Abstract
Gene expression profiling was used to compare the gene expression patterns of human germinal center (GC) T helper (Th) cells with other CD4+ T-cell subsets (naive, central, and effector memory T cells). GC-Th cells, specifically localized in germinal centers to help B cells, are distantly related to central and effector memory T cells in global gene expression profiles. GC-Th cells displayed substantial differences in mRNA for adhesion molecules, chemoattractant receptors, and cytokines compared with other populations. Distinct expression of transcriptional factors by GC-Th cells is consistent with the hypothesis that they may be different from other T cells in cell lineage. Interestingly, CXCL13, a critical chemokine for B-cell entry to lymphoid follicles, is one of the most highly up-regulated genes in GC-Th cells. GC-Th cells (but not other T cells) produce and secrete large amounts of functional CXCL13 upon T-cell receptor activation, a process that is dependent on costimulation, requires translation and transcription, and is dramatically enhanced by activation in the presence of GC-B cells. This study revealed for the first time the unique gene expression program of GC-Th cells.
Introduction
Heterogeneous subsets of CD4+ T cells are master regulators of immune responses. Diverse memory and effector CD4+ T-cell subsets originate from naive T cells. Naive T cells, generated in the thymus, keep circulating the blood and secondary lymphoid tissues in search of antigens.1-3 In secondary lymphoid tissues, naive T cells undergo activation, clonal expansion, and differentiation to become memory and effector cells in response to antigenic signals provided by antigen presenting cells.4 Many memory and effector T cells migrate to peripheral nonlymphoid tissues for immune surveillance.5,6 Depending on their effector function and cytokine production capacity, conventional memory and effector T cells are divided into nonpolarized T cells, T helper 1 (Th1), Th2, and regulatory T cells.7-10 They are also classified into “central memory cells” best characterized as CXCR5+CCR7+ cells and “effector memory cells” enriched among CCR7– T cells.11 CCR7– memory T cells contain a higher frequency of cells producing Th1 and Th2 cytokines than CXCR5+CCR7+ memory T cells upon T-cell receptor (TCR) stimulation, thus closer to effector T cells in cytokine production.12
Recently, another CD4+ T-cell subset, distinct from polarized Th1 and Th2 cells in cytokine production, migration, and effector function, has been characterized.13 In human lymphoid tissues, CD4+ T cells expressing CXCR5 and CD57 are specifically localized in germinal centers (termed GC-Th cells hereafter).13,14 These T cells are related to other CXCR5+ memory T cells15,16 in that they are mostly nonpolarized (lacking interleukin 4 [IL-4] and interferon γ [IFN-γ] production) but are efficient in inducing B-cell production of immunoglobulin.13 In mice, a similar type of T cells exists with efficient B-cell helping capacity but no inflammatory immune response.17 GC-Th cells lack CCR7, a T-cell area localization receptor, and do not migrate to ELC/CCL19 but migrate toward the B-cell area chemokine CXCL13, a feature consistent with their specific localization in germinal centers.13
In a search for the molecular basis for the differences between GC-Th cells and other T-cell subsets (CXCR5+CCR7+ early or central memory and CCR7– effector memory cells), we compared their gene expression profiles with a cDNA microarray technique using naive cells as reference cells. GC-Th cells display unique gene expression patterns not only on the global scale but also in several major gene groups such as effector molecules (cytokine and chemokine), cell cycle and apoptosis, transcription factors, and migration. Most striking is the selective and high-level expression of a potent B-cell– and CXCR5+ T-cell–attracting chemokine, CXCL13, in GC-Th cells. CXCL13 production by GC-Th cells is induced by TCR activation and is dependent on the costimulation signal through CD28. The significance of the unique gene expression profile of GC-Th cells in their biology and function in germinal centers is discussed herein.
Materials and methods
Cell isolation
Mononuclear cells from human peripheral blood and tonsils obtained from young patients (3-10 years old) were prepared by density gradient centrifuge on histopaque 1077 (Sigma-Aldrich, St Louis, MO). CD4+ T cells (purity > 97%) were isolated by depleting non-CD4+ T cells using a magnetic bead depletion method (Miltenyi Biotec, Auburn, CA). After staining of the isolated CD4+ T cells with appropriate antibodies, CD57+CD4+ GC-Th cells, CD45RA+CD4+ naive, CCR7–CD45RA–CD4+ effector memory and CXCR5+CCR7+CD45RA– memory cells were sorted using the FACSVantage SE (Becton Dickinson, San Jose, CA). T cells with a purity more than 99% were used for microarray experiments. Naive and CD57+ T cells were also sorted by magnetic bead labeling using antibodies to CD45RO or CD57 for cell culture experiments. Monocyte-derived mature dendritic cells were prepared as previously described.18 GC-B cells were isolated from tonsils as described elsewhere19 using anti-CD44 and anti-IgD antibodies (purity > 95% based on CD38, IgD, and CD19 expression).
To determine the levels of CXCL13 messages after culture (Figure 5D-E), GC-Th cells were resorted using antibodies to CD57 and CD3, and anti–mouse immunoglobulin microbeads (Miltenyi Biotec). B cells were positively isolated using anti-CD19 magnetic beads (Dynal, Oslo, Norway) or negatively isolated by depleting T cells as described in the paragraph above. Dendritic cells were either positively isolated using anti-CD1a microbeads (Miltenyi Biotec) or negatively selected by depleting T cells. Isolated cells were checked for purity by flow cytometry and dissolved in Trizol solution (Invitrogen, Carlsbad, CA) for RNA preparation.
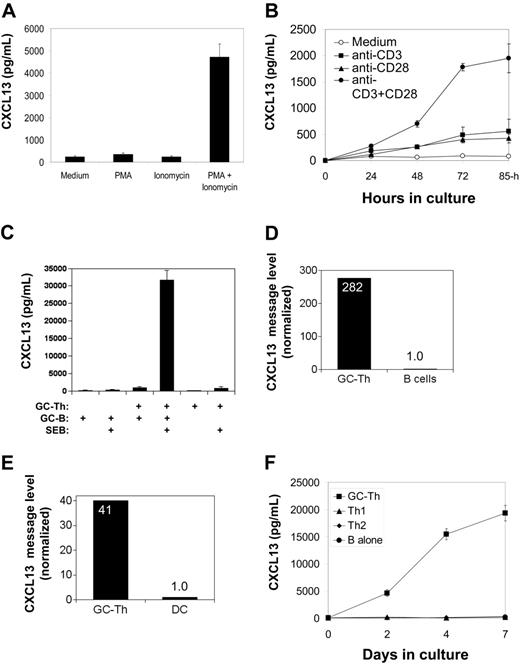
CXCL13 production by GC-Th cells is regulated by TCR activation and CD28 costimulation. (A) Regulation of CXCL13 production in GC-Th cells by PMA and/or ionomycin. (B) Efficient production of CXCL13 requires both TCR activation and CD28 costimulation. (C) GC-B cells support GC-Th cells in production of CXCL13. GC-B cells were cocultured with equal numbers of GC-Th cells in the presence of superantigen SEB (1 μg/mL) for 3 to 4 days. (D) High levels of CXCL13 mRNA were detected in GC-Th cells but not in B cells reisolated after culture. (E) High levels of CXCL13 messages were detected in GC-Th cells but not in cultured dendritic cells reisolated after culture. Expression of CXCL13 message was determined by quantitative real-time PCR assay (D-E). The CXCL13 message number of each sample was divided by the β-actin message number for normalization. Relative expression levels (when the normalized CXCL13 copy level in B cells or dendritic cells equals one) are shown. (F) GC-Th cells but not Th1 or Th2 cells produce CXCL13. GC-Th cells, Th1, or Th2 cells were in vitro generated and cultured with GC-B cells in the presence of SEB. CXCL13 production was examined by ELISA (A-C, F). Data shown are representatives of at least 3 separate experiments. The error bars are standard deviations of triplicate experiments.
CXCL13 production by GC-Th cells is regulated by TCR activation and CD28 costimulation. (A) Regulation of CXCL13 production in GC-Th cells by PMA and/or ionomycin. (B) Efficient production of CXCL13 requires both TCR activation and CD28 costimulation. (C) GC-B cells support GC-Th cells in production of CXCL13. GC-B cells were cocultured with equal numbers of GC-Th cells in the presence of superantigen SEB (1 μg/mL) for 3 to 4 days. (D) High levels of CXCL13 mRNA were detected in GC-Th cells but not in B cells reisolated after culture. (E) High levels of CXCL13 messages were detected in GC-Th cells but not in cultured dendritic cells reisolated after culture. Expression of CXCL13 message was determined by quantitative real-time PCR assay (D-E). The CXCL13 message number of each sample was divided by the β-actin message number for normalization. Relative expression levels (when the normalized CXCL13 copy level in B cells or dendritic cells equals one) are shown. (F) GC-Th cells but not Th1 or Th2 cells produce CXCL13. GC-Th cells, Th1, or Th2 cells were in vitro generated and cultured with GC-B cells in the presence of SEB. CXCL13 production was examined by ELISA (A-C, F). Data shown are representatives of at least 3 separate experiments. The error bars are standard deviations of triplicate experiments.
Gene expression analysis by microarray technology
cDNA microarray experiments were performed as described elsewhere.20 Briefly, total RNA was isolated from T cells with Trizol solution (Invitrogen), amplified by a T7 linear amplification system,21 labeled with CY-3 (reference cells: naive T cells) or CY-5 dyes (naive, GC-Th cells, and other memory or effector T cells), and used to hybridize cDNA microarrays. These arrays contain 38 432 cDNA spots corresponding to human sequence verified genes, and were purchased from the Microarray Core Facility at the Stanford School of Medicine. Fluorescent intensity for CY-3 and CY-5 was detected by using a GenePix 4000b microarray scanner (Axon Instruments, Foster City, CA). The data files were processed so that the red signal was normalized by applying a single multiplicative factor to all intensities measured for the red dye Cy5. The normalization factor was computed so that the median Cy3/Cy5 fluorescent ratio of nonflagged spots on each array was 1.0. Selection and filtering of high-quality genes were performed as described elsewhere22 using the Stanford Microarray Database (SMD). To analyze levels of up- or down-regulation, we applied a hierarchical clustering algorithm implemented by the software Cluster as described by Eisen et al.23 This software clusters genes according to their similarity in the pattern of gene expression and displays the data in a dendrogram that resembles a tree (TreeView). Average linkage clustering based on Pearson correlation was performed to compare similarities among the T-cell subsets.
Cell culture
Cell culture was performed in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), gentamycin, streptomycin, and penicillin. T and B cells were cultured at a 1-to-1 ratio in 96-well plates or 24-well plates. For generation of conditioned medium for chemotaxis, 0.5 × 107 to 1 × 107 GC-Th cells were cultured with equal numbers of tonsil B cells in the presence of staphylococcal enterotoxin B (SEB; 1 μg/mL; Sigma-Aldrich). For T-cell receptor activation, immobilized anti-CD3 antibodies (one μg/well) along with anti-CD28 antibody (10 μg/mL; Pharmingen, San Diego, CA) were used. Other stimulants (Sigma-Aldrich) used in this study were: phytohemagglutinin (PHA; 10 μg/mL), phorbol 12-myristate 13-acetate (PMA; 1 μg/mL), and ionomycin (one μM). Cells were incubated in 5% CO2 incubators at 37° C for 3 to 5 days. In some experiments, GC-Th cells were cultured in the presence of T-cell stimulators with or without actinomycin D (1 μg/mL) or cycloheximide (8 μg/mL) to inhibit transcription or translation, respectively. Th1 and Th2 cells were generated by stimulating CD45RA+ T cells with anti-CD3 and anti-CD28 in the presence of IL-12 (2 ng/mL) and neutralizing anti–IL-4 antibodies (200 ng/mL) for Th1 cells, or IL-4 (200 U/mL) and neutralizing anti–IL-12 antibodies (2 μg/mL) for Th2 cells, respectively (all antibodies were from Pharmingen). Cells were further expanded for 4 days in medium containing IL-2 (100 U/mL).
In situ fluorescent immunohistochemistry
Frozen sections of tonsils were acetone-fixed and stained using directly labeled antibodies to CD11C (phycoerythrin [PE]), CD57 (fluorescein isothiocyanate [FITC]), CD38 (PE), and/or CD19 (FITC). To detect CXCL13 expression, acetone-fixed and goat serum–blocked (20%; 30 minutes; 25° C) tonsil sections were stained with anti-CXCL13 antibody (mouse IgG1, clone 53610; R&D Systems, Minneapolis, MN; 30 minutes at 10 μg/mL) or isotype control antibody (mouse IgG1; 30 minutes at 5 μg/mL; Caltag, Burlingame, CA) and then by anti–mouse immunoglobulin goat antibody (PE-conjugated; BioSource, Camarillo, CA). Sections were blocked in mouse serum (5% in phosphate-buffered saline [PBS]) and further stained with anti–CD57-FITC. Stained sections were analyzed using a confocal microscopy system (Bio-Rad MRC 1024UV [Bio-Rad, Hercules, CA] and Diaphot 300 microscope [Nikon, Melville, NY]) at Purdue Cytometry Lab.
ELISA
Isolated T cells were cultured for 3 to 5 days in various conditions, and supernatants were assayed by a 3-step sandwich enzyme-linked immunosorbent assay (ELISA) kit for CXCL13 (R&D Systems) according to the manufacturer's protocols. The sensitivity of this ELISA kit was more than 10 pg/mL.
Quantitative real-time PCR analysis
Total RNA was extracted from isolated tonsil cell populations with Trizol reagent (Invitrogen), and was reverse-transcribed into cDNAs with SuperScript First-Strand Synthesis System for reverse transcriptase–polymerase chain reaction (RT-PCR; Invitrogen) according to the manufacturer's protocol. Numbers of CXCL13 and β-actin messages in the cDNA samples were determined by real-time PCR assays using SYBR Green PCR Master Mix system (PE Applied Biosystems, Foster City, CA) on an ABI PRISM 5700 sequence detection system (PE Applied Biosystems). For standard curves, known amounts of universal β-actin and CXCL13 cDNA were amplified. Real-time PCR reactions were performed on serially diluted cDNA samples by pretreating the reaction samples for 2 minutes at 50° C and then for 10 minutes at 95° C followed by 40 cycles of PCR reaction (denaturation at 95° C for 15 seconds, annealing at 55° C for 45 seconds, and extension at 72° C for 30 seconds). The specificity of the amplified products was verified based on unique dissociation curves and sizes of the amplified CXCL13 and β-actin products. The CXCL13 message number of each sample was divided by the β-actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) message number for normalization. The primer sequences used for the real-time PCR experiments are shown in Table S3 (see the Supplemental Tables link at the top of the online article on the Blood website).
Chemotaxis assay
Chemotaxis was assayed by a 2-chamber transwell cell migration system (5 μm pores; Corning, Corning, NY).24 Large numbers (0.5 × 107-1 × 107) of GC-Th cells were activated with a superantigen SEB and GC-B cells. Highly conditioned media were harvested after 7 days and used for chemotaxis assays. Fresh uncultured and conditioned media from GC-B cell culture were used for negative controls. Fresh tonsil mononuclear cells were incubated 3 hours to overnight to resensitize them from potential desensitization effects in vivo. Percent cell migration was calculated by dividing the numbers of cells migrated to the bottom wells by those of input cells. All chemotaxis was performed in duplicate.
Results
GC-Th cells and other T-cell subsets examined in this study
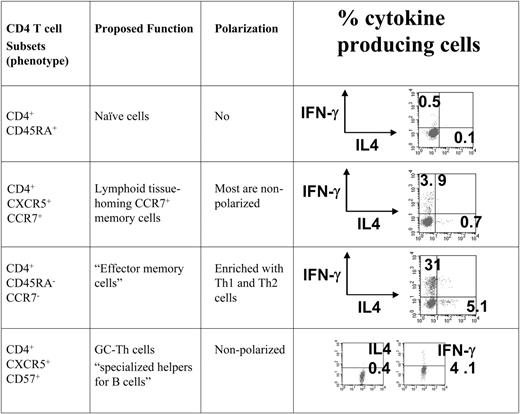
To compare the gene expression patterns of GC-Th cells and other T-cell subsets we used a cDNA microarray method. All of the CD4+ T-cell subsets examined in this study were freshly isolated from peripheral blood or tonsils in the absence of any artificial stimulation in vitro. We included GC-Th, naive, “early or central memory,” and “effector memory” CD4+ T cells in our array study. The phenotypes of these T cells are shown in Figure 1. GC-Th cells are localized in germinal centers, and are efficient in helping B cells produce antibodies.13 It has been shown that the early memory CXCR5+ CCR7+ T cells are rapidly generated from naive T cells upon activation with dendritic cells within 2 to 3 days, and are mostly composed of nonpolarized cells.25,26 The CD4+CCR7– T-cell population lacks a lymphoid homing receptor CCR7 and is enriched with Th1 and Th2 cells.11,12 These cell subsets were highly purified (> 99%) for cDNA microarray experiments, where naive T cells were used as reference cells to focus our interest on genes differentially expressed in memory/effector populations.
The T-cell subsets that were compared with GC-Th cells in the microarray study. All T-cell subsets used in the study were highly purified cells from peripheral blood (naive, CXCR5+CCR7+ memory, and CCR7-effector memory cells) and tonsils (CD57+ GC-Th cells). Cells were isolated by a combination of magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS). The surface markers used for cell isolation, proposed function, and typical polarization status of T-cell subsets are shown.
The T-cell subsets that were compared with GC-Th cells in the microarray study. All T-cell subsets used in the study were highly purified cells from peripheral blood (naive, CXCR5+CCR7+ memory, and CCR7-effector memory cells) and tonsils (CD57+ GC-Th cells). Cells were isolated by a combination of magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS). The surface markers used for cell isolation, proposed function, and typical polarization status of T-cell subsets are shown.
GC-Th cells are unique in overall gene expression
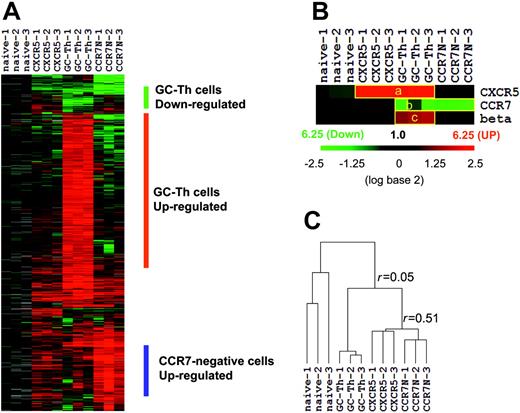
We performed microarray experiments using RNA isolated from the 4 T-cell subsets. After filtering out the genes that are of low intensity, poor quality, or not differentially expressed compared with naive T cells, we identified approximately 1600 genes that are differentially expressed in any of the memory/effector cells versus naive CD4+ T cells. The expression patterns of these genes are shown in Figure 2A. Three independent array experiments were performed for each T-cell subset using different samples. The overall pattern of gene expression in the 3 experiments for each cell subset is reproducible as evidenced by an average regression correlation of r = 0.73. Genes down-regulated and up-regulated in GC-Th cells were identified. A list of these genes and their up- and down-regulation fold ratios compared with naive, CCR7+CXCR5+ early memory, and CCR7– cells are shown in Table S1 and Table S2. β-1,3-glucuronyl transferase 1 is the enzyme responsible for the transfer of the HNK-1glycosyl epitope to CD57.27 Its gene along with those of CCR7 and CXCR5 can serve as controls for the microarray experiments. CXCR5 is up-regulated in GC-Th cells and in CCR7+CXCR5+ early memory T cells as expected (Figure 2B). CCR7 is down-regulated in GC-Th cells and in CCR7– T cells as most (GC-Th cells) or all (CCR7– T cells) of these cells lack CCR7 where the naive reference T cells are CCR7-positive. β-1,3-glucuronyl transferase 1 was up-regulated only in GC-Th cells, consistent with their unique expression of CD57. The expression patterns of the control genes show that the array experiments indeed represent the gene expression patterns of the cell subsets of interest. On the basis of the Pearson correlation method, we draw a hierarchical dendrogram showing the relationship of their gene expression profiles (Figure 2C). GC-Th cells were distinct from CCR7+CXCR5+ early memory (r = 0.06 ± 0.08) and CCR7– T cells in gene expression (r = –0.01 ± 0.14). Interestingly, the CCR7+CXCR5+ early memory T cells and CCR7– T cells were more closely related to each other (r = 0.51 ± 0.08) than to GC-Th cells. One difference between the 2 cell subsets was that CCR7– T cells expressed a group of genes (∼200) at higher levels than did early memory T cells (Figure 2A).
The overall gene expression profile of GC-Th cells (GC-Th) versus naive CD4+CD45RA+ (naive), CD4+CXCR5+CCR7+ “early memory” (CXCR5), and CD4+CCR7– “effector memory” (CCR7N) CD4+ cells. (A) Depicted are the 20 000 measurements of gene expression filtered from 12 microarray analyses of the 4 T-cell subsets (3 independent arrays per subset using cells from different donors, indicated as 1-3). Hierarchical clustering was performed to arrange genes based on their patterns of expression. Each row represents a separate cDNA clone on the microarray and each column a separate mRNA or T-cell subset. The results presented represent the ratio of hybridization of fluorescent cDNA probes prepared from each mRNA sample isolated from naive or memory/effector cells to reference mRNA samples from naive CD4+ T cells. These ratios are measures of relative gene expression in each mRNA sample and were depicted according to the color scale shown at the right. As indicated, the scale extends from fluorescence ratios of 0.25 to 6.25 (–2.5 to +2.5 in log base 2 units). Gray indicates missing or excluded data. (B) Individual expression patterns of several marker genes in the T-cell subsets. beta indicates beta-1,3-glucoronyltransferase 1. (C) Relationship between T-cell subsets as a function of similarity of gene expression programs shown in hierarchical dendrogram. Average linkage clustering based on Pearson correlation was performed to compare similarities among the T-cell subsets. The r values represent the correlation between gene expression profiles of T-cell subsets (+1 being a perfect positive linear relationship, 0 meaning no linear relationship, and –1 being a perfect negative linear relationship).
The overall gene expression profile of GC-Th cells (GC-Th) versus naive CD4+CD45RA+ (naive), CD4+CXCR5+CCR7+ “early memory” (CXCR5), and CD4+CCR7– “effector memory” (CCR7N) CD4+ cells. (A) Depicted are the 20 000 measurements of gene expression filtered from 12 microarray analyses of the 4 T-cell subsets (3 independent arrays per subset using cells from different donors, indicated as 1-3). Hierarchical clustering was performed to arrange genes based on their patterns of expression. Each row represents a separate cDNA clone on the microarray and each column a separate mRNA or T-cell subset. The results presented represent the ratio of hybridization of fluorescent cDNA probes prepared from each mRNA sample isolated from naive or memory/effector cells to reference mRNA samples from naive CD4+ T cells. These ratios are measures of relative gene expression in each mRNA sample and were depicted according to the color scale shown at the right. As indicated, the scale extends from fluorescence ratios of 0.25 to 6.25 (–2.5 to +2.5 in log base 2 units). Gray indicates missing or excluded data. (B) Individual expression patterns of several marker genes in the T-cell subsets. beta indicates beta-1,3-glucoronyltransferase 1. (C) Relationship between T-cell subsets as a function of similarity of gene expression programs shown in hierarchical dendrogram. Average linkage clustering based on Pearson correlation was performed to compare similarities among the T-cell subsets. The r values represent the correlation between gene expression profiles of T-cell subsets (+1 being a perfect positive linear relationship, 0 meaning no linear relationship, and –1 being a perfect negative linear relationship).
Comparison of GC-Th cells with other memory/effector T cells in expression of the genes in 5 functional groups
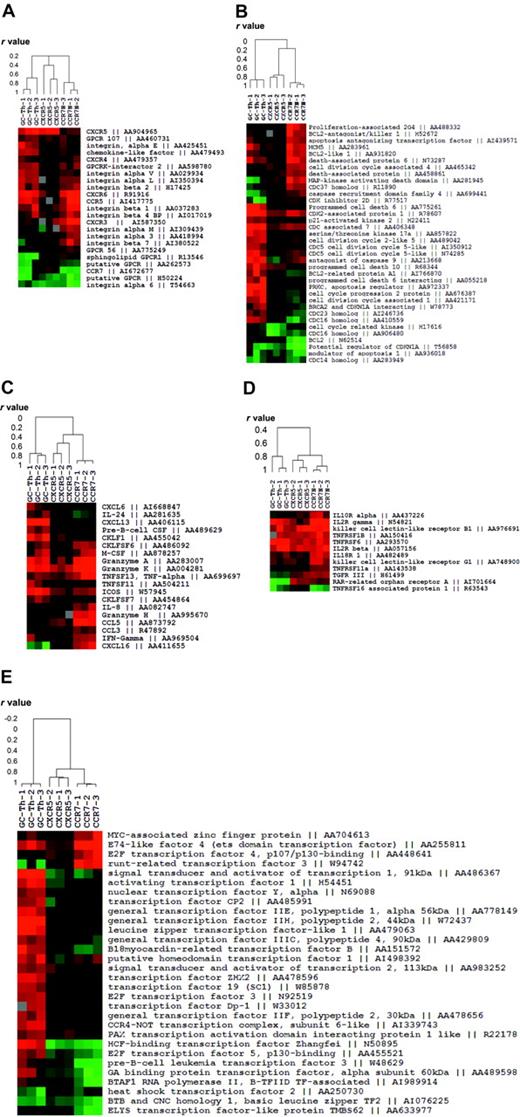
We fractionated genes into 5 functional groups and examined the relationship of GC-Th cells with other T-cell subsets in gene expression. The first group contains the genes that are related to migratory function such as chemokine receptors and adhesion molecules. GC-Th cells and CCR7+CXCR5+ early memory T cells shared the expression of CXCR5 but they were quite different in expression of other genes (Figure 3A). Whereas GC-Th cells and CCR7– T cells shared expression of a few genes such as CCR5 and CXCR6, they were very different from each other. Generally, the early memory T cells were similar to naive T cells in expression of these cell trafficking–related genes.
Expression patterns of the genes classified into 5 functional groups in GC-Th cells versus other memory/effector T-cell subsets. (A) Migration-related genes. (B) Cell-cycle– and apoptosis-related genes. (C) Chemokine- and cytokine-related genes. (D) Cytokine receptors. (E) Transcription-related genes. Each clustered image accompanies a dendrogram showing the relationship (in r values from Pearson correlation) among the T-cell subsets or 12 experiments (3 per subset) in gene expression. Genbank accession numbers are shown along with gene names.
Expression patterns of the genes classified into 5 functional groups in GC-Th cells versus other memory/effector T-cell subsets. (A) Migration-related genes. (B) Cell-cycle– and apoptosis-related genes. (C) Chemokine- and cytokine-related genes. (D) Cytokine receptors. (E) Transcription-related genes. Each clustered image accompanies a dendrogram showing the relationship (in r values from Pearson correlation) among the T-cell subsets or 12 experiments (3 per subset) in gene expression. Genbank accession numbers are shown along with gene names.
The second group includes a collection of the genes involved in cell proliferation and death (Figure 3B). In this case, GC-Th cells were extremely different from CCR7+CXCR5+ early memory and CCR7– T cells. GC-Th cells up-regulated a number of genes related to cell cycle and cell death such as programmed cell death 6, antagonist of caspase 9 and prostate apoptosis response regulator 4 (PAR-4). CCR7– T cells also express a group of cell-cycle– and death-related genes such as Bcl2 antagonist and mitogen-activated protein kinase (MAPK)–activating death domain, a pattern different from that of GC-Th cells and CCR7+CXCR5+ early memory T cells. However, the genes expressed by GC-Th cells and CCR7– T cells were distinct from each other. In contrast, CCR7+CXCR5+ early memory cells did not up-regulate this group of genes, again most resembling naive T cells.
The third and fourth groups of genes contain cytokine receptors and cytokines. In expression of cytokine receptors the 3 T-cell subsets were not very different from each other except a few genes such IL-2Rγ, IL-10R, and IL-18R (Figure 3D). In contrast, the 3 cell subsets were very different from each other in expression of some cytokines and chemokines, molecules potentially important for their effector function (Figure 3C). GC-Th cells uniquely express CXCL13, a chemokine critical for B-cell entry to lymphoid follicles. They up-regulated another molecule called CKLF1, a relatively uncharacterized chemotactic molecule.28 ICOS, an important costimulation molecule in formation of humoral immune response and germinal center formation,29,30 was highly up-regulated in GC-Th cells as well. GC-Th cells share the expression of granzymes A and K with CCR7– T cells, while some genes such as granzyme H, CCL5, and IFN-γ were up-regulated only in CCR7– T cells. The most striking difference in gene expression among the 3 cell subsets was observed in expression of transcriptional factors (Figure 3E). The genes highly up-regulated in GC-Th cells include activating transcription factor 1, host cell factor (HCF)–binding transcription factor Zangfei and transcription factor ZH3 among many others. CCR7– T cells showed up-regulated expression levels of some transcription factors such as runt-related transcription factor 3 and E2F transcription factor 4, which GC-Th cells had not up-regulated. The CCR7+CXCR5+ early memory T cells did not up-regulate the transcription factors shown in GC-Th cells or CCR7– T cells, in many respects resembling naive T cells.
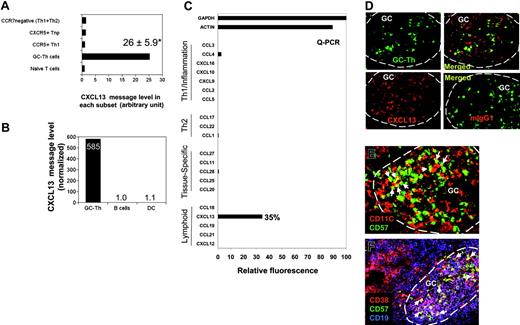
Specific and high-level expression of CXCL13 messages by GC-Th cells
Expression of CXCL13 messages was not expected (Figure 3C), as there has been a substantial body of evidence that non-T cells (including follicular dendritic cells) produce CXCL13 in secondary lymphoid tissues. CXCL13 is one of the most highly up-regulated genes in GC-Th cells versus other T-cell subsets along with other uncharacterized genes such as hypothetical protein FLJ23309 and early growth response 2 (a zinc finger protein; Table S1). The expression levels of CXCL13 mRNA in GC-Th cells were on average 26 times higher than in other T-cell subsets (Figure 4A). We examined expression of 20 chemokines including CXCL13 by GC-Th cells using a real-time PCR method, because the arrays used in this study did not include the cDNAs for many chemokines and we wanted to confirm the CXCL13 expression with a different method. Strikingly, CXCL13 was the only chemokine significantly expressed at the RNA level by the GC-Th cells (Figure 4C). The expression levels of CXCL13 mRNA were very high in GC-Th cells in that 0.05-0.5 copies of CXCL13 messages were expressed in GC-Th cells per copy of β-actin. Freshly isolated tonsil B cells and monocyte-derived mature dendritic cells, however, did not express any significant level of CXCL13 message (Figure 4B). The possibility of CXCL13 production by GC-Th cells is intriguing in that T cells may play a more active role in recruitment of follicle-homing B and T lymphocytes than previously thought. Therefore we further investigated the regulation of CXCL13 in GC-Th cells.
The unique expression of CXCL13 in human CD4+ GC-Th cells. (A) Expression of CXCL13 in GC-Th cells and other T-cell subsets measured by a cDNA array technique. Data shown are in relative units (naive T cells equal one). *Average up-regulation of CXCL13 mRNA in GC-Th cells versus naive T cells plus or minus standard deviation from 3 independent measurements. (B) Quantitative RT-PCR examination of CXCL13 in freshly isolated GC-Th cells, monocyte-derived mature dendritic cells, and freshly isolated CD19+ tonsil B cells. The copy numbers of B-lymphocyte chemoattractant (BLC) were normalized for copy numbers for GAPDH. Relative expression levels are shown (B cells equal 1). (C) CXCL13 is the major chemokine expressed by GC-Th cells. Real-time RT-PCR was used to examine 20 chemokines (in 4 groups: “Th1/inflammation,”“Th2,”“tissue-specific,” and “lymphoid” based on their expression sites or functions); GAPDH and β-actin were used as internal controls. Expression levels are shown in relative SYBR green fluorescence of the gene products to GAPDH products (GAPDH intensity = 100%). (D) CXCL13 expression in GC-Th cells was visualized by an in situ immunohistology technique. Frozen tonsil sections were stained with anti-CXCL13 antibody or control mouse IgG1 and then with secondary antibody conjugated with PE (red). Sections were further stained with anti–CD57-FITC for GC-Th cells (green). A confocal microscope system (Bio-Rad MRC 1024UV and Nikon Diaphot 300 microscope) was used to analyze the sections. (E-F) Many GC-Th cells are in physical contact with dendritic cells and GC-B cells. Frozen tonsil sections were stained with fluorescent monoclonal antibodies to CD11C (red for dendritic cells and some macrophages), CD57 (green for GC-Th cells), CD38 (red), and CD19 (blue). Please note that green (CD57+) cells are GC-Th cells and red cells are dendritic cells in panel E. Purple (CD38+CD19+) cells are GC-B cells and red (CD38+CD19–) cells are plasma cells in panel F. Arrows indicate several examples of GC-Th cells that are in direct contact with dendritic cells or GC-B cells. The images were taken at × 200 magnification.
The unique expression of CXCL13 in human CD4+ GC-Th cells. (A) Expression of CXCL13 in GC-Th cells and other T-cell subsets measured by a cDNA array technique. Data shown are in relative units (naive T cells equal one). *Average up-regulation of CXCL13 mRNA in GC-Th cells versus naive T cells plus or minus standard deviation from 3 independent measurements. (B) Quantitative RT-PCR examination of CXCL13 in freshly isolated GC-Th cells, monocyte-derived mature dendritic cells, and freshly isolated CD19+ tonsil B cells. The copy numbers of B-lymphocyte chemoattractant (BLC) were normalized for copy numbers for GAPDH. Relative expression levels are shown (B cells equal 1). (C) CXCL13 is the major chemokine expressed by GC-Th cells. Real-time RT-PCR was used to examine 20 chemokines (in 4 groups: “Th1/inflammation,”“Th2,”“tissue-specific,” and “lymphoid” based on their expression sites or functions); GAPDH and β-actin were used as internal controls. Expression levels are shown in relative SYBR green fluorescence of the gene products to GAPDH products (GAPDH intensity = 100%). (D) CXCL13 expression in GC-Th cells was visualized by an in situ immunohistology technique. Frozen tonsil sections were stained with anti-CXCL13 antibody or control mouse IgG1 and then with secondary antibody conjugated with PE (red). Sections were further stained with anti–CD57-FITC for GC-Th cells (green). A confocal microscope system (Bio-Rad MRC 1024UV and Nikon Diaphot 300 microscope) was used to analyze the sections. (E-F) Many GC-Th cells are in physical contact with dendritic cells and GC-B cells. Frozen tonsil sections were stained with fluorescent monoclonal antibodies to CD11C (red for dendritic cells and some macrophages), CD57 (green for GC-Th cells), CD38 (red), and CD19 (blue). Please note that green (CD57+) cells are GC-Th cells and red cells are dendritic cells in panel E. Purple (CD38+CD19+) cells are GC-B cells and red (CD38+CD19–) cells are plasma cells in panel F. Arrows indicate several examples of GC-Th cells that are in direct contact with dendritic cells or GC-B cells. The images were taken at × 200 magnification.
GC-Th cells express CXCL13 protein in germinal centers where these cells are surrounded by antigen presenting cells
To detect CXCL13 expression at the protein level in situ, frozen tonsil sections were stained with anti-CXCL13 antibody. CXCL13 expression (Figure 4D; red) was detected in GC-Th cells stained in green (Figure 4D). As expected, CXCL13 expression was detected in other cell types in germinal centers in addition to CD57+ GC-Th cells. This is consistent with the finding that follicular dendritic cells and GC dendritic cells can express CXCL13 in germinal centers of secondary lymphoid tissues and chronically inflamed tissues in human and mice.31-35
Germinal centers are filled with activated GC-B cells undergoing differentiation to memory and plasma B cells. Dendritic cells are also present in large numbers in germinal centers.36 We examined whether these antigen presenting cells localize close to GC-Th cells. GC-Th cells were found in the areas where CD11C+ cells (GC dendritic cells and Tingible body macrophages) were abundant (Figure 4E). As pointed with arrows, many GC-Th cells are in direct contact with CD11C+ cells. Many GC-Th cells are also in direct contact with CD38+CD19+ GC-B cells (Figure 4F), suggesting that GC-Th cells are present in microenvironments where they can readily receive antigenic signals from dendritic cells and GC-B cells.
CXCL13 production by GC-Th cells is positively regulated by TCR activation
Freshly isolated GC-Th cells secrete low levels of CXCL13 (typically 50-100 pg/106 cells/day) in the absence of any stimulation. T-cell activation with small molecule agonists PMA (a protein kinase C activator) and ionomycin (a calcium ionophore), which induce signaling similar to T-cell receptor activation, increased CXCL13 expression by 20- to 40-fold (Figure 5A). Although anti-CD3 alone was a weak inducer of CXCL13 production, it efficiently stimulated CXCL13 production by GC-Th cells when combined with anti-CD28 (Figure 5B).
We examined whether interaction of GC-B cells induces CXCL13 production by GC-Th cells. GC-B cells and GC-Th cells were isolated from the same donors and cocultured (Figure 5C). When staphylococcal enterotoxin B (SEB) was used to conjugate the major histocompatibility complex (MHC) class II molecules of GC-B cells and the TCR of GC-Th cells, GC-Th cells produced CXCL13 at very high levels (5-10 ng/106 cells/day). In this culture system, B cells themselves did not produce any detectable level of CXCL13 in the conditions tested (the sensitivity of CXCL13 ELISA is > 10 pg/mL). It was still possible that CXCL13 produced in GC-Th cell–B-cell cocultures could be from GC-B cells in response to activation by GC-Th cells. To rule out this possibility, we reisolated the 2 cell subsets after culture and individually examined them for expression of CXCL13 and β-actin by quantitative real-time PCR. Whereas high levels of CXCL13 message expression were observed in the reisolated GC-Th cells, no significant expression of CXCL13 message was detected in the reisolated B cells (Figure 5D). Mature dendritic cells prepared from blood monocytes were also efficient in induction of CXCL13 production by GC-Th cells. CXCL13 concentrations of 20 ng/mLto 30 ng/mLwere achieved after 3 to 4 days in culture with allogenic dendritic cells in the presence of SEB (not shown). When cocultured GC-Th cells and dendritic cells were reisolated and individually examined for CXCL13 message levels, only the GC-Th cells had high levels of CXCL13 message (Figure 5E).
GC-Th cells were further compared with in vitro generated Th1 and Th2 cells. When activated with GC-B cells as antigen presenting cells in the presence of SEB, only the cultures with GC-Th cells but not with Th1 or Th2 cells produced high amounts of CXCL13 (Figure 5F). CXCL13 production by GC-Th cells is dependent on transcription and translation processes: when treated with actinomycin D or cycloheximide, CXCL13 synthesis by GC-Th cells in response to TCR activation was almost completely suppressed (not shown).
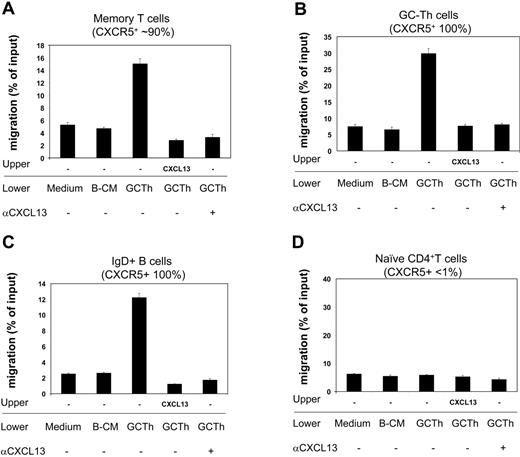
GC-Th cell–derived CXCL13 induces migration of specialized T- and B-cell subsets
Next we examined the biologic activity of the CXCL13 secreted by GC-Th cells during interaction with GC-B cells. Using the culture media as a chemoattractant, we performed chemotaxis assays on fresh tonsil lymphocytes, and identified responsive cell subsets (Figure 6). Conditioned media from the cultures of GC-Th cells, activated by SEB-pulsed GC-B cells, efficiently attracted memory CD4+ T cells, GC-Th cells, and IgD+ naive B cells. However, it did not have any chemotactic effect on naive CD4+ T cells. B-cell–conditioned media did not have any chemotactic effect on the cell types tested. In order to determine whether this chemotaxis was mediated by CXCL13 but not by other chemokines, we performed the chemotaxis assays in the presence of CXCL13 in the upper chamber, thus under a negative gradient of CXCL13. In this condition, no migration of any cell type has been observed to the GC-Th cell–conditioned media. We further performed chemotaxis assays in the presence of neutralizing anti-CXCL13 in the lower chambers. Addition of anti-CXCL13 abolished the chemotaxis of memory T cells, GC-Th cells, and naive B cells to the GC-Th cell–conditioned media, demonstrating that CXCL13 is responsible for the chemotactic activity of the conditioned media.
GC-Th cell–derived chemoattractants attract CXCR5+ T and B cells in a CXCL13-dependent manner. Conditioned media obtained from culture of GC-Th cells (shown as GC-Th) were used for chemotaxis assays. Fresh uncultured (medium) and conditioned media from GC-B cell cultures (B-CM) in the presence of SEB were used for negative controls. CXCL13 was added to upper chambers at 5 μg/mL. Anti-CXCL13 (or control antibody) was added at 25 μg/mL to lower chambers. The isotype control antibody did not inhibit the chemotaxis (not shown). After chemotaxis, the migrated cells were stained with fluorescent antibodies (anti–CD57-FITC, anti–IgD-PE, anti–CD4-PerCP, and anti–CD45RA-APC) to identify and numerate various tonsil lymphocyte subsets: CD4+CD45RA–CD57– memory T cells, CD4+CD57+ GC-Th cells, IgD+ naive B cells, and CD4+CD45RA+ naive T cells. The numbers of cells in the lower chambers (migrated cells) and in input cells were determined by FACS. Data are shown as percent migration of the indicated subsets. The error bars are the differences of duplicated experiments. Representative data from 3 independent experiments are shown.
GC-Th cell–derived chemoattractants attract CXCR5+ T and B cells in a CXCL13-dependent manner. Conditioned media obtained from culture of GC-Th cells (shown as GC-Th) were used for chemotaxis assays. Fresh uncultured (medium) and conditioned media from GC-B cell cultures (B-CM) in the presence of SEB were used for negative controls. CXCL13 was added to upper chambers at 5 μg/mL. Anti-CXCL13 (or control antibody) was added at 25 μg/mL to lower chambers. The isotype control antibody did not inhibit the chemotaxis (not shown). After chemotaxis, the migrated cells were stained with fluorescent antibodies (anti–CD57-FITC, anti–IgD-PE, anti–CD4-PerCP, and anti–CD45RA-APC) to identify and numerate various tonsil lymphocyte subsets: CD4+CD45RA–CD57– memory T cells, CD4+CD57+ GC-Th cells, IgD+ naive B cells, and CD4+CD45RA+ naive T cells. The numbers of cells in the lower chambers (migrated cells) and in input cells were determined by FACS. Data are shown as percent migration of the indicated subsets. The error bars are the differences of duplicated experiments. Representative data from 3 independent experiments are shown.
Discussion
In this study, we examined the gene expression pattern of human CD57+ GC-Th cells in an effort to understand the molecular basis of their unique function in germinal centers. We determined the overall differences in gene expression patterns between CD57+ GC-Th cells and other memory/effector T-cell subsets. More specific differences in expression of individual genes were observed in several key gene groups. We further studied the regulation of CXCL13 production, one of the most highly up-regulated genes in GC-Th cells versus naive, CXCR5+CCR7+ early memory, and CCR7– effector memory cells.
Based on their migratory characteristics, general lack of Th1 or Th2 cytokine production and function in helping B cells, it was proposed that GC-Th cells may represent a novel functional lineage of CD4+ effector T cells.10 They are mostly nonpolarized, express CXCR5 but not CCR7, and are localized in the GC. They are efficient in inducing B-cell antibody production compared with other T-cell subsets.13 GC-Th cells proliferate poorly compared with other naive and memory T cells in response to TCR activation.37 This information is consistent with the hypothesis that GC-Th cells may be terminally differentiated effector T cells in germinal centers, specialized in helping GC-B cells differentiate to memory and plasma B cells. The microarray data in this study show that GC-Th cells are indeed very different in gene expression from the rest of CD4+ T-helper subsets. Especially, GC-Th cells and CCR7– effector memory cells show highly distinctive patterns of gene expression from each other. Whereas the CXCR5+CCR7+ early memory and CCR7– “effector memory” CD4+ T cells are more closely related to each other, GC-Th cells are distant from these T-cell subsets in overall gene expression. CXCL13 is produced when GC-Th cells are activated. Therefore, the fact that freshly isolated GC-Th cells expressed high levels of CXCL13 mRNA suggests that they had been stimulated by antigen presenting cells in germinal centers. In contrast, CXCR5+CCR7+ early memory and CCR7– effector memory cells, isolated from peripheral blood, were mostly resting cells. Therefore, the unique global gene expression of GC-Th cells may be the result, in part, of their activation status. On the other hand, CXCL13 is produced by GC-Th cells but not by Th1 or Th2 cells regardless of their activation status (Figure 5F). Therefore, the unique gene expression of GC-Th cells may also be the result of their cell type–specific gene expression program.
GC-Th cells differed from the other subsets in expression of numerous transcription factors. In this regard, GC-Th cells and CCR7– effector memory T cells displayed almost opposite expression patterns of transcription factors from each other, a finding consistent with the hypothesis that GC-Th cells are distinct from polarized effector T cells in cell lineage. Some of the transcription factors up-regulated in GC-Th cells are signal transducer and activator of transcription 1 (STAT1), STAT2, Zhangfei, B-cell chronic lymphocytic leukemia (CLL)/lymphoma 6 (BCL6), general transcription factor IIH, and leucine zipper transcription like factor 1. STAT1 and STAT2 play important roles in T-cell differentiation and interferon-mediated responses.38,39 An increase in expression of general transcription factor component, which forms the preinitiation transcription complex,40 suggests that general transcription capacity is enhanced in GC-Th cells. Zhangfei (ZF) regulates the host transcription factor (HCF) required for transcriptional activation of herpes simplex virus (HSV) immediate-early (IE) genes by the virion protein VP16.41 A number of transcription factors such as leucine zipper transcription like factor 1 and BCL6 with their functions yet to be characterized in GC-Th cells were up-regulated (Figure 3). It is possible that combinations of these transcription factors confer on GC-Th cells their unique gene expression pattern.
The gene expression profile of GC-Th cells in this study provides some clues on their effector function, migration, cell cycle, and activation state. Among the genes uniquely expressed in GC-Th cells, some may directly account for their function in germinal centers. The chemokine receptor CXCR5 is important for their localization; ICOS on GC-Th cells may provide a costimulation signal necessary for B-cell differentiation30,42,43 ; and CXCL13 could attract the appropriate immune cells into germinal centers. Additionally, however, GC-Th cells express genes whose functions in the germinal centers are unclear: for example, they contain high message levels of granzyme A and granzyme K, enzymes whose functions are typically associated with cytotoxic activity.
The microarray data showed that GC-Th cells express CXCL13, a chemokine specifically expressed in lymphoid follicles and required for B-cell localization. We verified with real-time PCR analysis that CXCL13 is the only chemokine highly expressed by GC-Th cells among the 20 chemokines examined. Mice deficient in CXCL13 or its receptor CXCR5 are defective in B-cell homing to follicles in lymph nodes as well as in spleen.44,45 In addition to its chemotactic activity toward B cells, it has been shown in mice that CXCL13 activates naive B cells and up-regulates membrane LTα1β2, which promotes follicular dendritic cell development46-49 and production of CXCL13 itself.31,44 It has been reported that several non–T-cell types such as follicular dendritic cells, dendritic cells, epithelial cells, and endothelial cells express CXCL13.31-35 Our study provides new evidence that T cells in germinal centers can produce large amounts of CXCL13, and the CXCL13 production by GC-Th cells is regulated by TCR activation. Moreover, costimulation through the CD28 receptor along with TCR activation is necessary for induction of CXCL13 in GC-Th cells (Figure 5). Freshly isolated GC-Th cells express high levels of CXCL13 mRNA. Without persistent T-cell activation in culture, however, GC-Th cells would not maintain the initial levels of CXCL13 mRNA and are unable to produce significant levels of CXCL13 protein. The CXCL13 secreted by GC-Th cells can induce chemotaxis of memory T cells, IgD+ B cells, and GC-Th cells themselves, but not naive T cells (Figure 6). It would also attract as-yet-unactivated GC-Th cells, focusing helper activity to active sites of stimulation. Therefore, GC-Th cells, in addition to their strategic localization in germinal centers to help B-cell differentiation, are in a position to recruit more CXCR5+ immune cells into germinal centers. The fact that GC-Th cells can produce CXCL13 in response to TCR activation may be strategically important for the immune system to produce the right amount of CXCL13 according to the amount of antigen presented during immune responses.
Additional information on the genes highly up-regulated or down-regulated in GC-Th cells compared with naive, CXCR5+CCR7+, and CCR7–CD4+ T cells is shown in descending order in Table S1 and Table S2. One of the genes specifically down-regulated in GC-Th cells versus other T-cell subsets is sphingolipid G-protein–coupled receptor 1 (also called S1P1 receptor or Edg-1; down-regulated 5- to 7-fold). Interestingly, this is the receptor required for exit of lymphocytes from secondary lymphoid organs and thymus.50,51 Therefore, the down-regulation of this gene is consistent with the fact that CD57+ GC-Th cells are trapped in germinal centers and don't circulate in the blood system. Taken together, we conclude that GC-Th cells have a unique gene expression program that is distinct from that of other memory and effector T-cell subsets.
Prepublished online as Blood First Edition Paper, June 22, 2004; DOI 10.1182/blood-2004-03-1206.
Supported by grants from the Leukemia and Lymphoma Society, the American Cancer Society (IRG-58-006-44), the Leukemia Research Foundation, and Eli and Edythe L. Broad Foundation (C.H.K.), and from the National Institutes of Health (E.C.B.).
The online version of the article includes a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We appreciate the critical comments from Drs Harm HogenEsch and Ramesh Vemulapalli at Purdue University. We thank Jennie Sturgis at Purdue Cytometry Lab for her excellent assistance in confocal microscopy image analyses.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal