we still have little information concerning the regulation of their antitumor activity in vivo or ex vivo.16,17 Therefore, several questions remain to be addressed to understand the antineoplastic potential of human NK lymphocytes:
. HLA class I molecules are reported to be down-regulated during solid tumor progression.18 Is this a general paradigm in tumor disease development? What happens to MHC class I surface expression in hematologic malignancy, where a high number of NK cells are present in the tumor environment during disease progression?
. Which is the role, if any, for tumor-associated MICA, MICB, and ULBP molecules, and for NK-activating receptors (NKG2D and NCR) in the recognition of hematopoietic-derived tumors?
. Can freshly ex vivo human NK cells recognize explanted autologous tumor cells?
Human NK cells recognize healthy B cells and regulate their activation and differentiation.19,20 Other hematopoietic-derived cells can stimulate NK lymphocytes as described for dendritic cells (DCs).21 The B-cell membrane-associated proteins CD40 and CD1 regulate natural killer cell cytotoxicity.22-25 Furthermore, NK lymphocytes are specifically activated after bone marrow graft but not by other tissue transplantations.26 They localize in lymph nodes and spleen, mainly in B-cell follicles and in the marginal zone.27 Blood, spleen, and bone marrow are the anatomic districts where the highest number and activity of NK cells are present.1
Taking together these considerations, hematologic malignancies (B-cell-derived tumors in particular) could be considered an appealing system to investigate the potential role of NK cells in the control of tumor progression.
Multiple myeloma (MM) is a plasma cell-derived tumor. It is characterized by accumulation of plasma cells in the bone marrow (BM); it is often an incurable disease with a median survival of a few years, only recently improved with the wider use of autotransplant procedures. Three progression and localization stages can be envisaged in MM28 : 1) inactive phase, in which the tumor cells are scarcely proliferating intramedullary plasma cells; 2) active phase, in which plasma cells proliferate in BM, with some spilling in the circulating blood; and 3) in some cases an advanced phase, characterized by increase in the plasmablastic compartment, with BM and extramedullary proliferation.
So far, most studies analyzed allogeneic NK recognition of in vitro long-term cultured myeloma lines or fresh myeloma cells,29,30 or autologous natural cytotoxicity mediated by interleukin 2 (IL-2)-activated peripheral blood mononuclear cells (PBMCs) against multiple myeloma targets31 ; one report described myeloma as a NK-resistant tumor.32 Therefore, no general agreement exists on NK susceptibility of MM cells.
The aims of our study were 1) to challenge, in vitro and ex vivo, NK cell recognition of myeloma cells; 2) to dissect the molecular mechanisms governing NK recognition of myeloma cells; and 3) to verify plasma cell immunophenotype expressed in different myeloma localizations and disease stages.
To address these issues, we used a newly established MM cell system composed by relatively short-term in vitro passages of myeloma cell lines derived from different disease localizations: bone marrow (BM), peripheral blood (PB), and pleural effusion (PE) cells.33 So far, only 57 cases of PE have been reported, 4 of them included in this study. In our system, BM and PE myeloma cells were highly related, since they had been generated from the same individuals (KMS12 and KMS21). Indeed, BM and PE belonged to the same clone, since KMS12BM and KMS12PE shared the same karyotypic abnormality, and the KMS21BM and KMS21PE had the same immunoglobulin (Ig) production.33
Patients, materials, and methods
Cell lines
The human myeloma cell lines were established at the Kawasaki Medical School, Okayama, Japan. They were generated from bone marrow (KMS12BM, KMS21BM), pleural effusion (KMS12PE, KMS21PE, KMS11PE, KMS26PE), and peripheral blood (KMS18PB, KMS27PB). KMS12BM and KMS12PE were established from a single patient, as were KMS21BM and KMS21PE. KMS cells were established in vitro as described33,34 ; to avoid in vitro selection, all KMS cell lines were studied within the third/fifth in vitro culture passage. The cells were grown in RPMI 1640 (Invitrogen, Milan, Italy) supplemented with 10% fetal bovine serum (FBS; Invitrogen) at 37°C in a humidified atmosphere with 5% CO2. In the course of the study, KMS21PE was lost accidentally. The cervical carcinoma cell line HeLa (CCL-2; ATCC, Manassas, VA) and the T-B lymphoblastoid hybrid T2 cell line were grown in RPMI 1640 (Invitrogen) supplemented with 5% fetal calf serum (FCS).
Monoclonal antibodies and immunofluorescence procedures
The myeloma cell lines were analyzed by direct or indirect staining and flow cytometry analysis using the following antibodies: anti-HLA class I monoclonal antibodies (mAbs): anti-pan HLA class I clone W6/32HL (IgG2a; Cymbus Biotechnology, Chandlers Ford, England); anti-HLA-G clone G7 (IgG2a; a kind gift of Dr D. Gerarghy, Fred Hutchinson Cancer Research Center, Seattle, WA). To analyze CD38 and CD138 the fluorescein isothiocyanate (FITC) directly conjugated clone HB-7 (IgG1) and phycoerythrin (PE)-conjugated BB-4 (IgG1; Becton Dickinson, Mountain View, CA) were used, respectively. To evaluate the following surface molecules these mAbs were used: anti-CD40 clone B-B20 (IgG1), anti-CD80 clone BB-1 (IgM), anti-CD86 clone BU63 (IgG1), anti-CD9 clone MM 2/57-PE (IgG2b), anti-CD1b clone M-T 101-FITC (IgG1), all available from Cymbus Biotechnology. To analyze MIC expression on tumor cell membrane we used the anti-MICA clone BAM 195 (IgG1),8 anti-MICA-MICB-specific monoclonal antibody 6D4 (IgG1).9 The anti-ULBP1-3 mAbs used were M295 (anti-ULBP1, IgG1), M310 (anti-ULBP2, IgG1), and M550 (anti-ULBP3, IgG1) (Amgen, Seattle, WA).
In the indirect staining, cells were incubated with the appropriate mAb followed by the PE-conjugated goat anti-mouse secondary reagent (Jackson Immuno Research, Baltimore, MD). In all experiments, as first step the cells were incubated with human serum for 15 minutes, and isotype-matched controls were used to set up the negative values. Samples were analyzed by the fluorescence-activated cell sorter FACS VANTAGE (Becton Dickinson). In MHC class I induction experiments, myeloma cells were cultured at cell concentration of 5 × 105/mL for 48 hours in the presence of 500 UI/mL exogenous IFNγ (PeproTech, Rocky Hill, NJ). Then, the cells were stained with either W6-32 or with G7 antibodies plus PE-conjugated secondary antibodies and analyzed by FACS.
Patient recruitment
A cohort of 14 patients with cytologic or histologic diagnosis of multiple myeloma was investigated in this study. The patients were cared at the Hematology Clinic of the University of Naples “Federico II” Medical School. All patients gave informed consent, according to the ethical regulations of our institutions and the Declaration of Helsinki. Only patients with bone marrow CD38+-infiltrating plasma cells greater than 30% entered in this study. Patient's relevant clinical parameters are reported in Table 1.
Patient characteristics
Patient no. . | Age, y/Sex . | Clinical stage . | Monoclonal Ig . | % PC in BM . |
|---|---|---|---|---|
| 1 | 62/M | IIIA | IgG κ | 45 |
| 2 | 59/F | IIIA | IgD λ | 70 |
| 3 | 60/F | IIA | IgG λ | 36 |
| 4 | 76/M | IIIA | IgA κ | 40 |
| 5 | 65/M | IIA | IgA κ | 45 |
| 6 | 72/F | IIA | IgA κ | 40 |
| 7 | 79/F | IIIB | IgA κ | 80 |
| 8 | 65/M | IIIA | IgA κ | 35 |
| 9 | 51/F | IIA | IgG κ | 37 |
| 10 | 63/M | IIIB | IgA κ | 60 |
| 11 | 61/M | IIIA | IgG κ | 34 |
| 12 | 73/F | IIB | IgA κ | 54 |
| 13 | 73/M | IIB | IgG κ | 30 |
| 14 | 72/M | IIIB | IgG λ | 51 |
Patient no. . | Age, y/Sex . | Clinical stage . | Monoclonal Ig . | % PC in BM . |
|---|---|---|---|---|
| 1 | 62/M | IIIA | IgG κ | 45 |
| 2 | 59/F | IIIA | IgD λ | 70 |
| 3 | 60/F | IIA | IgG λ | 36 |
| 4 | 76/M | IIIA | IgA κ | 40 |
| 5 | 65/M | IIA | IgA κ | 45 |
| 6 | 72/F | IIA | IgA κ | 40 |
| 7 | 79/F | IIIB | IgA κ | 80 |
| 8 | 65/M | IIIA | IgA κ | 35 |
| 9 | 51/F | IIA | IgG κ | 37 |
| 10 | 63/M | IIIB | IgA κ | 60 |
| 11 | 61/M | IIIA | IgG κ | 34 |
| 12 | 73/F | IIB | IgA κ | 54 |
| 13 | 73/M | IIB | IgG κ | 30 |
| 14 | 72/M | IIIB | IgG λ | 51 |
PC indicates plasma cells; BM, bone marrow.
Circulating and bone marrow-derived CD38+ plasma cells were detected by direct immunofluorescence and FACS analysis using the anti-CD38 FITC or anti-CD138 PE directly conjugated monoclonal antibodies (clone HB-7 and BB-4; Becton Dickinson) gating on CD38bright or on CD38+/CD138+ cells. The immune phenotype of the myeloma cells was obtained by double staining of either CD38+bright or CD38/CD138+ cells with the pertinent monoclonal antibody.
NK cell and MM cell generation ex vivo
NK cells were obtained from healthy donor buffy coats. Briefly, PBMCs were isolated by Ficoll-Paque (Biochrom, Berlin, Germany) density gradient centrifugation. For the isolation of human peripheral blood NK cells, we used the NK Cell Isolation Kit and VarioMACS for the depletion of non-NK cells (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) according to manufacturer's recommendations; NK cell purity was above 95%. NK cells were suspended in Iscoves modified Dulbecco medium (IMDM) culture medium (Life Technology, Milan, Italy) supplemented with penicillin (100 IU/mL), streptomycin (100 μg/mL), 10% fetal bovine serum; (Invitrogen), and 3% human serum in the presence of 200 U/mL recombinant interleukin 2 (rIL-2; Chiron, Emeryville, CA) and cultured for 2 days to obtain activated polyclonal NK cells.
From each patient, a sample of 20 mL heparinized peripheral blood and 10 mL bone marrow aspirate (in the occasion of diagnostic procedures) was obtained, in sterile condition. Bone marrow myeloma cells were isolated by Ficoll-Paque gradients; once resuspended in culture medium (RPMI 1640) supplemented with penicillin (100 IU/mL), streptomycin (100 μg/mL), 10% fetal bovine serum, NK cells were purified as described above.
For each patient, the range of myeloma cells present in the explanted tumor sample was between 35% and 95% of the bone marrow mononuclear cells. To normalize the number of myeloma cells among the different patients, mononuclear cells were cultured for 48 hours in the presence of human recombinant IL-6, 10 ng/mL (PeproTech). The IL-6 treatment yielded to a specific CD38+ tumor cell proliferation; therefore, in each ex vivo cytotoxicity experiment CD38+ myeloma target cells were always 95% to 100% of the labeled target cells. In some patients, myeloma cells were purified by immunomagnetic approach using the CD138 purification kit (Miltenyi).
Cytotoxicity assay
Cytotoxicity assays were performed using the fluorescent carboxyfluorescein diacetate (c′FDA) NK assay35 ; the results obtained were confirmed using the standard 51Cr-release assay.
In the fluorescent c′FDA NK assays, cytotoxicity was analyzed by flow cytometry using the protocol described elsewhere.35 Briefly, the target cells were labeled with carboxy-fluorescein diacetate (c'FDA; Molecular Probes, Eugene, OR). The incubation was performed in 96-well plates at 37°C in a humidified 5% CO2 incubator. The specific lysis of target cells was calculated as follows: % specific lysis = (CT - TE/CT) × 100 (where CT = mean number of fluorescent target cells in control tubes and TE = mean number of fluorescent cells in target + effectors tubes).
In the standard 51Cr release assay, effector cells were mixed with 5 × 103 target cells. Myeloma cells were labeled with 100 μCi (3.7 MBq) 51Cr sodium chromate (Amersham Biosciences Europe GmbH, Freiburg, Germany) and then washed extensively. In all experiments the effector-to-target (E/T) ratios were 25:1, 12:1, 6:1, or 3:1, except when differently indicated. The percentage of specific lysis was calculated as follows: % specific lysis = (cpm experimental - cpm spontaneous release)/(cpm maximum release - cpm spontaneous release) × 100.
The spontaneous release was determined by incubation of the labeled target cells alone in RPMI 1640 medium supplemented with 10% FBS. Maximum release was obtained by target cells incubation with 0.5% Triton X-100. In all experiments, spontaneous release was less than 20% of the total uptake.
In the experiments in which mAbs were used to prevent the interactions between HLA class I, or MICA molecules with their receptor counterpart, the target cells were preincubated for 30 minutes at room temperature with A6-136 (IgM), or BAM 195. As control, we used the anti-CD57 clone TIB200 (IgM) (ATCC). The mAbs were present in the culture medium during the whole assay period. For NK receptor blocking experiments, we incubated NK cells for 15 minutes at room temperature with various mAbs before addition of target cells. For anti-NKG2D, the clone 1D11 (IgG1; Becton Dickinson) was used. For masking NCR, the following mAb of IgM isotype were used: anti-NKp46 clone KL247, anti-NKp44 clone KS38, both kindly provided by S. Parolini (University of Brescia, Brescia, Italy) and anti-NKp30 clone F252.7
We also used a control mAb, T345, recognizing a lymphocyte membrane-associated vaccinia virus growth factor (VGF) glycoprotein (kind gift of Soldano Ferrone, Rosewell Cancer Park, Buffalo, NY). All mAbs were used at the final concentration of 10 μg/mL.
Statistic analysis
Results of experimental points obtained from multiple experiments were reported as mean ± SEM. Significance levels were determined by Student t test analysis; P value of .05 or less was considered significant.
RNA preparation, RT-PCR, and Northern blotting procedure
For RNA preparation, 1 × 106 cells were lysed in 800 μL TRIzol reagent (Invitrogen) followed by phenol/chloroform extraction and ethanol precipitation. The samples obtained were quantified by absorbance at 260 nm; RNA integrity was assessed by electrophoresis on 1.5% agarose gel. cDNAs were synthesized from 1 μg total cellular RNA using 0.5 U Superscript II reverse transcriptase (Invitrogen) and 2.5 μM random hexamers (Promega, Milan, Italy) at 42°C for 60 minutes. cDNA aliquots were amplified in duplicate in a Robocycler (Stratagene, Italy) in a total volume of polymerase chain reaction (PCR) buffer of 50 μL, in the presence of 1 U Taq DNA Polymerase (Eppendorf, Milan, Italy), 200 μM deoxyribonucleoside triphosphates (dNTPs; Promega), and 20 pmol primers. To exclude amplification of genomic DNA contaminating the samples, experiments were also performed using RNA as substrate for reverse transcriptase (RT)-PCR assay. Oligonucleotide sequences were 1) glyceraldehyde-3-phosphate dehydrogenase (GAPDH) fwd,0 5′-CACCATCTTCCAGGAGCGAG-3′, and rev, 5′-TCACGCCACAGTTTCCCGGA-3′; 2) MHC class I (HLA-A, -B, -C) fwd, 5′-CCTTGTGTGGGACTGAGAGG-3′, and rev, 5′-CAGAGATGGAGACACCTCAGC-3′ (UniSTS, RH80274); 3) MICA fwd, 5′-AGCGTAGTCCTGAGGAGAACAGTGCC-3′, and rev, 5′-CCTGTGGTCACTCGTCCCAACTGGG-3′; and 4) transporter associated with antigen processing (TAP) fwd, 5′-CGCCTCACTGACTGGATTCT-3′, and rev, 5′-TGGCTGGACTTTGCCAGAGA-3′.
For the Northern blot procedure, 10 μg total RNA was size-fractionated by 1.5% agarose/formaldehyde electrophoresis and transferred to a Nytran membrane (Sigma, Milan, Italy). The probes were labeled with α-32P deoxycytidine triphosphate (dCTP) with a random primer DNA-labeling Kit (Amersham). The filters were prehybridized in Church buffer36 for 1 hour and then hybridized overnight at 65°C with heat-denaturated double-stranded DNA probes (2 × 105 cpm/mL).37
Results
NK recognition of BM- and PE-derived myeloma cell lines
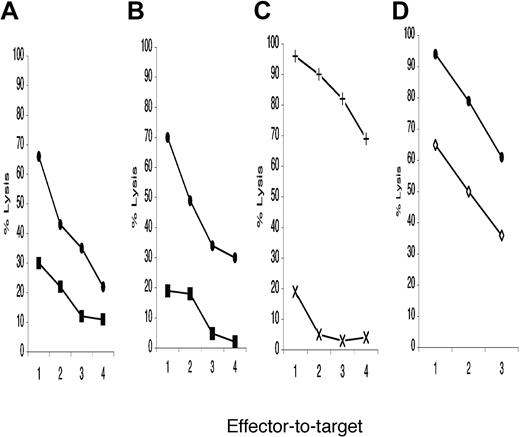
To evaluate NK recognition of MM cells derived from different sites, cytotoxicity tests were performed using as targets the lines KMS12BM, KMS21BM, and the related PE. BM-derived myeloma cells were readily eliminated, while the related PE-derived cells were spared. Figure 1A-B depicts 1 representative of 5 experiments carried out. PB-derived (KMS27PB) and PE-derived (KMS11PE) myeloma cell lines were exposed to NK cell lysis (Figure 1C). The KMS11PE line was resistant to NK cytotoxicity, while KMS27PB-derived myeloma cells were readily eliminated. When KMS26PE cells were compared with KMS12BM, the former were more resistant (Figure 1D), although they were recognized better than the other 3 PE myeloma lines. The NK susceptibility differences between BM and PE myeloma lines were significant (P < .01).
NK recognition of bone marrow, peripheral blood, and pleural effusion-derived myeloma cell line by purified allogeneic NK cells activated with IL-2. Close circles represent the percentage of specific lysis of distinct myeloma cell targets: (A and D) KMS12BM, (B) KMS21BM, (C) with cross KMS27PB. Close squares indicate the lysis levels of pleural effusion-derived myeloma target cells: (A) KMS12PE, (B) KMS21PE. In panel C and D with X and open diamond the percentage of specific lysis of KMS11PE and KMS26PE, respectively. The ratios used were 25:1, 12:1, 6:1, 3:1 indicated in abscissa as 1, 2, 3, and 4, respectively. Each panel described 1 representative experiment of at least 3 performed. NK cells preferentially killed BM myeloma lines, while PE-derived cells were less NK susceptible.
NK recognition of bone marrow, peripheral blood, and pleural effusion-derived myeloma cell line by purified allogeneic NK cells activated with IL-2. Close circles represent the percentage of specific lysis of distinct myeloma cell targets: (A and D) KMS12BM, (B) KMS21BM, (C) with cross KMS27PB. Close squares indicate the lysis levels of pleural effusion-derived myeloma target cells: (A) KMS12PE, (B) KMS21PE. In panel C and D with X and open diamond the percentage of specific lysis of KMS11PE and KMS26PE, respectively. The ratios used were 25:1, 12:1, 6:1, 3:1 indicated in abscissa as 1, 2, 3, and 4, respectively. Each panel described 1 representative experiment of at least 3 performed. NK cells preferentially killed BM myeloma lines, while PE-derived cells were less NK susceptible.
MHC class I, MICA, and ULBP1-3 expression on myeloma cell lines
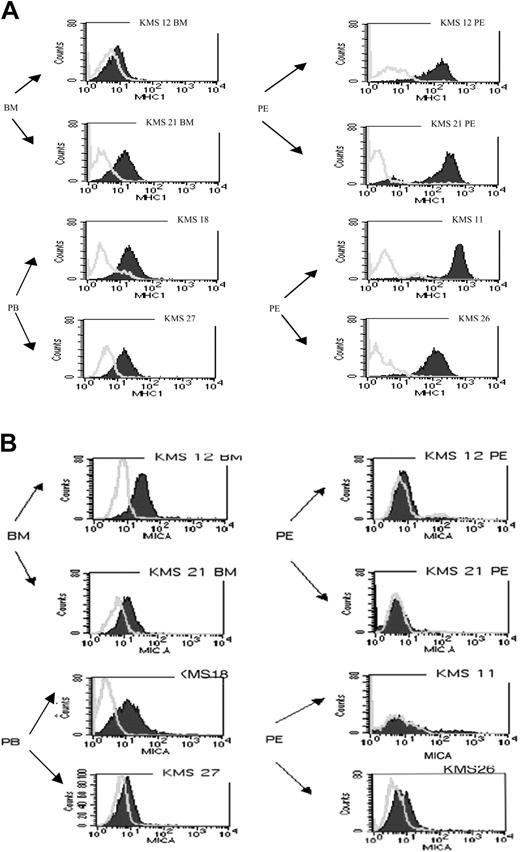
To test the mechanism behind the different NK susceptibility of the myeloma cell lines the MHC class I, MICA, MICB, and ULBP1-3 expression was evaluated on the myeloma cell surface. Two BM (KMS12BM, KMS21BM), 2 PB (KMS18PB, KMS27PB), and 4 PE (KMS12PE, KMS21PE, KMS11PE, KMS26PE) derived cell lines were stained with pan-anti-MHC class I (Figure 2A), anti-MICA (Figure 2B), and anti-MICA/B (data not shown). ULBP expression was measured on the 2 BM and 3 PE myeloma cell lines (data not shown). MHC class I surface expression was higher on the PE cell lines compared with the BM and PB cell lines (P ≤ .05 in 5 independent experiments) (Figure 2A). It is worth noting that in some experiments BM and PE pairs were both derived from the same patient (KMS12 and KMS21). MICA expression (Figure 2B) was readily detectable on both BM cell lines and on the KMS18 PB cell line, while the other PB cell line (KMS27) had lower levels. The anti-MICA and the anti-MICA/B antibody did not stain 3 of 4 PE cell lines (Figure 2B and data not shown). The difference in MICA levels comparing BM with PE cell lines was significant (P ≤ .05) except for the difference between KMS21BM/KMS26PE (P = .06). Thus, we found that MHC class I expression was low in BM- and high in PE-derived cell lines; conversely, MICA expression was high in BM cell lines and low in PE cell lines. ULBPs analysis showed no clear correlation among the various cell lines (data not shown).
Reciprocal surface expression of MHC class I and MICA between bone marrow- and pleural effusion-derived myeloma cell lines. (A) BM, PB, and PE myeloma cell lines were stained with isotypic mAb control (open curves) or with anti-MHC class I antibody (filled area). A clear overexpression of MHC class I molecule is detected in PE cell lines (1 of 5 representative experiments). (B) Staining obtained using BAM195 anti-MICA antibodies: MICA expression is detectable in BM cell lines but not in PE cell lines. Similar results were obtained using the anti-MICA-MICB antibody 6D4.
Reciprocal surface expression of MHC class I and MICA between bone marrow- and pleural effusion-derived myeloma cell lines. (A) BM, PB, and PE myeloma cell lines were stained with isotypic mAb control (open curves) or with anti-MHC class I antibody (filled area). A clear overexpression of MHC class I molecule is detected in PE cell lines (1 of 5 representative experiments). (B) Staining obtained using BAM195 anti-MICA antibodies: MICA expression is detectable in BM cell lines but not in PE cell lines. Similar results were obtained using the anti-MICA-MICB antibody 6D4.
Molecular analysis of MHC class I and MICA expression in myeloma cell lines
MHC class I up-regulation in myeloma cells from different sites and during disease progression is a novel finding. Given the limited knowledge available on the role of MICA gene expression on tumor progression, we decided to investigate further the molecular bases of the different immunophenotype of BM- and PE-derived cell lines.
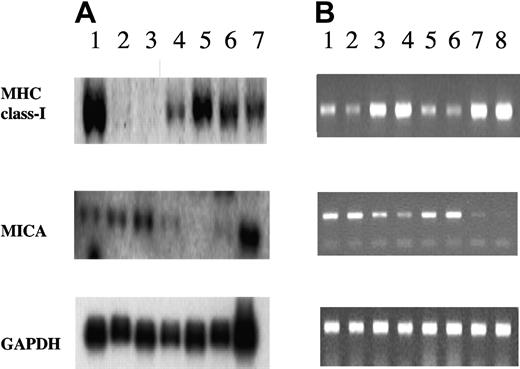
The differential expression of MHC class I and MICA molecules between BM- and PE-derived myeloma cell membrane was investigated at transcriptional level by Northern blot and RT-PCR (Figure 3). By Northern analysis, we detected significant differences in the RNA amount for MHC class I heavy chain between the 2 BM-derived myeloma lines (KMS12BM, KMS21BM) and the PE-derived lines (KMS12PE, KMS26PE, KMS11PE) (Figure 3A). Unfortunately, in this experimental set we were unable to include the KMS21PE cells, which had been lost during the study. TAP1 transcripts were equally represented in all myeloma lines (data not shown). Thus, PE myeloma cells showed a more active transcription of MHC class I heavy chain genes, when compared with bone marrow lines. Identical results were obtained analyzing the MHC class I heavy-chain transcripts by RT-PCR; in this experimental setting, RNA extracted from KMS21PE could be included, confirming the intense and specific MHC class I heavy-chain transcriptional activity in all PE lines tested (Figure 3B).
Northern blot and RT-PCR analysis of MHC-I and MICA expression in myeloma cell lines. (A) Northern blot analysis. Total RNA (10 μg) from T2 (lane 1), KMS21BM (lane 2), KMS12BM (lane 3), KMS12PE (lane 4), KMS26PE (lane 5), KMS11PE (lane 6), and HeLa (lane 7) cell lines was hybridized with MHC-I and MICA cDNA fragments. For the evaluation of gel load and transfer, the same filters were rehybridized with a fragment of GAPDH cDNA. Reciprocal amounts of MHC class I heavy chains and MICA transcripts levels were observed between BM and PE myeloma cell lines. (B) RT-PCR was performed from RNA preparations in duplicate, using specific primers for MHC class I, and MICA (40 cycles). cDNAs were obtained from: KMS12BM (lanes 1-2), KMS12PE (lanes 3-4), KMS21BM (lanes 5-6), and KMS21PE (lanes 7-8). The products were analyzed by agarose gel and were normalized using the levels of GAPDH (20 cycles) as an internal control. The RT-PCR analysis confirmed the inverse correlation between the MHC class I and MICA transcript levels found in BM and PE cell lines.
Northern blot and RT-PCR analysis of MHC-I and MICA expression in myeloma cell lines. (A) Northern blot analysis. Total RNA (10 μg) from T2 (lane 1), KMS21BM (lane 2), KMS12BM (lane 3), KMS12PE (lane 4), KMS26PE (lane 5), KMS11PE (lane 6), and HeLa (lane 7) cell lines was hybridized with MHC-I and MICA cDNA fragments. For the evaluation of gel load and transfer, the same filters were rehybridized with a fragment of GAPDH cDNA. Reciprocal amounts of MHC class I heavy chains and MICA transcripts levels were observed between BM and PE myeloma cell lines. (B) RT-PCR was performed from RNA preparations in duplicate, using specific primers for MHC class I, and MICA (40 cycles). cDNAs were obtained from: KMS12BM (lanes 1-2), KMS12PE (lanes 3-4), KMS21BM (lanes 5-6), and KMS21PE (lanes 7-8). The products were analyzed by agarose gel and were normalized using the levels of GAPDH (20 cycles) as an internal control. The RT-PCR analysis confirmed the inverse correlation between the MHC class I and MICA transcript levels found in BM and PE cell lines.
In the RNA extracted from KMS12BM and KMS21BM, the MICA transcripts were found increased when compared with the related PE cell lines (Figure 3A-B).
To further understand the nature of the MHC class I transcriptional down-regulation in BM myeloma lines, we investigated whether IFNγ could increase it. IFNγ pretreatment of BM myeloma cells restored MHC class I surface levels (data not shown). These data strongly suggest that the difference in MHC class I expression between BM- and PE-derived myeloma lines is due to a different transcriptional regulation.
Involvement of HLA-class I, NKG2D, and NCR in myeloma cell targeting by fresh NK cells
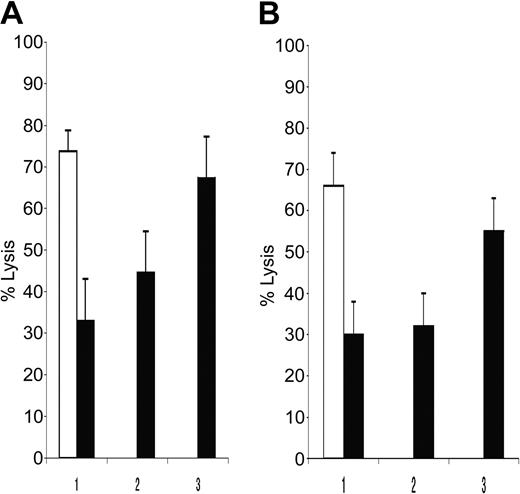
To verify whether PE myeloma resistance to NK cytotoxicity was due to high MHC class I molecules expression, KMS12PE and KMS21PE target cells were pretreated with either anti-MHC class I IgM antibodies or isotypic control (Figure 4A-B). Anti-MHC class I antibody restored NK susceptibility of these cells, increasing their lysis up to 40% to 80% of the lysis of the related BM myeloma cells.
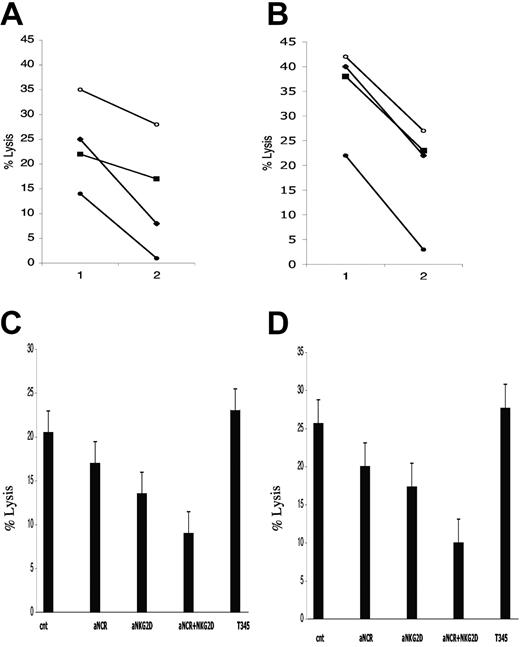
MHC class I inhibits NK cell recognition of pleural effusion-derived myeloma cell lines. Mean NK lysis percentage of KMS12BM and of KMS12PE are reported in panel A, while NK lysis of KMS21BM and KMS21PE are reported in panel B. BM and PE myeloma cell lysis are indicated with white and black columns, respectively. Antibody pretreatment of the PE targets are reported on abscisses: 1) complete medium, 2) anti-CD57 (TIB200) isotype control, 3) anti-MHC class I mAb A6 to 136 (IgM). The percentage of lysis reported was calculated with E/T ratio of 12:1. MHC class I masking restored myeloma susceptibility to NK recognition.
MHC class I inhibits NK cell recognition of pleural effusion-derived myeloma cell lines. Mean NK lysis percentage of KMS12BM and of KMS12PE are reported in panel A, while NK lysis of KMS21BM and KMS21PE are reported in panel B. BM and PE myeloma cell lysis are indicated with white and black columns, respectively. Antibody pretreatment of the PE targets are reported on abscisses: 1) complete medium, 2) anti-CD57 (TIB200) isotype control, 3) anti-MHC class I mAb A6 to 136 (IgM). The percentage of lysis reported was calculated with E/T ratio of 12:1. MHC class I masking restored myeloma susceptibility to NK recognition.
To explore which of the NK-activating receptors was involved in myeloma cell killing, we performed receptor blockade experiments. KMS12BM and KMS21BM were used as target. Anti-NCR (NKp46, NKp30, NKp44) antibodies were used alone or in combination either with anti-MICA or anti-NKG2D to prevent NK receptor engagement of the cognate ligands expressed on myeloma targets. In all experiments on both myeloma cell lines the strongest and most reproducible lysis inhibition was observed using the combinations of either anti-NCR + anti-NKG2D or anti-NCR + anti-MICA (Figure 5A-B). Using fresh NK cells, the simultaneous NCR and MICA blockade gave an inhibition range of 60% to 100% at 12:1 effector/target ratio (Figure 5C-D).
NCR and NKG2D are the main activating receptors involved in NK recognition of bone marrow-derived myeloma cells. KMS12BM (A) and KMS21BM (B) killing by resting purified NK cells. Target cells were treated with control antibody (○), anti-NCR (▪), anti-NKG2D (▴), and anti-NCR + NKG2D (•). One representative experiment of 4 performed is reported. E/T ratio indicated on abscissa are 12:1 (1) and 6:1 (2). (C-D) Average NK lysis and SEM are calculated from 4 experiments with KMS12BM and KMS21BM, respectively. The target treatment with different antibodies is reported on the abscissa: untreated target cell percentage of killing is indicated as cnt. Separately, anti-NCR and anti-NKG2D treatment of NK cells inhibited BM myeloma cells recognition; when used together, a clear synergistic inhibitory effect emerged.
NCR and NKG2D are the main activating receptors involved in NK recognition of bone marrow-derived myeloma cells. KMS12BM (A) and KMS21BM (B) killing by resting purified NK cells. Target cells were treated with control antibody (○), anti-NCR (▪), anti-NKG2D (▴), and anti-NCR + NKG2D (•). One representative experiment of 4 performed is reported. E/T ratio indicated on abscissa are 12:1 (1) and 6:1 (2). (C-D) Average NK lysis and SEM are calculated from 4 experiments with KMS12BM and KMS21BM, respectively. The target treatment with different antibodies is reported on the abscissa: untreated target cell percentage of killing is indicated as cnt. Separately, anti-NCR and anti-NKG2D treatment of NK cells inhibited BM myeloma cells recognition; when used together, a clear synergistic inhibitory effect emerged.
MHC class I and MICA expression on freshly explanted myeloma cells
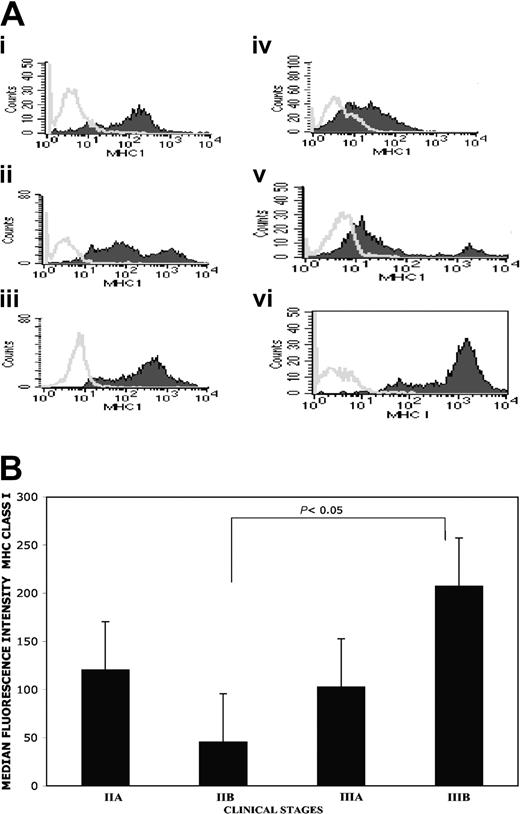
The expression level of MHC class I molecules was analyzed on freshly explanted CD38bright or CD38+/CD138+ myeloma cells obtained by bone marrow aspirate in 14 patients suffering with MM. A heterogeneous staining pattern emerged in the neoplastic cell population: bright, dim, and, in some case, intermediate MHC class I expression was found (Figure 6A), while MHC class I expression on healthy cells present in the same patient's bone marrow was homogeneous (data not shown). A direct correlation between myeloma MHC class I expression and clinical stage was observed (Figure 6B), and a statistically significant (P < .05) difference was found comparing MHC class I expression between patients with clinical stage IIB and IIIB.
Heterogeneous MHC class I molecule expression on freshly isolated CD38+ and CD138+/CD38+ bone marrow myeloma cells. Staining profile of MHC class I expression on patients 2, 3, 4, 5, 6, and 10 derived CD38+ cells are reported in panels i, ii, iii, iv, v, and vi, respectively (A). It should be noted that in patients 2 and 3 the myeloma cells were stained equally well by either anti-CD38 or anti-CD138 mAbs; for p10 MHC class I expression was tested on immunomagnetic-purified CD38+/CD138+ myeloma cells, and for patients 4, 5, and 6 CD38bright cells were gated. At least 10 000 events were analyzed within the CD38+bright or the CD38+/CD138+ gated cells. Open curves are the isotypic control; close curves are W6-32 staining. The correlation between myeloma clinical stages and MHC class I expression was analyzed (B). Stage IIA (n.4), stage IIB (n.2), stage IIIA (n.5), and stage IIIB (n.3). The differences between stage IIB and IIIB reached statistical significance with P of 05 or less. The analysis was performed via gating on living MHC class I-positive myeloma cells.
Heterogeneous MHC class I molecule expression on freshly isolated CD38+ and CD138+/CD38+ bone marrow myeloma cells. Staining profile of MHC class I expression on patients 2, 3, 4, 5, 6, and 10 derived CD38+ cells are reported in panels i, ii, iii, iv, v, and vi, respectively (A). It should be noted that in patients 2 and 3 the myeloma cells were stained equally well by either anti-CD38 or anti-CD138 mAbs; for p10 MHC class I expression was tested on immunomagnetic-purified CD38+/CD138+ myeloma cells, and for patients 4, 5, and 6 CD38bright cells were gated. At least 10 000 events were analyzed within the CD38+bright or the CD38+/CD138+ gated cells. Open curves are the isotypic control; close curves are W6-32 staining. The correlation between myeloma clinical stages and MHC class I expression was analyzed (B). Stage IIA (n.4), stage IIB (n.2), stage IIIA (n.5), and stage IIIB (n.3). The differences between stage IIB and IIIB reached statistical significance with P of 05 or less. The analysis was performed via gating on living MHC class I-positive myeloma cells.
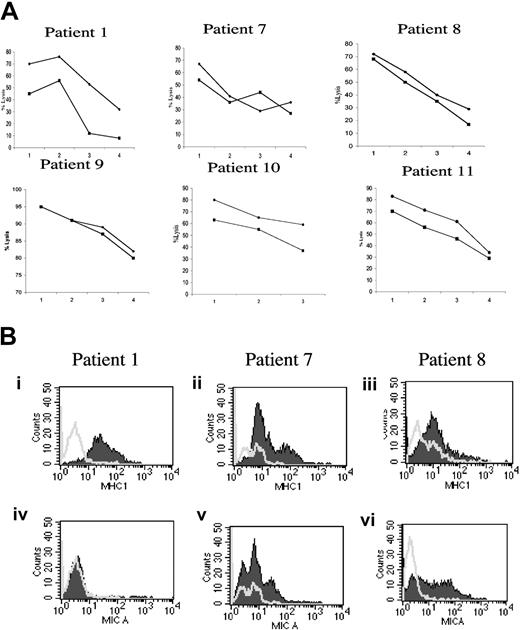
In 3 patients (1, 7, and 8) it was possible to stain bone marrow myeloma cells for MHC class I and MICA molecules and perform autologous NK cytotoxicity assays. Freshly explanted myeloma cells from these 3 patients were exposed to purified autologous NK cells either fresh or activated by IL-2. In patient 1, NK cells recognized autologous myeloma cells, and their activation by IL-2 improved the autologous killing (Figure 7A). The experiments performed with cells from patients 7 and 8 confirmed that autologous NK cells could recognize bone marrow-infiltrating myeloma cells; in these patients IL-2 activation had only a marginal effect, but it has to be noted that NK cells from these patients showed maximal autologous myeloma cell killing even before IL-2 activation. Moreover, NK cells derived from all 3 patients efficiently eliminated allogeneic myeloma cells (data not shown). Similar results were obtained when autologous NK antimyeloma cytotoxicity was analyzed using CD138+ myeloma targets purified by an immunomagnetic technique. MHC class I and MICA staining of myeloma cells from patients 1, 7, and 8 are reported in Figure 7B.
Autologous NK cells recognize freshly explanted bone marrow myeloma cells. (A) Freshly explanted autologous NK and myeloma cells were used in cytotoxicity assays. The results obtained in patients 1, 7, 8, 9, 10, and 11 are shown. ♦ indicates recognition by autologous fresh NK cells; •, percentage of lysis obtained using as effectors IL-2 activated autologous NK cells. The E/T ratios used were 25:1 (1), 12:1 (2), 6:1 (3), and 3:1 (4). Myeloma cells from patient 10 were obtained by immunomagnetic selection using BB-4 (anti-CD138) mAb and kept without exogenous IL-6 treatment. Five of 6 patients showed a very high autologous NK recognition of BM-derived myeloma cells, while patient 1 had less efficient autologous myeloma elimination. In patient 1, 10, and 11, IL-2 activation of NK cells had a beneficial effect on autologous myeloma recognition. (B) CD38+ cells from bone marrow aspirates were double stained with anti-MHC class I, W6-32 (i-iii) or anti-MICA, BAM195 (iv-vi) antibodies. Open areas are isotype controls; filled areas are specific antibody staining. Patient 1 had higher MHC class I expression than patient 7 and patient 8, while MICA was not detectable. Patient 7- and patient 8-derived myeloma cells were readily stained with anti-MICA antibody. After subtracting the background value the MHC class I surface expression median value for the patients 1, 7, and 8 were, respectively, 30, 8, and 10, while the MICA median values were 3, 10, and 8. For patients 9, 10, and 11 it was not possible to evaluate the complete phenotype because of the limited number of cells obtained.
Autologous NK cells recognize freshly explanted bone marrow myeloma cells. (A) Freshly explanted autologous NK and myeloma cells were used in cytotoxicity assays. The results obtained in patients 1, 7, 8, 9, 10, and 11 are shown. ♦ indicates recognition by autologous fresh NK cells; •, percentage of lysis obtained using as effectors IL-2 activated autologous NK cells. The E/T ratios used were 25:1 (1), 12:1 (2), 6:1 (3), and 3:1 (4). Myeloma cells from patient 10 were obtained by immunomagnetic selection using BB-4 (anti-CD138) mAb and kept without exogenous IL-6 treatment. Five of 6 patients showed a very high autologous NK recognition of BM-derived myeloma cells, while patient 1 had less efficient autologous myeloma elimination. In patient 1, 10, and 11, IL-2 activation of NK cells had a beneficial effect on autologous myeloma recognition. (B) CD38+ cells from bone marrow aspirates were double stained with anti-MHC class I, W6-32 (i-iii) or anti-MICA, BAM195 (iv-vi) antibodies. Open areas are isotype controls; filled areas are specific antibody staining. Patient 1 had higher MHC class I expression than patient 7 and patient 8, while MICA was not detectable. Patient 7- and patient 8-derived myeloma cells were readily stained with anti-MICA antibody. After subtracting the background value the MHC class I surface expression median value for the patients 1, 7, and 8 were, respectively, 30, 8, and 10, while the MICA median values were 3, 10, and 8. For patients 9, 10, and 11 it was not possible to evaluate the complete phenotype because of the limited number of cells obtained.
Discussion
We evaluated the expression of MHC class I, MICA, and ULBPs in an experimental MM cell system that allowed us to correlate the myeloma immunophenotype with NK recognition at different disease localizations within the same patient.
The anatomic site of the disease may be indicative of the disease stage: BM-derived cell lines are representative of early disease, while myeloma pleural effusions are related with advanced stage. PE myeloma is a rare and extremely aggressive tumor with a prognosis of less than 4 months.38 CD28 was described as a key landmark to differentiate between slowly progressing, early-stage myeloma (dim CD28) and advanced disease with high tumor malignancy (bright CD28).39 We confirmed this observation in our experimental system: all the 4 PE-derived myeloma lines expressed high membrane levels of CD28 while the BM lines showed low CD28 levels on their cell membrane (data not shown).
Our data demonstrate that PE myeloma lines express higher levels of MHC class I compared with BM and PB myeloma cells. Intriguingly, MHC overexpression was mirrored by the loss or drastic reduction of MICA molecules. Low MHC class I- and high MICA-expressing KMS12 and KMS21 BM cells were readily eliminated by NK cells, while KMS12 PE, KMS21PE, and KMS11PE myeloma lines were NK resistant. It is worthy to note that, among the PE lines KMS26PE express the lowest MHC class I levels and the highest NK susceptibility. The MHC class I protection from NK recognition was directly proven by antibody masking experiments in the KMSPE myeloma lines.
Therefore, from in vitro studies we conclude the following:
BM myeloma cell lines have reduced membrane levels of MHC class I molecules, while PE myeloma cells have high amount of MHC class I.
A reciprocal MICA expression was found between the 2 disease sites.
BM-derived myeloma cell lines were susceptible to NK recognition, while the PE-derived myeloma cell lines were spared. It was possible to restore NK susceptibility in PE-derived cells by MHC class I antibody masking. Thus, NK recognition of myeloma cells seems to be negatively regulated by MHC class I molecules.
The increased MHC class I expression of PE cell lines is in contrast with previous observations made in solid tumors, where disease progression led to MHC class I loss or down-regulation.18
MHC class I molecules are reduced on several tumors, and it is widely accepted that MHC loss leads to tumor escape by CTL recognition. MHC class I molecule down-regulation is an event correlated with cell transformation. Several reviews summarize allele, locus, haplotype, or even complete MHC class I deficiency in primary tumors, especially in metastatic lesions.40-42 However, some exception to this rule was reported. In the uveal melanoma, Blom et al43 observed that liver metastatic lesions expressed high MHC class I levels and that NK cell depletion worsened the disease course. Those researchers demonstrated in a mouse system that NK cells were involved in uveal melanoma progression control and that the higher MHC class I expression found on the metastases was due to a selection process based on NK activity, regulated by MHC class I inhibitory receptors.43 It should be noted that uveal melanoma spread occurs through blood circulation, where NK activity and cell number are high.1 It is conceivable to propose that in tumors exposed to high numbers of NK cells, such as myeloma and uveal melanoma, the immune editing is operated by cytotoxic lymphocytes regulated by MHC class I inhibitory receptors. The comparative analysis of myeloma cells MHC class I expression levels with the clinical stage gives further support to our hypothesis.
While the molecular mechanisms leading to MHC class I down-regulation are well known, how a tumor could increase its MHC class I expression is less understood. In melanoma, cutaneous lymphoma, glioma, and in some lung carcinoma it was reported that the ectopic expression of HLA-G allelic products could contribute to the overall MHC class I levels.44 In our myeloma cell system HLA-G was not expressed and not IFNγ inducible (data not shown).
MHC class I heavy-chain transcription was analyzed by Northern blot and RT-PCR in our myeloma system. The transcripts for MHC class I were reduced in BM cell lines, while high amount of MHC class I heavy-chain mRNA was detected in PE-derived cells. In the same experiments, when the transcript levels for TAP1 were measured, no changes were found between BM and PE myeloma. Intriguing reciprocal results were obtained by hybridizing our filters with the MICA probe. High levels of MICA transcripts were found by Northern blot and RT-PCR analysis in BM myeloma cells while very little if any MICA mRNA was found in the related PE cell lines (KMS12PE, KMS21PE). These findings suggest that, during myeloma progression, a clonal variant with defective MICA transcription could emerge, resulting in tumor cell escape from NKG2D driven immune surveillance, a mechanism distinct from the previously described MICA shedding.45
So far, ULBP expression was studied mainly on solid tumors and long-term in vitro cultured cell lines8 ; here we evaluated their expression for the first time on myeloma cells. ULBP1-3 was expressed on myeloma cells from different disease sites, and MICA down-regulation during myeloma progression did not prevent tumor cells to express ULBPs; however, ULBP1-3 distribution was not clearly correlated with MM sites and stage. Perhaps, the recently suggested differences in the association kinetics of NKG2D with MICA and ULBP46 and the demonstrated different binding affinity among murine NKG2D ligands47 could be invoked to justify the discrepancy found in our study among MICA and ULBP expression in different MM sites or stages.
The blocking experiments with NCR and NKG2D antibodies showed that both receptors are critically involved in resting NK cell recognition of myeloma cells. We analyzed the expression of MHC class I and MICA on primary CD38+ myeloma cells at diagnosis. All tumor populations showed a simultaneous presence of cells expressing high and low MHC class I levels. We speculate that in the early stage of MM the heterogeneous MHC class I expression could be explained as an intrinsic instability of the tumor gene expression and/or as a back-migration of MHC class I bright myeloma clones from the peripheral blood to the bone marrow. The observed discrepancy between the low MHC class I expression on KMSBM cell lines and the heterogeneous pattern found in patients could be explained by an in vitro selection of a dominant cell population, or with the small number of BM myeloma lines available in our study. According with our data, we can predict that in myeloma the initial tumor population is controlled by NK cells; then, myeloma subclones with low MICA and high MHC class I levels could emerge. The residual MHC class Ibright, MICAdim cells will dampen a possible further recognition by NKG2D expressing CTL, γδ T, and NK cells. Our data are in agreement with the NK role in controlling myeloma progression suggested in previous reports.29,48,49 Our work nicely complements a previous study by Frohn et al,29 in which a clear recognition of MHC class I-expressing fresh myeloma cells by allogeneic NK cells was demonstrated. The MICA, NKG2D, and NCR key role in myeloma recognition described here may offer an explanation for these observations.29 We provide direct evidence that in patients with myeloma NK cells recognize autologous tumor targets, and NK cells from patients appeared to be activated in vivo. Thalidomide has been used with success in myeloma therapy. In one study the beneficial effect of thalidomide was demonstrated to be dependent on IL-2 induction of natural cytotoxicity.50 Our data strengthen and develop further these observations. NK cells and the knowledge available on their regulation could represent an intriguing possibility for innovative biologically based immunotherapy approach of multiple myeloma.
Prepublished online as Blood First Edition Paper, August 24, 2004; DOI 10.1182/blood-2004-04-1422.
Supported by Associazione Italiana per la Ricerca sul Cancro (AIRC) and Strategic research center for studies of Integrative Recognition in Immune System-Stiftelsen för Strategisk Forskning.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Klas Kärre for data discussion and critical reading of the manuscript, Petter Höglund for data discussion, and Dan Geraghty for providing us with reagents.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal