Abstract
The overexpanded clone in most B-cell-type chronic lymphocytic leukemia (BCLL) patients expresses an immunoglobulin (Ig) heavy chain variable (VH) region gene with some level of mutation. While it is presumed that these mutations were introduced in the progenitor cell of the leukemic clone by the canonical somatic hypermutation (SHM) process, direct evidence of such is lacking. Nucleotide sequences of the Ig VH genes from 172 B-CLL patients were analyzed. Previously described VH gene usage biases were noted. As with canonical SHM, mutations found in B-CLL were more frequent in RGYW hot spots (mutations in an RGYW motif = 44.1%; germ line frequency of RGYW motifs = 25.6%) and favored transitions over transversions (transition-transversion ratio = 1.29). Significantly, transition preference was also noted when only mutations in the wobble position of degenerate codons were considered. Wobble positions are inherently unselected since regardless of change an identical amino acid is encoded; therefore, they represent a window into the nucleotide bias of the mutational mechanism. B-CLL VH mutations concentrated in complementarity-determining region 1 (CDR1) and CDR2, which exhibited higher replacement-to-silent ratios (CDR R/S, 4.60; framework region [FR] R/S, 1.72). These results are consistent with the notion that VH mutations in B-CLL cells result from canonical SHM and select for altered, structurally sound antigen receptors. (Blood. 2004;103:3490-3495)
Introduction
B-cell-type chronic lymphocytic leukemia (B-CLL) is a disease of clonally expanded CD5+ lymphocytes that express intact antigen receptors. Roughly half of B-CLL patients have a clone that expresses an immunoglobulin (Ig) variable (V) region gene that differs from the germ line sequence by at least 2% in the V segment of the heavy chain (VH).1 These patients have a significantly better clinical outcome than patients whose B-CLL clone expresses a relatively unmutated VH gene.2,3 The biologic basis for the difference in clinical outcomes is not understood, although gene expression and surface membrane phenotypic differences have been described.4-6
Normal B cells accumulate mutations in their Ig V genes during their affinity maturation in the germinal center by a T-cell-dependent process termed somatic hypermutation (SHM).7 These mutations alter the structure of the antibody molecule and allow for the selection of variants with greater affinity for the driving antigen. As such, the presence of Ig V gene mutations produced by SHM in B-CLL cells would indicate that the progenitor cell was antigen experienced. While the precise mechanistic basis for the SHM process is currently being elucidated,8 the characteristics of the resulting mutations have been delineated in a number of systems. The final spectrum of mutations observed in VH sequences deviates from randomness because of the intrinsic mutability of the germ line VH genes and the effects of selection for the antibody molecule's structure.
The intrinsic mutability of VH genes has been the subject of much theoretical and empirical study9-11 but in essence resolves to 2 aspects. The first is the relative likelihood that a given nucleotide will be mutated and the second is the relative chance that it will mutate into the 3 other nucleotides. The latter is firmly established as a transition (purine-to-purine or pyrimidine-to-pyrimidine mutation) bias.7,12 The former involves a role for sequence context and hot spots such as RGYW (R = A or G, Y = C or T, W = A or T)10,13,14 and is probably also influenced by position15 and local DNA or RNA structure.16 Since precise mechanistic models for SHM are still evolving, empirical descriptions of mutational spectra remain the best available method for establishing the likelihood that a given set of sequences were affected by the same or similar processes and selections.
It is often assumed or inferred that the mutations present in VH genes of B-CLL cells developed by the standard SHM process that is initiated during a T-cell-dependent germinal center (GC) reaction. However, this has not been formally proven and may be suspect. Ig V gene mutations can develop in the absence of T-cell help17 and outside of classical germinal centers.18 The finding of V gene mutations in B-CLL cells was initially surprising because most CD5+ B lymphocytes in the blood or follicular mantles of humans lack V gene changes. For this and other reasons, it has been suggested that some or all B-CLL cells are derived from a subset of B cells that can accumulate Ig V gene mutations in a T-cell-independent fashion (eg, marginal zone [MZ] B cells).19 The characteristics of the mutations acquired in this manner and their similarities or differences with those occurring during a T-cell-dependent process are not yet defined.
Therefore, we undertook an empirical study on a large database of B-CLL VH sequences to determine whether the mutations in the Ig V gene exhibited features that were similar or different from canonical SHM. The targeting preference for RGYW motifs, base change bias for transitions, and focusing of replacement mutations in the complementarity-determining regions (CDRs) and away from the framework regions (FRs) in B-CLL sequences were consistent with SHM. Thus, regardless of the T-cell or GC dependence of these mutations, our data do not support a necessity to invoke a mutational process other than the normal SHM mechanism as the mediator of Ig V gene mutations in B-CLL cells.
Patients, materials, and methods
Patients
The Institutional Review Boards of North Shore University Hospital (Manhasset, NY) and Long Island Jewish Medical Center (New Hyde Park, NY) approved these studies. Informed consent was provided by all participants in accordance with the Declaration of Helsinki. The patients in this study were diagnosed with typical B-CLL based on clinical criteria and laboratory features. Peripheral blood mononuclear cells (PBMCs) were separated from heparinized venous blood of B-CLL patients by density gradient centrifugation using Ficoll-Paque (Amersham Biosciences, Piscataway, NJ). PBMCs were cryopreserved with a programmable cell-freezing machine (Cryomed, Mt Clemens, MI).
Ig VH gene sequencing
VH cDNA sequences were determined by the previously described method.1 Briefly, mRNA was purified from cryopreserved PBMCs of B-CLL patients. The cDNA was synthesized by reverse transcription and used as template in polymerase chain reaction (PCR) reactions with VH family FR-specific forward primers and μ, γ, and α chain reverse primers. Once the VH family was determined, the PCR was repeated using a leader-specific forward primer, and this product was either directly sequenced or cloned and sequenced. Only complete (beginning of FR1 through FR4) B-CLL VH sequences were considered here.
Other VH sequences
A collection of mutated VH sequences from the public GenBank database was obtained. All of these sequences were entered by Dr P. E. Lipsky and used the VH genes 3-07, 3-23, 4-34, or 4-59.20-23
Sequence analysis
The closest germ line VH for each B-CLL VH sequence was assigned using DNAPLOT (http://www.mrc-cpe.cam.ac.uk/DNAPLOT.php?menu=901) or IgBlast (http://www.ncbi.nlm.nih.gov/igblast/). Other sequence manipulations were done using DNASTAR v5.06 (DNASTAR, Inc, Madison, WI) and Microsoft Excel 2002. Nucleotide differences that were assignable to known alleles or that occurred independently among multiple B-CLL sequences, indicating allelic origin, were not scored as mutations. Deletions or insertions were not considered. CDRs and FRs were defined by the rules of Kabat et al.24
Average mutability calculations
The average mutability of the “wobble” nucleotides was calculated for each germ line gene as follows. For a given gene, the redundant codon positions were identified and, for each one, the mutability scores of the 3 triplets in which the redundant nucleotide resides were averaged. The nucleotide mutability scores were then averaged for all positions containing the same nucleotide to create an average mutability score for every A, G, C, and T in each germ line gene. Finally the overall average mutability was determined by weighting the score for each nucleotide in each germ line gene by summing the products of each gene's mutability values and the number of mutations in VH sequences of that gene and dividing the total by the overall number of mutations considered.
Statistical analysis
The percentage of mutations in RGYW motifs, the transition-to-transversion ratios, and the replacement-to-silent ratios were evaluated by the χ2 test using the non-CLL dataset as the expected values. When the number of observed events was too low for the χ2 test to be considered accurate, the Fisher exact test was employed. All calculations were performed using GraphPad InStat version 3.05 (GraphPad Software, San Diego, CA).
Results
VH gene usage and mutation frequencies
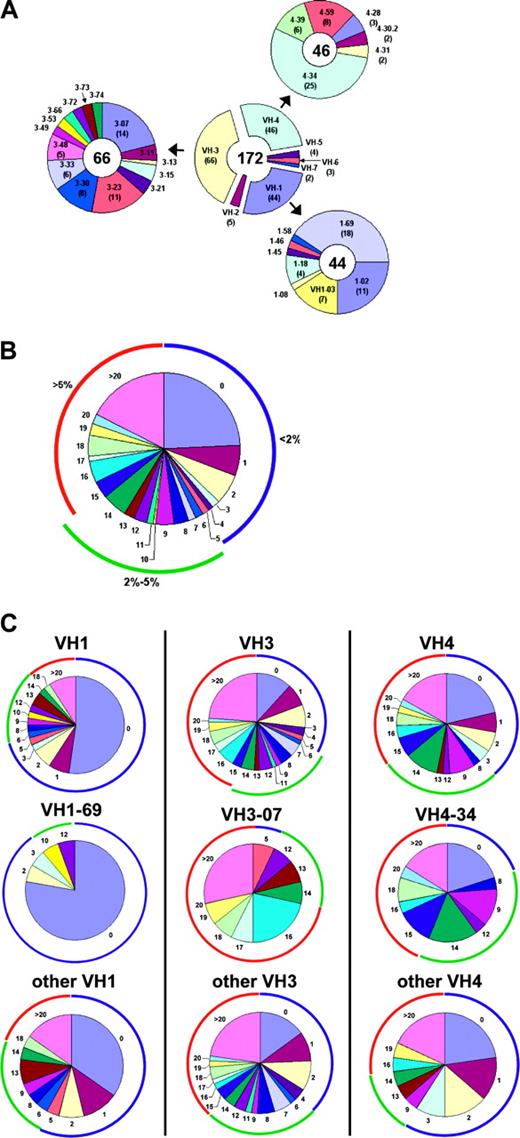
Complete VH sequences were obtained from 172 leukemic clones of B-CLL patients. Each sequence was assigned a germ line counterpart using the public tools (IgBlast and DNAplot). VH3 family genes were the most commonly used, with VH1 and VH4 used at somewhat lower frequencies. VH4-34, 1-69, 3-07, and 3-23 and their alleles were the most commonly used VH genes (Figure 1A), as expected.1,3,25,26 The frequency of cases using VH4-34 and 1-69 was greater than the frequency of those genes reported in a normal CD5+ B-cell repertoire,21 while the frequency of VH3-30- and 3-09 gene-using cases was lower than the frequency of those genes in the normal repertoire (P < .5; data not shown). These specific VH gene frequencies are consistent with those described previously.1,27,28 However, these frequency comparisons may be specious if the cell of origin of B-CLL cells is not the circulating CD5+ B cell found in most healthy subjects.
VH gene usage and mutation distribution. (A) The number of B-CLL VH sequences using genes of each VH family are shown in the center donut and the number of sequences using particular VH1, 3, and 4 genes are depicted peripherally. The VH gene used by each sequence was assigned using DNAPLOT and IgBlast. (B) Overall distribution of mutations per sequence. The colored arcs encompass sequences with less than 2% mutation (blue), 2% to 5% mutation (green), or more than 5% mutation (red). (C) The distribution of mutations in sequences using the 3 most commonly used VH families: VH1 (left), VH3 (middle), and VH4 (right). The distributions are shown when all genes of the family (top), the most commonly used gene of the families (middle), and the other genes of that family (bottom) are considered.
VH gene usage and mutation distribution. (A) The number of B-CLL VH sequences using genes of each VH family are shown in the center donut and the number of sequences using particular VH1, 3, and 4 genes are depicted peripherally. The VH gene used by each sequence was assigned using DNAPLOT and IgBlast. (B) Overall distribution of mutations per sequence. The colored arcs encompass sequences with less than 2% mutation (blue), 2% to 5% mutation (green), or more than 5% mutation (red). (C) The distribution of mutations in sequences using the 3 most commonly used VH families: VH1 (left), VH3 (middle), and VH4 (right). The distributions are shown when all genes of the family (top), the most commonly used gene of the families (middle), and the other genes of that family (bottom) are considered.
A database of the differences from the germ line sequences was compiled, with attention paid to eliminating possible and probable allelic variants from subsequent inclusion in mutation analysis. The total number of mutations in each B-CLL sequence was determined. Approximately 41% of the Ig VH genes analyzed had less than 2% difference from the most similar germ line gene, the commonly used threshold for considering a case “unmutated,” and 24% had no changes whatsoever (Figure 1B). Approximately 22% of the leukemic clones had between 2% and 5% mutations and 37% had more than 5% mutations. These mutations occurred in all VH families, although within each family the distribution of mutations among the family members differed significantly.
For example, of the 3 most used VH genes, 1-69 genes were predominantly completely unmutated (78%), VH3-07 genes were predominantly mutated (93%), and VH 4-34 were usually mutated (80%), but the remainder were completely the same as the germ line. Interestingly, these biases in either the presence or absence of VH mutation are much more pronounced in these specific genes than the family as a whole. The other VH1 genes are unmutated more than (58%) the overall frequency (41%) but not nearly as much as the VH1-69 gene. Similarly, although approximately 90% of the 3-07 genes exhibit at least 2% mutations, the other VH3 genes have almost the same percentage of unmutated genes as the overall (38%). Finally, while VH4-34 genes are deficient in unmutated sequences, the remaining VH4 family genes are slightly enriched in unmutated sequences (59%; Figure 1C).
Nucleotide characteristics of B-CLL VH mutations
Targeting of mutations to specific sequences and areas within VH genes. The targeting and base change preferences associated with canonical SHM are nonrandom and have been empirically determined.29 A sampling of non-B-CLL mutated VH sequences from the literature was analyzed and found to be consistent with expected biases toward mutations in RGYW hot spots, increased replacement-to-silent (R/S) ratios in the CDR and depressed ratios in the framework, and for transitions (A>G, G>A, C>T, and T>C) over transversions (Table 1). Overall, the mutations in the B-CLL sequences exhibited quantitatively similar biases that were not statistically different from the representative non-CLL sequences. There was an increased frequency of mutation in RGYW motifs, with a preference for transitions over transversions regardless of the nucleotide that was mutated (Figure 2A). Interestingly, the characteristics of the mutations occurring in “unmutated” cases (those with < 2%) were also not significantly different from non-CLL, although the lower level of replacement mutations in the CDR was almost significant (P = .0501). A larger number of B-CLL-derived VH gene sequences will be needed to answer this question definitively.
Targeting and base change characteristics of mutations in B-CLL, unmutated B-CLL, and non-B-CLL VHgenes
. | No. of sequences . | No. of mutations . | R/S ratio . | . | . | Germ line RGYW, % . | Mutation in RGYW, % . | Transitions-to-transversions ratio . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Type . | . | . | Total . | CDR . | FR . | . | . | . | ||
| B-CLL | 172 | 1789 | 2.35* | 4.60* | 1.72* | 25.6 | 43.9* | 1.294* | ||
| B-CLL with less than 2% mutation | 71 | 71 | 1.86* | 3.50† | 1.40* | 24.7 | 53.5* | 1.152* | ||
| Non-B-CLL | 67 | 452 | 2.23 | 4.27 | 1.50 | 24.9 | 49.1 | 1.294 | ||
. | No. of sequences . | No. of mutations . | R/S ratio . | . | . | Germ line RGYW, % . | Mutation in RGYW, % . | Transitions-to-transversions ratio . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Type . | . | . | Total . | CDR . | FR . | . | . | . | ||
| B-CLL | 172 | 1789 | 2.35* | 4.60* | 1.72* | 25.6 | 43.9* | 1.294* | ||
| B-CLL with less than 2% mutation | 71 | 71 | 1.86* | 3.50† | 1.40* | 24.7 | 53.5* | 1.152* | ||
| Non-B-CLL | 67 | 452 | 2.23 | 4.27 | 1.50 | 24.9 | 49.1 | 1.294 | ||
Not a significant difference compared with non-CLL (P > .4).
Not a significant difference compared with non-CLL (P > .05).
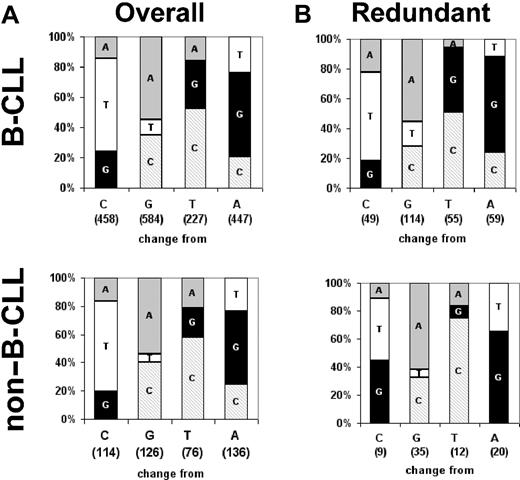
A transition bias is apparent in all mutated nucleotides. Graphs show the relative frequency with which each nucleotide mutates into each of the other 3. The number of mutations occurring in each nucleotide is shown in parentheses on the x-axis. (A) All mutations. (B) Mutations occurring at the third position of completely redundant codons. Data are from 172 B-CLL sequences from our internal laboratory database and 67 non-B-CLL sequences from the public databases.
A transition bias is apparent in all mutated nucleotides. Graphs show the relative frequency with which each nucleotide mutates into each of the other 3. The number of mutations occurring in each nucleotide is shown in parentheses on the x-axis. (A) All mutations. (B) Mutations occurring at the third position of completely redundant codons. Data are from 172 B-CLL sequences from our internal laboratory database and 67 non-B-CLL sequences from the public databases.
Nucleotides biases in VH gene mutations. In order to assure that the transition/transversion biases were not the result of amino acid selection for antigen binding or undefined structural requirements, we analyzed the transition-to-transversion ratios at redundant wobble positions in the VH sequences (Figure 2B). Wobble positions are the third nucleotide of redundant codons. Since any nucleotide at these positions encodes the same amino acid, these nucleotides are useful for deconvoluting the potential role of amino acid selection because they cannot be selected at the protein level. As such, these positions offer a window into the targeting and base change preferences of the mutational mechanism with no interference from protein structure-based selective pressures (although selection at the level of transcription or translation cannot be formally excluded). These analyses also indicated that nucleotide changes that are found in B-CLL VH favored transitions.
Bias against changes in C nucleotides at wobble positions of redundant codons. One surprising result of the analysis of these redundant codons was an unexpectedly strong bias against mutations at C nucleotides that is not present when the mutations at all sites are considered. This bias was also found in redundant codons of the non-B-CLL VH sequences analyzed here (Figure 3).
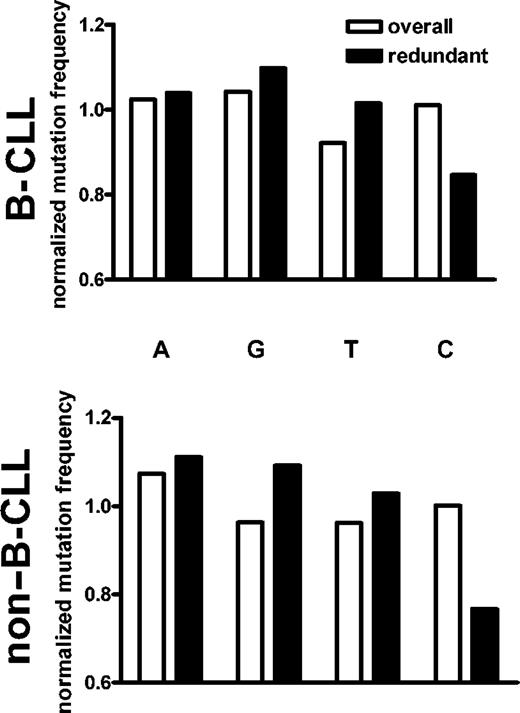
Nucleotide biases of the mutational mechanism. Normalized mutation frequencies were calculated by comparing the fraction of mutations in a given nucleotide to the fraction of that nucleotide in the germ line sequences. If mutations occurred in a nucleotide at the same relative rate that it appeared in the germ line VH sequences, it would have a normalized mutation rate of 1. When all mutations are considered, there is a slight bias away from mutating T nucleotides. However, when only the silent redundant positions are considered, there is a pronounced bias against mutations in C nucleotides in both B-CLL (top) and normal (bottom) sequences.
Nucleotide biases of the mutational mechanism. Normalized mutation frequencies were calculated by comparing the fraction of mutations in a given nucleotide to the fraction of that nucleotide in the germ line sequences. If mutations occurred in a nucleotide at the same relative rate that it appeared in the germ line VH sequences, it would have a normalized mutation rate of 1. When all mutations are considered, there is a slight bias away from mutating T nucleotides. However, when only the silent redundant positions are considered, there is a pronounced bias against mutations in C nucleotides in both B-CLL (top) and normal (bottom) sequences.
One possible explanation for such a bias could invoke sequence context effects. Other studies have demonstrated that the conventional SHM process has a hierarchy of preference for various dinucleotide and trinucleotide motifs.14 The difference between the most mutated and least mutated motifs can be significant. Therefore, the flanking sequences of each redundant codon were considered. The average mutability of the 3 trinucleotide windows (upstream, centered, and downstream) that contained the particular wobble nucleotide was calculated for each germ line redundant codon position using the mutability index of Shapiro et al,30 as described in “Patients, materials, and methods.” While C nucleotides had the lowest relative mutability, the difference was not as pronounced as the actual frequencies observed here (average mutability: A, 1.08; G, 0.84; C, 0.77; T, 0.98). Furthermore, the average mutability calculated for G nucleotides was only marginally greater than that for C, yet the normalized mutation frequency of G was slightly elevated in both the B-CLL and non-B-CLL sequences analyzed (Figure 3). Similar results were obtained when dinucleotide contexts were considered (data not shown).
The rate of mutation drops off exponentially toward the 3′ end of the Ig V gene sequence,15 and therefore a clustering of redundant C nucleotides in one portion of the VH gene could lead to a lowered mutation rate as a function of their position. However, the various redundant nucleotides were evenly distributed throughout the germ line genes, excluding a gross positional effect (data not shown).
B-CLL VH mutations are selected
Selection for improved antigen binding sites or for the maintenance of antibody structure typically causes localized shifts in the R/S ratios of mutations during canonical SHM. The B-CLL mutations analyzed here follow a similar pattern (Table 1), with an elevated R/S in CDRs (mean, 4.60) and decreased R/S in FRs (mean, 1.72). This pattern mirrored that seen in the normal sequences (4.27 and 1.50, respectively).
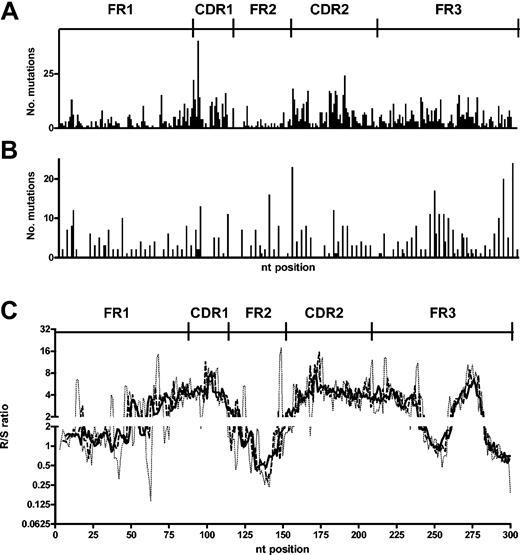
When the mutations are mapped onto the gene, there is an apparent targeting of replacement mutations toward the CDR1 and CDR2 (Figure 4A). Although such mapping is helpful in identifying nucleotides of particular vulnerability, it is less valuable when considering targeting of larger stretches of DNA sequence. Therefore, to gain more resolution as to which areas were enriched or suppressed for replacement mutations, a “moving window” approach was applied to the positional R/S data. In this analysis, we determined the R/S ratio that surrounds each nucleotide by using the cumulative R/S of stretches of 5, 6, and 7 nucleotides upstream and downstream of the central position (moving windows of 11, 13, and 15 nucleotides). These windows make the differences from individual nucleotide to individual nucleotide (Figure 4A-B) less pronounced, and the larger the window the greater the “smoothing” becomes. This analysis clearly localized the CDR1 and CDR2 as having elevated R/S ratios and the FR1 and FR2 as having lowered R/S ratios. Interestingly an increased R/S ratio at the end of FR3 was also clearly identified with this approach (Figure 4C).
CDRs of B-CLL sequences contain more replacement mutations. (A) Number of replacement mutations at each position in the VH genes of B-CLL. (B) Number of silent mutations observed at each position in the VH of B-CLL. (C) A “moving window” R/S ratio for each position was calculated by considering the cumulative R/S ratio of the centered position and the 5 (dotted line), 6 (dashed line), or 7 (solid line) positions on either side. CDR and FR are as defined by Kabat et al.24
CDRs of B-CLL sequences contain more replacement mutations. (A) Number of replacement mutations at each position in the VH genes of B-CLL. (B) Number of silent mutations observed at each position in the VH of B-CLL. (C) A “moving window” R/S ratio for each position was calculated by considering the cumulative R/S ratio of the centered position and the 5 (dotted line), 6 (dashed line), or 7 (solid line) positions on either side. CDR and FR are as defined by Kabat et al.24
Discussion
The results presented here strongly support the hypothesis that the clonal VH mutations seen in B-CLL cells are the product of the classical antigen-driven SHM process. In addition, previously observed mutation distribution and gene usage biases were confirmed and extended. While these observations are not unexpected, the large sample size analyzed here creates a more firm footing for models of B-CLL development that suggest that B-CLL cases with mutated VH genes derived from cells whose V segments were somatically mutated (Table 1; Figure 1) via a process that strongly resembles or is (Table 1; Figures 2, 3, 4) the conventional SHM machinery in response to antigen stimulation.
The targeting preference and base change biases of the mutational mechanism responsible for the B-CLL VH mutations were distinguished from the effects of selection by focusing on intrinsically unselectable wobble positions in redundant codons (Figure 2B). This approach confirmed that the overall pattern of transition bias was not excessively influenced by selecting for protein structure that provides advantages in antigen binding or eliminates the danger of destabilizing the B-cell-receptor scaffolding. In addition, this approach revealed an unexpected aspect of mutation targeting. Cytidine nucleotides at redundant wobble positions were mutated at approximately half the frequency of the other bases in both B-CLL and canonical SHM-generated sequences (Figure 3). Curiously, there was no overall bias against mutations in cytidine, suggesting that the wobble positions are somehow distinct in their mutation profile.
An inherent limitation of this analysis is the restriction to B-CLL cases with VH mutations, the degree of which can differ from case to case. Sequences with higher numbers of mutations had a greater influence on the results, although this was unavoidable. The data reported here, however, in no way rule out a role for antigen selection in the unmutated cases, since many germ line or natural antibodies can have moderate affinity to defined antigens.31-33 Recently, sets of Ig V unmutated B-CLL cases that express certain VHDJH and VLJL gene segments with stereotyped CDR3s have been defined.34,35 Such patterned variable region gene structures are characteristic of antibodies to defined epitopes in several experimental and vaccine systems,36-39 suggesting that at least some Ig V unmutated B-CLL cases may have a common antigen specificity.
The few mutations observed in sequences with less than 2% differences from the most similar germ line gene (“unmutated”) also resembled the canonical SHM spectra, although a depressed R/S ratio in the CDR approached a statistically significant difference. This may indicate that the unmutated Ig V sequences occurred in cells that began, but quickly aborted, the SHM process, perhaps due to an already avid antigen receptor or impermissible autoreactivity. However, these are speculations in the absence of defined antigens and affinities.
It has been proposed that B-CLL is derived from marginal-zone (MZ) B cells.19 Although MZ B cells can display somatic mutations,40 these may occur via a T-independent mechanism outside of classical GCs.17,18 If MZ cells are the precursors of B-CLL cells, then the mechanism that generates mutations in MZ cells would presumably be mechanistically similar to the SHM that takes place in GCs. The results of the current study would be inconsistent with a model that attributes B-CLL mutations to a distinct mechanism with a different mutational spectrum.
Ongoing mutations in Ig V genes41 and BCL-642 have been observed in some B-CLL cases. At this time, there are an insufficient number of sequences of emerging variants available to perform the analyses similar to those in this study, although one study did suggest that targeting might differ in these ongoing.41 However, such an analysis would be valuable, as it would indicate whether the ongoing mutations are consistent with continued SHM, as has been postulated from demonstrations that activation-induced cytidine deaminase (AID) is expressed in some B-CLL cells.43-45 The alternative, that the ongoing mutations are not the product of SHM, would indicate that at least some B-CLL cells have a more elevated mutation rate. Elevated mutation rates are often conducive to increased aggressiveness46 and would be cause for clinical concern.
Prepublished online as Blood First Edition Paper, December 24, 2003; DOI 10.1182/blood-2003-10-3407.
Supported in part by grants from the National Institutes of Health National Cancer Institute (NIH NCI; CA 81554 and CA 87956), the Joseph Eletto Leukemia Research Fund, the Jean Walton Fund for Lymphoma & Myeloma Research, and the S.L.E. Foundation, Inc.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank past and former members of the laboratory for their contributions toward compiling the sequence database used here. We thank David Thaler, Manlio Ferrarini, and Federico Caligaris-Cappio for useful discussions of the data.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal