Abstract
Recently, overlapping molecular phenotypes of hematopoietic and neuropoietic cells were described in mice. Here, we examined primary human CD34+ hematopoietic stem and progenitor cells applying specialized cDNA arrays, real-time reverse-transcriptase–polymerase chain reaction (RT-PCR), and fluorescent-activated cell sorter (FACS) analysis focusing on genes involved in neurobiologic functions. We found expression of vesicle fusion and motility genes, ligand- and voltage-gated ion channels, receptor kinases and phosphatases, and, most interestingly, mRNA as well as protein expression of G protein–coupled receptors of neuromediators (corticotropin-releasing hormone 1 [CRH 1] and CRH 2 receptors, orexin/hypocretin 1 and 2 receptors, GABAB receptor, adenosine A2B receptor, opioid κ1 and μ1 receptors, and 5-HT 1F receptor). As shown by 2-color immunofluorescence, the protein expression of these receptors was higher in the more immature CD38dim than in the CD38bright subset within the CD34+ population, and completely absent in fully differentiated blood cells, suggesting that those receptors play a role in developmentally early CD34+ stem and progenitor cells. The intracellular concentration of cyclic adenosine monophosphate (cAMP) in CD34+ cells was diminished significantly upon stimulation of either CRH or orexin receptors, indicating that those are functionally active and coupled to inhibitory G proteins in human hematopoietic cells. In conclusion, these findings suggest a molecular interrelation of neuronal and hematopoietic signaling mechanisms in humans.
Introduction
Human CD34+ hematopoietic stem and progenitor cells ensure lifelong production of mature blood cells according to the varying needs of the individual. Hematopoiesis is a precisely regulated process based upon a balance of self-renewal and commitment to differentiation along the different hematopoietic lineages. The restorative capacity of human CD34+ cells is clinically used in the autologous and allogeneic transplantation setting to reconstitute hematopoiesis following cytotoxic therapy for the treatment of patients with malignant or autoimmune diseases.1-3 Beyond that, data of recent studies suggest that hematopoietic progenitors might also be able to transdifferentiate into nonhematopoietic cells, which could open novel therapeutic avenues in the treatment of diseases such as myocardial or cerebral infarction as well as other degenerative disorders.4-7 However, novel data have challenged the transdifferentiation model by suggesting cell fusion rather than plasticity of stem cells.8-10 Abetter molecular understanding of the signal perception pathways of hematopoietic stem and progenitor cells seems to be required to understand the conditions under which transdifferentiation of hematopoietic cells may occur.11 Studies in animal models showed the presence of sensory and autonomic nerves in the bone marrow as a morphologic correlate of a possible neural regulation of hematopoiesis.12-14 However, the idea that neuromediators might directly influence hematopoietic progenitors is controversially discussed.15-19 Recently, several investigators described partly overlapping genetic programs of hematopoietic and neuropoietic cells in mice.20,21 Those findings prompted us to examine human hematopoietic cells by means of specialized cDNA arrays, quantitative real-time reversetranscription–polymerase chain reaction (RT-PCR), and fluorescent-activated cell sorter (FACS) analysis focusing on gene expression known to be involved in neurobiologic functions. We particularly concentrated on receptors and signaling molecules that have not been described in human hematopoietic stem cells so far. We found that primary human CD34+ cells express numerous genes that are primarily assigned to the nervous system, among them G protein–coupled receptors of neuromediators, receptor tyrosine/serine kinases, receptor tyrosine phosphatases, ligand- and voltage-gated ion channels, as well as genes involved in vesicle fusion, motility, and adhesion.
Materials and methods
Cells
After informed consent, human peripheral blood mononuclear cells (PB-MNCs) were obtained from 4 healthy volunteers who donated hematopoietic stem cells for allografting. The donors received 12 μg human recombinant granulocyte colony-stimulating factor (G-CSF; NEUPOGEN; Amgen, Thousand Oaks, CA) per kilogram body weight for 5 days. Afterward, cells were harvested using a CobeSpectra Apheresis System (Gambro BCT, Planegg-Martinsried, Germany). Donors mobilized between 5 × 106 and 11 × 106 CD34+ cells per kilogram body weight, as determined by flow cytometry. PB-MNCs were obtained by density centrifugation using the lymphocyte separation medium lymphoprep (Nycomed Pharma, Oslo, Norway) as previously described.22 CD34+ cells were positively selected using the midiMACS immunomagnetic separation system (Miltenyi Biotec, Bergisch Gladbach, Germany) as described before.23 Purity of separated CD34+ cells ranged between 96% and 99% as determined by flow cytometry.
Immunofluorescence staining and flow cytometry
Immunomagnetically selected cells were stained with a phycoerythrin (PE)–conjugated monoclonal anti-CD34 antibody (clone 8G12; Becton Dickinson, Heidelberg, Germany) in 50 μL phosphate-buffered saline (PBS) at 4° C for 30 minutes. An isotype-identical monoclonal antibody (immunoglobulin G1 [IgG1]–PE; Becton Dickinson) served as control. For detection of receptors, indirect immunofluorescence was used. Highly enriched CD34+ cells were incubated for 30 minutes at 4° C with primary goat polyclonal IgG antibodies raised against opioid κ1 receptor, opioid μ1 receptor, GABA B receptor, serotonin (5-HT) 1F receptor, adenosine A2B receptor, corticotropin-releasing hormone (CRH) receptors, and orexin/hypocretin receptors (Santa Cruz Biotechnology, Heidelberg, Germany). Normal goat IgG served as a control for the primary staining. After washing with PBS, secondary staining was performed for 15 minutes at 4° C using a rhodamine (R)–conjugated donkey anti–goat IgG antibody (Santa Cruz Biotechnology). Rhodamine-conjugated normal donkey IgG was used as a control for the secondary staining. For 2-color immunofluorescence staining of CD34+ cells, we used fluorescein isothiocyanate (FITC)–conjugated monoclonal anti-CD38 (clone HB7; Becton Dickinson) or anti–HLA-DR (clone TU36; Becton Dickinson) antibodies. For staining of monocytes and granulocytes, FITC-conjugated monoclonal anti-CD14 (clone Mϕ9; Becton Dickinson) or anti-CD15 (clone 80H5; Immunotech, Marseilles, France) was applied. After antibody staining, cells were washed and suspended in PBS buffer. The cells were analyzed using a Becton Dickinson FACScan with a 2-W argon ion laser. Resultant data were analyzed using the CellQuest software (Becton Dickinson) after gating on viable cells.
RNA isolation, reverse transcription, and hybridization to nylon cDNA arrays
Isolation of total RNA from 1 × 106 to 4 × 106 highly enriched CD34+ cells was performed with the Rneasy Mini Kit (Qiagen AG, Hilden, Germany) according to the manufacturer's instructions. RNA yield and purity were measured photometrically. RNA samples were dissolved in 2 μL RNase-free water. Atlas Human Neurobiology arrays (BD Biosciences Clontech, Heidelberg, Germany) were used for hybridization experiments. Between 2 to 4 μg total RNA from each sample was reversely transcribed and radioactively labeled with 32P according to a modified protocol of the manufacturer's instructions as previously described.24 Reverse transcription was performed using gene-specific primers instead of random primers, which increased the sensitivity by diminishing the complexity of the resultant cDNA pool. This method permitted the examination of gene expression profiles of CD34+ cells derived from single rather than pooled samples. Both pooling of samples from different donors and amplification of cDNA would have hampered the reliability of the gene expression data. Double-spotted cDNA arrays were used to further enhance validity.
Quantification, normalization
Radioactive signals were assessed using a Phosphorimager (Fuji FLA-3000; FujiFilm, Tokyo, Japan) controlled by the BAS-Reader 3.01 Software (Raytest, Straubenhardt, Germany). We measured dot intensities with the AIDA-Software (Raytest). After background subtraction, raw data were normalized using a standard global intensity-based normalization strategy.25 Relative expression level of each gene was expressed as the ratio of dot intensity to median intensity of 20% highest expressed genes.
Quantitative real-time reverse-transcription–polymerase chain reaction
Total RNA was extracted using the Absolute RNA Microprep system (Stratagene, La Jolla, CA), which includes DNase treatment of the samples for prevention from contamination with genomic DNA. Total RNA was reversely transcribed using random hexamer primers as previously described.26 For detection and quantification, LightCycler technology (Roche Molecular Biochemicals, Mannheim, Germany) was used.27 Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA served as external control for relative quantification. PCR reactions were performed using the LightCycler-FastStart DNA Master SYBR Green kit (Roche Molecular Biochemicals). PCR was carried out in a final volume of 20 μL using 0.5 μM of each primer, 4 mM MgCl2, 2 μL of the supplied enzyme mix containing the reaction buffer, FastStart Taq DNA polymerase, and DNA double strand–specific SYBR Green I dye for detection of PCR products. PCR was performed with a 480-second preincubation at 95° C followed by 45 cycles of 15 seconds at 95° C, 5 seconds at 60° C, and 20 seconds at 72° C. PCR products were verified by melting curve analysis, agarose gel electrophoresis, and DNA sequencing (SEQLAB, Goettingen, Germany). The following primers were used: GAPDH specific: forward primer, 5′-TCCATGACAACTTTGGTATCG-3′ and reverse primer, 5′-CTAATTCTAGTGGGTCAAGATGTAGC-3′ (product, 380 bp); opioid μ1 receptor specific: forward primer, 5′-TCCGTTGCCCTAACAG-3′ and reverse primer, 5′-CCTCGGTGTGGTATATT-3′ (product, 233 bp); GABA B receptor specific: forward primer, 5′-GTGGGCATGGCTATCTACAAT-3′ and reverse primer, 5′-ACGCTCCTCTTTCTCAGCAAT-3′ (product, 312 bp); serotonin 1F receptor specific: forward primer, 5′-TGGTGTCCCTCACTCT-3′ and reverse primer, 5′-ACCACTTGCCCCATAA-3′ (product, 211 bp); adenosine A2B receptor specific: forward primer, 5′-TTCAATTTCTTTGGGTGT-3′ and reverse primer 5′-GTTCCGGTAAGCATAG-3′ (product, 330 bp); purinergic P2X4 receptor specific: forward primer, 5′-CTACGTCATCGGGTGG-3′ and reverse primer, 5′-CGTTGAAAGCTACGCA-3′ (product, 338 bp); voltage-gated potassium channel SRS4 (shaker related subfamily, member 4) specific: forward primer, 5′-TGACCCTTTGCGCAATGAGT-3′ and reverse primer 5′-AGATGACCAGGACGGACACAA-3′ (product, 317 bp); voltage-gated potassium channel SRS6 specific: forward primer, 5′-CACGGTAGGTTACGGG-3′ and reverse primer, 5′-CCAAGTCCGTTGTCAG-3′ (product, 228 bp); neurotrophin 3 specific: forward primer, 5′-ACGGTACGCGGAGCATAAGA-3′ and reverse primer, 5′-CTCGGACGTAGGTTTGGGAT-3′ (product, 264 bp); synaptobrevin 1 specific: forward primer, 5′-CCTCCTCCTAACATGACCAGTAA-3′ and reverse primer, 5′-AGCATGATCATCATCTTGCAGT-3′ (233 bp); and dystroglycan α specific: forward primer, 5′-CTTACAGCAGTTTGTACG-3′ and reverse primer, 5′-CAGTCGATATGGCTTAG-3′ (product, 384 bp). Corticotropin-releasing hormone 1 (CRH 1) and CRH 2 receptors were detected using TaqMan technology as previously described.28,29 To exclude genomic DNA contamination, RNA extracts that had not undergone reverse transcription prior to PCR served as control. Primers were designed using the LightCycler Probe Design software 1.0 (Roche Molecular Biochemicals). The crossing points (CPs) of real-time PCR curves were determined by the LightCycler 3.5 software using the second derivative maximum method.
Enzyme immunoassay
Intracellular cyclic adenosine monophosphate (cAMP) levels of CD34+ cells were measured by a competitive enzyme immunoassay (EIA; Sigma, Taufkirchen, Germany). Immunomagnetically selected CD34+ cells were cultured at 1 × 106 cells per milliliter in RPMI 1640 medium (Sigma) containing 10% heat-inactivated fetal calf serum (PAA Laboratories, Coelbe, Germany), 100 U/mL penicillin (Sigma), 100 μg/mL streptomycin (Sigma), 2 mM l-glutamine (PAA Laboratories), 20 ng/mL interleukin-3, 20 ng/mL interleukin-6, and 50 ng/mL stem cell factor, in 5% CO2 at 37° C for 15 hours. Cytokines were obtained from PromoCell (Heidelberg, Germany). Subsequently, the cells were incubated with either 1 μM CRH, 1 μM orexin A, 0.1 μM orexin B, or PBS alone (control) for 30 minutes at 37° C. After centrifugation, the supernatant was completely removed and the cell pellets were treated with 100 mM HCl for 20 minutes to lyse the cells and decrease endogenous phosphodiesterase activity. After centrifugation at 600g at room temperature, cell lysates were acetylated by adding 5% of acetic anhydrate in order to prevent cAMP degradation and thereby improving the sensitivity of the assay. There were 5 samples containing defined amounts of cAMP (0.078 pmol/mL, 0.312 pmol/mL, 1.25 pmol/mL, 5 pmol/mL, or 20 pmol/mL; Sigma) that served as standards and were treated in the same manner with HCl followed by acetylation as described. Following acetylation, standards as well as samples were incubated in separate wells of a microtiter plate coated with goat anti–rabbit IgG (Sigma). Alkaline phosphatase (50 μL) covalently linked to cAMP as well as 50 μL of a polyclonal rabbit anti-cAMP antibody (Sigma) were added. The sealed microtiter plate was incubated at room temperature on a shaker at 300 rpm for 2 hours to allow competitive binding of the anti-cAMP antibody, and afterward wells were treated with 200 μL wash buffer (Sigma) 3 times. Then 200 μL of a solution of p-nitrophenyl phosphate (Sigma), serving as a substrate for alkaline phosphatase, was added to every well and incubated at room temperature without shaking. After one hour, the enzyme reaction was stopped and the optical density at 405 nm, being inversely proportional to the concentration of cAMP in either standards or samples, was determined using the Wallac 1420 Victor2 multilabel plate reader (EG&G; Wallac, Turku, Finland). The concentration of cAMP in the samples was calculated using the standards as a reference and expressed as percentage of the unstimulated controls.
Results
Gene expression analysis using specialized cDNA arrays
We examined primary human hematopoietic stem and progenitor cells by means of specialized cDNA arrays covering the expression of 588 human genes primarily assigned to the nervous system. We found expression of motility genes, ligand- and voltage-gated ion channels, receptor tyrosine/serine kinases, receptor tyrosine phosphatases, and G protein–coupled receptors, which had not been described in CD34+ cells so far (Table 1; Figure 1). The complete data of our array experiments are available in the database of the German Resource Center for Genome Research at http://www.rzpd.de according to the MIAME (Minimum Information About a Microarray Experiment) standards.30
Neurobiologic genes expressed in human CD34+ cells
Name . | GenBank accession no. . |
|---|---|
| Receptors | |
| GABA B receptor | Y11044 |
| Adenosine A2B receptor | M97759 |
| Opioid κ1 receptor | U11053 |
| Opioid μ1 receptor | L25119 |
| 5-HT 1F receptor | L05597 |
| Prostaglandin E4 receptor | L25124 |
| Somatostatin 4 receptor | D16826 |
| Thrombin receptor (PAR1) | M62424 |
| CRH 1 receptor | X72304 |
| Ephrin A1 receptor (EphA1) | M18391 |
| Insulin-like growth factor 2 receptor | Y00285 |
| Activin A receptor, type 2b | X77533 |
| Postsynaptic density protein 95 (PSD-95) | U83192 |
| Collagen type I receptor (CD36) | D12676 |
| Transforming growth factor β receptor 1 | L11695 |
| Protein tyrosine phosphatase, receptor type A | M34668 |
| Protein tyrosine phosphatase, receptor type E | X54134 |
| Protein tyrosine phosphatase, receptor type J | U10886 |
| Protein tyrosine phosphatase, receptor type M | X58288 |
| Ion channels | |
| Purinergic P2X4 receptor | Y07684 |
| Purinergic P2X7 receptor | Y09561 |
| Voltage-gated potassium channel, SRS4 | M55514 |
| Voltage-gated potassium channel, SRS6 | X17622 |
| Voltage-gated chloride channel 3 | X78520 |
| Voltage-gated chloride channel 4 | X77197 |
| Voltage-gated chloride channel 7 | Z67743 |
| Cytoskeleton, motility, adhesion | |
| Synaptobrevin 1 (VAMP1) | Z48924 |
| Syntaxin 1A | L37792 |
| Syntaxin 3A | U32315 |
| Syntaxin 4A | U07158 |
| Syntaxin 7 | U77942 |
| Syntaxin-binding protein 2 | U63533 |
| Synaptosomal-associated protein 23 (SNAP23) | Y09567 |
| N-ethylmaleimide-sensitive factor (NSF) | U03985 |
| α-SNAP (NSF attachment protein) | U39412 |
| Membrane protein of cholinergic synaptic vesicles (VAT1) | U18009 |
| RAB 1 | M28209 |
| RAB 11 | X79780 |
| Dynein, light chain | U32944 |
| Dynein, heavy chain | L23958 |
| Dynamin 2 | L36983 |
| Dynactin 1 | X98801 |
| Actinin alpha 1 | X15804 |
| Drebrin 1 | U00802 |
| Dystroglycan α | L19711 |
Name . | GenBank accession no. . |
|---|---|
| Receptors | |
| GABA B receptor | Y11044 |
| Adenosine A2B receptor | M97759 |
| Opioid κ1 receptor | U11053 |
| Opioid μ1 receptor | L25119 |
| 5-HT 1F receptor | L05597 |
| Prostaglandin E4 receptor | L25124 |
| Somatostatin 4 receptor | D16826 |
| Thrombin receptor (PAR1) | M62424 |
| CRH 1 receptor | X72304 |
| Ephrin A1 receptor (EphA1) | M18391 |
| Insulin-like growth factor 2 receptor | Y00285 |
| Activin A receptor, type 2b | X77533 |
| Postsynaptic density protein 95 (PSD-95) | U83192 |
| Collagen type I receptor (CD36) | D12676 |
| Transforming growth factor β receptor 1 | L11695 |
| Protein tyrosine phosphatase, receptor type A | M34668 |
| Protein tyrosine phosphatase, receptor type E | X54134 |
| Protein tyrosine phosphatase, receptor type J | U10886 |
| Protein tyrosine phosphatase, receptor type M | X58288 |
| Ion channels | |
| Purinergic P2X4 receptor | Y07684 |
| Purinergic P2X7 receptor | Y09561 |
| Voltage-gated potassium channel, SRS4 | M55514 |
| Voltage-gated potassium channel, SRS6 | X17622 |
| Voltage-gated chloride channel 3 | X78520 |
| Voltage-gated chloride channel 4 | X77197 |
| Voltage-gated chloride channel 7 | Z67743 |
| Cytoskeleton, motility, adhesion | |
| Synaptobrevin 1 (VAMP1) | Z48924 |
| Syntaxin 1A | L37792 |
| Syntaxin 3A | U32315 |
| Syntaxin 4A | U07158 |
| Syntaxin 7 | U77942 |
| Syntaxin-binding protein 2 | U63533 |
| Synaptosomal-associated protein 23 (SNAP23) | Y09567 |
| N-ethylmaleimide-sensitive factor (NSF) | U03985 |
| α-SNAP (NSF attachment protein) | U39412 |
| Membrane protein of cholinergic synaptic vesicles (VAT1) | U18009 |
| RAB 1 | M28209 |
| RAB 11 | X79780 |
| Dynein, light chain | U32944 |
| Dynein, heavy chain | L23958 |
| Dynamin 2 | L36983 |
| Dynactin 1 | X98801 |
| Actinin alpha 1 | X15804 |
| Drebrin 1 | U00802 |
| Dystroglycan α | L19711 |
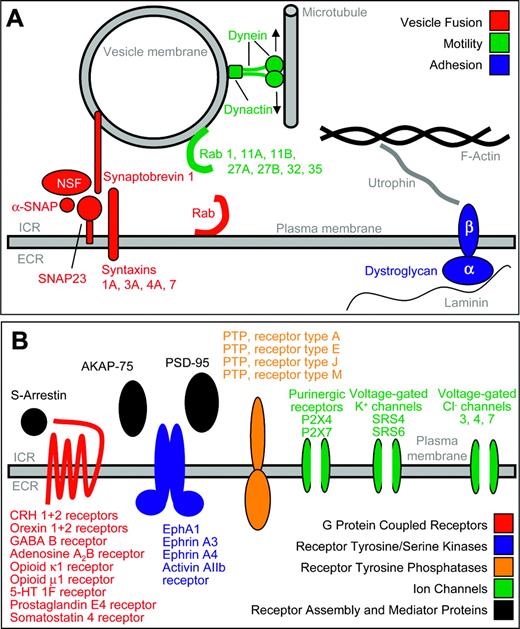
Schematic display of neurobiologic genes expressed in primary human CD34+ hematopoietic stem and progenitor cells. All genes, the expression of which we newly demonstrated in human CD34+ cells, are colored. Intracellular rooms (ICR) and extracellular rooms (ECR) are indicated. (A) Motility and cytoskeletal genes. (B) Receptors and ion channels.
Schematic display of neurobiologic genes expressed in primary human CD34+ hematopoietic stem and progenitor cells. All genes, the expression of which we newly demonstrated in human CD34+ cells, are colored. Intracellular rooms (ICR) and extracellular rooms (ECR) are indicated. (A) Motility and cytoskeletal genes. (B) Receptors and ion channels.
Corroboration by real-time RT-PCR and immunofluorescence
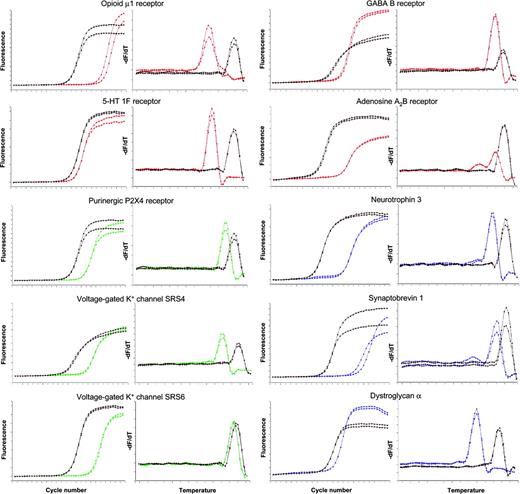
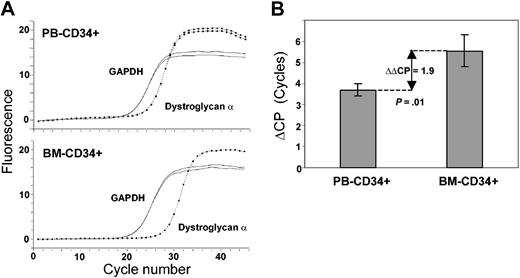
To corroborate mRNA expression data obtained by cDNA array technology, we performed real-time RT-PCR for 12 genes. The results of real-time RT-PCR were in line with the cDNA array data. In detail, we demonstrated mRNA expression of GABA B receptor, adenosine A2B receptor, corticotropin-releasing hormone 1 (CRH1) and CRH2 receptors, opioid μ1 receptor, serotonin 1F receptor, purinergic P2X4 receptor, voltage-gated potassium channels SRS4 and SRS6, neurotrophin 3 (NT-3), synaptobrevin 1 (VAMP1), and dystroglycan α (Figures 2, 3). Furthermore, we examined differential expression of dystroglycan α in CD34+ cells from peripheral blood (PB-CD34+) and bone marrow (BM-CD34+) using quantitative real-time RT-PCR. We found that PB-CD34+ cells expressed dystroglycan α 3.8-fold higher than BM-CD34+ cells (Figure 4).
Detection of mRNA expression by real-time RT-PCR. Real-time RT-PCR curves (left) and respective melting curves (right) of the PCR products are displayed. Curves of the 10 target genes are colored; curves of the respective GAPDH controls are displayed in black. The analysis of 1 representative of 3 CD34+ cell samples is shown. Each analysis was performed in duplicate.
Detection of mRNA expression by real-time RT-PCR. Real-time RT-PCR curves (left) and respective melting curves (right) of the PCR products are displayed. Curves of the 10 target genes are colored; curves of the respective GAPDH controls are displayed in black. The analysis of 1 representative of 3 CD34+ cell samples is shown. Each analysis was performed in duplicate.
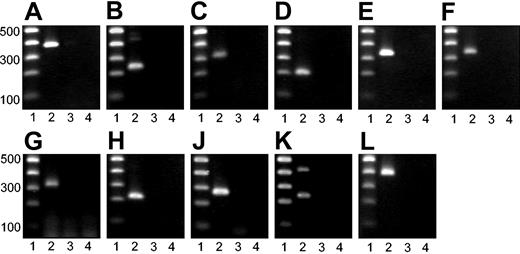
Agarose gel electrophoresis of PCR products. PCR products were subjected to agarose gel electrophoresis. Lane 1: 100–base pair (bp) DNA ladder. Base pairs are indicated. Lane 2: specific RT-PCR product. Lane 3: PCR analysis of RNA without RT (no-RT control). Lane 4: water control. (A) GAPDH. (B) Opioid μ1 receptor. (C) GABA B receptor. (D) 5-HT 1F receptor. (E) Adenosine A2B receptor. (F) Purinergic P2X4 receptor. (G) Voltage-gated K+ channel SRS4. (H) Voltage-gated K+ channel SRS6. (J) Neurotrophin 3. (K) Synaptobrevin 1. (L) Dystroglycan α.
Agarose gel electrophoresis of PCR products. PCR products were subjected to agarose gel electrophoresis. Lane 1: 100–base pair (bp) DNA ladder. Base pairs are indicated. Lane 2: specific RT-PCR product. Lane 3: PCR analysis of RNA without RT (no-RT control). Lane 4: water control. (A) GAPDH. (B) Opioid μ1 receptor. (C) GABA B receptor. (D) 5-HT 1F receptor. (E) Adenosine A2B receptor. (F) Purinergic P2X4 receptor. (G) Voltage-gated K+ channel SRS4. (H) Voltage-gated K+ channel SRS6. (J) Neurotrophin 3. (K) Synaptobrevin 1. (L) Dystroglycan α.
Differential dystroglycan α expression in human CD34+ cells of the blood and marrow. (A) Real-time RT-PCR curves of dystroglycan α (dotted line) and the GAPDH control (solid line) of 1 representative of 4 experiments are shown. (B) Average differences of crossing points (ΔCP) of dystroglycan α and GAPDH for all examined PB-CD34+ (n = 4) and BM-CD34+ (n = 4) samples are displayed. The lines in each bar indicate one standard deviation. The average difference of ΔCPs (ΔΔCP) of PB and BM is indicated by the double-headed arrow (P = .01).
Differential dystroglycan α expression in human CD34+ cells of the blood and marrow. (A) Real-time RT-PCR curves of dystroglycan α (dotted line) and the GAPDH control (solid line) of 1 representative of 4 experiments are shown. (B) Average differences of crossing points (ΔCP) of dystroglycan α and GAPDH for all examined PB-CD34+ (n = 4) and BM-CD34+ (n = 4) samples are displayed. The lines in each bar indicate one standard deviation. The average difference of ΔCPs (ΔΔCP) of PB and BM is indicated by the double-headed arrow (P = .01).
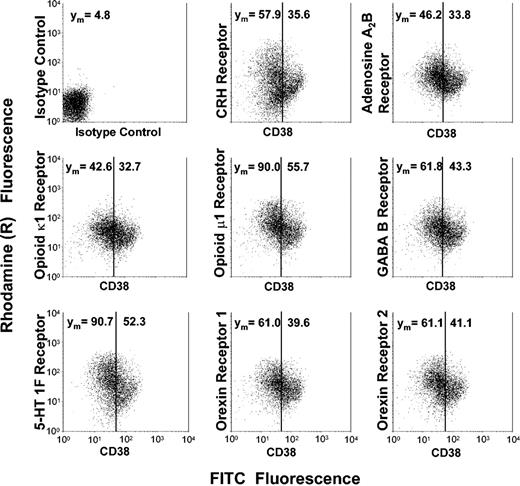
To confirm that the transcripts were also expressed at the protein level, we measured expression of 8 of the newly found receptors by indirect immunofluorescence and FACS analysis of highly enriched CD34+ cells. Protein expression of orexin/hypocretin 1 and 2 receptors, CRH receptors, GABA B receptor, adenosine A2B receptor, opioid κ1 and μ1 receptors, and serotonin 1F receptor was found on CD34+ cells, confirming the mRNA expression data obtained by cDNA array technology and real-time RT-PCR (Figure 5).
Protein expression of neurobiologic surface receptors dependent on the developmental stage of human CD34+ cells. Highly enriched CD34+ cells were subjected to 2-color immunofluorescence using a FITC-conjugated anti-CD38 antibody and surface receptor–specific antibodies, the binding of which was detected by a rhodamine (R)–conjugated secondary antibody. In each dot plot, cell populations are divided into a developmentally early CD38dim subset and a CD38bright subset representing more mature hematopoietic progenitors. Mean rhodamine fluorescence values, which reflect the surface receptor expression, are indicated for each subset. Displayed is 1 representative of 4 experiments.
Protein expression of neurobiologic surface receptors dependent on the developmental stage of human CD34+ cells. Highly enriched CD34+ cells were subjected to 2-color immunofluorescence using a FITC-conjugated anti-CD38 antibody and surface receptor–specific antibodies, the binding of which was detected by a rhodamine (R)–conjugated secondary antibody. In each dot plot, cell populations are divided into a developmentally early CD38dim subset and a CD38bright subset representing more mature hematopoietic progenitors. Mean rhodamine fluorescence values, which reflect the surface receptor expression, are indicated for each subset. Displayed is 1 representative of 4 experiments.
Development-dependent receptor expression
After having demonstrated mRNA as well as protein expression of the receptors, we wondered whether receptor expression was related to the developmental stage of human CD34+ cells. We used 2-color immunofluorescence to measure receptor protein expression dependent on CD38 expression in CD34+ cells. We found that expression of CRH receptors, orexin receptors, GABA B receptor, adenosine A2B receptor, opioid κ1 and μ1 receptors, as well as 5-HT 1F receptor was greater among the more immature CD34+ CD38dim subset in comparison with the lineage-determined CD34+ CD38bright progenitors (Figure 5). The same results were obtained when we used HLA-DR staining to subdivide cells within the CD34+ population (data not shown). Fully differentiated CD14+ monocytes or CD15+ granulocytes showed no expression of CRH receptors, orexin receptors, GABA B receptor, adenosine A2B receptor, opioid receptors, or 5-HT 1F receptor (data not shown).
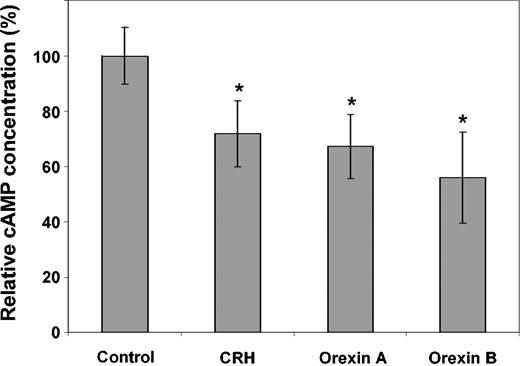
Functional activity of CRH and orexin receptors
In order to assess whether the G protein–coupled receptors are functionally active in CD34+ cells, we measured intracellular concentration of the second messenger cAMP upon stimulation with CRH, orexin A, or orexin B using a competitive enzyme immunoassay. Intracellular cAMP decreased by 28% (SD: 12%), 33% (SD: 11%), and 44% (SD: 16%) following stimulation of highly enriched human CD34+ cells with CRH, orexin A, or orexin B, respectively (Figure 6). This decrease was statistically highly significant (P < .001).
Enzyme immunoassay (EIA) of intracellular cAMP in human CD34+ cells. Highly enriched human CD34+ cells were stimulated with CRH (1 × 10–6 M), orexin A (1 × 10–6 M), or orexin B (1 × 10–7 M). Afterward, intracellular cAMP concentration was measured by EIA and compared with untreated controls. CD34 cell samples of 6 patients were examined in duplicate. The lines in each bar represent one standard deviation. Significant changes (P < .001) are indicated (*).
Enzyme immunoassay (EIA) of intracellular cAMP in human CD34+ cells. Highly enriched human CD34+ cells were stimulated with CRH (1 × 10–6 M), orexin A (1 × 10–6 M), or orexin B (1 × 10–7 M). Afterward, intracellular cAMP concentration was measured by EIA and compared with untreated controls. CD34 cell samples of 6 patients were examined in duplicate. The lines in each bar represent one standard deviation. Significant changes (P < .001) are indicated (*).
Discussion
Recently, several investigators detected overlapping gene expression profiles of neuropoietic and hematopoietic cells in mice.20,21 Here, we examined primary human hematopoietic stem and progenitor cells using specialized cDNA arrays measuring the expression of 588 human genes known to represent the functional repertoire of neuronal cells. Particular emphasis was put on the examination of expression of genes that mediate communication of cells within the nervous system such as receptors and ion channels. The cDNA arrays also included components of the specialized cytoskeleton involved in vesicle trafficking, synaptic transmission, and adhesion of neuronal cells. We found expression of members of different categories of genes such as vesicle fusion and motility genes, ligand- and voltage-gated ion channels, receptor tyrosine/serine kinases, receptor tyrosine phosphatases, and G protein–coupled receptors, which had not been observed in CD34+ cells so far. For all experiments performed in this study, we used highly enriched CD34+ cells of a purity between 96% and 99%. However, it is not impossible that in some instances gene expression detected by array analysis or RT-PCR might at least partly derive from other blood cells, although FACS analysis established expression of 8 genes on CD34+ cells on a single-cell level.
Specialized cytoskeleton, vesicle transport, and adhesion
In the nervous system, transmission of receptor-mediated signals or messenger uptake requires a specialized cytoskeleton allowing endocytosis and exocytosis as well as directed intracellular transport of vesicles and molecules. The molecular elements of this machinery are well characterized in polar cell types such as neuronal or epithelial cells, while human hematopoietic stem and progenitor cells have not been examined so far. We found expression of a variety of genes necessary for directed transport of molecules. Synaptobrevin (vesicle-associated membrane protein 1, VAMP1), synaptosomal-associated protein 23 (SNAP23), N-ethylmaleinimide-sensitive factor (NSF), and α-SNAP (NSF attachment protein) were expressed in CD34+ cells as well as other essential elements of the vesicle fusion apparatus such as the syntaxins 1A, 3A, 4A, and 7, and syntaxin binding protein 2. There was also expression of genes coding for proteins known to be part of vesicle membranes such as the membrane protein of cholinergic synaptic vesicles (VAT1), clathrin, clathrin-associated proteins, and clathrin coat assembly proteins. A variety of genes of the RAB (Ras-related in brain) family were also expressed (RAB 1, 11A, 11B, 27A, 27B, 32, and 35, and guanosine diphosphate (GDP) dissociation inhibitor 1 and 2). The respective proteins facilitate directed transport of vesicles along cytoskeletal structures and mediate tethering of vesicles to target membranes, which is an early step in vesicle fusion.
Genes for members of the cytoskeletal motility apparatus expressed in CD34+ cells were dynein, dynamin 2, dynactin 1, alpha-actinin, and drebrin 1. In conclusion, CD34+ hematopoietic stem and progenitor cells apparently have the molecular equipment available to exert active and directed molecular transport.
Dystroglycan is a heterodimeric transmembrane molecule binding laminin-1 and -2, perlecan, and agrin. It is known to link the cytoskeleton to the extracellular matrix in nerve and muscle cells.31,32 Dystroglycan α was strongly expressed in human CD34+ cells, suggesting that it might play a role in adhesion of hematopoietic stem and progenitor cells. As assessed by real-time RT-PCR, circulating CD34+ cells from peripheral blood expressed dystroglycan significantly higher than CD34+ cells residing in the bone marrow. This finding implies a role for dystroglycan in the trafficking and homing of CD34+ cells, possibly by using dystroglycan as a molecular anchor. Another interesting aspect is that dystroglycan serves as a major receptor for arenaviruses, lymphocytic choriomeningitis (LCM) virus for instance.33 It has been recognized for a long time that the hematopoietic stem cell compartment is suppressed upon systemic LCM virus infection without understanding the molecular causes of this observation.34,35 Our data point out the possibility of arenaviral infection of CD34+ cells, which might result in the infection-associated hematopoietic failure.
Ion channels
Recently, it was reported that intracellular ion concentrations play a crucial role for signaling and development of neuronal cells.36 Shirihai et al have shown that expression of 2 inward rectifying potassium channels is necessary for cytokine-induced differentiation of hematopoietic progenitors.37 Here, we describe expression of the ligand-gated ion channels P2X4 and P2X7 as well as expression of the voltage-gated potassium channels SRS4 and SRS6 and voltage-gated chloride channels 3, 4, and 7 in CD34+ cells. P2X4 and P2X7 are purinergic receptors that can be activated by adenosine triphosphate (ATP) and result in opening of nonselective cation channels permeable for Na+,K+, and Ca2+ ions.38 Voltage-gated channels may lead to prolonged changes of intracellular ion concentration. Our data may provide a molecular basis for functional studies of the electrophysiologic properties of CD34+ cells.
Receptors
In this study, we demonstrate that primary human CD34+ cells express mRNA and protein of receptors primarily ascribed to the nervous system.
We found that the G protein–coupled opioid receptors κ1 as well as μ1 are expressed on human CD34+ cells. From previous studies, it is known that opioid receptor signaling exerts functional effects in certain types of blood cells. Maestroni et al reported that hematopoiesis is stimulated via κ1 opioid receptor–mediated signaling.39 On the other hand, the loss of a particular opioid receptor as shown in opioid μ1 receptor–deficient mice was associated with increased proliferation of granulocyte-macrophage, erythroid, and multipotential progenitor cells.40 However, the molecular causes of those observations are unclear. From our data, we assume that the natural ligands of opioid receptors may act on the level of CD34+ cells.
The adenosine A2B receptor represents a stimulatory G protein–coupled receptor, which we also found to be expressed in CD34+ cells on the transcriptional as well as on the proteomic level. Adenosine A2B receptors play an important role for axonal outgrowth and hypoxic preconditioning in the heart and the nervous system.41-43 The presence of this receptor in CD34+ cells could explain the recent observation that an increase in the concentration of extracellular adenosine enhances cell cycling of hematopoietic progenitors.44
Another G protein–coupled receptor with potentially growth mediating activity is the 5-HT 1F receptor. Our finding of mRNA and surface protein expression of the 5-HT 1F receptor in CD34+ cells might explain the observation of Skurikhin et al who found that hematopoiesis is influenced by the serotoninergic system in vivo.45
We also detected expression of the prostaglandin E4 receptor, another G protein–coupled receptor. In a burn sepsis model, it could be shown that the natural ligand prostaglandin E2 of this receptor augments monocytopoiesis and decreases granulopoiesis in mice.46 Furthermore, prostaglandin E2 seems to suppress B lymphopoiesis.47 Our data suggest that the differential effects observed in both lineages are mediated via prostaglandin E4 receptor on the level of CD34+ progenitor cells.
GABA B receptor mRNA as well as surface protein was also expressed in human CD34+ cells. Within the nervous system, GABA plays a central role as a neurotransmitter mediating inhibitory postsynaptic potentials either by increasing the permeability of potassium channels or by blocking voltage-dependent calcium channels. Since we observed the expression of several genes encoding for potassium channels, GABA could be viewed as a mediator between neuronal and hematopoietic cells.
Most interestingly, we detected mRNA as well as protein expression of 4 receptors of hypothalamic peptides, the CRH 1 and 2 receptors and the orexin 1 and 2 receptors, in human CD34+ cells. CRH receptors are involved in systemic stress response via the hypothalamus-pituitary-adrenal axis and in topic stress-related effects.28,29 The orexin receptors were thought to be almost exclusively expressed in the nervous system up to now and play a role in regulation of sleep-wake-rhythm and energy homeostasis.48,49 The physiologic functions of those receptors in human CD34+ hematopoietic stem and progenitor cells are not known.
We also found expression of a couple of novel non–G protein–coupled receptors in CD34+ cells. We report that ephrin A1 receptor (EphA1) as well as its membrane-anchored ligands ephrin A3 and ephrin A4 are expressed in CD34+ hematopoietic cells. Ephrin receptors (Eph) are a large family of receptor tyrosine kinases that interact with ephrin proteins.50 Interactions of ephrins and ephrin receptors play a central role for developmental patterning in the nervous system by guiding migration and repulsion of axons.51 Further reports suggest a functional relevance for angiogenesis, tumor formation, and adhesion.52 Another special feature of the ephrin receptor/ligand system is bidirectional signaling mechanisms, which means that the membrane-bound ephrin proteins not only serve as ligands for Eph receptors but also transduce afferent signals upon receptor binding.53 Our data suggest that signaling via EphA1 receptor, and the ligands ephrin A3 and ephrin A4, may also play a role for hematopoietic stem and progenitor cells—maybe through a homotypic interaction of CD34+ cells.
Beyond surface receptors, we also found expression of genes that act as intracellular organizers of receptor assembly and integrators of signals, thus representing a second level of signal processing following receptor activation. The postsynaptic density protein 95 (PSD-95), expression of which we found in human CD34+ cells, is known to be linked to receptors in the postsynaptic density of neuronal cells.54 PSD-95 seems to function as an organizer of receptor complexes and as a mediator of signal transduction.55 Additionally, we detected expression of protein kinase A anchor protein 5 (AKAP75) in CD34+ cells. AKAP75 anchors protein kinase A to postsynaptic densities and increases cAMP signaling to the nucleus.56 Further, AKAP75 works as an integrator of signals from multiple transduction pathways in neuronal cells.57 Our findings imply complex signal processing mechanisms in CD34+ cells similar to those in neuronal cells.
Development-dependent expression of neuromediator receptors
Wondering whether the neuromediator receptor expression was related to the developmental stage of human CD34+ cells, we found that expression of CRH receptors, orexin receptors, GABA B receptor, adenosine A2B receptor, opioid κ1 and μ1 receptors, as well as 5-HT 1F receptor was greater among the more immature CD34+ CD38dim subset in comparison with the lineage-determined progenitors staining brightly positive for CD38. On the other hand, these receptors were not expressed at all by fully differentiated blood cells such as monocytes or granulocytes. Apparently, expression of the receptors is restricted to immature developmentally early hematopoietic stem and progenitor cells.
Functional activity of CRH and orexin receptors in human CD34+ cells
We found that the intracellular cAMP decreased significantly following stimulation of highly enriched human CD34+ cells with either CRH, orexin A, or orexin B. This finding indicates that CRH as well as orexin receptors are actively signaling in human CD34+ cells and that they are coupled to inhibitory G proteins. These observations are in line with previous studies having shown the coupling of CRH and orexin receptors to inhibitory G proteins in the nervous system.58,59 Further studies will be necessary to elucidate the exact signaling pathways of these and the other newly described neuromediator receptors in human CD34+ cells.
In conclusion, primary human CD34+ hematopoietic cells express mRNA as well as protein of numerous genes that so far were thought to exert their functions predominantly in the nervous system, among them receptors, receptor assembly molecules, ion channels, and components of a specialized cytoskeleton mediating vesicle trafficking and adhesion. Therefore, the functional role of those genes and their products might be broader and more universal than assumed up to now. Alternatively, the expression of neuronal marker genes might not be as specific as previously believed. Our finding that early CD34+ cells express several functionally active receptors primarily assigned to the nervous system demonstrates a potential molecular interrelation of neuronal and hematopoietic signaling mechanisms, and supports molecularly the model of a possible neuronal regulation of immature hematopoietic progenitors. The higher expression of neurobiologic receptors in immature, developmentally early hematopoietic cells and their undetectable expression in fully differentiated blood cells might furthermore suggest a developmental affinity of human hematopoietic and neural cells. Our data provide a basis for studies examining the functional role of the newly detected receptors in human CD34+ cells and their significance in hematopoietic regulation, stem cell signaling, differentiation, and perhaps transdifferentiation.
Prepublished online as Blood First Edition Paper, March 11, 2004; DOI 10.1182/blood-2004-01-0373.
Supported by the Leukämie Liga e.V. Düsseldorf and the Deutsche Krebshilfe (grant D/03/41221; U.S.).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Anke Boeckmann, Maren Free, Hildegard Gaussmann, Baerbel Junge, Sabrina Pechtel, and Elke Rosenbaum-Koenig for expert technical support.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal