Abstract
Fludarabine, the current standard treatment for B-cell chronic lymphocytic leukemia (CLL), can induce apoptosis in CLL cells in vitro, and a number of molecular mechanisms contribute to its cytotoxicity. Using gene expression profiling, we investigated the molecular consequences of fludarabine treatment of patients with CLL in vivo. In 7 patients with CLL, a consistent gene expression signature of in vivo fludarabine exposure was identified. Many of the fludarabine signature genes were known p53 target genes and genes involved in DNA repair. In vitro treatment of CLL cells with fludarabine induced the same set of genes as observed in vivo, and many of these genes were also induced by in vitro exposure of CLL cells to ionizing radiation. Using isogenic p53 wild-type and null lymphoblastoid cell lines, we confirmed that many of the fludarabine signature genes were also p53 target genes. Because in vivo treatment with fludarabine induces a p53-dependent gene expression response, fludarabine treatment has the potential to select p53-mutant CLL cells, which are more drug resistant and associated with an aggressive clinical course. These considerations suggest that fludarabine treatment should be given in strict accordance to the current National Cancer Institute (NCI) guidelines that have established criteria of disease activity that warrant treatment.
Introduction
Curative treatments for B-cell chronic lymphocytic leukemia (CLL) are not yet available.1 Treatment with the purine analog fludarabine has been shown to increase the complete remission rate, enhance progression-free survival, and increase the median duration of the clinical response but not the survival in previously untreated patients with CLL, as compared with treatment with chlorambucil alone or combination chemotherapy.2,3 Treatment with fludarabine induces apoptosis in quiescent lymphocytes and CLL cells,4 and a variety of in vitro studies have addressed the mechanisms underlying this cell death. Although CLL cells are predominantly nondividing, the triphosphate of fludarabine can be incorporated into the DNA of the CLL cells, possibly during repair DNA synthesis.5,6 Nucleoside analog-induced DNA strand breaks lead to the activation of p53 and p53-dependent target genes,7 and p53-mediated induction of apoptosis has been suggested to play a central role in the killing of CLL cells. Whether an intact p53 pathway is required for fludarabine-induced killing is controversial,8-10 because, at least in some cases, killing can occur in a p53-independent fashion.11-13
The presence of p53 mutations in CLL cells is associated with decreased survival and clinical resistance to fludarabine treatment.14,15 However, because p53 loss can promote genomic instability, it is unclear whether this fludarabine resistance is a direct effect of p53 inactivation or an indirect effect of other genomic alterations in p53-deficient CLL cells.
Although the effects of fludarabine on CLL cells have been well studied in vitro,6,13,16 the molecular consequences of fludarabine treatment on CLL cells in vivo have not been fully investigated. In this study, we used genomic-scale gene expression profiling to monitor changes in gene expression in leukemic cells from CLL patients during their first course of fludarabine treatment and focused on the molecular changes during fludarabine treatment. We observed that fludarabine induced the expression of a characteristic and discrete set of genes that largely overlaps with p53-responsive genes, providing evidence that the p53 pathway is triggered in vivo during fludarabine treatment of patients with CLL.
Patients, materials, and methods
Clinical study
All patients with CLL were entered on a protocol approved by the National Cancer Institute (NCI) Institutional Review Board and gave informed consent. CLL was diagnosed and treated according to the revised NCI working group criteria.17 All 7 patients in the in vivo gene expression profiling study were previously untreated and received intravenous fludarabine (25 mg/m2) daily for 5 days, repeated every 28 days for up to 6 cycles. All 7 patients achieved a partial remission according to NCI criteria.17 In patient CLL 39, the sixth cycle of fludarabine could not be given because of an intercurrent myocardial infarction, and in patient CLL 45 fludarabine treatment had to be stopped after 4 cycles because of Coombs+ mild hemolysis. Clinical details of all patients are summarized in Table 1.
Clinical characteristics of patients with CLL studied
. | Before Tx . | . | . | . | . | . | Tx response . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | Sex/age, y . | IgVH status . | Cytogenetics/FISH . | RAI at Tx . | Mo to Tx . | Blood count before Tx . | Blood count after Tx . | Bone marrow after Tx . | Adenopathy before and after Tx . | Spleen CT before/after Tx, cm . | ||||||||
| CLL 15 | M/58 | Unmutated VH1-69 | del 13q, del 6q, del 11q | IV | 28 | ALC 388 × 109/L, Hb 138 g/L, plts 93 × 109/L | ALC 2 × 109/L, Hb 152 g/L, plts 78 × 109/L | Hypercellular, 9% lymphocytes | Clinically resolved, CT > 50% ↓ | 16/13 | ||||||||
| CLL 39 | M/83 | Mutated VH4-61 | del 13q | II | 115 | ALC 82 × 109/L, Hb 130 g/L, plts 133 × 109/L | ALC 5 × 109/L, Hb 125 g/L, plts 118 × 109/L | ND | Clinically and CT resolved | 14/12 | ||||||||
| CLL 55 | M/81 | Unmutated VH1-02 | + 12, del 11q | III | 14 | ALC 225 × 109/L, Hb 94 g/L, plts 130 × 109/L | ALC 8 × 109/L, Hb 128 g/L, plts 146 × 109/L | ND | Clinically resolved, CT > 50% ↓ | 18/14 | ||||||||
| CLL 62 | F/50 | Unmutated VH5-51 | del 13q, del 11q | II | 43 | ALC 115 × 109/L, Hb 135 g/L, plts 269 × 109/L | ALC 7 × 109/L, Hb 138 g/L, plts 171 × 109/L | Normocellular, 45% lymphocytes | Clinically resolved, CT > 50% ↓ | 13/10 | ||||||||
| CLL 52 | M/54 | Unmutated VH2-05 | t(1;14)(p32;q22) | II | 27 | ALC 161 × 109/L, Hb 119 g/L, plts 160 × 109/L | ALC 5 × 109/L, Hb 137 g/L, plts 201 × 109/L | Hypercellular, 95% lymphocytes | Clinically resolved CT > 50% ↓ | 16/14 | ||||||||
| CLL 45 | M/43 | Unmutated VH3-30 | +12 | III | 5 | ALC 124 × 109/L, Hb 69 g/L, plts 170 × 109/L | ALC 7 × 109/L, Hb 77 g/L, plts 88 × 109/L (AIHA) | Normocellular, 50% lymphocytes | Clinically and CT > 50% ↓ | 21/17 | ||||||||
| CLL 50 | M/56 | Unmutated VH3-39 | +12, del 11q | II | 27 | ALC 17 × 109/L, Hb 125 g/L, plts 57 × 109/L | ALC 1 × 109/L, Hb 142 g/L, plts 97 × 109/L | Hypocellular, lymphoid nodules | Clinically and CT resolved | 23/16 | ||||||||
| CLL 31 | F/77 | Mutated VH3-23 | del 13q | II | 10 | NA | NA | NA | NA | NA | ||||||||
| CLL 53 | F/46 | Mutated VH3-33 | del 13q | NA, stable | NA | NA | NA | NA | NA | NA | ||||||||
. | Before Tx . | . | . | . | . | . | Tx response . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | Sex/age, y . | IgVH status . | Cytogenetics/FISH . | RAI at Tx . | Mo to Tx . | Blood count before Tx . | Blood count after Tx . | Bone marrow after Tx . | Adenopathy before and after Tx . | Spleen CT before/after Tx, cm . | ||||||||
| CLL 15 | M/58 | Unmutated VH1-69 | del 13q, del 6q, del 11q | IV | 28 | ALC 388 × 109/L, Hb 138 g/L, plts 93 × 109/L | ALC 2 × 109/L, Hb 152 g/L, plts 78 × 109/L | Hypercellular, 9% lymphocytes | Clinically resolved, CT > 50% ↓ | 16/13 | ||||||||
| CLL 39 | M/83 | Mutated VH4-61 | del 13q | II | 115 | ALC 82 × 109/L, Hb 130 g/L, plts 133 × 109/L | ALC 5 × 109/L, Hb 125 g/L, plts 118 × 109/L | ND | Clinically and CT resolved | 14/12 | ||||||||
| CLL 55 | M/81 | Unmutated VH1-02 | + 12, del 11q | III | 14 | ALC 225 × 109/L, Hb 94 g/L, plts 130 × 109/L | ALC 8 × 109/L, Hb 128 g/L, plts 146 × 109/L | ND | Clinically resolved, CT > 50% ↓ | 18/14 | ||||||||
| CLL 62 | F/50 | Unmutated VH5-51 | del 13q, del 11q | II | 43 | ALC 115 × 109/L, Hb 135 g/L, plts 269 × 109/L | ALC 7 × 109/L, Hb 138 g/L, plts 171 × 109/L | Normocellular, 45% lymphocytes | Clinically resolved, CT > 50% ↓ | 13/10 | ||||||||
| CLL 52 | M/54 | Unmutated VH2-05 | t(1;14)(p32;q22) | II | 27 | ALC 161 × 109/L, Hb 119 g/L, plts 160 × 109/L | ALC 5 × 109/L, Hb 137 g/L, plts 201 × 109/L | Hypercellular, 95% lymphocytes | Clinically resolved CT > 50% ↓ | 16/14 | ||||||||
| CLL 45 | M/43 | Unmutated VH3-30 | +12 | III | 5 | ALC 124 × 109/L, Hb 69 g/L, plts 170 × 109/L | ALC 7 × 109/L, Hb 77 g/L, plts 88 × 109/L (AIHA) | Normocellular, 50% lymphocytes | Clinically and CT > 50% ↓ | 21/17 | ||||||||
| CLL 50 | M/56 | Unmutated VH3-39 | +12, del 11q | II | 27 | ALC 17 × 109/L, Hb 125 g/L, plts 57 × 109/L | ALC 1 × 109/L, Hb 142 g/L, plts 97 × 109/L | Hypocellular, lymphoid nodules | Clinically and CT resolved | 23/16 | ||||||||
| CLL 31 | F/77 | Mutated VH3-23 | del 13q | II | 10 | NA | NA | NA | NA | NA | ||||||||
| CLL 53 | F/46 | Mutated VH3-33 | del 13q | NA, stable | NA | NA | NA | NA | NA | NA | ||||||||
Tx indicates treatment; IgVH, immunoglobulin gene heavy chain variable region; unmutated, sequence homology of IgVH region > 98% to germ line; del, cytogenetic deletion: ALC, absolute lymphocyte count (normal range, 0.46-4.7 × 109/L [460-4700/μL]); Hb, hemoglobin (normal range, 127-167 g/L [12.7-16.7g/dL] for men, 111-150 g/L (11.1-15 g/dL) for women); Plts, platelet count (normal range, 154-345 × 109/L [154 000-345 000/μL]); CT, computed tomography of relevant disease sites; Mutated, sequence homology of IgVH region < 98% to germ line; ND, not done; AIHA, autoimmune hemolytic anemia; and NA, not applicable.
For the in vitro studies, cells were obtained from one untreated (CLL 53) and one previously treated patient (CLL 31). CLL 31 was diagnosed with CLL in 1988 and required treatment in 1989 (chlorambucil and prednisone). In 1996, the patient progressed to stage IV disease and received 3 cycles of fludarabine with a partial response. Six cycles of fludarabine were again administered in 1997 and in 1998. Following further clinical progression, the patient enrolled in a clinical study at the National Cancer Institute of the investigational agent, 506U78, in fludarabine refractory patients. Cells from this patient were obtained prior to the start of the study medication.
CLL purification from blood samples
Mononuclear cells were separated using density gradient centrifugation (LSM Lymphocyte Separation Medium; ICN Biomedicals, Aurora, OH), and leukemic cells were purified by magnetic selection for CD19+ at 4° C (Miltenyi Biotec, Bergisch Gladbach, Germany). CD19+ cells were stored as a pellet at –80° C until RNA was isolated with use of the Fast-Track kit 2.0 (Invitrogen, Carlsbad, CA).
Cell culture
Dimethyl sulfoxide (DMSO)–preserved mononuclear cells from patients 15, 31, and 53 were thawed and immediately taken through negative selection by using equipment and reagents from StemCell Technologies (Vancouver, BC, Canada). Our previous study of gene expression profiles in CLL that included DMSO-preserved specimens demonstrated no effect of the freezing procedure on gene expression patterns compared with fresh CLL cells.18 CD19+ cells (purity > 98%) were cultured at 37° C at a concentration of 1 × 107 cells/mL in alpha-modified minimum essential medium supplemented with 100 U/mL penicillin, 0.1 mg/mL streptomycin, interleukin 4 (IL-4) at 50 ng/mL, and 10% heat-inactivated fetal bovine serum. Cells were treated with 9-β-d-arabinosyl-2-fluoroadenine (fludarabine) monophosphate at a concentration of 1 μM, a concentration that is comparable to that achieved in blood during treatment of patients with CLL.19 Alternatively, CLL cells were exposed to γ radiation (10 Gy). CLL cells were harvested for RNA preparation 24 hours or 48 hours after these treatments were initiated.
The isogenic human lymphoblast cell lines TK6 (wild-type p53) and NH32 (p53-null) were maintained as exponentially growing cultures in RPMI 1640 media supplemented with 10% horse serum, 100 U/mL penicillin, and 0.1 mg/mL streptomycin.20 These cells were irradiated with 10 Gy, and total RNA was extracted by using the Trizol reagent (Invitrogen) before irradiation and at 1, 3, 7, and 24 hours following irradiation.
Immunoglobulin (Ig) VH sequencing
To generate oligo-deoxythymidine (dT)–primed cDNA with use of Superscript (Life Technologies, Bethesda, MD), 500 ng mRNA or 1 to 5 μg total RNA from purified CLL cells was used. Amplification of the immunoglobulin VH sequence was performed essentially as described.21 Briefly, cDNA was amplified by polymerase chain reaction (PCR) by using a mixture of 5′ oligonucleotides specific for each leader sequence of the VH1 to VH7 families (VH1 and VH7, 5′-CCA TGG ACT GGA CCT GGA-3′; VH2, 5′-ATG GAC ATA CTT TGT TCC AC-3′; VH3, 5′-CCA TGG AGT TTG GGC TGA GC-3′; VH4, 5′-ATG AAA CAC CTG TGG TTC TT-3′; VH5, 5′-ATG GGG TCA ACC GCC ATC CT-3′; VH6, 5′-ATG TCT GTC TCC TTC CTC AT-3′) as forward primers and either a 3′ oligonucleotide complementary to the JH consensus sequence (5′-ACC TGA GGA GAC GGT GAC C-3′) or the constant region of the IgM locus (5′-AGG AGA AAG TGA TGG AGT CG-3′) as reverse primers. Patients with leukemic VH sequences that had 98% or greater identity to a germline-encoded VH gene were categorized as Ig-unmutated patients and the remainder of the patients were categorized as Ig-mutated patients.
CD38 expression
Whole blood was stained within 24 hours of collection with a panel of antibodies as previously described.22 Six-parameter, 4-color flow cytometry was performed with a BD FACSCalibur flow cytometer. The sensitivity of fluorescent detectors was set and monitored using Calibrite Beads (BD, San Jose, CA) according to manufacturer's recommendations. Data (collected in List mode) was analyzed with CellQuest software (BD). For analysis, lymphocytes were gated by forward and side scatter, or by CD45 staining versus side scatter. CD19, CD20, and CD3 populations were back-gated to determine appropriateness of analysis gates. Isotype controls were run with each patient specimen. CD38+ cells were determined as the percentage of lymphocytes staining more intensely with anti-CD38 than with isotype controls. CLLs for which CD38 was expressed in 30% or more of the leukemic cells were considered CD38+.
Cytogenetics
Buffy coats from fresh, heparinized peripheral blood were cultured in duplicate with each of the following mitogens: phytohemagglutinin, phorbol 12-myristate 13-acetate, pokeweed mitogen, and Escherichia coli lipopolysaccharide. After 96 hours in a humidified 5% CO2 incubator at 37° C, cells were harvested and fixed in 3:1 methanol-glacial acetic acid. For G-banded karyotype analysis, slides were stained with Wright stain and, if present, at least 10 metaphase cells from each culture condition were fully analyzed at the microscope. Two karyotypes of each cell line were prepared, and findings were designated according to the International System for Human Cytogenic Nomenclature (ISCN).23 For interphase fluorescence in situ hybridization (FISH), fresh slides were made from the fixed cell pellets, and hybridization was performed by using commercially available probes (Vysis, Downers Grove, IL) to detect genomic losses in 13q14 (D13S319, D13S25), 11q23 (MLL), 11q22 (ATM), and 17p13.1 (p53), and to detect trisomy 12 (CEP12). Minimums of 200 interphase nuclei were scored for hybridization signals for each probe. On the basis of results of healthy controls, losses and gains were interpreted as negative if they occurred in 4% or fewer of the nuclei; however, none of the positive results were equivocal as all were present in 12% or more of the nuclei.
Microarray procedures
Lymphochip DNA arrays prepared from 17 856 cDNA clones were used for analysis of gene expression as described.24,25 For the analysis of gene expression in CLL cells obtained during in vivo fludarabine treatment, fluorescent cDNA probes were prepared from CLL mRNA with incorporation of Cy5-labeled nucleotides. A Cy3-labeled cDNA probe was prepared from a previously described reference pool of RNA25 and cohybridized with the Cy3-labeled experimental probe to the Lymphochip DNA microarray. The use of the reference probe in each hybridization allowed us to compare the relative expression of a given gene across all of the experiments.24 For analysis of gene expression in in vitro–cultured cells, Cy5-labeled cDNA probes prepared from fludarabine- or γ radiation–treated cells were compared with Cy3-labeled cDNA probes prepared from untreated control cells. Microarrays were scanned at 10-μm resolution using a GenePix scanner (Axon Instruments, Union City, CA) at variable photomultiplier tube (PMT) voltage (< 1% saturated spots), and data files were entered into a custom database maintained at the National Cancer Institute. Expression data used for analysis fulfilled the following criteria: spot size of at least 25 μm, minimum intensities of at least 50 relative fluorescent units (RFUs) in the Cy3 and Cy5 channels or minimum intensity of at least 500 RFU in either channel alone. Hierarchical clustering and display of the gene expression data were performed with use of the Cluster and TreeView software packages.26 The complete dataset can be downloaded at http://lymphochip.nih.gov/Fludarabine.
Results
Cytogenetics results
Combined results from G-banded karyotype and interphase FISH analyses are summarized in Table 1. Analyses were performed just prior to starting fludarabine therapy in all patients except CLL 52 who was studied 2 months after completing treatment. G-banded analysis was inadequate (< 20 normal metaphase cells) in CLL 53 only. Clonal chromosome abnormalities were found by G-banding prior to treatment in 4 patients (CLL 15, 62, 45, and 50) and following treatment in CLL 52. Interphase FISH confirmed the G-band findings in CLL 15, 62, 45, and 50 and also revealed the deletion 11q and trisomy 12 clones in the sample from CLL 55. Deletions in band 13q14 not detected by G-banding were found by using interphase FISH in CLL 15, 39, 62, 31, and 53. Interphase FISH was performed by using the p53 probe on all pretreatment samples; no losses of p53 were detected.
In vivo gene expression response of CLL cells to fludarabine treatment
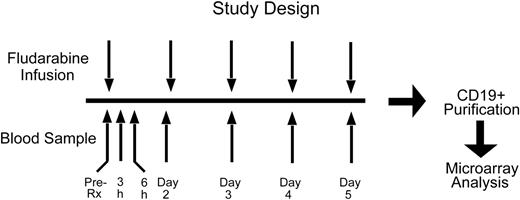
We designed a clinical protocol aimed at elucidating the gene expression changes that are induced in vivo in the leukemic cells of patients receiving fludarabine treatment (Figure 1). Seven previously untreated patients with CLL who required fludarabine treatment were studied. Blood samples were taken immediately prior to the start of the first cycle of fludarabine (time 0), at 3 and 6 hours after the first dose of fludarabine, and immediately before each subsequent dose on days 2, 3, 4, and 5. The CD19+ leukemic cells from each sample were purified by magnetic sorting. In all instances, purity of CD19+ B-CLL cells was more than 98% (data not shown). For subsequent DNA microarray analysis of gene expression mRNA was extracted from the tumor specimens.
Design of the study. Gene expression profiles of leukemic cells from patients with CLL were studied at 7 time points immediately before and after initiation of the first cycle of fludarabine treatment.
Design of the study. Gene expression profiles of leukemic cells from patients with CLL were studied at 7 time points immediately before and after initiation of the first cycle of fludarabine treatment.
The clinical characteristics of the patients are summarized in Table 1, along with analysis of their leukemic cells for immunoglobulin gene mutations and CD38 expression. At diagnosis, the patients had a median age of 56 years (range, 43-83 years) and had Rai stage I or II disease. The median time to treatment was 27 months with a range of 5 to 115 months from diagnosis. Six of these patients had unmutated immunoglobulin (Ig) VH genes and required treatment within 5 to 43 months of initial diagnosis, as expected given the more progressive nature of this subtype of CLL.1 The one patient with mutated Ig VH genes required treatment at 115 months from diagnosis.
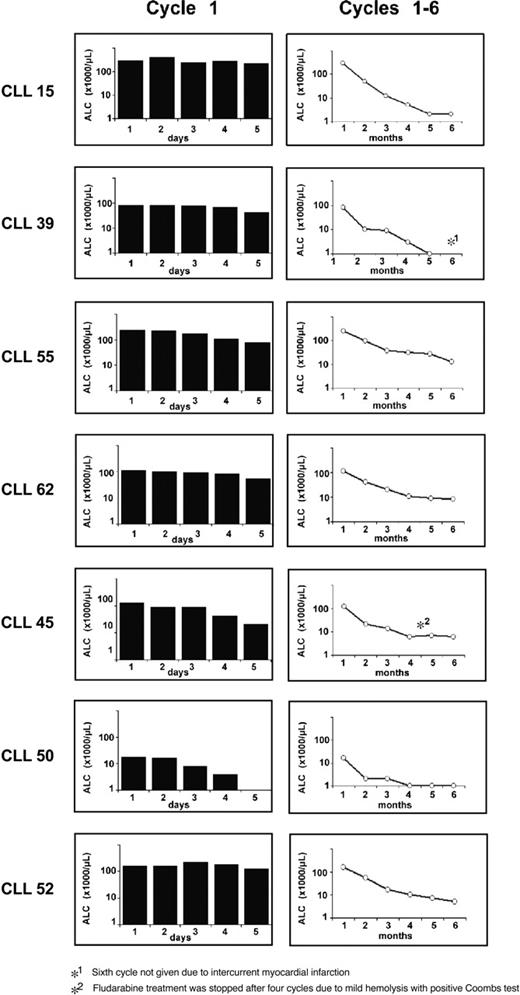
Following fludarabine treatment, all patients responded well with decreases in peripheral white blood cell (WBC) counts and achieved partial remissions (Figure 2). Following the first cycle of fludarabine, patients had only modest decreases in white counts, whereas 2 patients (CLL 45 and CLL 50) had more dramatic reductions.
Clinical response to fludarabine treatment. Shown are the absolute lymphocyte counts (ALCs) for each of the 7 patients with CLL during days 1 to 5 of the first cycle of fludarabine (left column) and during the fludarabine cycles 1 to 6 (right column).
Clinical response to fludarabine treatment. Shown are the absolute lymphocyte counts (ALCs) for each of the 7 patients with CLL during days 1 to 5 of the first cycle of fludarabine (left column) and during the fludarabine cycles 1 to 6 (right column).
We profiled gene expression in the CLL leukemic samples obtained before and during fludarabine administration by using Lymphochip DNA microarrays.25 Initial exploration of the dataset with use of a hierarchical clustering algorithm26 revealed that most of the genes represented on the microarray were not altered in expression by fludarabine treatment (data not shown). Nonetheless, this approach identified one cluster of genes that was up-regulated during fludarabine treatment. No change in the expression of these genes was evident at 3 and 6 hours after treatment, but increased and consistent expression occurred at days 3 to 5 after treatment.
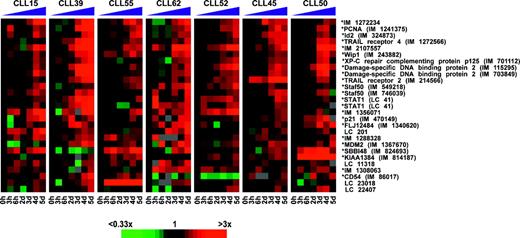
On the basis of these observations, we searched the dataset by using mathematical selection criteria aimed at identifying genes that were increased in expression by fludarabine in most patients. Specifically, we identified microarray elements for which the expression levels were higher in the day 3 to 5 samples than in the time 0, 3-hour, and 6-hour samples in at least 5 of the 7 patients. This procedure selected 27 microarray elements, 18 of which represented named genes and 9 of which represented novel genes of unknown function. Figure 3 depicts the change in expression of each gene during fludarabine treatment relative to its pretreatment level. The data are displayed by using a color scheme in which the pretreatment expression level is black, and expression levels greater or less than this level are represented by shades of red and green, respectively. Notably, 7 of these microarray elements represent genes that are documented target genes of the p53 transcription factor, including p21, MDM2, DDB2 (damage-specific DNA binding protein 2, 48 kDa), TNFRSF10B (TRAIL receptor 2/DR5), PCNA (proliferating cell nuclear antigen), and PPM1D (protein phosphatase 1D magnesium-dependent, delta isoform/Wip1). In addition, several of the other genes that were induced by in vivo fludarabine treatment were experimentally confirmed to be p53 targets (described in “In vitro gene expression response of CLL cells to fludarabine and ionizing radiation”). These data suggest that in vivo administration of fludarabine induces a p53-dependent transcriptional response in the CLL cells. In addition, we searched the dataset for sets of genes that were consistently down-regulated in response to fludarabine treatment, but none were detected.
DNA microarray analysis identifies a homogeneous p53-dependent gene expression response of CLL cells to treatment with fludarabine in vivo. Changes in gene expression are depicted for each gene relative to its pretreatment level (black squares). Shades of red and green indicate up- or down-regulation of a given gene according to the color scheme shown below. Sequence-verified Lymphochip clones are labeled with an asterisk (*). For unnamed genes IMAGE clone numbers (IM) are given. Unnamed genes without sequence verification are labeled with a Lymphochip (LC) number.
DNA microarray analysis identifies a homogeneous p53-dependent gene expression response of CLL cells to treatment with fludarabine in vivo. Changes in gene expression are depicted for each gene relative to its pretreatment level (black squares). Shades of red and green indicate up- or down-regulation of a given gene according to the color scheme shown below. Sequence-verified Lymphochip clones are labeled with an asterisk (*). For unnamed genes IMAGE clone numbers (IM) are given. Unnamed genes without sequence verification are labeled with a Lymphochip (LC) number.
In vitro gene expression response of CLL cells to fludarabine and ionizing radiation
To determine whether the gene expression changes observed in vivo were a direct effect of fludarabine treatment, we treated CLL cells in vitro with a pharmacologically relevant concentration of fludarabine (1 μM) and studied gene expression at 24 and 48 hours after treatment. CLL cells were obtained from 2 previously untreated patients: patient CLL 15 who subsequently required fludarabine treatment and patient CLL 53 who had stable disease and did not require treatment. In addition, we studied CLL cells from patient CLL 31 who had received several cycles of fludarabine treatment and was clinically resistant to fludarabine. In patients CLL 15 and CLL 53, fludarabine treatment was cytotoxic and cell viability decreased significantly after 48 hours, whereas no cytotoxic effect was observed in the cells obtained from patient CLL 31 (data not shown). This differential sensitivity to fludarabine was mirrored in the gene expression changes induced in the CLL cells by fludarabine (Figure 4A). Fludarabine treatment of cells from patients CLL 15 and CLL 53 up-regulated many of the same genes that were induced by in vivo fludarabine administration, including the known p53 target genes as well as several novel genes. In contrast, fludarabine had little if any effect on the expression of these genes in cells from the fludarabine-resistant patient CLL 31 (Figure 4). Interestingly, the tumor cells from this patient showed strong positivity for p53 by immunohistochemistry, suggesting inactivation of the p53 pathway (Figure 4B).
p53-dependent gene expression. (A) p53-dependent gene expression response of CLL cells in vitro following treatment with fludarabine or irradiation. Gene expression changes at 24 hours (1 day) and 48 hours (2 days) are displayed for each gene relative to its expression in control cultures. In CLL patients 15 and 53, fludarabine treatment and irradiation were cytotoxic, whereas no effect of either treatment was observed in CLL patient 31, who was clinically resistant to fludarabine. (B) Immunohistochemical staining for p53 in CLL 31, demonstrating overexpression in approximately 30% of tumor cells. The image was obtained using a Zeiss Axioplan 2 microscope, 100 ×/1.4 N.A. planar apochromatic oil immersion objective, and MicroMAX digital camera (Princeton Instruments, Trenton, NJ), and then processed with IPLab Spectrum (Signal Analytics, Vienna, VA) and Adobe Photoshop Software.
p53-dependent gene expression. (A) p53-dependent gene expression response of CLL cells in vitro following treatment with fludarabine or irradiation. Gene expression changes at 24 hours (1 day) and 48 hours (2 days) are displayed for each gene relative to its expression in control cultures. In CLL patients 15 and 53, fludarabine treatment and irradiation were cytotoxic, whereas no effect of either treatment was observed in CLL patient 31, who was clinically resistant to fludarabine. (B) Immunohistochemical staining for p53 in CLL 31, demonstrating overexpression in approximately 30% of tumor cells. The image was obtained using a Zeiss Axioplan 2 microscope, 100 ×/1.4 N.A. planar apochromatic oil immersion objective, and MicroMAX digital camera (Princeton Instruments, Trenton, NJ), and then processed with IPLab Spectrum (Signal Analytics, Vienna, VA) and Adobe Photoshop Software.
One of the proposed mechanisms by which fludarabine induces apoptosis in CLL cells is by the induction of DNA strand breaks. We, therefore, exposed CLL cells in vitro to ionizing radiation and compared the gene expression response with this stimulus with the response to fludarabine. As expected, exposure to γ radiation was toxic in the tumor cells from patients CLL 15 and CLL 53, whereas the viability of the cells from the fludarabine-resistant patient CLL 31 was unaffected (data not shown). The gene expression response to γ radiation was highly similar to the response to fludarabine (Figure 4). Virtually all of the genes that were induced by fludarabine were also induced by γ radiation, and γ radiation did not induce any genes that were not induced by fludarabine.
Finally, we tested whether the genes that were up-regulated by fludarabine and γ radiation were dependent on p53 for their induction. We used 2 isogenic lymphoblastoid cell lines for these experiments, which differed only with respect to their p53 loci. The parental TK6 cell line has wild-type p53 alleles, whereas the TK6-derived NH32 cells have 2 null p53 alleles that were created by homologous recombination.20 We subjected these cell lines to γ radiation and assessed gene expression changes occurring within 24 hours of treatment (Figure 5). Many of the p53 target genes that were induced in CLL cells by fludarabine and γ radiation were also induced in the p53 wild-type cell line but were not induced in the p53-null cell line.
Genes that are induced in CLL cells after fludarabine treatment or irradiation are dependent on p53. Two isogenic cell lines that differ in their p53 loci only (TK6 with p53 wild-type alleles and NH32 with 2 null p53 alleles) were irradiated, and gene expression changes for selected genes are depicted relative to their pretreatment levels (black squares) according to the color scheme shown in Figure 4.
Genes that are induced in CLL cells after fludarabine treatment or irradiation are dependent on p53. Two isogenic cell lines that differ in their p53 loci only (TK6 with p53 wild-type alleles and NH32 with 2 null p53 alleles) were irradiated, and gene expression changes for selected genes are depicted relative to their pretreatment levels (black squares) according to the color scheme shown in Figure 4.
Discussion
Although fludarabine is the most effective single drug at inducing clinical responses in CLL,2 the molecular changes that occur in CLL cells during treatment have not been extensively investigated. In general, the in vivo effects of a chemotherapeutic agent may not be fully modeled by in vitro cultures for several reasons. First, the concentration of a therapeutic agent during in vivo administration is not constant and determined by multiple factors, including dose, schedule, and pharmacokinetics. In particular, pharmacokinetic-dependent fluctuations in blood concentrations are difficult to accurately mimic in vitro. Second, cancer cells in vivo are subject to various pro-proliferative and antiapoptotic stimuli, many of which are not fully characterized, and these cannot be faithfully reproduced under in vitro culture conditions. Furthermore, under standard in vitro cultures of purified CLL cells, the cells die of apoptosis over a several-day period, which is not a characteristic of such cells in vivo. Thus, the in vitro response of CLL cells to drug treatment may be influenced by nonphysiologic intracellular mechanisms that promote spontaneous apoptosis in vitro.
For these reasons, we undertook the present clinical trial to identify molecular changes that occur in CLL cells during actual treatment. The gene expression profiles of CLL cells sampled during fludarabine administration in vivo revealed a stereotypical and relatively discrete molecular response. In all 7 patients studied, we observed a consistent up-regulation of known transcriptional targets of p53, including p21, MDM2, DDB2, TNFRSF10B, PCNA, and PPM1D.27-29 Furthermore, many of the other genes induced by fludarabine in vivo were also induced in vitro by γ irradiation in a p53-dependent manner. Similarly, fludarabine treatment in vitro induced many of the same genes induced in vivo, demonstrating that the in vivo gene expression changes were due to direct effects of fludarabine on the CLL cells. Further, CLL cells from a patient with clinical fludarabine resistance failed to show either a cytotoxic or gene expression response to fludarabine in vitro. Taken together, our data suggest that at standard therapeutic doses, fludarabine induces a p53-mediated gene expression response.
Although our study may have included more patients with refractory CLL (based on the low complete response (CR) rate and the inclusion of 4 patients with 11q deletions), it appears unlikely that the observed p53-mediated gene expression response is specific to this clinical subset of patients with CLL, because the CLL cases 15, 45, 50, 52, 53, and 55 all had a typical CLL gene expression profile in our previous gene expression study.18
Previous in vitro studies investigating the effect of nucleoside analogs on CLL cells have led to varying conclusions about the role of p53 in the apoptotic response to purine analogs such as fludarabine. In a study of 2-chlorodeoxyadenosine, a purine analog closely related to fludarabine, in vitro apoptosis in CLL cells was preceded by induction of p53 and its downstream target p21.8 Likewise, another study found that fludarabine treatment in vitro up-regulated p53 and its target gene MDM2, and further showed that fludarabine induced apoptosis in CLL cells with wild-type p53 but not in p53-mutated cells.9 In contrast, other studies report that fludarabine can also induce apoptosis of CLL cells in vitro in a p53-independent fashion.10-12 For example, spleen cells from p53 knock-out mice can be killed by fludarabine treatment in vitro, although they were somewhat more resistant than p53 wild-type cells.12 Although our results are consistent with the hypothesis that p53 is involved in the apoptosis of CLL cells during in vivo fludarabine administration, it is also possible that p53-independent mechanisms may play a role. It is noteworthy, however, that we did not detect prominent gene expression changes in vivo that could not be associated with either a p53-dependent or DNA damage–dependent response. Future studies will have to show whether the observed p53-dependent gene expression changes are specific for the response to fludarabine treatment or whether they can also occur in other therapies (eg, with chlorambucil).
Our finding that fludarabine activated a p53 response in all of our study patients has implications for the clinical management of CLL. Our results suggest that treatment of patients with CLL with fludarabine has the potential to select for outgrowth of p53 mutant subclones that would be cross-resistant to several other chemotherapeutic agents. It is also possible that p53 mutant CLL cells could cause a more aggressive disease on the basis of recent evidence that deletion of the p53 locus is associated with poor prognosis. These considerations suggest that fludarabine treatment should be given according to the current NCI guidelines that establish criteria of disease activity that warrant treatment. Further, the use of fludarabine could also be guided by prognostic data on the basis of the immunoglobulin mutation status of the leukemic cells,21,30 zeta-associated protein 70 (ZAP70) expression by the CLL cells31,32 and cytogenetic abnormalities.33 Ultimately, clinical trials will be needed to determine the optimal use of fludarabine in the treatment of patients with CLL. Finally, our data suggest that the combination of fludarabine with other therapeutic agents that may have a p53-independent mode of action, such as alemtuzumab (Campath-1H), may be synergistic.34
Prepublished online as Blood First Edition Paper, May 11, 2004; DOI 10.1182/blood-2003-09-3236.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Lisa Kruger, MS; Shannon Skarshaug, MS; and April Tos, MT, CLsp(CG) for performing the cytogenetic analyses.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal