Abstract
Patients with Philadelphia chromosome-positive (Ph+) acute lymphoblastic leukemia (ALL) generally have a poor prognosis and would benefit from the development of new therapeutic approaches. We previously demonstrated that an allosterically controllable ribozyme, maxizyme (Mz), can induce apoptosis in chronic myelogenous leukemia (CML) cells. Ph+ ALL cells harbor a bcrabl fusion gene (e1a2) encoding a 190-kDa fusion protein (p190) involved in disease pathogenesis. In this study, we have designed a Mz that specifically cleaves e1a2 mRNA and transduced this e1a2Mz into Ph+ ALL cells using a third-generation lentiviral vector system. In 3 of 5 Ph+ ALL cell lines, e1a2Mz transduction resulted in a significant decrease in viability and increased cell apoptosis. We observed a decrease in e1a2 mRNA in all Ph+ ALL cells transduced with e1a2Mz, and the e1a2 mRNA level was higher in e1a2Mz-resistant cells than in e1a2Mz-sensitive cells. All samples of primary Ph+ ALL cells tested showed e1a2Mz-induced growth inhibition and apoptosis. Importantly, e1a2Mz did not influence the colony formation of normal CD34+ cord blood cells. These results indicate that e1a2Mz kills Ph+ ALL cells specifically, suggesting that it may be used as a novel gene therapy strategy for Ph+ ALL. (Blood. 2004;104:356-363)
Introduction
The Philadelphia chromosome (Ph) can harbor one of several kinds of fusion genes between bcr and c-abl and plays a major pathogenetic role for chronic myelogenous leukemia (CML) and acute lymphoblastic leukemia (ALL).1 The p190 bcr-abl fusion gene (e1a2) is detectable in 20% to 35% of patients with ALL.2,3 Patients shown to express the mRNA transcript encoding e1a2 have a poor prognosis despite intensive treatment such as high-dose chemotherapy followed by hematopoietic stem cell transplantation.2-4 In addition, almost all of them become resistant quickly even to imatinib mesylate (STI571), a newly developed bcr-abl tyrosine kinase inhibitor.5-8 Therefore, development of ribozyme therapy targeting the p190 form of bcr-abl is considered a promising new therapeutic strategy.
A hammerhead ribozyme discovered initially in a plant viroid is known to be a strong RNA enzyme that only digests RNA at a specific target sequence, NUX (N indicates any ribonucleotide; X indicates A, C, or U).9-11 Therefore, we could design a ribozyme for the b3a2-type bcr-abl fusion gene, one of the major fusion genes in CML, which contains an NUX (GUU) sequence at its particular fusion junction, but not for other types of bcr-abl fusion genes such as b2a2 and e1a2, which are found in CML and ALL, respectively. To overcome this limitation, we previously developed a novel ribozyme, which we named maxizyme (Mz), that does not require the cleavage site at the junction site.12-14 In our previous study, we demonstrated that a Mz targeting the b2a2-type bcr-abl induced apoptosis in the BV173 CML cell line12 and prevented the death of mice that received BV173 cell transplants,15 and we proposed that it might provide a new strategy for future treatment of CML.
In our previous reports, we used a Moloney murine leukemia virus (MLV) vector to transduce the Mz that targets the b2a2-type bcr-abl (b2a2Mz), which resulted in very low transduction efficiencies into primary leukemia cells from CML patients; therefore, we could not demonstrate the effect of this Mz on these cells.16 As a high level of gene transduction was required to demonstrate the effect of Mz on primary leukemia cells, we examined various kinds of vector systems, including retroviral and adenoviral vectors, but could not obtain a sufficient level. Moreover, gene transduction of Ph+ ALL cells with the MLV vector was very difficult, even when we used established cell lines as target cells. Recently, we and others reported that gene transduction efficiencies of vesicular stomatitis virus (VSV)-pseudotyped lentiviral vectors, which were derived from human immunodeficiency virus type-1 (HIV), into patients' leukemia cells were much higher than those obtained using the MLV vector.17,18
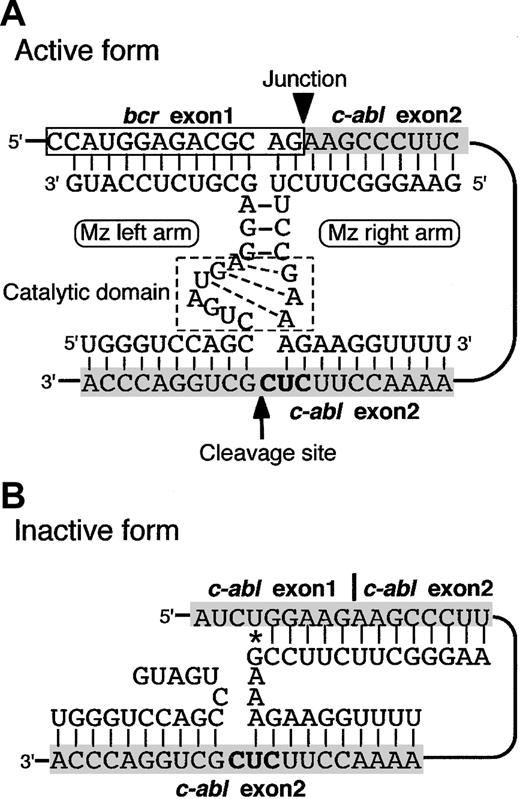
Therefore, we have developed a Mz-expressing HIV vector targeting the p190-type bcr-abl (e1a2Mz) for the treatment of Ph+ ALL. This e1a2Mz vector was designed to form an active secondary structure only in the presence of the p190-type bcr-abl fusion gene and not in its absence (Figure 1A-B, reproduced from Tanabe et al16 ) and was found to successfully digest synthetic e1a2 oligonucleotide.16 In this study, we have successfully transduced the e1a2Mz into Ph+ ALL cells with a third-generation HIV vector system and examined the effect of this Mz on Ph+ ALL cells and normal human hematopoietic progenitor cells.
Secondary structures of a maxizyme targeting the p190-type bcr-abl fusion gene. The e1a2Mz forms active and inactive structures when it recognizes and binds to p190-type bcr-abl mRNA (A) and normal c-abl mRNA (B), respectively.16 The gray shaded areas represent the squence of c-abl mRNA, and the bold letters “CUC” are the NUX sequence. *Non-Watson-Crick base pair.
Secondary structures of a maxizyme targeting the p190-type bcr-abl fusion gene. The e1a2Mz forms active and inactive structures when it recognizes and binds to p190-type bcr-abl mRNA (A) and normal c-abl mRNA (B), respectively.16 The gray shaded areas represent the squence of c-abl mRNA, and the bold letters “CUC” are the NUX sequence. *Non-Watson-Crick base pair.
Materials and methods
Cell lines
Human cervical carcinoma cell line HeLa cells were cultured in Eagle minimal essential medium supplemented with 10% fetal bovine serum (FBS). The 293T cells, a human embryonic kidney cell line transduced with simian virus 40 T antigen, were cultured in Dulbecco modified Eagle medium (DMEM) with 10% FBS. Ph+ ALL cell line OM9;22 cells were kindly supplied by Dr K. Ohyashiki (Tokyo Medical College, Tokyo, Japan),19 and KOPN-30, -57, and -72 cells were established as previously reported.20 Leukemia cell lines consisting of SUP-B15 Ph+ ALL cells or K562 CML-blastic crisis (CML-BC) cells were obtained from the American Type Culture Collection (Rockville, MD). The NB4 acute promyelocytic leukemia cell line was kindly provided by Dr M. Lanotte (Hôpital Saint-Louis, Paris, France).21 All cells except for SUP-B15 cells, which were cultured in Iscove modified Dulbecco medium (IMDM) supplemented with 20% FBS, were cultured in RPMI 1640 medium supplemented with 10% FBS.
Primary leukemia cells and cobblestone area formation
Primary leukemia cells were obtained from peripheral blood (PB) or bone marrow (BM) and used in the following experiments after obtaining informed consent, and this study was approved by the institutional review board of the Institute of Medical Science, The University of Tokyo. These cells were purified by Ficoll-Hypaque centrifugation, and more than 98% of the cells in the population were identified microscopically as leukemia cells. After e1a2Mz transduction, 5 × 103 to 5 × 104 cells were cocultured with a mouse stroma feeder cell line consisting of HESS-5 cells (a generous gift from Dr T. Tsuji, Science University of Tokyo, Chiba, Japan),22 which were seeded in 6-well plates with IMDM and 20% FBS 2 weeks before coculture and were formed into a confluent monolayer by the start of the coculture. Cobblestone-like areas, which consisted of more than 5 leukemia cells, were counted 14 to 17 days after transduction. A fraction of the cells were also cultured in IMDM with 20% FBS without feeder cells up to 7 days after transduction to determine the transduction efficiencies and apoptosis by flow cytometry (FCM).
Preparation and culture of cord blood CD34+ cells
Umbilical cord blood was drawn from the umbilical cord of healthy volunteers using a heparinized syringe after obtaining informed consent and the approval of the institutional review board of The Institute of Medical Science, The University of Tokyo, based on the Declaration of Helsinki. Mononuclear cells were purified from these samples by Ficoll-Hypaque methods as previously reported.18 These cells were processed further to isolate CD34+ cells (CD34+ cord blood cells [CBCs]) using a magnetic-activated cell separation (MACS) direct CD34 progenitor cell isolation kit (Miltenyi Biotech, Bergisch Gladbach, Germany) according to the manufacturer's instructions. After 2 rounds of isolation, more than 95% of cells were CD34+, as assessed by FCM. After transduction with the lentiviral vectors, CD34+ CBCs were cultured in IMDM supplemented with 10% BIT9500 (a serum substitute; Stem Cell Technologies, Vancouver, BC, Canada), 100 ng/mL stem cell factor (SCF; Genzyme, Cambridge, MA), 100 ng/mL thrombopoietin (Chemicon, Temecula, CA), and 100 ng/mL granulocyte colony-stimulating factor (G-CSF; kindly provided by Chugai Pharmaceuticals, Tokyo, Japan) for 14 days. To determine the colony-forming abilities of e1a2Mz-transduced CD34+ CBCs, a sample of cells (300 cells) was obtained immediately after the start of gene transduction and plated in IMDM containing 30% FBS, 1% bovine serum albumin, 1.2% methylcellulose, and the following hematopoietic factors: 25 ng/mL SCF, 10 ng/mL interleukin-3 (kindly provided by Chugai Pharmaceuticals), 100 ng/mL interleukin-6 (kindly provided by Ajinomoto, Tokyo, Japan), 10 ng/mL G-CSF, and 2 U/mL erythropoietin (kindly provided by Chugai Pharmaceuticals). Colony numbers were counted 14 days after transduction was performed.
Construction of e1a2Mz expression plasmids
The sequences and secondary structure of e1a2Mz were reported previously16 and are shown in Figure 1. The e1a2Mz-encoding DNA was synthesized by polymerase chain reaction (PCR) using Platinum Pfx DNA polymerase (Invitrogen, Carlsbad, CA) and synthetic oligonucleotides. Template and primer sequences were as follows: left arm template, 5′-CCCGGTTCGAAACCGGGCACTACAAAAACCAACTTTGGTCCAGCCTGATGAGGAGCGTCTCCATGGAAAAAGGTACCCCGGA-3′; right arm template, 5′-CCCGGTTCGAAACCGGGCACTACAAAAACCAACTTTTCTCAATGGCTGCATCGAAACCTCACTTGGTACCCCGGA-3′; a common sense primer for the right and left arms of e1a2Mz, 5′-TCCCCGGTTCGAAACCGGGCACTAC-3′; antisense primer for the right arm, 5′-TATCCGGGGTACCTTTTTCCATGGAGACGC-3′; and antisense primer for the left arm, 5′-TATCCGGGGTACCTTTAAACCTTCTTTCGGAAG-3′. The PCR products were digested with Csp45I and KpnI and cloned downstream of the tRNAVal promoter of ptRNA-KE, which is a plasmid derived from tRNAVal/pUC-dt with a minor modification in the 3′ end of its cloning site (a SalI site was replaced with a KpnI site), as previously described.13 Finally, we confirmed the sequence of the e1a2Mz expression cassettes.
Construction and preparation of e1a2Mz expression lentiviral vector
We embedded genes encoding e1a2Mz downstream of genes for human tRNAVal. Our e1a2Mz expression unit consisted of 2 tandemly arranged tRNAVal-driven expression cassettes corresponding to each component of the e1a2Mz heterodimer, similar to previous reports.12,23 This e1a2Mz expression unit was inserted into a self-inactivating lentiviral vector plasmid, pHIV-ECD2, which was derived from pCS-CDF-EG-PRE and contains human CD2 (hCD2; provided by Health Science Research Resources Bank, Osaka, Japan) as a marker gene (Figure 2A). We also constructed control plasmids to make control HIV vectors as described in Table 1. A b2a2Mz expression unit, which was used in our previous study,12 was inserted into the pHIV-ECD2 as the e1a2Mz expression unit. Lentiviral vector particles were produced as described previously.18 Briefly, the pHIV-e1a2Mz or control plasmids were cotransfected with the third-generation packaging plasmid pMDLg/p.RRE (kindly provided by Cell Genesys, South San Francisco, CA), pRSV-rev, and pMD.G into 293T cells. Viral supernatants were harvested 48 and 72 hours after transfection, passed through 0.22-μm filters (Millipore, Bedford, MA), and concentrated by 2 rounds of ultracentrifugation. The viral pellet was resuspended in serum-free IMDM to obtain a 10 000-fold concentrate. Viral stocks were stored at -80°C until assay, and viral titers were determined by the transduction of HeLa cells with serial dilutions followed by FCM, ranging from 8 × 108 to 2 × 109 HeLa-transducing units per milliliter.
Transduction of the e1a2Mz expression vector into Ph+ ALL cells. (A) Schematic diagrams of the 4 constructs used to produce the e1a2Mz expression HIV(VSV) vector. pHIV-e1a2Mz, pMDLg/p.RRE, pRSV-rev, and pMD.G indicate the e1a2Mz-encoding transfer vector, packaging plasmid, rev expression plasmid, and VSV G-protein expression plasmid, respectively. P indicates tRNAVal (pol III promoter of the e1a2Mz); MzL, e1a2Mz left arm; MzR, e1a2Mz right arm; CMV, cytomegalovirus promoter; LTR, long terminal repeat; ψ, packaging signal; SD, splice donor; SA, splice acceptor; RRE, Rev-responsive element; R, repeat region; ΔU3, promoter/enhancer sequence-deleted U3 (▨); cPPT, central polypurine tract; CTS, central termination sequence; EF-1α, elongation factor 1α subunit promoter; hCD2, human CD2; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element; GAG, gag region of HIV-1; POL, pol region of HIV-1; pA, poly A; RSV, respiratory syncytial virus promoter; and VSV.G, vesicular stomatitis virus G protein. (B) The hCD2 expression in the Ph+ ALL cell line, KOPN-30. The hCD2 expression in KOPN-30 cells 3 days after transduction with control vector (ECD2; Table 1) or e1a2Mz expression vector (e1a2Mz; Table 1) at an MOI of 10 was determined by FCM. Histograms in solid lines indicate untransduced cells and the area under the solid lines represents vector-transduced cells. *The numbers above the bars indicate the percentage of hCD2-positive cells. (C) Transduction efficiencies into human leukemia cell lines, including 5 Ph+ ALL cell lines, with the ECD2 (▪) or e1a2Mz (▨) vector. The results represent means of triplicate experiments.
Transduction of the e1a2Mz expression vector into Ph+ ALL cells. (A) Schematic diagrams of the 4 constructs used to produce the e1a2Mz expression HIV(VSV) vector. pHIV-e1a2Mz, pMDLg/p.RRE, pRSV-rev, and pMD.G indicate the e1a2Mz-encoding transfer vector, packaging plasmid, rev expression plasmid, and VSV G-protein expression plasmid, respectively. P indicates tRNAVal (pol III promoter of the e1a2Mz); MzL, e1a2Mz left arm; MzR, e1a2Mz right arm; CMV, cytomegalovirus promoter; LTR, long terminal repeat; ψ, packaging signal; SD, splice donor; SA, splice acceptor; RRE, Rev-responsive element; R, repeat region; ΔU3, promoter/enhancer sequence-deleted U3 (▨); cPPT, central polypurine tract; CTS, central termination sequence; EF-1α, elongation factor 1α subunit promoter; hCD2, human CD2; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element; GAG, gag region of HIV-1; POL, pol region of HIV-1; pA, poly A; RSV, respiratory syncytial virus promoter; and VSV.G, vesicular stomatitis virus G protein. (B) The hCD2 expression in the Ph+ ALL cell line, KOPN-30. The hCD2 expression in KOPN-30 cells 3 days after transduction with control vector (ECD2; Table 1) or e1a2Mz expression vector (e1a2Mz; Table 1) at an MOI of 10 was determined by FCM. Histograms in solid lines indicate untransduced cells and the area under the solid lines represents vector-transduced cells. *The numbers above the bars indicate the percentage of hCD2-positive cells. (C) Transduction efficiencies into human leukemia cell lines, including 5 Ph+ ALL cell lines, with the ECD2 (▪) or e1a2Mz (▨) vector. The results represent means of triplicate experiments.
Mz expression and control HIV(VSV) vectors
. | . | Genes inserted in HIV(VSV) vectors* . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|
| Vector . | Abbreviation . | CD2 . | tRNAVal . | e1a2Mz left arm . | e1a2Mz right arm . | |||
| HIV-ECD2 | ECD2 | + | − | − | − | |||
| HIV-tRNAVal | tRNA | + | + | − | − | |||
| HIV-e1a2MzL | e1a2MzL | + | + | + | − | |||
| HIV-e1a2MzR | e1a2MzR | + | + | − | + | |||
| HIV-e1a2Mz | e1a2Mz | + | + | + | + | |||
. | . | Genes inserted in HIV(VSV) vectors* . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|
| Vector . | Abbreviation . | CD2 . | tRNAVal . | e1a2Mz left arm . | e1a2Mz right arm . | |||
| HIV-ECD2 | ECD2 | + | − | − | − | |||
| HIV-tRNAVal | tRNA | + | + | − | − | |||
| HIV-e1a2MzL | e1a2MzL | + | + | + | − | |||
| HIV-e1a2MzR | e1a2MzR | + | + | − | + | |||
| HIV-e1a2Mz | e1a2Mz | + | + | + | + | |||
+ indicates inserted; and −, not inserted.
Indicated genes were inserted in parental HIV vector.
Inoculation of viral vectors
Human leukemia cell lines, primary leukemia cells, or CD34+ CBCs (2 × 105) were inoculated with e1a2Mz vector or control vectors for 2 hours at 37°C in a CO2 incubator. After inoculation at a multiplicity of infection (MOI) of 0.1 to 80, cells were washed and cultured in RPMI 1640 with 10% FBS for leukemia cell lines except SUP-B15 cells, in IMDM with 20% FBS for SUP-B15 cells and primary leukemia cells, or in IMDM with 10% BIT9500, 100 ng/mL SCF, 100 ng/mL TPO, and 100 ng/mL G-CSF for CD34+ CBCs. To obtain CD2-expressing cells after transduction at low titers, cells were selected by the magnetic beads method using a MACS CD2 MicroBead kit (Miltenyi Biotech) 3 days after transduction.
Determination of transduction efficiency
Transduction efficiencies of target cells were determined by examining hCD2 expression. Cells transduced with vectors were stained with fluorescein isothiocyanate (FITC)-labeled anti-hCD2 antibody (Becton Dickinson Pharmingen, San Diego, CA) and analyzed by FCM, using a FACS Calibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ). Primary leukemia cells were also stained with biotin-labeled antihuman CD19 antibody (Becton Dickinson Pharmingen) and then stained with allophycocyanin-labeled streptavidin (Becton Dickinson Pharmingen). CD34+ CBC cells were also stained with phycoerythrin-labeled antihuman CD34 antibody (Becton Dickinson Pharmingen). These double-stained cells were analyzed by 2-color FCM.
Determination of viability and apoptosis
To determine the viability of cells transduced with e1a2Mz or control vectors, we counted live and dead cell numbers every 2 or 3 days up to 14 days by the trypan blue exclusion method. Apoptosis of cells was determined using the annexin V-apoptosis detection kit I (Becton Dickinson Pharmingen) according to the manufacturer's instruction. We stained cells with FITC-labeled annexin V and propidium iodide (PI) and analyzed them by 2-color FCM.
Determination of copy number of integrated viral DNA
The copy number of the integrated lentiviral DNA in the host cells was determined by the real-time Alu-PCR method, which amplifies and quantifies the sequences between genomic Alu repeat sequences of the host cells and the LTR of integrated HIV vectors, as reported previously.24 Briefly, DNA extracted from Ph+ ALL cells transduced with HIV(VSV) vectors was amplified with the following PCR primers and probes using TaqMan Universal PCR Master Mix and ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Branchburg, NJ). PCR primer and probe sequences were as follows: MH535 (Alu-PCR sense), 5′-AACTAGGGAACCCACTGCTTAAG-3′; MH704 (Alu-PCR antisense), 5′-TGCTGGGATTACAGGCGTGAG-3′; and MH603 (Alu-PCR probe), 5′-FAMACACTACTTGTTGCACTCAAGGCAAGCTTT-TAMRA-3′. The copy number of integrated vector DNA was calculated from a standard curve drawn by the copy number of vector DNA and the threshold cycle number of Alu-PCR for stably transduced 293T cells cultured for 1 month after transduction with the ECD2 vectors. The copy number of vector DNA in stably transduced 293T cells was determined by Alu-PCR for the HIV-long terminal repeat (HIV-LTR) as described previously.18 To determine the copy number of integrated vectors per cell, the copy number of β-actin DNA was also determined by real-time PCR with TaqMan β-actin Control Reagents (Applied Biosystems).
Detection of the e1a2Mz expression
On day 7 after transduction with e1a2Mz or control vector, total RNA was isolated from leukemia cells or CD34+ CBCs using a Qiagen RNA/DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol, and the RNA was treated with amplification-grade DNase I (Invitrogen) to digest contaminating DNA. The cDNA was synthesized using a SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) with the following reverse transcription (RT) primers. To detect the right or left arm of e1a2Mz, RT-PCR was performed on 300 ng RNA using AmpliTaq Gold DNA polymerase (Applied Biosystems) and the following PCR primers in a final volume of 50 μL, with 35 cycles of thermal cycling at 95°C for 30 seconds, 57°C for 30 seconds, and 72°C for 30 seconds. RT-PCR for glyceraldehyde phosphate dehydrogenase (GAPDH) was performed using an oligo (dT) RT primer and the following primers as a control for the RT-PCR of e1a2Mz. PCR primer sequences were as follows: RT primer, 5′-AAAAAAA-3′ (complementary sequence of the transcriptional stop sequence in the e1a2Mz expression cassettes); a common sense primer for the right and left arms of e1a2Mz, 5′-ACCGTTGGTTTCCGTAGTGTAGTGGTTATC-3′; antisense primer for the right arm, 5′-TAAACCTTCTTTCGGAAGAAGCCCTTCAGC-3′; antisense primer for the left arm, 5′-TTTCCATGGAGACGCTCCTCATCAGGCT-3′; and GAPDH, 5′-ACCACAGTCCATGCCATCAC-3′ (sense) and 5′-TCCACCACCCTGTTGCTGTA-3′ (antisense). PCR products were examined by electrophoresis in a 2% agarose gel.
Detection of bcr-abl mRNA
To quantify p190-type bcr-abl mRNA, cDNA synthesized from total RNA of Ph+ ALL cells using an oligo (dT) primer as described in the preceding section was analyzed by real-time PCR using TaqMan Universal PCR Master Mix and ABI PRISM 7700 Sequence Detection System (Applied Biosystems). Copy numbers calculated from the threshold cycle number and a standard curve were normalized against values determined for GAPDH. Probe and primer sequences were as follows: bcr-abl, 5′-FAM-CGCCCTCGTCATCGTTGGGCCAGATCTAMRA-3′ (probe), 5′-ATCGTGGGCGTCCGCAAGAC-3′ (sense primer), 5′-GCTCAAAGTCAGATGCTACTG-3′ (antisense primer); and GAPDH, 5′-VIC-CAAGCTTCCCGTTCTCAGCC-TAMRA-3′ (probe), 5′-GAAGGTGAAGGTCGGAGTC-3′ (sense primer), and 5′-GAAGATGGTGATGGGATTTC-3′ (antisense primer).
Results
Effect of the e1a2Mz on Ph+ ALL cell lines
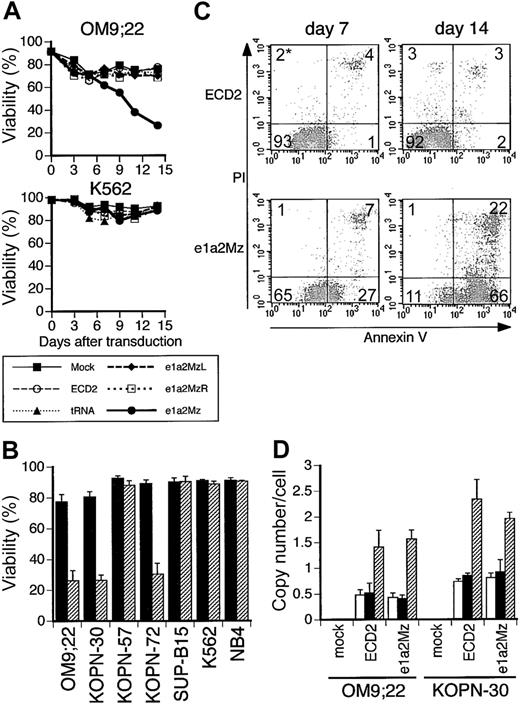
We transduced e1a2Mz into Ph+ ALL cell lines with the third-generation lentiviral vector system at an MOI of 10. To determine the transduction efficiencies of the e1a2Mz expression vector and control vectors, we examined the transgenic expression of human CD2 in Ph+ ALL cells by FCM. As shown in Figure 2B, 100% of KOPN-30 Ph+ ALL cells were transduced with the e1a2Mz expression vector or ECD2 vector. In addition, the transduction efficiency of these vectors in all cell lines tested was 100% (Figure 2C). The transduction efficiency of all of the remaining vectors (Table 1) was also 100% in all cell lines (data not shown). Expression of the CD2 transgene was maintained over the entire period of the assay (data not shown). After transduction of the e1a2Mz vector, the survival rate of OM9;22 Ph+ ALL cells began to decrease from day 7 and was about 30% on day 14 (Figure 3A). The viability of the other Ph+ ALL cell lines that were tested, KOPN-30 and KOPN-72, also decreased after e1a2Mz vector transduction (Figure 3B). In addition, as for those cells, there were no live cells 21 days after transduction with the e1a2Mz vector (data not shown). Ph+ ALL cells transduced with control vectors at an MOI of 10, however, did not exhibit this decrease in survival rate (Figure 3A-B). To further determine the specificity of the e1a2Mz-induced cytotoxicity, we transduced cells with the b2a2Mz expression HIV vector. This vector contains the Mz expression unit that has been reported to digest b2a2-type bcr-abl RNA and to induce the apoptosis of BV173 CML-BC cells12 and did not affect the viability of Ph+ ALL cell lines (data not shown).
Effect of e1a2Mz transduction on the survival of Ph+ ALL cells. (A) Death of OM9;22 Ph+ ALL cells (p190-type bcr-abl positive) and K562 CML-BC cells (p190-type bcr-abl negative) transduced with the indicated vectors, or untransduced, was determined by the trypan blue exclusion test every 2 or 3 days. The results represent means of triplicate experiments. (B) Viabilities of leukemia cell lines transduced with the ECD2 (▪) or e1a2Mz (▨) vector on day 14. These results represent means ± SDs of triplicate experiments. (C) Apoptosis of the Ph+ ALL cell line KOPN-30 transduced with e1a2Mz. KOPN-30 cells transduced with the ECD2 or e1a2Mz vector were stained with FITC-labeled annexin V and PI on days 7 and 14 and were analyzed by 2-color FCM. Cells in each quadrant correspond to the following conditions: bottom left, live; bottom right, in early apoptosis; top right, in late apoptosis or dead. *The numbers in the plots indicate the percentage of cells in that quadrant. (D) Copy numbers of integrated lentiviral vector per cell transduced with the indicated vectors at an MOI of 0.1 (□), 1 (▪) or 10 (▨). As for the cells transduced at an MOI of 0.1, the results of CD2+ cells selected by magnetic bead separation are shown. These results represent mean ± SD of triplicate experiments.
Effect of e1a2Mz transduction on the survival of Ph+ ALL cells. (A) Death of OM9;22 Ph+ ALL cells (p190-type bcr-abl positive) and K562 CML-BC cells (p190-type bcr-abl negative) transduced with the indicated vectors, or untransduced, was determined by the trypan blue exclusion test every 2 or 3 days. The results represent means of triplicate experiments. (B) Viabilities of leukemia cell lines transduced with the ECD2 (▪) or e1a2Mz (▨) vector on day 14. These results represent means ± SDs of triplicate experiments. (C) Apoptosis of the Ph+ ALL cell line KOPN-30 transduced with e1a2Mz. KOPN-30 cells transduced with the ECD2 or e1a2Mz vector were stained with FITC-labeled annexin V and PI on days 7 and 14 and were analyzed by 2-color FCM. Cells in each quadrant correspond to the following conditions: bottom left, live; bottom right, in early apoptosis; top right, in late apoptosis or dead. *The numbers in the plots indicate the percentage of cells in that quadrant. (D) Copy numbers of integrated lentiviral vector per cell transduced with the indicated vectors at an MOI of 0.1 (□), 1 (▪) or 10 (▨). As for the cells transduced at an MOI of 0.1, the results of CD2+ cells selected by magnetic bead separation are shown. These results represent mean ± SD of triplicate experiments.
In contrast, the viability of KOPN-57 and SUP-B15 Ph+ ALL cells did not decrease upon e1a2Mz vector transduction nor did that of the p190-type bcr-abl-negative K562 and NB4 cells (Figure 3A-B). To determine the mechanism of e1a2Mz-induced cell death, we performed an annexin V assay to look for signs of apoptosis. In e1a2Mz-sensitive KOPN-30 cells, apoptotic cells began to appear 7 days after e1a2Mz vector transduction and increased in number until day 14 (Figure 3C). In other e1a2Mz-sensitive Ph+ ALL cells, but not in e1a2Mz-resistant Ph+ ALL or p190-type bcr-abl-negative cells, apoptosis was induced similarly by e1a2Mz vector transduction (data not shown).
Integration of the e1a2Mz vector
Recently, X-linked severe combined immunodeficiency (SCIDX1) patients whose lymphocytes were transduced with an MLV vector have been reported to develop leukemia, and insertional mutagenesis is an important problem in the gene therapy using retroviral vectors.25 In retroviral transduction, the number of integration sites/cell was reported to be increased at higher MOI.26 Hence, we tried to transduce the e1a2Mz vector to OM9;22 and KOPN-30 cells at a low titer to reduce the integration of the vector DNA. At an MOI of 1, although the transduction efficiency of these cells was 96% to 98%, the viability of OM9;22 cells and KOPN-30 cells on day 7 was about 50% and 60%, respectively, indicating that the cytotoxic effect under these conditions was less than that at an MOI of 10. Moreover, the viability of KOPN-30 cells recovered to the initial level of transduction on day 14. We also examined the effect of e1a2Mz in OM9;22 or KOPN-30 cells when transduced with one infectious particle. Those cells were transduced at an MOI of 0.1 followed by magnetic sorting for CD2, and results similar to those seen in nonsorted cells transduced at an MOI of 1 were obtained. To clarify the relation between the effects of e1a2Mz and the transduction titer, we measured the number of chromosomally integrated vectors in those cells. As shown in Figure 3D, the copy number of integrated DNA in OM9;22 or KOPN-30 cells transduced at an MOI of 1 and 10 was about 0.5 and 1.5 per cell for OM9;22 cells and 0.9 and 2 per cell for KOPN-30, respectively. In addition, similar to the copy numbers of integrated vector DNA at an MOI of 1, those in CD2+ OM9;22 and KOPN-30 cells transduced at an MOI of 0.1 were about 0.5 and 0.8 per cell, respectively (Figure 3D). These results suggested that the integration of vector DNA into the chromosome of leukemia cells is important to realize the cytotoxic effects of e1a2Mz, and relatively low copy number of integrated DNA, which were obtained by the transduction at an MOI of 10, was considered to be enough.
The e1a2Mz expression in Ph+ ALL cells
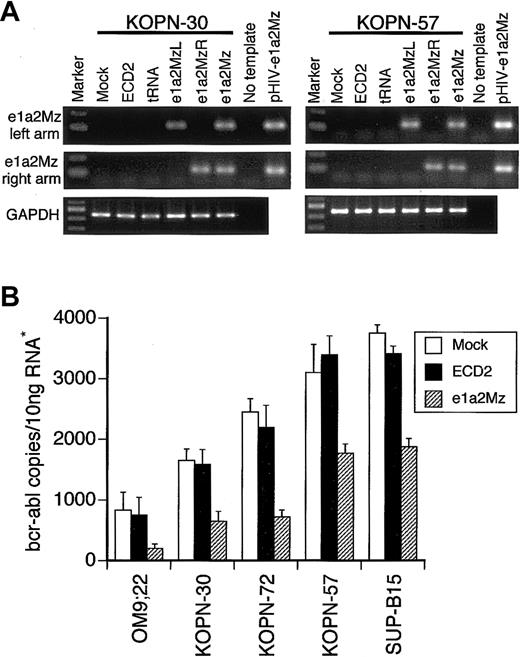
To confirm the expression of e1a2Mz RNA in e1a2Mz-transduced cells, we performed an RT-PCR assay for the left and right arms of the e1a2Mz gene. In both e1a2Mz-sensitive KOPN-30 and e1a2Mz-resistant KOPN-57 cells, we detected both arms of e1a2Mz 7 days after transduction with the e1a2Mz vector (Figure 4A). In all other cell lines transduced with the e1a2Mz vector, both arms of e1a2Mz RNA were also detected (data not shown). As e1a2Mz expression was confirmed in both e1a2Mz-sensitive and -resistant cells, one or more mutations in the e1a2Mz-targeting sites of the bcr-abl fusion gene were thought to underlie the e1a2Mz resistance. Analysis of the e1a2Mz-recognition and -digestion sequences of the bcr-abl mRNA in both e1a2Mz-resistant and e1a2Mz-sensitive Ph+ ALL cells, however, revealed no such mutation. Therefore, an alternative mechanism, such as failure of the bcr-abl mRNA reduction or the contribution of oncogenes other than bcr-abl to cell survival, could underlie the resistance of KOPN-57 and SUP-B15 cells to e1a2Mz.
Expression of the e1a2Mz and p190-type bcr-abl mRNA. (A) Expression of the left and right arms of e1a2Mz in KOPN-30 and KOPN-57 cells transduced with the indicated vectors was examined by RT-PCR using specific primer pairs. RT-PCR for GAPDH was performed as an internal control. The pHIV-e1a2Mz plasmid was used as a positive control for the PCR. The RT-PCR products were electrophoresed through a 2% agarose gel and were stained with ethidium bromide. (B) Quantification of p190-type bcr-abl mRNA in Ph+ ALL cell lines transduced with or without the e1a2Mz by real-time RT-PCR using specific primer pairs and probe. The copy number of bcr-abl mRNA was calculated from the standard curve. The results represent means ± SDs of triplicate experiments. *The amount of RNA was normalized by the copy number of GAPDH mRNA.
Expression of the e1a2Mz and p190-type bcr-abl mRNA. (A) Expression of the left and right arms of e1a2Mz in KOPN-30 and KOPN-57 cells transduced with the indicated vectors was examined by RT-PCR using specific primer pairs. RT-PCR for GAPDH was performed as an internal control. The pHIV-e1a2Mz plasmid was used as a positive control for the PCR. The RT-PCR products were electrophoresed through a 2% agarose gel and were stained with ethidium bromide. (B) Quantification of p190-type bcr-abl mRNA in Ph+ ALL cell lines transduced with or without the e1a2Mz by real-time RT-PCR using specific primer pairs and probe. The copy number of bcr-abl mRNA was calculated from the standard curve. The results represent means ± SDs of triplicate experiments. *The amount of RNA was normalized by the copy number of GAPDH mRNA.
Decrease of p190-type bcr-abl expression in Ph+ ALL cells
To evaluate the effect of e1a2Mz on the expression level of bcr-abl, we measured the copy number of bcr-abl mRNA by quantitative real-time RT-PCR using specific primers and probes. A decrease in bcr-abl mRNA was observed not only in e1a2Mz-sensitive OM9;22, KOPN-30, and KOPN-72 Ph+ ALL cells but also in KOPN-57 and SUP-B15 cells (Figure 4B). The bcr-abl mRNA copy number in e1a2Mz-resistant cells, however, was higher than that in e1a2Mz-sensitive cells after e1a2Mz transduction (Figure 4B).
Titer escalation study
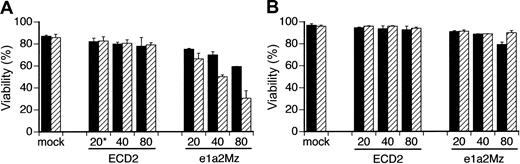
Because the insufficient decrease of bcr-abl mRNA was a possible mechanism of e1a2Mz resistance, we transduced e1a2Mz into KOPN57 and SUP-B15 cells at a higher MOI to observe the extent of the resistance by titer escalation. In KOPN57 cells, growth inhibition and a decrease in viability were observed in a dose-dependent manner without any increase in nonspecific toxicity (Figure 5A). In contrast, SUP-B15 cells were still resistant to e1a2Mz, even after transduction of the e1a2Mz vector at an MOI of 80, although a transient decrease in viability was observed 7 days after transduction (Figure 5B)
Effect of titer escalation on e1a2Mz transduction into Ph+ ALL cell lines. KOPN-57 (A) and SUP-B15 (B) cells were transduced with the ECD2 or e1a2Mz vector at the indicated MOI. Cell death was determined by the trypan blue exclusion test on days 7 (▪) and 14 (▨). The results represent means ± SDs of triplicate experiments. *Numbers along the x-axis indicate the MOI of transduction.
Effect of titer escalation on e1a2Mz transduction into Ph+ ALL cell lines. KOPN-57 (A) and SUP-B15 (B) cells were transduced with the ECD2 or e1a2Mz vector at the indicated MOI. Cell death was determined by the trypan blue exclusion test on days 7 (▪) and 14 (▨). The results represent means ± SDs of triplicate experiments. *Numbers along the x-axis indicate the MOI of transduction.
Effect of e1a2Mz on primary Ph+ ALL cells
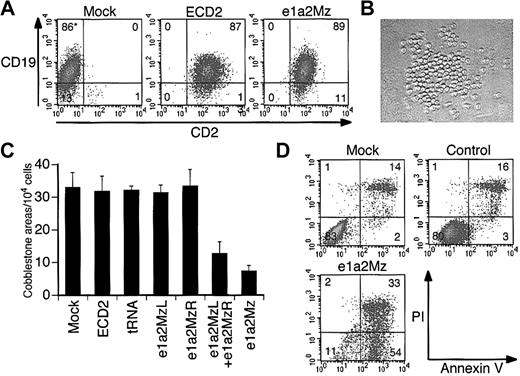
Since primary leukemia cells from Ph+ ALL patients are the proposed targets of gene therapy, we examined the effect of e1a2Mz on such cells. We transduced the e1a2Mz or control vectors into these cells at an MOI of 20. Similar to the results seen for Ph+ cell lines, the transduction efficiency into these cells was almost 100% in all 4 samples 3 days after transduction (Figure 6A). At first, we examined the viability of primary leukemia cells in the same culture system as that used for cell lines. We could not determine clearly, however, whether e1a2Mz had any inhibitory effects because it was very difficult to culture these cells in our conventional liquid culture system: the number of live cells decreased continuously after the start of culture regardless of whether or not the e1a2Mz vectors were transduced. Therefore, we used a coculture system reported previously in which a murine bone marrow stroma cell line was used as a source for feeder cells and the primary leukemia cells formed cell clusters upon the feeder cells (Figure 6B cobblestone area [CA]).27 We counted the number of cobblestone formations on days 14 to 17 after transduction. In all of the experiments, a significant decrease of the size of CAs was observed upon transduction with the e1a2Mz vector. In addition, the number of CAs decreased in number after transduction with the e1a2Mz left arm (e1a2MzL) and e1a2Mz right arm (e1a2MzR) vectors together at an MOI of 10 for each vector (total MOI = 20) but not with the other control vectors (Figure 6C; Table 2). However, we could not determine whether the CA formation would be inhibited completely by e1a2Mz transduction because all of the CAs from untransduced or control vector-transduced cells also eventually disappeared after approximately 21 days from the start of the assay as those from the e1a2Mz-transduced cells did in 14 to 17 days as described above. As observed in the cell lines, e1a2Mz transduction of primary leukemia cells induced an increase in apoptosis (Figure 6D).
Effect of e1a2Mz transduction on primary leukemia cells from Ph+ ALL patients. (A) Transduction efficiencies of primary leukemia cells from a Ph+ ALL-1 patient. Expression of CD2 (transgene) and CD19 (natively expressed gene) in cells transduced with or without the indicated vectors was determined by FCM on day 3. (B) Cobblestone area formation of primary leukemia cells from Ph+ ALL-1 patients on day 17. Primary leukemia cells were cocultured with mouse stroma cells, and cobblestone areas were formed about 2 weeks after transduction (original magnification, × 100). The image was acquired using an inverted microscope (Diaphot TMD 300; Nikon, Tokyo, Japan) with a digital camera (DXM 1200; Nikon). The acquired image was processed using ACT-1 software (Nikon). The objective lenses used were Nikon NCF Plan ELWD DM 20×/0.40. (C) Number of cobblestone areas derived from primary leukemia cells of the Ph+ ALL-4 patient. Cells were transduced with the indicated vectors at an MOI of 20, and cobblestone areas were counted 17 days after transduction. The results represent means ± SDs of duplicate experiments. (D) Apoptosis of primary leukemia cells from Ph+ ALL-1 patients. Apoptosis was determined by annexin V assay on day 7 as performed for the cell lines. *In panels A and D, the numbers in the plot indicate the percentage of cells in that quadrant.
Effect of e1a2Mz transduction on primary leukemia cells from Ph+ ALL patients. (A) Transduction efficiencies of primary leukemia cells from a Ph+ ALL-1 patient. Expression of CD2 (transgene) and CD19 (natively expressed gene) in cells transduced with or without the indicated vectors was determined by FCM on day 3. (B) Cobblestone area formation of primary leukemia cells from Ph+ ALL-1 patients on day 17. Primary leukemia cells were cocultured with mouse stroma cells, and cobblestone areas were formed about 2 weeks after transduction (original magnification, × 100). The image was acquired using an inverted microscope (Diaphot TMD 300; Nikon, Tokyo, Japan) with a digital camera (DXM 1200; Nikon). The acquired image was processed using ACT-1 software (Nikon). The objective lenses used were Nikon NCF Plan ELWD DM 20×/0.40. (C) Number of cobblestone areas derived from primary leukemia cells of the Ph+ ALL-4 patient. Cells were transduced with the indicated vectors at an MOI of 20, and cobblestone areas were counted 17 days after transduction. The results represent means ± SDs of duplicate experiments. (D) Apoptosis of primary leukemia cells from Ph+ ALL-1 patients. Apoptosis was determined by annexin V assay on day 7 as performed for the cell lines. *In panels A and D, the numbers in the plot indicate the percentage of cells in that quadrant.
Cobblestone area formation of primary Ph+ ALL cells
. | Transduced vectors . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample . | CD2 . | tRNA . | e1a2MzL . | e1a2MzR . | e1a2MzL + e1a2MzR . | e1a2Mz . | |||||
| Ph+ ALL-1 | 90.8 | 88.2 | 85.7 | 92.0 | NT | 32.4 | |||||
| Ph+ ALL-2 | 98.2 | 95.5 | 93.8 | 96.2 | NT | 25.4 | |||||
| Ph+ ALL-3 | 95.6 | 94.3 | 90.8 | 88.6 | 43.3 | 28.8 | |||||
| Ph+ ALL-4 | 96.4 | 97.6 | 95.2 | 101.2 | 38.8 | 22.4 | |||||
. | Transduced vectors . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample . | CD2 . | tRNA . | e1a2MzL . | e1a2MzR . | e1a2MzL + e1a2MzR . | e1a2Mz . | |||||
| Ph+ ALL-1 | 90.8 | 88.2 | 85.7 | 92.0 | NT | 32.4 | |||||
| Ph+ ALL-2 | 98.2 | 95.5 | 93.8 | 96.2 | NT | 25.4 | |||||
| Ph+ ALL-3 | 95.6 | 94.3 | 90.8 | 88.6 | 43.3 | 28.8 | |||||
| Ph+ ALL-4 | 96.4 | 97.6 | 95.2 | 101.2 | 38.8 | 22.4 | |||||
Values presented in the table are the percentage of cobblestone area number derived from mock-transduced cells. Data represent the mean of duplicated assay. NT indicates not tested.
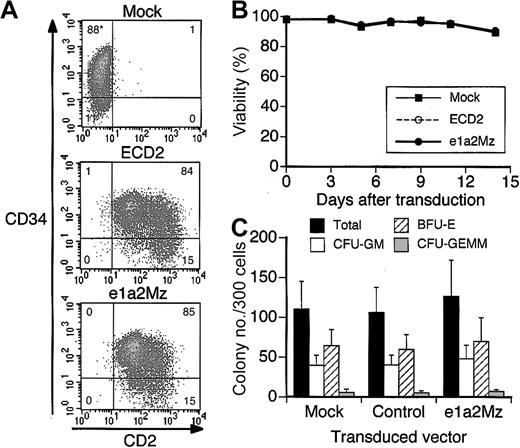
Effect of e1a2Mz on CD34+CBCs
In K562 and NB4 cells, whose survival is not dependent upon p190-type bcr-abl, transduction with e1a2Mz did not affect viability and proliferation (Figure 3A-B), suggesting that there is little nonspecific cytotoxicity associated with the procedure. To evaluate further the safety of the e1a2Mz vector, we transduced it into normal human CD34+ CBCs. Almost all cells transduced with the e1a2Mz or control vectors expressed CD2 (Figure 7A). In these cells, we also confirmed e1a2Mz expression by RT-PCR (data not shown). The viability of CD34+ CBCs did not decrease after transduction with the e1a2Mz or control vectors (Figure 7B). In addition, the number and composition of colonies were not affected by e1a2Mz transduction (Figure 7C).
Effect of e1a2Mz transduction on normal human CD34+ CBCs. (A) Transgenic expression of hCD2 in CD34+ CBCs was determined by FCM 3 days after gene transduction. Cells were also stained with anti-CD34 antibody. Representative data from 3 samples are shown. *The numbers in the plots indicate the percentage of cells in that quandrant. (B) Death of CD34+ CBCs transduced with the indicated vectors, or untransduced, was determined by the trypan blue exclusion test up to 14 days after transduction. The results represent means of data from 3 CD34+ CBC samples. (C) Colony formation analysis of CD34+ CBCs transduced with the indicated vectors or untransduced. Number and types of colonies formed in semisolid media were assessed 14 days after transduction. The results represent means ± SDs of data from 3 CD34+ CBC samples. CFU-GM indicates granulocyte-macrophage colony-forming unit; BFU-E, erythroid burst-forming unit; and CFUGEMM, granulocyte-erythrocyte-megakaryocyte-macrophage colony-forming unit.
Effect of e1a2Mz transduction on normal human CD34+ CBCs. (A) Transgenic expression of hCD2 in CD34+ CBCs was determined by FCM 3 days after gene transduction. Cells were also stained with anti-CD34 antibody. Representative data from 3 samples are shown. *The numbers in the plots indicate the percentage of cells in that quandrant. (B) Death of CD34+ CBCs transduced with the indicated vectors, or untransduced, was determined by the trypan blue exclusion test up to 14 days after transduction. The results represent means of data from 3 CD34+ CBC samples. (C) Colony formation analysis of CD34+ CBCs transduced with the indicated vectors or untransduced. Number and types of colonies formed in semisolid media were assessed 14 days after transduction. The results represent means ± SDs of data from 3 CD34+ CBC samples. CFU-GM indicates granulocyte-macrophage colony-forming unit; BFU-E, erythroid burst-forming unit; and CFUGEMM, granulocyte-erythrocyte-megakaryocyte-macrophage colony-forming unit.
Discussion
We have already reported that a Mz targeting the p190-type bcr-abl fusion gene could digest synthetic chimeric oligonucleotide RNA in vitro.16 Due to the difficulty of obtaining 100% transduction of e1a2Mz into leukemia cells using conventional vector systems, however, we could not demonstrate the effect of the e1a2Mz on live cells until a VSV-pseudotyped lentiviral vector system was developed. Recent reports, including one from our group, demonstrated that such a lentiviral vector is the most efficient method for transducing human blood cells, including leukemia cells.17,18,28,29 In addition, long-term expression of a transgene with this vector system was demonstrated in human leukemia cells, including primary cells from leukemia patients.18 Therefore, we adopted a third-generation HIV(VSV) vector system and successfully transduced an e1a2Mz vector into 5 different Ph+ ALL cell lines and 2 control cell lines with 100% efficiency (Figure 2B-C). Moreover, we could transduce those vectors into primary leukemia cells from Ph+ ALL patients with the same efficiency (Figure 6A). This is the first report of such a high transduction efficiency into Ph+ ALL cells, although several studies have demonstrated relatively high transduction efficiencies using second-generation VSV-pseudotyped HIV vectors with human leukemia cells from patients with B-precursor ALL (2.2%-43.35%),30 ALL (40.3%-90.3%), and acute myeloblastic leukemia (61.7%-87.6%).17 One cause of the higher transduction efficiencies observed in this study compared with previous reports is likely to be the insertion into our vector of the central DNAflap sequence of HIV, which facilitates the integration into the genome of host cells.31-33 In addition, however, the transduction efficiencies observed in this study were higher than those obtained in a previous report from our laboratory (52%-95%),18 which also employed a third-generation HIV vector with the central DNA flap sequence. This difference was probably due to the use of the elongation factor 1 α (EF-1α) promoter in this study, which has been reported to work more strongly in lymphoid cells than the cytomegalovirus (CMV) promoter used in the previous study.34
We successfully demonstrated that the e1a2Mz efficiently induced cell death in the Ph+ ALL cell lines OM9;22, KOPN-30, and KOPN-72 (Figure 3). These cytotoxic effects were thought to be e1a2Mz dependent because transduction of any control vectors, including vectors harboring only left or right arm of the e1a2Mz or b2a2Mz, did not affect the survival of these cells. In addition, we demonstrated that transduction of the e1a2Mz vector inhibited cobblestone area formation of primary Ph+ ALL cells (Figure 6C; Table 2). Furthermore, the growth of these cells was inhibited not only upon transduction of the e1a2Mz vector, which encoded both the left and right arms of the e1a2Mz, but also with cotransduction of the e1a2MzL and e1a2MzR vectors, which encoded only the left or right arm, respectively (Figure 6C; Table 2), further suggesting that the cytotoxicity demonstrated in this study was the effect of the e1a2Mz ribozyme itself. The effect of cotransduction of the e1a2MzL and e1a2MzR vectors was weaker than that of transduction of the e1a2Mz vector, probably because the titer of e1a2MzL and e1a2MzR vectors used in cotransduction (MOI = 10 for each vector) was half of the e1a2Mz vector titer (MOI = 20), resulting simply in a lesser amount of functional ribozyme in transduced cells.
Ph+ ALL cell lines of KOPN-57 and SUP-B15, however, were resistant to e1a2Mz transduction (Figure 3B), although e1a2Mz was expressed efficiently in these cells (Figure 4A) and no mutation existed in the e1a2Mz-targeting sequence of their bcr-abl fusion genes. We confirmed a decrease in bcr-abl mRNA in these cells, but the amount of residual bcr-abl mRNA in these cells was higher than that in e1a2Mz-sensitive cells (Figure 4B), suggesting that the amount of undigested bcr-abl mRNA was high enough to make the cells functionally e1a2Mz resistant. Therefore, we transduced the cells with a higher titer of e1a2Mz vector to increase the intracellular digestion of bcr-abl mRNA. As expected, the viability of KOPN-57 cells decreased in a titer-dependent manner (Figure 5A) but that of SUP-B15 cells did not decrease, despite the escalation of viral titer (Figure 5B). We could not elucidate the cause of e1a2Mz resistance in SUP-B15 cells; it is possible, however, that the expression of other oncogenes besides bcr-abl contributes autonomously to the proliferation of this cell line.
Importantly, the e1a2Mz was expressed in but did not affect the survival of both leukemia cell lines not harboring a p190-type bcr-abl fusion gene (Figure 3B) and normal hematopoietic progenitor cells (Figure 7B-C). These results strongly suggest that transduction of e1a2Mz with our lentiviral vector system would be a secure as well as effective system for use in the clinical treatment of Ph+ ALL.
Of course, long-term safety should be confirmed in vivo before the introduction of this new technology to a clinical setting. Recently, 2 SCID-X1 patients were reported to develop leukemia after gene therapy using an MLV vector. In these cases, vector integration in the proximity of the LIM domain only 2 (LMO2) protooncogene promoter was thought to cause aberrant transcription and expression of LMO2 and induce leukemia. Insertional mutagenesis is a frequently discussed problem of using retroviral vectors in a clinical setting.25 In our study, the copy number of integrated vector DNA in OM9;22 and KOPN-30 cells was only about 1.5 and 2, respectively, when transduced at an MOI of 10 (Figure 3D). The results suggested that e1a2Mz transduction using our HIV(VSV) vector is effective with a minimum integration of the vector DNA and potentially useful for the treatment of Ph+ ALL. It may eventually be necessary, however, to exclude possible lentiviral integration around vital genes by nucleotide sequencing before administration to each patient. The future development of gene targeting technology is, of course, another important issue.
The future application of this new technology for the treatment of ALL includes the in vitro purging of leukemia cells from patients' autologous peripheral blood mononuclear cells (PBMCs) or BM cells using our lentiviral-mediated e1a2Mz transduction system followed by autologous stem cell transplantation. Since the level of minimal residual disease has been reported as an important predictor of relapse for Ph+ ALL after imatinib mesylate treatment35 and some cell lines were resistant or less sensitive to e1a2Mz, the bcr-abl level in residual leukemia cells in the PBMCs or BM must be determined before administration of the Mz-treated cells. For in vivo use, selective transduction of the e1a2Mz into target cells would be ideal because the long-term safety of e1a2Mz transduction into normal somatic cells, including hematopoietic progenitor cells, has not yet been proven as described above. Recently, a lentiviral vector system using an alphavirus envelope containing the Fc-binding site of protein A has been reported to target CD4-expressing cells specifically.36 We have also developed a targeted drug delivery system for CD19-expressing Ph+ ALL cells using an anti-CD19 antibody-binding polyethylene glycol liposome and have demonstrated its efficient and specific transduction into these specific cells.37 These reagents and vectors will be useful in the development of a selective transduction system to deliver e1a2Mz into target cells, potentially allowing the establishment of effective in vivo gene therapy using e1a2Mz.
In conclusion, we have demonstrated the specific cytocidal effect of e1a2Mz on Ph+ ALL cells using a third-generation lentiviral vector system. This is the first report demonstrating the cytotoxic effect of a ribozyme on Ph+ ALL cells without any cytotoxicity to normal hematopoietic progenitor cells during in vitro observation period. Therefore, the e1a2Mz is considered to be a useful tool for gene therapy of Ph+ ALL.
Prepublished online as Blood First Edition Paper, March 23, 2004; DOI 10.1182/blood-2003-06-1948.
Supported in part by Taisho Pharmaceutical Co, Ltd (Tokyo, Japan) and a grant-in-aid from the Ministry of Education, Science, Sports and Culture and the Ministry of Health, Labor and Welfare of Japan. Y.B. is an awardee of a Research Fellowship of the Japan Society of the Promotion of Science for Young Scientists.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors would like to thank Dr I. M. Verma (Salk Institute, La Jolla, CA) and Cell Genesys for providing HIV vector constructs. We also would like to express our gratitude to Drs K. Ohyashiki and T. Tsuji for kindly providing us OM9;22 cells and HESS-5 cells, respectively. We are grateful to Dr H. Urano (Ikuryo Clinic, Tokyo, Japan) for providing us with cord blood samples. We also appreciate Drs T. Iseki, F. Nagamura, J. Ooi, and A. Tomonari and the hospital staff of the Research Hospital of The Institute of Medical Science, The University of Tokyo for their support in sample collection from patients. We are indebted to Dr Y. Nakazaki (Kyushu University), Dr N. Satoh, Ms N. Yusa, Mr K. Takahashi, Ms M. Oiwa, and Ms S. Suzuki (The University of Tokyo) for technical support.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal