Abstract
BCR/ABL is the causative genetic aberration in chronic myelogenous leukemia (CML). Mice lacking expression of the interferon (IFN) consensus sequence binding protein (ICSBP), an IFNγ-inducible transcription factor of the interferon regulatory factor (IRF) family, develop a disease similar to human CML. Mounting evidence suggests a role for ICSBP in the pathogenesis of CML. However, the underlying mechanisms are largely unknown. By stable and conditional expression of ICSBP in wild-type and BCR/ABL-transformed 32D cells (32D/wt and 32D/BA), we found that ICSBP inhibited BCR/ABL-mediated leukemogenesis in vivo. Moreover, ICSBP also overrode BCR/ABL-mediated morphology changes, chemotherapy, and imatinib resistance, as well as BCR/ABL-induced repression of differentiation. Some of these ICSBP effects may be explained in part by an ICSBP-mediated repression of bcl-2, a major antiapoptotic target of BCR/ABL, on transcriptional and protein level. Using reporter gene assays and electrophoretic mobility shift assays we identified that the bcl-2 promoter activity was inhibited by ICSBP by way of a fragment containing 2 characteristic ICSBP-responsive elements. An inverse correlation between ICSBP and bcl-2 expression was confirmed in vivo. Collectively, our findings suggest that ICSBP antagonizes BCR/ABL by down-regulation of bcl-2 and implicates a central role for ICSBP in the pathogenesis of CML, as well as a therapeutic target to overcome drug resistance in bcl-2-dependent tumors. (Blood. 2004;103:3480-3489)
Introduction
The reciprocal gene translocation t(9;22)(q34;q11) creates the Philadelphia chromosome (Ph+), which is the causative genetic aberration of chronic myelogenous leukemia (CML).1 Translation of this leukemia-specific hybrid gene generates a 210-kDa fusion protein (p210 BCR/ABL),2 which constitutively activates various signal transduction pathways that result in malignant transformation.3-5
Even though BCR/ABL has been established as the unique cause of CML, the clinical heterogeneity of CML cannot entirely be explained by activation of BCR/ABL. Several lines of evidence suggest that interferon consensus sequence binding protein (ICSBP/IRF-8), a member of the interferon γ (IFNγ)-inducible transcription factor family may be involved in the pathogenesis of CML.6-10 Interferon regulatory factors (IRFs) are generally characterized by homologous DNA binding domains that recognize specific consensus sequences present in IFNs and IFN-inducible genes (reviewed in Tamura and Ozato11 and Levi et al12 ). ICSBP regulates immune responses7,13-15 by controlling the transcription of important genes of the innate and adoptive immunity, such as major histocompatibility complex (MHC) class I and II genes, IFNγ, interleukin-1β (IL-1β), IL-12 p40, and IL-18.6,16-19 However, recent findings also suggested a role for ICSBP as a tumor suppressor and regulator of apoptosis.20,21
Still, little is known exactly how ICSBP exerts antitumor effects. By using stable and conditional systems coexpressing BCR/ABL and ICSBP, we were seeking to uncover mechanisms of the tumor suppressor activity of ICSBP. To this end, we found that ICSBP down-regulated bcl-2, one of the key antiapoptotic cancer genes, known to be specifically required for BCR/ABL-mediated transformation.22-24
Materials and methods
Mice, mice injections, and analyses
Male mice 6 to 9 weeks old were injected intravenously into mouse lateral tail vein with 1 × 106 32Dcl3-derived cells (hereafter referred to as 32D). Mice were closely monitored and killed if moribund. Dead mice were checked for weight and size of their livers and spleens. Peripheral blood and bone marrow smears were prepared, and Giemsa stainings were analyzed for cell morphology.
Plasmids, retroviral constructs, and retroviral infection
For pBABE-ICSBP-Hygro we subcloned murine ICSBP cDNA from mICSBP-pcDNA3 (kindly provided by K. Ozato [National Institute of Child Health and Human Development, Bethesda, MD]) into pBABEHygro. The retroviral vector Mig-p210, containing a full-length 7.2-kb EcoRI BCR/ABL cDNA fragment was a kind gift from W. Pear.25 For pTRE2hyg-ICSBP, murine ICSBP was cloned into the BamHI site of pTRE2hyg (Clontech Laboratories, Heidelberg, Germany).
For retroviral infections of parental 32D cells and 32D/BA cells with ICSBP virus or control virus, and of 32D/ICSBP on/off with BCR/ABL virus, the Phoenix-eco packaging cell line (G. Nolan, Stanford University, Palo Alto, CA) was transiently transfected with the retroviral plasmids, and 48 hours later the infectious supernatant was used to infect the target cell lines as previously described.26 Stable ICSBP-expressing clones were selected beginning 72 hours after infection by using 800 μg/mL hygromycin. Stable BCR/ABL-expressing cells were obtained after IL-3 withdrawal and outgrowth of factor-independent cell lines.
Stable cell lines and transfection
The IL-3-dependent, murine myeloid precursor cell line 32Dcl3 (hereafter referred to as 32D) was obtained from the DSMZ cell culture collection (DSMZ, Braunschweig, Germany) and maintained in RPMI 1640 medium supplemented with 1% glutamine, 10% fetal calf serum (FCS) (all Gibco/BRL, Eggenstein, Germany), 1% penicillin/streptomycin (Biochrom KG, Berlin, Germany), and 10% WEHI-conditioned media as a source for IL-3. The oligoclonal cell lines 32D-v and 32D/BA were generated by electroporating the empty pcDNA3 vector (Invitrogen, Karlsruhe, Germany) or pcDNA3 containing a 7.2-kb EcoRI BCR/ABL cDNA fragment. Cell lines transduced with the BCR/ABL were factor independent, eg, grew in absence of IL-3. 32D/ICSBP cells and 32D/BA-ICSBP were generated by retroviral infection of the full-length murine ICSBP cDNA into 32D-v or 32D/BA cells. From the factor-independent 32D/BA-ICSBP cells clone 3E (32D/BA-ICSBP-3E) was generated by limiting dilution technique and used for most of the experiments.
Development of 32D-ICSBP/tet-off cell systems
32D-ICSBP on/off and 32D/BA-ICSBP on/off were generated by using the Tet-Off gene expression system (Clontech Biotechnology), as described by the manufacturer. Briefly, 32D cells were electroporated with the pTet-Off vector and selected with genecitin (G418) at 800 μg/mL. Stable clones were subsequently generated by limiting dilution. Presence of pTet-off in these clones was confirmed by polymerase chain reaction (PCR). 32D-pTet-off clones were then electroporated with pTRE2-hyg-ICSBP and selected with hygromycin at 2 μg/mL to obtain stable 32D/ICSBP on/off cells. Clone C4 of 32D/ICSBP on/off (C4) was selected for further experiments because of its high baseline ICSBP expression levels and very low ICSBP background expression in the presence of doxycycline (1-2 μg/mL). C4 was then infected with Mig-p210 viral supernatant and selected by IL-3 withdrawal to obtain a triple stable-expressing cell line. Limiting dilution of this cell line yielded in 32D/BA-ICSBP on/off cells.
RT-PCR
Messenger RNA was isolated by using the Qiagen-RNA extraction kit (Qiagen, Hilden, Germany) as recommended by the manufacturer. Total RNA (1 μg) was reverse transcribed into cDNA by using 140 U Superscript reverse transcriptase (RT) (GIBCO-BRL, Eggenstein, Germany) as previously described.27 PCR amplification was performed under the following reaction conditions: 1 × reaction buffer, 2.5 mM MgCL2, 200 μM dNTP (deoxynucleotide triphosphate), 5 μM of each primer, 0.625 U Taq-DNA polymerase (Roche Diagnostics, Mannheim, Germany) in a total volume of 25 μL. Primer sequences, annealing temperatures, and number of amplification cycles are specified in Table 1. The remaining cycling conditions were as follows: 94°C for denaturation for 30 seconds, annealing for 1 minute, extension at 72°C for 40 seconds, followed by a final cycle of 90°C for 1 minute and 60°C for 10 minutes. The products were electrophoresed on a 3% agarose gel. Gels were stained with ethidium bromide and photographed.
Sequences of primers and PCR conditions
Genes . | Primers . | Annealing temperature, °C . | Cycles . |
|---|---|---|---|
| BCR/ABL | Sense, 5′-AGCATTCCGCTGACCATCA-3′ | 62 | 30 |
| Antisense, 5′-AGCGGCTTCACTCAGACCC-3′ | |||
| ICSBP | Sense, 5′-AGAGGGAGACAAAGCTGAACC-3′ | 59 | 30 |
| Antisense, 5′-TGAATGGTGTGTGTCATAGGC-3′ | |||
| G-CSFR | Sense, 5′-CTCAAACCTATCCTGCCTCATG-3′ | 60 | 35 |
| Antisense, 5′-TCCAGGCAGAGATGAGCGAATG-3′ | |||
| c-fms | Sense, 5′-GCGATGTGTGAGCAATGGCAGT-3′ | 62 | 35 |
| Antisense, 5′-AGACCGTTTTGCGTAAGACCTG-3′ | |||
| GM-CSFR-α | Sense, 5′-GAGGTCCACAGGTCAAGGTG-3′ | 60 | 35 |
| Antisense, 5′-GATTGACAGTGGCAGGCTTC-3′ | |||
| GM-CSFR-β | Sense, 5′-TTTCCATCACAAACGAAGACT-3′ | 60 | 35 |
| Antisense, 5′-AATGAATGAGTAAGCCATCTT-3′ | |||
| PU.1 | Sense, 5′-TCTGATGGAGAAGCTGATGG-3′ | 55 | 31 |
| Antisense, 5′-CTTGACTTTCTTCACCTCGC-3′ | |||
| MPO | Sense, 5′-ATGCAGTGGGGACAGTTTCTG-3′ | 55 | 30 |
| Antisense, 5′-GTCGTTGTAGGATCGGTACTG-3′ | |||
| CEBP-α | Sense, 5′-GAACAGCAACGAGTACCGGGTA-3′ | 55 | 35 |
| Antisense, 5′-GCCATGGCCTTGACCAAGGAG-3′ | |||
| CEBP-ϵ | Sense, 5′-AGCCCCCGACACCCTTGATGA-3′ | 68 | 31 |
| Antisense, 5′-GTCCCCTTCTCAAGGCACCCT-3′ | |||
| Egr-1 | Sense, 5′-CAACTACCCCAAACTGGAGGAG-3′ | 56 | 32 |
| Antisense, 5′-GAGGATTGGTCATGCTCACGAG-3′ | |||
| SR | Sense, 5′-GTACTAATACCTGTTGTTGGA-3′ | 55 | 35 |
| Antisense, 5′-CGTGCGCTTGTTCTTCTTTCA-3′ | |||
| β-Actin | Sense, 5′-TGGCACCACACCTTCTACAA-3′ | 60 | 25 |
| Antisense, 5′-AGGAGCCAGAGCAGTAATCT-3′ |
Genes . | Primers . | Annealing temperature, °C . | Cycles . |
|---|---|---|---|
| BCR/ABL | Sense, 5′-AGCATTCCGCTGACCATCA-3′ | 62 | 30 |
| Antisense, 5′-AGCGGCTTCACTCAGACCC-3′ | |||
| ICSBP | Sense, 5′-AGAGGGAGACAAAGCTGAACC-3′ | 59 | 30 |
| Antisense, 5′-TGAATGGTGTGTGTCATAGGC-3′ | |||
| G-CSFR | Sense, 5′-CTCAAACCTATCCTGCCTCATG-3′ | 60 | 35 |
| Antisense, 5′-TCCAGGCAGAGATGAGCGAATG-3′ | |||
| c-fms | Sense, 5′-GCGATGTGTGAGCAATGGCAGT-3′ | 62 | 35 |
| Antisense, 5′-AGACCGTTTTGCGTAAGACCTG-3′ | |||
| GM-CSFR-α | Sense, 5′-GAGGTCCACAGGTCAAGGTG-3′ | 60 | 35 |
| Antisense, 5′-GATTGACAGTGGCAGGCTTC-3′ | |||
| GM-CSFR-β | Sense, 5′-TTTCCATCACAAACGAAGACT-3′ | 60 | 35 |
| Antisense, 5′-AATGAATGAGTAAGCCATCTT-3′ | |||
| PU.1 | Sense, 5′-TCTGATGGAGAAGCTGATGG-3′ | 55 | 31 |
| Antisense, 5′-CTTGACTTTCTTCACCTCGC-3′ | |||
| MPO | Sense, 5′-ATGCAGTGGGGACAGTTTCTG-3′ | 55 | 30 |
| Antisense, 5′-GTCGTTGTAGGATCGGTACTG-3′ | |||
| CEBP-α | Sense, 5′-GAACAGCAACGAGTACCGGGTA-3′ | 55 | 35 |
| Antisense, 5′-GCCATGGCCTTGACCAAGGAG-3′ | |||
| CEBP-ϵ | Sense, 5′-AGCCCCCGACACCCTTGATGA-3′ | 68 | 31 |
| Antisense, 5′-GTCCCCTTCTCAAGGCACCCT-3′ | |||
| Egr-1 | Sense, 5′-CAACTACCCCAAACTGGAGGAG-3′ | 56 | 32 |
| Antisense, 5′-GAGGATTGGTCATGCTCACGAG-3′ | |||
| SR | Sense, 5′-GTACTAATACCTGTTGTTGGA-3′ | 55 | 35 |
| Antisense, 5′-CGTGCGCTTGTTCTTCTTTCA-3′ | |||
| β-Actin | Sense, 5′-TGGCACCACACCTTCTACAA-3′ | 60 | 25 |
| Antisense, 5′-AGGAGCCAGAGCAGTAATCT-3′ |
Patient samples
The cDNA of 7 healthy donors, 10 patients in chronic phase of CML at diagnosis, and 10 patients with CML in major cytogenetic remission under a standard IFNα-based treatment regimen was studied.27 All patients and healthy donors gave written informed consent for the use of their samples.
Quantitative real-time PCR
Quantitative real-time PCR was performed on an ABI Prism 7700 by using the QuantiTect SYBR Green kit according to the instructions from the manufacturer (Qiagen). In each reaction 1 μL cDNA synthesized from total RNA was used. The ICSBP primers were 5′-GTC CCA ACT GGA CAT TTC CG-3′ (forward) and 5′-CAT TCA CGC AGC CAG CAG-3′(reverse). The bcl-2 primers were 5′-TGC GGC CTC TGT TTG ATT TC-3′ (forward) and 5′-CTT GTG GCC CAG ATA GGC AC-3′ (reverse). The GAPDH primers were 5′-CTC CTC CAC CTT TGA CGC TG-3′ (forward) and 5′-ACC ACC CTG TTG CTG TAG CC-3′ (reverse) (all primers were from TIB-Mol Biol, Berlin, Germany). The amplification conditions for all reactions were 1 × 95°C for 15 minutes followed by 45 × 94°C for 15 seconds, 60°C for 30 seconds, 72°C for 30 seconds. ICSBP and bcl-2 expression was normalized by comparison with the expression of the housekeeping gene GAPDH. Standard curves were constructed for each run by using serial dilutions of ICSBP, bcl-2, and GAPDH plasmids with known copy numbers. Each standard and sample was measured in triplicates. Quantitative results of real-time PCR were assessed by determination of the cycle threshold (CT) value. A calibration curve was derived by plotting the CT values, obtained for each dilution, against the logarithm of the plasmid copy number. The calibration curves showed a correlation coefficient (R2) between 0.98 and 0.99. The estimated amount of ICSBP and bcl-2 mRNA was normalized to GAPDH mRNA expression to compensate for variations in quantity or quality of starting mRNA and for differences in reverse transcriptase efficiency. ICSBP/ or bcl-2/GAPDH ratio was expressed in percentages.
Northern blotting
Total RNA (5 μg) was separated on a 1.5% (wt/vol) agarose/formaldehyde gel and subsequently transferred to a nylon membrane (Hybond N+; Amersham Buchler, Braunschweig, Germany) by capillary blotting. A 529-bp murine bcl-2 cDNA fragment was generated by RT-PCR from total mRNA of 32D cells by using the oligonucleotide primers 5′-AGATCGTGAAGTACATACATTA-3′ and 5′-TTATCCTGGATCCAGGTGTGCAGAT-3′, subsequently cloned into the pCR2.1-TOPO vector (Invitrogen) and sequenced to confirm its identity to published gene sequence (GenBank accession no. M16506). The EcoRI insert was purified radiolabeled by using a random primer DNA labeling kit. The random primer labeling kit (Decaprime II) was from Ambion (Austin, TX) and α-32P-dCTP was from DuPont-NEN (Boston, MA). The human β-actin cDNA insert and the ExpressHyb solution were purchased from Clontech (Palo Alto, CA). Hybridization procedures were carried out according to the manufacturer's instructions. Control hybridizations with human β-actin cDNA verified that equal amounts of RNA were loaded.
Assessment of apoptosis by annexin/propidium iodide staining
For assessment of apoptosis 1 × 106 cells at 2 × 105/mL were cultured in 6-well plates (Greiner, Solingen, Germany) and treated with 100 μg/mL cytarabine (AraC), 10 μg/mL etoposide (VP-16), or 1 μM imatinib (kindly provided by Novartis, Basel, Switzerland), respectively, for indicated times (24-72 hours). Alternatively, phosphate-buffered saline (PBS) (GIBCO-BRL) washed cells were starved for 18 and 36 hours either in IL-3-free RPMI 1640 plus 10% FCS for factor-dependent 32D cells or in serum-free RPMI 1640 for BCR/ABL-transformed cells. After incubation, cells were stained by using the Annexin V-FITC (fluorescein isothiocyanate) Apoptosis detection kit (Calbiochem, Schwalbach, Germany) essentially as recommended. Briefly, after cytotoxic treatment, cells were washed with cold PBS, and 1 × 105 cells were resuspended in 195 μL cold 1 × binding buffer plus 5 μL annexin V-FITC, followed by a 10-minute incubation at room temperature in the dark. Cells were again washed and resuspended with 190 μL binding buffer. Propidium iodide (PI) (10 μL of 20 μg/mL) was added immediately before analysis. Apoptosis was assessed by dual color flow cytometry on a FACScan cytofluorometer (Becton Dickinson, Mountain View, CA) by means of Cell Quest software package (Becton Dickinson).
Flow cytometric analysis
From a log-phase growing culture 5 × 105 cells were incubated with pretitrated amounts of either of the following phycoerythrin (PE)-labeled monoclonal antibodies (mAbs): anti-CD11b (Mac-1)-PE, clone M1/70, anti-B220 (CD45R)-PE, clone RA3-6B2, anti-CD80 (B7.1)-PE, clone 16-10A1, or irrelevant polyclonal antimouse immunoglobulin-PE as negative control (all from BD Pharmingen, San Diego, CA). To block nonspecific binding via Fc-receptors, 10 mg/mL mouse monoclonal immunoglobulin G1 (IgG1) mAb (clone 11711.11) was present throughout the incubation. After 30 minutes of incubation at 4°C, cells were washed and resuspended in fluorescence-activated cell sorting (FACS) buffer containing 0.5% bovine serum albumin (Behring, Marburg, Germany) in PBS. Finally, stained cells were washed and analyzed on a FACScan cytofluorometer by means of the Cell Quest software package (Becton Dickinson).
Immunoblotting analysis
Whole cell extracts were prepared by lysing cells for 30 minutes at 4°C with lysis buffer (20 mM Tris [tris(hydroxymethyl)aminomethane]-HCl pH 7.6, 100 mM NaCl, 1% NP40, 1 mM sodium vanadate [Na3VO4], 0.1% SDS (sodium dodecyl sulfate), 1 mM EGTA (ethylene glycol tetraacetic acid), 1 mM EDTA (ethylenediaminetetraacetic acid), 10 μg/mL PMSF (phenylmethylsulfonyl fluoride), 10 μg/mL leupeptin, 5 μg/mL aprotinin, 1.5 μg/mL Pepstain, 1 μg/mL Trypsin inhibitor, 10 mM sodium fluoride (NaF) (all from Sigma, St Louis, MO). Cell lysates were cleared by centrifugation at 4°C (12 000g for 10 minutes). Proteins (30 μg) were mixed with sample buffer, electrophoresed on a 10% SDS-polyacrylamide gel electrophoresis (PAGE), and subsequently electrotransferred to PVDF (polyvinylidene difluoride) membrane filters (Immobilin P; Millipore, Bedford, MA). Filters were probed with a goat polyclonal anti-ICSBP (C-19), specific to mouse and human ICSBP, and a rabbit polyclonal anticaspase3 (H-277) both from Santa Cruz Biotechnology, a mouse anti-Abl (8E9), a mouse anti-Bcl-xl (2H12), and a mouse anti-p53 (Pab122) (BD Pharmingen), a mouse monoclonal anti-Bcl-2 (BD Transduction Laboratories, Lexington, KY), a mouse anti-Bad, a mouse anti-Mcl-1, a mouse anti-Fas (all from BD Biosciences), and a mouse anti-β-actin mAb (AC-74) from Sigma Aldrich (Poole, United Kingdom) in 1% low-fat milk, 10 mM Tris-HCl, pH 7.6, 150 mM NaCl, and 0.025% Tween 20. Filters were washed in TBST (10 mM Tris-HCl, pH 7.6,150 mM NaCl, and 0.025% Tween 20) and TBS (10 mM Tris-HCl, pH 7.6, and 150 mM NaCl) and then reacted for 30 minutes at room temperature with the secondary, horseradish peroxidase-conjugated mouse antigoat, goat antirabbit, or goat antimouse antibody (all from Santa Cruz Biotechnology). Immunoreactive proteins were visualized by using the enhanced chemiluminescence (ECL) Western blotting detection kit (Amersham Pharmacia Biotech, Uppsala, Sweden) and BioMax-MR films (Kodak, Rochester, NY).
Bcl-2 reporter constructs and reporter gene assays
A 541-bp (long) and a 360-bp fragment (short) of the 3′-end of the 5′-untranslated region (UTR) of the mouse bcl-2 promoter were PCR-amplified from genomic DNA extracted from 32D cells and inserted into the pGL3-Basic firefly luciferase reporter vector (Promega, Madison, WI) using the following primer pairs: short fragment, forward primer 5′-TCACCTGAGCTCATTGCGGAGGAAGTAGACTG-3′ and reverse primer 5′-TGTACTCTCGAGCGATCTCCCGGTTATCATAC-3′; long fragment, forward primer 5′-ATATAAGAGCTCATGGACGCGTTTGAAATA-3′ and reverse primer 5′-TGTACTCTCGAGCGATCTCCCGGTTATCATAC-3′ at 57°C annealing temperature.
Bcl-2-promoter activation was measured using the dual luciferase assay (Promega) as described.27 Briefly, 5 nM respective bcl-2-reporter construct, containing the firefly luciferase gene under control of the human bcl-2 promoter, and the transfection control construct expressing the renilla luciferase gene were transiently coexpressed by electroporation. The control construct served as an internal reference for the efficiency of transfection and expression. Forty-eight hours after transfection luciferase activity was measured with a LB 96 P microlumat (EG&G Berthold, Bad Wildbad, Germany). Bcl-2-specific promoter activation was quantified as a ratio of measured firefly light units (flu) relative to renilla (rlu). Each experiment was done at least 3 times.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts from 32D/BA-v or 32D/BA-ICSBP(3E) cells were prepared with the TransFactor Extraction Kit (Clontech) according to instructions from the manufacturer. For EMSA, 50 ng double-stranded oligonucleotide DNA probes were labeled with α-32P]dCTP by Large Fragment of DNA Polymerase I (Invitrogen) and purified with NICKTM Columns (Amersham Biosciences). Subsequently, 14 μg nuclear extract was incubated (30 minutes, room temperature) with 1 ng radiolabeled probe in the presence of 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM dithiothreitol (DTT), 5% glycerol, 1 mM EDTA, 50 ng/μL bovine serum albumin (BSA), and 50 ng/μL polydeoxyinosinic deoxycytidylic acid (polydIdC). The samples were then separated on a nondenaturing 6% polyacrylamide gel and subjected to autoradiography. The sequences of the probes used in the EMSA are as follows: Ets/interferon regulatory factor (IRF)-composite element from the 5′-UTR of the bcl-2 promoter: (sense) 5′-ggggAAGCAAATTTCATTTCCAGA, (antisense) 5′-CTGTCTGGAAATGAAATTTGCTT; EICE mutated, (sense) 5′-ggggAAGCAAAATCAATTTGAAGA, (antisense) 5′-CTGTCTTCAAATTGATTTTGC-TT; Mock, (sense) 5′-AATGTAAACGGCGAGAATAGCAGAGTA, (antisense) 5′-TACTCTGCTATTCTCGCCGTTTACATT. The EICE sequence is underlined and mutations are shown in italics.
Statistical analysis
Analysis of the statistical significance of a difference between the apoptosis rate of ICSBP+ and ICSBP- cell lines was done using the Student t or Dunn multiple comparison t tests. Differences were considered significant in cases where P values were less than .05.
Results
Establishment of a stable and inducible expression system of ICSBP in 32D/wt and 32D/BA cell lines
The ICSBP-negative, IL-3-dependent myeloid precursor cells, 32D, or BCR/ABL-transformed IL-3-independent 32D cells, 32D/BA, were stable transfected with either ICSBP or an empty control vector to generate 32D/v, 32D/ICSBP, 32D/BA-ICSBP(3E), and 32D/BA-v. The expression of ICSBP and BCR/ABL in these stable cell lines is depicted in Figure 1A. The addition of doxycycline to the culture medium rapidly and durably repressed constitutive expression of ICSBP of 32D/ICSBP on/-off cells (clone C4) and 32D/BA-ICSBP on/-off cells (Figure 1B-C).
Generation of 32D/BA cells stably or inducibly coexpressing ICSBP. Protein lysates were harvested and immunoblotted with anti-ICSBP, anti-BCR/ABL, and anti-β-actin antibodies. β-actin immunoblotting was performed to assure equal protein loading. (A) 32D cells expressing BCR/ABL, ICSBP, or both, and appropriate vector (v) control cells. (B) ICSBP expression of 32D/ICSBP on/off cells 96 hours to 24 hours before, and 24 hours to 96 hours after addition of 1 μg/mL doxycycline. (C) ICSBP expression of 32D/ICSBP-BA on/off cells 96 hours to 24 hours before, and 24 hours to 96 hours after addition of 1 μg/mL doxycycline (top blot). (Bottom blots) BCR/ABL expression of 32D/ICSBP on/off and 32D/ICSBP-BA on/off cells in presence or absence of ICSBP.
Generation of 32D/BA cells stably or inducibly coexpressing ICSBP. Protein lysates were harvested and immunoblotted with anti-ICSBP, anti-BCR/ABL, and anti-β-actin antibodies. β-actin immunoblotting was performed to assure equal protein loading. (A) 32D cells expressing BCR/ABL, ICSBP, or both, and appropriate vector (v) control cells. (B) ICSBP expression of 32D/ICSBP on/off cells 96 hours to 24 hours before, and 24 hours to 96 hours after addition of 1 μg/mL doxycycline. (C) ICSBP expression of 32D/ICSBP-BA on/off cells 96 hours to 24 hours before, and 24 hours to 96 hours after addition of 1 μg/mL doxycycline (top blot). (Bottom blots) BCR/ABL expression of 32D/ICSBP on/off and 32D/ICSBP-BA on/off cells in presence or absence of ICSBP.
In both 32D cell systems ICSBP expression levels were in the range of physiologic ICSBP levels expressed, for example, in monocytic U937 or J774 cells (not shown). The BCR/ABL expression levels of 32D/BA-ICSBP on/-off cells remained high, when ICSBP was turned on (Figure 1C).
ICSBP expression inhibits BCR/ABL-mediated leukemogenesis in Balb/c mice
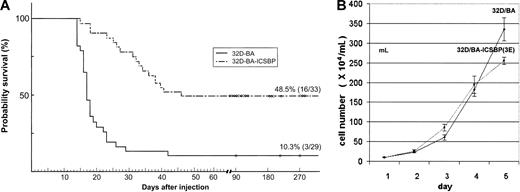
It has previously been demonstrated that ICSBP completely inhibited the manifestation of a BCR/ABL-induced lymphoid leukemia type in vivo.10 We here asked whether ICSBP also exerts its protective antileukemic effect when expressed in the BCR/ABL-transformed myeloid 32D cells, which may be a more CML-related cell type. When injected into healthy syngeneic Balb/c mice (n = 29), 32D/BA cells induced a rapid progressive leukemia that killed 89.7% of the hosts within 2 to 3 weeks (mean, 19 ± 4 days) (Figure 2A).
Survival of mice injected with 32D-BA/v and 32D-BA/ICSBP cells. (A) A total of 106 32D/BA-v or 32D/BA-ICSBP(3E) cells were injected intravenously into the tail veins. A total of 29 mice were injected in the 32D/BA-v arm, 33 in the 32D/BA-ICSBP(3E) arm. Mice were observed for survival and killed if moribund. (B) Growth characteristics of 32D/BA-v and 32D/BA-ICSBP cells under nonlimiting culture conditions. Cells were seeded at a density of 5 × 104 cell/mL and then counted daily in quadruplicate to generate proliferation curves from mean values ± SD. Each experiment was performed 3 times.
Survival of mice injected with 32D-BA/v and 32D-BA/ICSBP cells. (A) A total of 106 32D/BA-v or 32D/BA-ICSBP(3E) cells were injected intravenously into the tail veins. A total of 29 mice were injected in the 32D/BA-v arm, 33 in the 32D/BA-ICSBP(3E) arm. Mice were observed for survival and killed if moribund. (B) Growth characteristics of 32D/BA-v and 32D/BA-ICSBP cells under nonlimiting culture conditions. Cells were seeded at a density of 5 × 104 cell/mL and then counted daily in quadruplicate to generate proliferation curves from mean values ± SD. Each experiment was performed 3 times.
Spleens from 32D/BA-injected animals were enlarged, and all mice had a pathologic burden of 32D/BA cells in their peripheral blood, spleen, and bone marrow as evidenced by microscopy and detection of BCR/ABL by RT-PCR (data not shown). In contrast, a total of 16 (48.5%) of 33 mice that were intravenously injected with 32D/BA-ICSBP(3E) cells survived and lived without evidence of disease, that is, no organomegaly, no leukocytosis, or enlarged lymph nodes for more than 270 days. Thus, by using myeloid 32D cells these data confirm the potential of ICSBP to counteract the manifestation of a BCR/ABL-induced leukemia, albeit less effectively than previously reported for lymphoid BaF3-BA cells.10 Under nonlimiting culture conditions 32D/BA-ICSBP(3E) and 32D/BA cells proliferated at similar kinetics, making it unlikely that protection from BCR/ABL-mediated leukemia by ICSBP was due to decreased proliferation (Figure 2B).
ICSBP reverses BCR/ABL-mediated repression of surface antigen expression
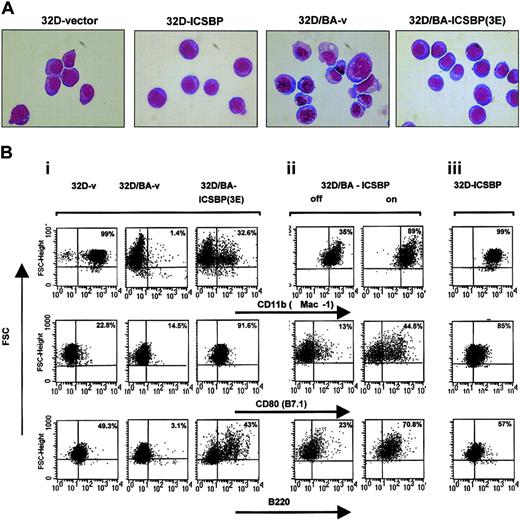
32D cells are dependent on IL-3 for cell growth and survival and do not spontaneously differentiate. However, expression of BCR/ABL in these cells induces growth factor independence.28 Although ICSBP had no effect on growth, cell cycle independence, or factor independence of 32D/v and 32D/BA cells (not shown), its expression reverted the transformed 32D/BA phenotype toward a typical 32D phenotype with normal cell morphology, fine chromatin structure, and loss of vacuolization (Figure 3A).
ICSBP reverses BCR/ABL-mediated morphology changes and disturbed antigen expression. (A) Wright-Giemsa staining. BCR/ABL expression in 32D cells induces a transformed phenotype characterized by vacuolization, anisocytosis, and irregular cell and chromatin morphology, which is reversed to a major extent by coexpression of ICSBP. Original magnification, × 600. (B) Representative dot plots of 32D and 32D/BA cells permanently (i,iii) or inducible (ii) expressing ICSBP. Cells were stained with anti-Mac-1, anti-CD80, or anti-B220 monoclonal antibodies. The staining intensity is plotted against the forward scatter (FSC) after gating tightly to live cells according to the scatter characteristics. For quantitation of antigen-positive cells identical quadrant gates were used for each of the tested antigens and for stable inducible systems. The percentage of positive cells of the right quadrants is indicated in each plot.
ICSBP reverses BCR/ABL-mediated morphology changes and disturbed antigen expression. (A) Wright-Giemsa staining. BCR/ABL expression in 32D cells induces a transformed phenotype characterized by vacuolization, anisocytosis, and irregular cell and chromatin morphology, which is reversed to a major extent by coexpression of ICSBP. Original magnification, × 600. (B) Representative dot plots of 32D and 32D/BA cells permanently (i,iii) or inducible (ii) expressing ICSBP. Cells were stained with anti-Mac-1, anti-CD80, or anti-B220 monoclonal antibodies. The staining intensity is plotted against the forward scatter (FSC) after gating tightly to live cells according to the scatter characteristics. For quantitation of antigen-positive cells identical quadrant gates were used for each of the tested antigens and for stable inducible systems. The percentage of positive cells of the right quadrants is indicated in each plot.
Whereas BCR/ABL decreased the expression of various 32D-specific differentiation antigens, such as Mac-1 (CD11b), a macrophage marker, CD80 (B7.1), a T-cell costimulatory receptor, and CD45R (B220), a pan B-cell marker (Figure 3Bi left and middle plots), ICSBP coexpression reversed this regulation to a major extent (Figure 3Bi right plots), indicating that ICSBP overcame the differentiation-repression by BCR/ABL.
This effect could be recapitulated in 32D/BA-ICSBP on/-off cells (Figure 3Bii), providing evidence for a direct cause-effect relationship between ICSBP expression and partial reversal of a BCR/ABL immunophenotype. Doxycycline itself did not cause these antigen regulations (data not shown). Interestingly, in 32D/BA-ICSBP on/off cells, which were generated by transduction of 32D/ICSBP on/off cells with BCR/ABL, the down-regulation of Mac-1 was less pronounced compared with 32D/BA-v, suggesting that pre-existing ICSBP prevented a BCR/ABL-mediated Mac-1 repression (Figure 3Bii left plot versus Figure 3Bi middle plot). Notably, ICSBP not only overcame BCR/ABL-mediated repression of antigen expression but also directly up-regulated Mac-1 and B7.1 in 32D/ICSBP cells (Figure 3Biii), suggesting these antigens as BCR/ABL-independent targets of ICSBP in 32D-cells.
ICSBP derepresses BCR/ABL-mediated inhibition of myelomonocytic gene expression
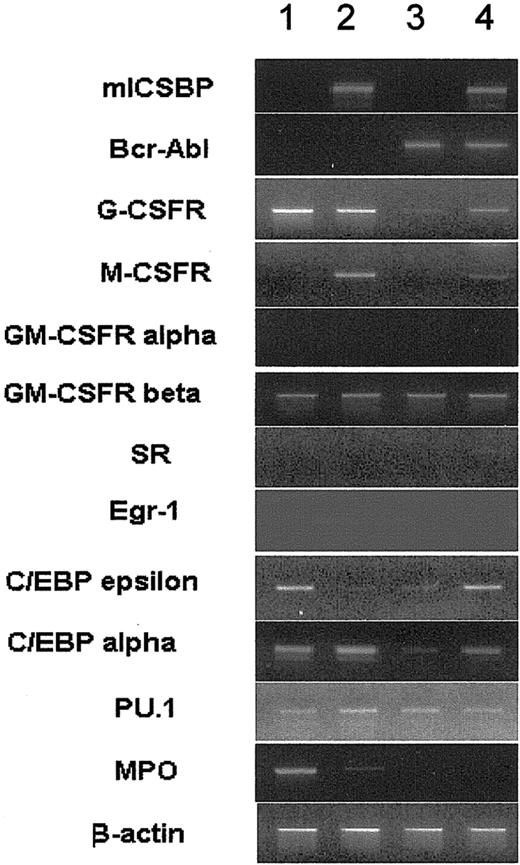
ICSBP overcame BCR/ABL-mediated repression of surface antigen expression. In line with this, ICSBP overrode BCR/ABL-mediated suppression of the expression of myeloid lineage marker genes, such as the granulocyte colony-stimulating factor receptor (G-CSFR), C/EBPα, C/EBPϵ, and the monocyte colony-stimulating factor receptor (M-CSFR, c-fms gene), a known target of ICSBP29 (Figure 4).
RT-PCR of mRNA derived from 32D/v, 32D/ICSBP, 32D/BA-v, and 32D/BA-ICSBP-3E cells. As a control, intron-spanning β-actin primers were used. Data from representative experiments are shown. Of note, ICSBP specifically restored the expression of the G-CSFR and C/EBPα, both of which were down-regulated by BCR/ABL. The expression of M-CSFR is specifically induced by ICSBP regardless of the presence of BCR/ABL. Lane 1 indicates 32D/v; lane 2, 32D/ICSBP; lane 3, 32D/BA-v; and lane 4, 32D/BA-ICSBP-3E.
RT-PCR of mRNA derived from 32D/v, 32D/ICSBP, 32D/BA-v, and 32D/BA-ICSBP-3E cells. As a control, intron-spanning β-actin primers were used. Data from representative experiments are shown. Of note, ICSBP specifically restored the expression of the G-CSFR and C/EBPα, both of which were down-regulated by BCR/ABL. The expression of M-CSFR is specifically induced by ICSBP regardless of the presence of BCR/ABL. Lane 1 indicates 32D/v; lane 2, 32D/ICSBP; lane 3, 32D/BA-v; and lane 4, 32D/BA-ICSBP-3E.
Intriguingly, C/EBPϵ, and, to a lesser extend, the macrophage-specific scavenger receptor, SR, were specifically induced by ICSBP in the presence of BCR/ABL. There was no difference between the transcription levels of the granulocyte macrophage colony-stimulating factor receptor (GM-CSFR) α and β, the transcription factor EGR-1, myeloperoxidase (MPO), and PU.1 in BCR/ABL- and ICSBP-expressing cells (Figure 4). Thus, ICSBP antagonized in part BCR/ABL-mediated transcriptional repression, causing the induction of differentiation genes.
ICSBP overcomes drug resistance
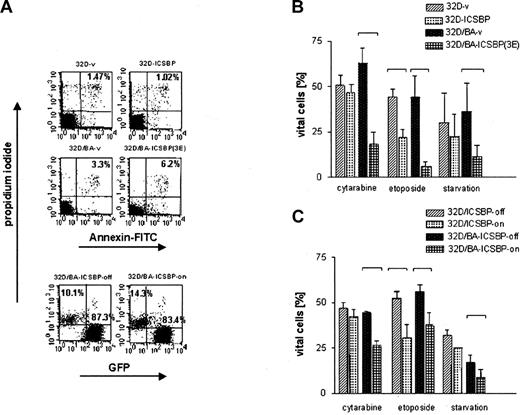
ICSBP antagonized BCR/ABL-mediated repression of transcription. We next asked whether ICSBP opposes apoptosis and drug resistance of BCR/ABL-transformed cells. Although ICSBP failed to increase the spontaneous apoptosis rate in any of the tested cell lines (Figure 5A), it caused a dramatic increase in the susceptibility of ICSBP-positive cells to die in response to cytotoxic stress such as serum starvation or exposure to the chemotherapeutic agents cytarabine and etopside (Figure 5B-C). The proapoptotic effect of ICSBP was less pronounced in BCR/ABL-negative 32D/ICSBP cells in which only exposure to VP16, but not IL-3 deprivation or AraC treatment, significantly increased apoptosis (Figure 5B). Augmented apoptosis was directly attributable to ICSBP expression, because turning off ICSBP expression by the addition of doxycycline in inducible cell systems decreased apoptotic responses (Figures 1 and 5C). Importantly, the proapoptotic effects of ICSBP could be recapitulated in other hematopoietic leukemia cell lines, namely K562 cells (BCR/ABL-positive erythroleukemic cells), KG1a cells (immature AML, BCR/ABL-negative), and Jurkat cells (BCR/ABL-negative T cells), implying that the proapoptotic effects of ICSBP are neither cell type nor oncogene specific (data not shown).
Assessment of apoptosis of 32D cells and BCR/ABL-transformed 32D cells permanently or inducibly expressing ICSBP. Apoptosis was determined by examining the uptake of annexin V-FITC and PI. Percentage of cells appearing as annexin- and PI-negative (annexin-/PI-) were counted as viable, whereas dead cells were defined as either annexin+/PI- (apoptotic), or annexin+/PI+ (necrotic). In the case of 32D/BA-ICSBP on/off cells, only PI+ cells could be counted as dead and PI- as vital, because these cells contained green fluorescent protein (GFP), which excluded the possibility of evaluation of annexin V-FITC staining. (A) Percentage of dead cells under nonlimiting culture conditions is indicated in the right upper quadrants. (B) Percentage of viable cells of ICSBP+ and ICSBP-stable cell lines cells after 18 hours of starvation without IL-3 (BA- cells) versus 36 hours of serum withdrawal (BA+ cells) or exposure to indicated chemotherapeutic drugs (24 hours versus 48 hours for BA- versus BA+ cells, respectively). (C) Percentage of viable cells of ICSBP-inducible cell lines (ICSBP off versus on) after 18 hours of starvation without IL-3 (BA- cells) versus 36 hours of serum withdrawal (BA+ cells) or exposure to indicated chemotherapeutic drugs (24 hours versus 48 hours for BA- versus BA+ cells, respectively). Bars represent mean values ± SEM of at least 3 independent experiments. Statistically significant differences in the apoptosis rates of cell line pairs were marked with brackets.
Assessment of apoptosis of 32D cells and BCR/ABL-transformed 32D cells permanently or inducibly expressing ICSBP. Apoptosis was determined by examining the uptake of annexin V-FITC and PI. Percentage of cells appearing as annexin- and PI-negative (annexin-/PI-) were counted as viable, whereas dead cells were defined as either annexin+/PI- (apoptotic), or annexin+/PI+ (necrotic). In the case of 32D/BA-ICSBP on/off cells, only PI+ cells could be counted as dead and PI- as vital, because these cells contained green fluorescent protein (GFP), which excluded the possibility of evaluation of annexin V-FITC staining. (A) Percentage of dead cells under nonlimiting culture conditions is indicated in the right upper quadrants. (B) Percentage of viable cells of ICSBP+ and ICSBP-stable cell lines cells after 18 hours of starvation without IL-3 (BA- cells) versus 36 hours of serum withdrawal (BA+ cells) or exposure to indicated chemotherapeutic drugs (24 hours versus 48 hours for BA- versus BA+ cells, respectively). (C) Percentage of viable cells of ICSBP-inducible cell lines (ICSBP off versus on) after 18 hours of starvation without IL-3 (BA- cells) versus 36 hours of serum withdrawal (BA+ cells) or exposure to indicated chemotherapeutic drugs (24 hours versus 48 hours for BA- versus BA+ cells, respectively). Bars represent mean values ± SEM of at least 3 independent experiments. Statistically significant differences in the apoptosis rates of cell line pairs were marked with brackets.
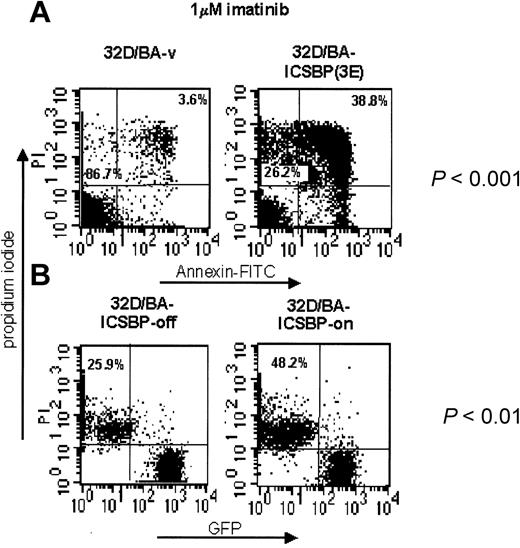
ICSBP ameliorates imatinib-induced apoptosis
We next asked whether ICSBP also promotes imatinib-induced apoptosis of BCR/ABL-transformed cells. In contrast to 32D/BA-v cells, imatinib treatment of 32D/BA-ICSBP(3E) cells caused a dramatic induction of early and late apoptosis (mean, 6.2-fold, P < .001, n = 4) (Figure 6A). Suppression of ICSBP by addition of doxycycline to 32D/BA-ICSBP on/off cells led to a 2-fold decreased killing efficacy of imatinib in these cells (P < .01, n = 3) (Figure 6B). This finding demonstrates a causal relationship between ICSBP expression and an increased sensitivity to imatinib. Notably, IL-3 rescue of imatinib-treated 32D/BA cells was independent from the presence of ICSBP (data not shown).
ICSBP overrides imatinib resistance in BCR/ABL-transformed 32D cells. ICSBP+ and ICSBP-cell lines were treated with 1 μM imatinib for 48 hours and subsequently stained with annexin V-FITC and PI. (A) The percentage of dead cells, defined as either annexin+/PI- (apoptotic) or annexin+/PI+ (necrotic) is indicated in the upper right quadrant of each plot. The percentage of live cells is indicated in the lower left quadrant. (B) Because of expression of GFP (Mig-p210) in 32D/BA-ICSBP on/off cells, the percentage of dead cells was calculated solely from the PI+ fraction, which is outlined in the upper left quadrant of each dot plot. The difference in the apoptosis rates of each pair of ICSBP+ and ICSBP-cell lines was statistically significant as indicated (P < .001 and P < .01 for stable and inducible ICSBP-expressing cell systems, respectively).
ICSBP overrides imatinib resistance in BCR/ABL-transformed 32D cells. ICSBP+ and ICSBP-cell lines were treated with 1 μM imatinib for 48 hours and subsequently stained with annexin V-FITC and PI. (A) The percentage of dead cells, defined as either annexin+/PI- (apoptotic) or annexin+/PI+ (necrotic) is indicated in the upper right quadrant of each plot. The percentage of live cells is indicated in the lower left quadrant. (B) Because of expression of GFP (Mig-p210) in 32D/BA-ICSBP on/off cells, the percentage of dead cells was calculated solely from the PI+ fraction, which is outlined in the upper left quadrant of each dot plot. The difference in the apoptosis rates of each pair of ICSBP+ and ICSBP-cell lines was statistically significant as indicated (P < .001 and P < .01 for stable and inducible ICSBP-expressing cell systems, respectively).
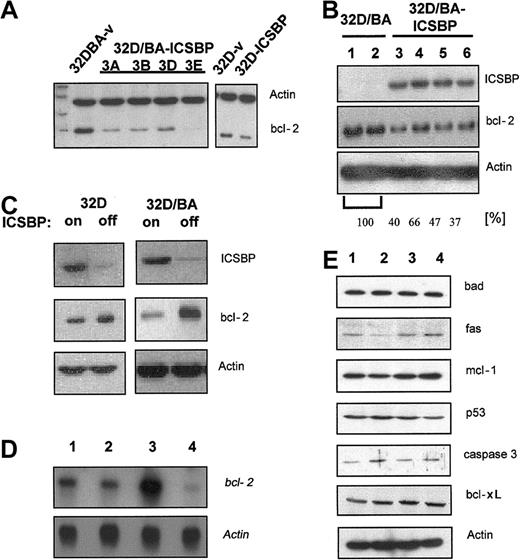
ICSBP suppresses bcl-2
ICSBP potently facilitated apoptosis of BCR/ABL-transformed cells in response to a variety of cytotoxic stimuli. Bcl-xl has been reported as a target of ICSBP and mediator of BCR/ABL-mediated apoptosis resistance.20 We here investigated whether bcl-xl or other antiapoptotic and/or proapoptotic genes were regulated by ICSBP. We found that 32D/BA-ICSBP cells and clonal derivative lines, especially clone 3E, had reduced bcl-2 expression levels compared with 32D/BA-v cells (Figure 7A). Transient expression of ICSBP in 32D/BA cells confirmed this ICSBP-mediated suppression of bcl-2. Gel-densitometric analysis revealed a mean of 47% reduction of bcl-2 expression in 32D/BA-ICSBP cells when compared with 32D/BA cells (Figure 7B). Bcl-2 down-regulation was a specific consequence of ICSBP expression, as it inversely correlated with ICSBP expression in ICSBP-inducible 32D/BA-ICSBP on/off cells (Figure 7C). Of note, stable or conditional expression of ICSBP in 32D cells caused a less remarkable down-regulation of bcl-2, indicating that the presence of BCR/ABL enhanced an ICSBP-dependent bcl-2 regulation (Figure 7A,C). Northern blotting confirmed this finding and indicated that bcl-2 was regulated on the transcriptional level (Figure 7D). The expression of other antiapoptotic target genes, namely mcl-1, fas, bcl-xl, p53, bad, and caspase 3, were unaffected by ICSBP (Figure 7E). Together, ICSBP was shown to be a transcriptional repressor of bcl-2.
ICSBP down-regulates bcl-2. For Western blot analysis protein lysates were harvested, separated on a 10% PAGE transferred to PVDF membranes, and immunoblotted using indicated antibodies. (A) Bcl-2 expression of clones derived from 32D/BA-v and 32D/BA-ICSBP (3A-D) by limited dilution (left) and 32D-v control and 32D-ICSBP cells (right). As a loading control the membrane was simultaneously also blotted with β-actin antibodies. (B) Transient expression of 32D cells with BCR/ABL (lanes 1-2) or 32D/BA cells with ICSBP (lanes 3-6). The percentage of reduction in bcl-2 expression was assessed by densitometric analysis as indicated. (C) Inverse correlation between the ICSBP and bcl-2 expression in 32D (left) as well as 32D-BA cells (right) with or without induction of ICSBP. ICSBP expression was turned off by addition of 1 μg/mL doxycycline for 48 hours. (D) Northern blotting: ICSBP causes a transcriptional down-regulation of bcl-2. Note the difference in the degree of bcl-2 regulation in BCR/ABL-positive and -negative cells; lane 1, 32D; lane 2, 32D/ICSBP; lane 3, 23D/BA; lane 4, 32D/BA/ICSBP-3E. (E) ICSBP does not affect the expression of other genes known to be involved in the regulation of apoptosis (lanes as described for panel D).
ICSBP down-regulates bcl-2. For Western blot analysis protein lysates were harvested, separated on a 10% PAGE transferred to PVDF membranes, and immunoblotted using indicated antibodies. (A) Bcl-2 expression of clones derived from 32D/BA-v and 32D/BA-ICSBP (3A-D) by limited dilution (left) and 32D-v control and 32D-ICSBP cells (right). As a loading control the membrane was simultaneously also blotted with β-actin antibodies. (B) Transient expression of 32D cells with BCR/ABL (lanes 1-2) or 32D/BA cells with ICSBP (lanes 3-6). The percentage of reduction in bcl-2 expression was assessed by densitometric analysis as indicated. (C) Inverse correlation between the ICSBP and bcl-2 expression in 32D (left) as well as 32D-BA cells (right) with or without induction of ICSBP. ICSBP expression was turned off by addition of 1 μg/mL doxycycline for 48 hours. (D) Northern blotting: ICSBP causes a transcriptional down-regulation of bcl-2. Note the difference in the degree of bcl-2 regulation in BCR/ABL-positive and -negative cells; lane 1, 32D; lane 2, 32D/ICSBP; lane 3, 23D/BA; lane 4, 32D/BA/ICSBP-3E. (E) ICSBP does not affect the expression of other genes known to be involved in the regulation of apoptosis (lanes as described for panel D).
ICSBP regulates bcl-2 promoter activity through an EICE response element
To delineate the mechanism of bcl-2 suppression by ICSBP, the bcl-2 promoter was inspected for IRF consensus binding sites. The 5′-UTR of the bcl-2 promoter revealed sequences characteristic for an Ets/IRF-composite element and an IFNγ activation site (GAS, pIRE element) (Figure 8A). Both sites are characteristic for interferon-regulated sites found in diverse ICSBP-responsive promoters (Figure 8B).11,12 By using luciferase gene reporter assays, we found that either the long fragment containing either response element or the short fragment containing only the GAS site repressed the bcl-2 promoter activity in 32D/BA-ICSBP cells, whereas an empty control vector had no effect (Figure 8C-D). In contrast to Western and Northern blot analyses, transient transfection the reporter constructs into 32D/ICSBP cells slightly activated the bcl-2 promoter (Figure 8D), indicating that BCR/ABL may alter IRS responsiveness in this part of the bcl-2 promoter. To investigate whether ICSBP could interact with the EICE in the 5′-UTR of the bcl-2 promoter, EMSA was performed by using this element as a 32P-labeled probe (Figure 8E). This investigation revealed an increased formation of a protein-DNA complex (shift I) in the presence of ICSBP using nuclear extracts from 32D/BA-ICSBP(3E) cells, as compared with 32D/BA cells, which lack ICSBP. The specificity of this protein-DNA complex was determined by competition both with unlabeled EICE-oligo and mock-oligo. Whereas the unlabeled EICE oligo competed away shift I (lanes 4-6), the mock oligo did not (lanes 7-9). The sequence selectivity of this protein-DNA interaction was further supported by the fact that a 32P-labeled mutated EICE probe did not give rise to shift I (lane 10). In contrast, another shift (shift II), was shown to be nonspecific, because it was competed away with unlabeled Mock oligo (lanes 4-6) as well as weakly detected also with the mutated EICE probe (lane 10). These data further support an important function for this regulatory element in ICSBP-mediated repression of the bcl-2 promoter. Which proteins that bind differentially to this EICE in the presence and absence of BCR/ABL are currently under investigation.
ICSBP regulates the bcl-2 promoter. (A) Sequence of the mouse bcl-2 promoter (5′-UTR). Two forward (right arrow) and one reverse (left arrow) primer for amplification of promoter fragments are underlined. Boxes indicate IRF-target sequences: IRF/Ets composite element and GAS element. (B) Homology analysis between IRF recognition sequences in promoters of known IRF-target genes and the putative bcl-2 sites. GAS and Ets/IRF sites from various genes are shown, and conserved sequences are highlighted in underlined bold letters. The GAS- and Ets/IRF-specific consensus sequences are depicted. Boxes indicate Ets/IRF and GAS sites of the murine bcl-2 promoter. (C) Schematic representation of the 5′-UTR containing 2 IRF target sites as indicated. The maps of the short and long fragment promoter constructs or the empty control construct are shown. (D) Luciferase expression 48 hours after transfection of cells with short (left) or long (middle) bcl-2 promoter firefly luciferase construct, or (right) empty vector construct. Renilla luciferase was used as internal control and the firefly-renilla ratio was depicted. The mean (± SE) of 3 experiments is shown. (E) Detection of an ICSBP-dependent increase in sequence-specific protein-DNA complex formation by EMSA. Nuclear extracts from 32D/BA or 32D/BA-ICSBP cells give rise to 2 shifts as indicated (shifts I and II), with the 32P-labeled EICE (EICE*) from the bcl-2 5′-UTR used as a probe. Competition with cold DNA oligos (EICE oligo cold) (lanes 4-6) or mock oligo (lanes 7-9) was performed with excess cold oligo compared with 32P-labeled probe (EICE*) (20 ×,50 ×, and 100 ×). Mutated 32P-labeled EICE (EICE* mutated) was also used as a probe, identifying shift II as unspecific (lanes 7-10).
ICSBP regulates the bcl-2 promoter. (A) Sequence of the mouse bcl-2 promoter (5′-UTR). Two forward (right arrow) and one reverse (left arrow) primer for amplification of promoter fragments are underlined. Boxes indicate IRF-target sequences: IRF/Ets composite element and GAS element. (B) Homology analysis between IRF recognition sequences in promoters of known IRF-target genes and the putative bcl-2 sites. GAS and Ets/IRF sites from various genes are shown, and conserved sequences are highlighted in underlined bold letters. The GAS- and Ets/IRF-specific consensus sequences are depicted. Boxes indicate Ets/IRF and GAS sites of the murine bcl-2 promoter. (C) Schematic representation of the 5′-UTR containing 2 IRF target sites as indicated. The maps of the short and long fragment promoter constructs or the empty control construct are shown. (D) Luciferase expression 48 hours after transfection of cells with short (left) or long (middle) bcl-2 promoter firefly luciferase construct, or (right) empty vector construct. Renilla luciferase was used as internal control and the firefly-renilla ratio was depicted. The mean (± SE) of 3 experiments is shown. (E) Detection of an ICSBP-dependent increase in sequence-specific protein-DNA complex formation by EMSA. Nuclear extracts from 32D/BA or 32D/BA-ICSBP cells give rise to 2 shifts as indicated (shifts I and II), with the 32P-labeled EICE (EICE*) from the bcl-2 5′-UTR used as a probe. Competition with cold DNA oligos (EICE oligo cold) (lanes 4-6) or mock oligo (lanes 7-9) was performed with excess cold oligo compared with 32P-labeled probe (EICE*) (20 ×,50 ×, and 100 ×). Mutated 32P-labeled EICE (EICE* mutated) was also used as a probe, identifying shift II as unspecific (lanes 7-10).
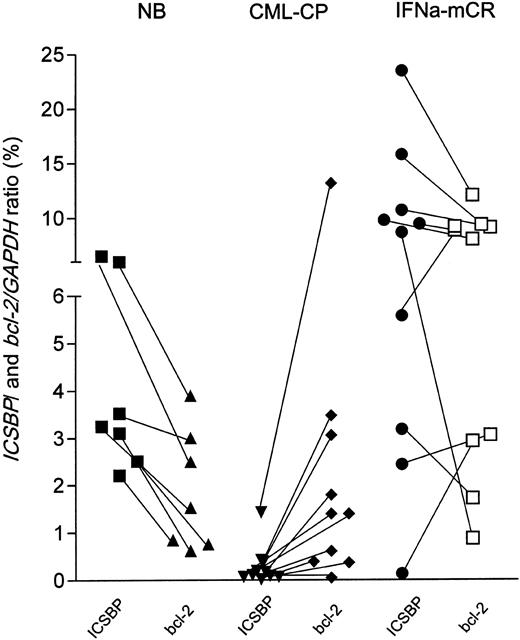
Inverse correlation between ICSBP and bcl-2 transcription in vivo
We then asked, using quantitative RT-PCR and cDNA from peripheral blood, whether the reverse regulation of ICSBP and bcl-2 transcription could be recapitulated in vivo. As shown in Figure 9, healthy blood donors (n = 7) displayed higher ICSBP than bcl-2 expression levels (represented by descending connecting lines between both values). In contrast, patients in chronic phase of CML (n = 10) demonstrated lower absolute ICSBP than corresponding bcl-2 transcripts levels (ascending connecting lines), and, accordingly, 7 of 10 patients in major cytogenetic remission under IFNα (n = 10) displayed higher ICSBP than correlating bcl-2 transcript levels (descending connecting lines). These in vivo data are in line with the finding that bcl-2 transcription is inversely regulated by ICSBP. However, the clinical relevance of the variation of the bcl-2 transcript levels and the ICSBP/bcl-2 ratios under IFNα therapy is currently being investigated.
ICSBP and bcl-2 expression in healthy donors and patients with CML. The expression levels of ICSBP and bcl-2 of each healthy donor or patient with CML was quantitated from peripheral blood cDNA by quantitative PCR and is outlined in the percentage of normalized to GAPDH. ICSBP and bcl-2 transcript levels are depicted by black (ICSBP) or open (bcl-2) symbols. Sample-specific expression pairs are indicated by connecting lines. NB (healthy donors, n = 7), CML-CP (chronic phase CML, untreated, n = 10), IFNα mcR (major cytogenetic responders with < 35% Ph-positive cells in bone marrow, n = 10).
ICSBP and bcl-2 expression in healthy donors and patients with CML. The expression levels of ICSBP and bcl-2 of each healthy donor or patient with CML was quantitated from peripheral blood cDNA by quantitative PCR and is outlined in the percentage of normalized to GAPDH. ICSBP and bcl-2 transcript levels are depicted by black (ICSBP) or open (bcl-2) symbols. Sample-specific expression pairs are indicated by connecting lines. NB (healthy donors, n = 7), CML-CP (chronic phase CML, untreated, n = 10), IFNα mcR (major cytogenetic responders with < 35% Ph-positive cells in bone marrow, n = 10).
Discussion
There is mounting evidence that ICSBP is involved in the pathogenesis of CML. Beyond this, ICSBP fulfills criteria of a tumor suppressor, as loss of ICSBP expression was documented also in AML patients30 and shown to permit AML1-ETO induced transformation in vivo.21 By using a BCR/ABL-transformed myeloid cell system we extended the evidence of a humoral, leukemia-protective effect of ICSBP and found that ICSBP up-regulates the bona fide costimulatory molecule B-7.1, which belongs to a class of molecules essentially required to elicit a T-cell response. However, in contrast to previous reports, we provide evidence that ICSBP counter-regulates features of transformation on the cellular level, namely BCR/ABL-mediated apoptosis, multidrug resistance, and a partial restoration of BCR/ABL-induced disturbances in the regulation of differentiation. Here, we describe for the first time bcl-2 as a down-regulated target of ICSBP, which provides a so far unknown mechanism for the anti-oncogenic effects of ICSBP on the cellular level.
ICSBP reversed BCR/ABL-induced morphology changes and restored or induced the expression of genes associated with differentiation. ICSBP physiologically directs macrophage differentiation and inhibits granulocytic maturation.29 In accordance to these observations from BCR/ABL-negative Tot2 cells, ICSBP blocked granulocytic differentiation (down-regulation of CEBPϵ and MPO) and induced a monocytic commitment (induction of c-fms and Mac-1) in 32D cells, but also in BCR/ABL-transformed 32D cells.29 This finding illustrates that the induction of a monocytic commitment by ICSBP dominated over a BCR/ABL-mediated block of maturation.
BCR/ABL is also known to inhibit G-CSF-induced granulocytic differentiation.31,32 This finding is in line with our experiments showing that BCR/ABL down-regulated granulocytic differentiation markers such as MPO, G-CSFR, and CEBPα and ϵ in 32D cells. However, in a BCR/ABL-positive cell context ICSBP also up-regulated granulocyte-specific differentiation markers, such as CEBPϵ and G-CSFR. Thus, we hypothesize that in a BCR/ABL-positive cell context ICSBP exerts 2 effects: it antagonizes a BCR/ABL-mediated block of differentiation, but simultaneously also induces monocytic commitment.
ICSBP prevented the development of a BCR/ABL-induced leukemia in about 50% of the challenged mice. An even better protection (100%) was reported from Deng and Daley10 using lymphoid BaF3 cells generated to express BCR/ABL and ICSBP. The reason for this difference is unclear. Notably, the regulation of a costimulatory antigen by an IRF member has previously not been reported but is in line with the role of ICSBP in inducing T-helper type-1 (Th-1) cytokines IL-12p40 and IFNγ,17,18 which are known to potently synergize with the B7/CD28 synapse formation during T-cell activation.33 In contrast to B7.1, the pan-lymphatic B220 antigen was dramatically up-regulated only in 32D/BA-ICSBP(3E) cells but not in 32D/ICSBP cells, indicating that BCR/ABL changed the outcome of an ICSBP-directed lineage commitment. Recently, Tamura et al34 found that ICSBP inhibits BCR/ABL-mediated mitogenic activity in Tot2 progenitor cells enabling differentiation. In these experiments ICSBP was also demonstrated to regulate c-myc.
Here, we show that ICSBP potently enhanced the sensitivity of BCR/ABL-transformed 32D cells to undergo apoptosis and overrode chemotherapy, and imatinib resistance (Figures 5-6). ICSBP potently facilitated apoptosis, but did neither alter growth nor the spontaneous apoptosis rate. There are only very few genes, such as the proapoptotic bcl-xs, which share this property with ICSBP.35,36 However, to the best of our knowledge, no other gene has been reported that reverts apoptosis and drug resistance of BCR/ABL-expressing cells.
By using a doxycycline inducible gene expression we found that a decrease of bcl-2 expression could be directly attributed to the induction of ICSBP, irrespective of the presence of BCR/ABL. However, the suppression of bcl-2 by ICSBP was pronounced in the presence of BCR/ABL (Figure 7). This may be due to a BCR/ABL- or differentiation-dependent expression of ICSBP binding partners, which are known to dictate the quality and degree of IRF-mediated regulations.18,19 This hypothesis is underscored by the fact that the inhibition of the bcl-2 family member, bcl-xl, by ICSBP has been described in U937 cells,20 but could not be recapitulated in our 32D systems. Because bcl-2 is essentially required for BCR/ABL-induced transformation, apoptosis, and drug resistance,22,23,37 we concluded that ICSBP mediates its anti-oncogenic effects by decreasing bcl-2. Consequently, high ICSBP expression levels in tumor cells may augment the efficacy of a cytotoxic therapy. This theory is supported by observations on the treatment of patients with CML,38 but also in vitro in lung cancer cell lines, which, after transfection with ICSBP, were prone to apoptosis (U. Wellner et al, manuscript in preparation, January 9, 2004). Thus, to improve therapeutic response rates or to overcome drug resistance it may be reasonable to use IFNγ, a potent stimulator of ICSBP expression6,39 in combination with chemotherapy or imatinib. The clinical use of IFNγ was feasible and effective.40-42 Intriguingly, in vitro IFNγ is known to sensitize tumor cells to apoptosis by up-regulation of caspases and down-regulation of bcl-2,43-46 both of which are targets of ICSBP. Thus, it is tempting to speculate that IFNγ mediates its proapoptotic effects in part by way of induction of ICSBP and subsequent bcl-2 suppression. This is also suggested by the observation of an inverse correlation of ICSBP and bcl-2 expression in patients with CML at diagnosis, and in major cytogenetic responders under IFNα. Interestingly, 4 of the latter had very low, whereas others had even higher bcl-2 transcript levels than healthy donors, irrespective of an increased ICSBP transcription. This finding indicates that IFNα may stimulate bcl-2 transcription in the peripheral blood and/or that the ICSBP-mediated bcl-2 regulation is partly impaired in some patients with CML, eg, those with less sustained response to IFNα. This hypothesis is currently being explored on a larger cohort of patients and by subfraction analysis of peripheral blood cells.
Together, by inducible and stable coexpression of ICSBP and BCR/ABL we provide evidence that ICSBP directly alters the biologic outcome of BCR/ABL signaling on the cellular level, presumably involving a down-regulation of bcl-2. ICSBP overcomes BCR/ABL-mediated apoptosis and drug resistance and reverts disturbances in the regulation of differentiation- and maturation-specific transcription factors. It is tempting to speculate that the manifestation of CML, as well as the kinetics of disease progression, could be significantly influenced by genes like ICSBP, which seem to have the potential to control the biology of leukemic clones either by way of strengthening the humoral antitumor immune surveillance or directly on the cellular level by maintaining the ability of an oncogene-bearing cell to undergo apoptosis. ICSBP may be a target to overcome multidrug and apoptosis resistance, for example, by induction with IFNγ.
Prepublished online as Blood First Edition Paper, December 4, 2003; DOI 10.1182/blood-2003-08-2970.
Supported by the Deutsche José Carreras Leukämie-Stiftung e.V. (A.B., A.N., M. Schmidt, and A.H.), by the P.E. Kempkes Stiftung (A.B.); the H. W. and J. Hector Stiftung (A.N., R.M., and M. Schmidt); the Wilhelm-Sander-Stiftung (A.N. and M. Schmidt); the Deutsche Forschungsgemeinschaft (R.M.); the Bundesministerium für Forschung und Bildung (BMBF); Kompetenznetz “Akute und chronische Leukämien” 01GI9974 (A.N.); and grants from Novartis GmbH and Hoffmann-La Roche AG (A.B. and A.N.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Mankel, M. Rehn, and E. Nalbatov for their excellent technical assistance.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal