Abstract
Hematopoietic stem progenitor cells (HSPCs) are highly enriched in a rare subset of Lin-CD34+CD38- cells. Independent of stage of human development, HSPC function segregates to the subset of Lin-CD34+CD38- cells. However, fetal-derived HSPCs demonstrate distinct self-renewal and differentiation capacities compared with their adult counterparts. Here, to characterize the molecular nature of fetal HSPCs, suppressive subtractive hybridization was used to compare gene expression of HSPCs isolated from fetal blood (FB-HSPCs) versus adult mobilized peripheral blood (MPB-HSPCs). We identified 97 differentially expressed genes that could be annotated into distinct groups that include transcription factors, cell cycle regulators, and genes involved in signal transduction. Candidate regulators, such as Lim only domain-2 (LMO2), nuclear factor–kappa B (NF-κB), tripartite motif 28 (Trim28), and N-myc protooncogene (MYCN), and a novel homeobox gene product were among transcripts that were found to be differentially expressed and could be associated with specific proliferation and differentiation properties unique to FB-HSPCs. Interestingly, the majority of genes associated with signal transduction belong to Ras pathway, highlighting the significance of Ras signaling in FB-HSPCs. Genes differentially expressed in FB-HSPCs versus adult MPB-HSPCs were verified using quantitative real-time polymerase chain reaction (Q-PCR). This approach also resulted in the identification of a transcript that is highly expressed in FB-HSPCs but not detectable in more differentiated Lin-CD34+CD38+ FB progenitors. Our investigation represents the first study to compare phenotypically similar, but functionally distinct, HSPC populations and to provide a gene profile of unique human HSPCs with higher proliferative capacity derived from early in utero human blood development.
Introduction
Proliferative and differentiation characteristics of human hematopoietic stem progenitor cells (HSPCs) allow the creation and maintenance of mammalian hematopoiesis.1 These rare cell types are operationally defined by their ability to reconstitute the hematopoietic system upon transplantation in vivo2 or give rise to large numbers of mature hematopoietic cells in vitro using long-term culture–initiating cells (LTC-ICs) or colony-forming unit (CFU) assays.3,4 Despite differences in readout assays used to define stem progenitors, human hematopoietic cells with these properties are highly enriched in the lineage-depleted (Lin-) population of hematopoietic cells with CD34+CD38- cell surface phenotype.5 The nonobese diabetic severe combined immunodeficient (NOD/SCID) mouse has been extensively used to evaluate the in vivo proliferation and differentiation characteristics of HSPCs2,5 that are mainly enriched in the Lin-CD34+CD38- subset. HSPCs are distinct from hematopoietic progenitor cells (HPCs) enriched in Lin-CD34+CD38+ subset, since HPCs are not able to repopulate in the NOD/SCID mouse and can only be detected using in vitro progenitor assays.5 To further illustrate the importance of this phenotype, HSPC function segregates to the subset of Lin-CD34+CD38- cells at all stages of human hematopoietic development, ranging from fetal to adult sources of HSPCs.6-9
Despite the ability to isolate human HSPCs, our understanding of mechanisms governing self-renewal or differentiation of these rare cells is limited. For example, the intrinsic and extrinsic factors that regulate the mitotic transitions of human HSPCs have yet to be determined.10,11 The dormant status of adult HSPCs may explain the inability to expand these cells without loss of HSPC function and why they are refractory to efficient retroviral gene transfer.1 However, fetal blood (FB)–derived HSPCs enriched in Lin-CD34+CD38- cells possess unique properties that include distinct proliferative and differentiation capacities compared with their adult counterparts. Over a series of investigations, a novel population of human HSPCs has been recently identified in the fetal circulation that possesses distinct proliferation and differentiation characteristics compared with adult circulating HSPCs harvested from mobilized peripheral blood (MPB).8,9 Lin-CD34+CD38- FB-HSPCs are distinct from adult Lin-CD34+CD38- MPB-HSPCs in their frequency among total mononuclear cells, number of human SCID repopulating cells (SRCs),8 enhanced retroviral transduction ability into HSPCs, in vivo myelopoietic reconstitution potential of FB-HSPCs, and the number of cycling HSPCs in FB that retains SRC capacity.9 Accordingly, FB-HSPCs provide a unique source of human HSPCs that are actively cycling8,9 with the opportunity to study and define the molecular mechanisms involved in HSPCs proliferation when self-renewal divisions are more frequent than in the adult hematopoietic system.
Recent advances in the area of gene expression studies have provided novel approaches, such as microarray,12 serial analysis of gene expression,13 differential display,14 and suppression subtractive hybridization (SSH),15 to study gene expression profiles in different populations of cells. Here, we aimed to evaluate differences in gene expression between FB-HSPCs and MPB-HSPCs that are likely responsible for distinct functional characteristics of FB-HSPCs using SSH. Our study identifies a gene profile for functionally distinct human HSPCs that differs from adult sources that possess limited proliferative potential and thereby provides the first molecular description of human HSPCs derived from early human blood development.
Materials and methods
Human cells
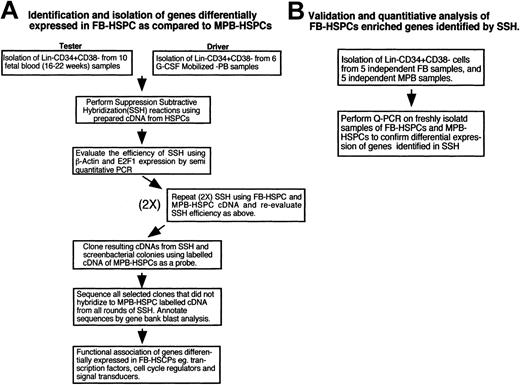
Samples of human FB (n = 10), from early gestation (16-22 weeks), and MPB (n = 6), mobilized by granulocyte colony-stimulating factor (G-CSF), were obtained in conjunction with local ethical and biohazard authorities of the University of Western Ontario and London Health Sciences Center. FB and MPB samples were diluted (1:4) with α-minimal essential medium (GibcoBRL, Grand Island, NY) or phosphate-buffered saline (PBS), and mononuclear cells (MNCs) were isolated as previously described.8,9 Figure 1 provides a schematic of our experimental approach using purified FB- and MPB-HSPCs. Given the individual variation between different human samples, FB and MPB samples from each population were pooled for SSH. Independent of this approach, genes identified to be differentially expressed in FB-HSPCs were confirmed using quantitative real-time polymerase chain reaction (Q-PCR) on freshly isolated samples of FB and MPB as indicated in Figure 1.
Experimental approach to identify genes differentially expressed in FB-HSPCs compared with adult MPB-HSPCs. (A) Development of cDNA libraries for identifying differentially expressed genes in FB-HSPCs. Isolation of Lin-CD34+CD38- cells from fetal blood (10 samples) and mobilized peripheral blood (6 samples) followed by performing SSH as explained in “Materials and methods.” SSH was performed 2 more times to ensure the quality and reproducibility of the cDNA library. Screening of the colonies in the subtracted library will detect differentially regulated transcripts in FB-HSPCs and BLAST analysis will identify the closest transcripts in the human genome. We further narrowed our analysis toward key regulatory genes including transcription factors, cell cycle regulators, and signal transduction genes presumably responsible for fate determination of HSPCs. (B) Verification and quantification of candidate transcripts identified in the SSH reactions. Lin-CD34+CD38- cells from fetal blood (5 samples) and mobilized peripheral blood cells (5 samples) and also Lin-CD34+CD38+ from several fetal blood (12 samples) were isolated and used for real-time Q-PCR. The Q-PCR experiments were done in 3 different concentrations and each concentration in duplicate. This analysis confirmed and validated our experimental approach to identify genes that are consistently differentially expressed in FB-HSPCs versus adult counterparts from M-PB-HSPCs.
Experimental approach to identify genes differentially expressed in FB-HSPCs compared with adult MPB-HSPCs. (A) Development of cDNA libraries for identifying differentially expressed genes in FB-HSPCs. Isolation of Lin-CD34+CD38- cells from fetal blood (10 samples) and mobilized peripheral blood (6 samples) followed by performing SSH as explained in “Materials and methods.” SSH was performed 2 more times to ensure the quality and reproducibility of the cDNA library. Screening of the colonies in the subtracted library will detect differentially regulated transcripts in FB-HSPCs and BLAST analysis will identify the closest transcripts in the human genome. We further narrowed our analysis toward key regulatory genes including transcription factors, cell cycle regulators, and signal transduction genes presumably responsible for fate determination of HSPCs. (B) Verification and quantification of candidate transcripts identified in the SSH reactions. Lin-CD34+CD38- cells from fetal blood (5 samples) and mobilized peripheral blood cells (5 samples) and also Lin-CD34+CD38+ from several fetal blood (12 samples) were isolated and used for real-time Q-PCR. The Q-PCR experiments were done in 3 different concentrations and each concentration in duplicate. This analysis confirmed and validated our experimental approach to identify genes that are consistently differentially expressed in FB-HSPCs versus adult counterparts from M-PB-HSPCs.
Cell purification
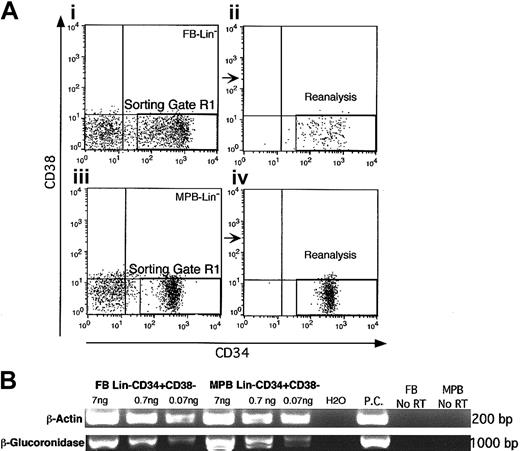
Subsets of Lin-CD34+CD38- from FB and MPB and Lin-CD34+CD38+ from FB were isolated as described.8 Briefly, the lineage-depleted (Lin-) cells were isolated from each population using a cocktail of monoclonal antibodies (Stem Cell Technologies, Vancouver, BC, Canada) against cell surface markers: CD2, CD3, CD14, CD16, CD19, CD24, CD33, CD38, CD41, CD56, CD66b, and glycophorinA. Next, the Lin- population was stained with antihuman CD34-allophycocyanin (APC) and antihuman CD38-phycoerithrin (PE; Becton Dickinson, San Jose, CA) antibodies. The stained cells were sorted on a FACS vantage SE (Becton Dickinson) using sorting gates illustrated in Figure 1A. Reanalysis of sorted Lin-CD34+CD38- cells from FB and MPB and Lin-CD34+CD38+ cells from FB demonstrated the isolation of the targeted populations with the highest purity (Figure 1A).
Synthesis and amplification of cDNA
The mRNA of pooled samples from each population was extracted using oligo(dT)-cellulose chromatography (Amersham Biosciences, Little Chalfont, United Kingdom) (Figure 1; experimental protocol). The mRNA underwent DNase-I digestion (GIBCO BRL, Burlington, ON, Canada) to eliminate the possibility of genomic DNA contamination. This was followed by the synthesis of full-length strand cDNA using 3′ and 5′ primers (Clontech, Palo Alto, CA) as previously described.16 The full-length strand cDNA was used as a template for exponential amplification in PCR using Advantage PCR Amplification kit (Clontech). The PCR conditions are as follows: first denaturation at 95°C for 1 minute followed by cycles of amplification at 95°C for 15 seconds (denaturation), 65°C for 30 seconds (annealing), and 68°C for 6 minutes (extension). The quality of amplified sample was evaluated on 1.2% agarose gel by observing the amplified products ranging from 7 kb to 0.5 kb. Amplified cDNA was purified (Clontech), quantified using Spectrophotometer (Fischer Scientific, Hampton, NH), and stored at -20°C until used.
Suppression subtractive hybridization (SSH)
Amplified cDNA of FB and MPB Lin-CD34+CD38- was used as tester and driver, respectively, for SSH as described.15 Briefly, the amplified cDNA of tester and driver underwent RsaI (Fermentas, Burlington, ON, Canada) restriction digestion and were purified (Clontech) followed by quantification using Spectrophotometer (Fisher Scientific). Next, in 2 separate tubes 100 ng of purified RsaI-digested tester cDNA was ligated to 10 mM of 2 different adaptor primers. Finally, in hybridization step, in 2 tubes, 15 ng of adaptor ligated tester cDNA was hybridized with 300 ng cDNA of driver population for 8 hours at 68°C. This was followed by mixing the content of both tubes along with adding 75 ng cDNA of driver population and overnight incubation at 68°C. The subtracted tester cDNA was amplified in suppression and nested PCR.
The efficiency of subtracted library was verified in semiquantitative PCR and Q-PCR, using the following primers: β-actin forward 5′-GATCCACATCTGCTGGAAGG-3′ and reverse 5′-AAGTGTGACGTTGACATCCG-3′; Tal-1 forward 5′-ATGGTGCAGCTGAGTCCTCC-3′ and reverse 5′-ATATACTTCATGGCCAGGCGG-3′; and elongation-2, factor-1 (E2F1) forward 5′-CCAACTCCCTCTACCCTTGA-3′ and reverse 5′-TGTCTCCCTCCCTCACTTTC-3′. The PCR condition was composed of primary denaturation (96°C for 5 minutes) followed by 40 cycles of 94°C (45 seconds), 60°C (45 seconds), and 72°C (2 minutes). The PCR components consisted of 2 mM MgCl2, 12.5 mm of each set of primers, and 2.5 U Taq DNA polymeRase (Invitrogen, Carlsbad, CA). As indicated in Figure 1A, SSH between FB-HSPC and MPB-HSPC was repeated 2 additional times to ensure the subtracted library was enriched for differentially expressed genes in FB-HSPCs (data not shown for these rounds of SSH).
Cloning and Southern blotting
The nested PCR products, from the last step of SSH, were ligated into TA vector (Invitrogen) and were transformed into TOP10 Escherichia coli cells (Invitrogen). Three hundred twenty colonies were screened using the RsaI-digested amplified cDNA of driver population as a probe. Briefly, the recombinant plasmids underwent EcoRI restriction digestion followed by fractionation on 1% agarose gel. The DNA was then transferred to the nylon membrane (Amersham Pharmacia) using capillary action.17 The membrane was hybridized with 20 × 106 α32P–deoxycytosine triphosphosphate (dCTP) random primed labeled (Roche, Mississauga, ON, Canada) cDNA probe for overnight at 42°C. The membrane was washed with sodium dodecyl sulfate (SDS; 0.1%) standard saline citrate (SSC; 2×) at room temperature for 30 minutes and SDS (0.1%) SSC (0.1×) at 68°C for 30 minutes. The membrane was exposed to BIOMAX MS film (Kodak, Rochester, NY) overnight at -80°C.
Quantitative real-time PCR (Q-PCR)
The differential expression of some of the transcripts was confirmed and quantified in Q-PCR (MX4000; Stratagene, La Jolla, CA) using SYBRGREEN (Stratagene) DNA binding dye. Freshly harvested populations of purified Lin-CD34+CD38- and Lin-CD34+CD38+ from 5 independent FB samples (ranging from 16-22 weeks of gestation) and 5 independent MPB samples of Lin-CD34+CD38- cells were used to isolate FB-HSPCs, FB-HPCs, and MPB-HSPCs, respectively, for real-time Q-PCR analysis. The Primer design program (http://www.broad.mit.edu/cgi-bin/primer/primer3_www.cgi) was used to design primers for the transcripts listed in Table 1. The regular PCR was performed for each gene to check the size of the amplicon followed by sequencing to ensure the authenticity of the amplified product. The Q-PCR condition was composed of 2 mM MgCl, 0.4 mM deoxynucleoside triphosphate (dNTP), 8% glycerol, 3% dimethyl sulfoxide (DMSO), 150 nM of each primer, 0.375 mL of 1:500 dilution of reference dye, and 2.5 mL of 1:2000 dilution of SYBRGREEN. The Q-PCR was performed along with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or hydroxymethyl bilane synthase gene (HMBS) as housekeeping genes (internal control) to ensure the quality and efficiency of PCR reaction. Each reaction was performed in duplicate for all samples and the comparative quantification, based on the cycle threshold (Ct) of the gene of interest (GOI) normalized to housekeeping gene (HKG; GAPDH or HMBS) in each population, was used to evaluate the relative gene expression in FB-HSPCs versus MPB-HSPCs and FB-HPCs. The following equation18 was applied to calculate the relative expression of candidate genes in FB-HSPCs versus MPB-HSPCs and FB-HPCs: 2-ΔΔCt = comparative expression of GOI in FB-HSPCs compared with MPB-HSPCs or FB-HPCs. ΔΔCt for FB-HSPCs versus MPB-HSPCs or FB-HPCs = ΔCt for FB-HSPCs - ΔCt for MPB-HSPCs or FB-HPCs. ΔCt of FB-HSPCs = Ct GOI - CtHKG. ΔCt of MPB-HSPCs = Ct GOI - CtHKG. ΔCt of FB-HPCs = Ct GOI - CtHKG.
Sequence of the forward and reverse primers used in Q-PCR for some of the transcripts identified in the subtracted library of FB-HSPCs
Transcript, gene . | Forward primer 5′ 3′ . | Reverse primer 5′ 3′ . | Amplicon size, bp . |
|---|---|---|---|
| CDK6 | CACGAACAGACAGAGAACCA | CGGTGTGAATGAAGAAAGTCC | 169 |
| CENPF | AGAACCCACCACGAAATCC | TGCCTTCACTGGACCTTACA | 169 |
| ETS2 | CCAAGAGAGGAGTGACCCAG | CAGGAGAAACTGCCACAGC | 114 |
| FB29 | CTTGGGTCAGAGCAGAGGTC | CAGCAGGTTTGGAACTGGA | 150 |
| FB83 | TCCAGCAACCTTCCATCTCT | TGAGCCAAGAAAGCCATC | 184 |
| FB136 | TTCCCAAAGTAACAGCCAGTC | TTCATCCTCAGTTTCACCAAA | 152 |
| FB248 | TCACTTTATCATCAGCCAGCA | TTCTTGGTCTCGCCTACTCC | 159 |
| GAPDH | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAC | 100 |
| HMBS | GGCAATGCGGCTGCAA | GGGTACCCACGCGAATCAG | 80 |
| LMO2 | TACTGGCACGAGGACTGC | CTTTCACCCGCATTGTCAT | 190 |
| LZTF1 | TCTACTCAGGGTTCAGGAGCA | TTGATTTGGTCATTCTTCTTGG | 120 |
| MTCO-1 | CCTGACTGGCATTGTATTAGC | TTTTGGCGTAGGTTTGGTCT | 179 |
| MYCN | TTGTGTGTTCCAAGTTTCCAA | CCACCTCTCATTACCCAGGA | 185 |
| NFKB1 | TGAGTCCTGCTCCTTCCAA | CTTCGGTGTAGCCCATTTGT | 105 |
| Rac1 | GTGAATCTGGGCTTATGGGA | GATGGGAGTGTTGGGACAGT | 189 |
| RALBP1 | GAAGCAGTATTTGCGAGACC | TGGCAGTTCTTTGAGTAAACG | 133 |
| RCC1 | TGGGTGAGAATGTGATGGAG | GCAGCCGAAGGAATAGACC | 137 |
| RNF4 | AAGGCACCAAAGAGCACAAT | TTCATCTCCAGCAGTTTCCA | 159 |
| TIF1 | AGTCATTCGTTGCCCAGTTT | CTTCTGCGTTGTCCTCACAG | 152 |
| TRIM28 | AATGATGCCCAGAAGGTGAC | TTGAGGTCCCACTGAAACTT | 245 |
| WEE1 | TGAAGAGGGCGATAGTCGTT | GCACTTGTGGTATCCGAGGT | 188 |
Transcript, gene . | Forward primer 5′ 3′ . | Reverse primer 5′ 3′ . | Amplicon size, bp . |
|---|---|---|---|
| CDK6 | CACGAACAGACAGAGAACCA | CGGTGTGAATGAAGAAAGTCC | 169 |
| CENPF | AGAACCCACCACGAAATCC | TGCCTTCACTGGACCTTACA | 169 |
| ETS2 | CCAAGAGAGGAGTGACCCAG | CAGGAGAAACTGCCACAGC | 114 |
| FB29 | CTTGGGTCAGAGCAGAGGTC | CAGCAGGTTTGGAACTGGA | 150 |
| FB83 | TCCAGCAACCTTCCATCTCT | TGAGCCAAGAAAGCCATC | 184 |
| FB136 | TTCCCAAAGTAACAGCCAGTC | TTCATCCTCAGTTTCACCAAA | 152 |
| FB248 | TCACTTTATCATCAGCCAGCA | TTCTTGGTCTCGCCTACTCC | 159 |
| GAPDH | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAC | 100 |
| HMBS | GGCAATGCGGCTGCAA | GGGTACCCACGCGAATCAG | 80 |
| LMO2 | TACTGGCACGAGGACTGC | CTTTCACCCGCATTGTCAT | 190 |
| LZTF1 | TCTACTCAGGGTTCAGGAGCA | TTGATTTGGTCATTCTTCTTGG | 120 |
| MTCO-1 | CCTGACTGGCATTGTATTAGC | TTTTGGCGTAGGTTTGGTCT | 179 |
| MYCN | TTGTGTGTTCCAAGTTTCCAA | CCACCTCTCATTACCCAGGA | 185 |
| NFKB1 | TGAGTCCTGCTCCTTCCAA | CTTCGGTGTAGCCCATTTGT | 105 |
| Rac1 | GTGAATCTGGGCTTATGGGA | GATGGGAGTGTTGGGACAGT | 189 |
| RALBP1 | GAAGCAGTATTTGCGAGACC | TGGCAGTTCTTTGAGTAAACG | 133 |
| RCC1 | TGGGTGAGAATGTGATGGAG | GCAGCCGAAGGAATAGACC | 137 |
| RNF4 | AAGGCACCAAAGAGCACAAT | TTCATCTCCAGCAGTTTCCA | 159 |
| TIF1 | AGTCATTCGTTGCCCAGTTT | CTTCTGCGTTGTCCTCACAG | 152 |
| TRIM28 | AATGATGCCCAGAAGGTGAC | TTGAGGTCCCACTGAAACTT | 245 |
| WEE1 | TGAAGAGGGCGATAGTCGTT | GCACTTGTGGTATCCGAGGT | 188 |
DNA sequencing and analysis
The recombinant DNA samples were sequenced (Sequencing Facility, Robarts Research Institute, London, ON, Canada) using M13 reverse and/or forward primers from the predetermined sequence in the vector. The sequences were analyzed using McVector 6.5.1 program (Oxford Molecular Group, Cambridge, United Kingdom).
Results
Development of cDNA libraries from purified Lin-CD34+CD38- cells derived from FB- and MPB-HSPCs
Distinct functional characteristics of FB-HSPCs compared with adult MPB-HSPCs8,9 prompted us to identify the molecular basis of these ontogenic differences. SSH has been shown by several investigations19-21 as one of the efficient approaches to study gene expression in closely related subpopulations of cells in which equally expressed transcripts are removed during the process of hybridization.15 However, one of the major difficulties in performing SSH in enriched HSPCs is the inability to obtain a sufficient number of cells to generate an adequate amount of cDNA using standard procedures. We overcame this limitation by amplification of first-strand cDNA, as was done in previous studies,16,22,23 in which the relative abundance of the primary transcripts is maintained during the amplification process. Given the distinct characteristics of FB-HSPC and its corresponding population of adult MPB-HSPC with identical cell surface phenotype, we used amplified cDNA of MPB-HSPCs as driver in SSH to remove genes that are equally expressed in both populations and enrich for transcripts that are differentially expressed in tester template generated from FB-HSPCs.
To ensure the purity of the populations of interest in FB and MPB, we used stringent selection gates to isolate the Lin-CD34+CD38- fraction from both sources (Figure 2Ai,iii). Upon reanalysis, these populations demonstrated as high as 99% purity (Figure 2Aii,iv), and preparations that were below 96% purity upon reanalysis were discarded and not used to generate cDNA template. Using an optimized cDNA amplification approach,16 we were able to generate up to 7 mg amplified cDNA, ranging from 7 kilobase (kb) to 0.5 kb in size (data not shown) from as few as 1 × 105 purified Lin-CD34+CD38- cells derived from FB or MPB sources. To evaluate the maintenance and representation of transcripts in the amplified preparations, expression of β-actin, as one of the highly expressed housekeeping genes,24 and β-glucoronidase, as a housekeeping gene expressed at a single transcript per cell,25 was determined by PCR. In addition, the size of β-glucoronidase amplicon (1.0 kb) is an indication of the integrity of amplified template. Using serial dilutions of amplified cDNA, β-actin and β-glucoronidase were detectable in both populations using as little as 0.07 ng of the amplified transcript (Figure 2B), indicating efficient cDNA amplification that has maintained the authenticity of the primary transcripts after several rounds of amplification. Based on these positive criteria, templates from MPB and FB Lin-CD34+CD38- cells representing HSPCs was used as driver and tester, respectively, in SSH reactions.
Purification of HSPC populations from FB and MPB. (A) Lineage-depleted population from several fetal blood (n = 10) and MPB (n = 6) samples were pooled separately and stained with CD34 antibody conjugated to APC and CD38 antibody conjugated to PE. Using a stringent sorting gate (R1), the CD34+CD38- population in FB (i) and MPB (iii) were sorted and reanalysis of the sorted populations ensured the high purity of FB Lin-CD34+CD38- (ii) and MPB Lin-CD34+CD38- (iv) cells. (B) Amplification of β-actin and β-glucoronidase to verify the integrity and maintenance of the abundant of primary transcripts after cDNA amplification in FB Lin-CD34+CD38- and MPB Lin-CD34+CD38-. Different concentrations (7 ng, 0.7 ng, and 0.07 ng) of amplified cDNA from FB Lin-CD34+CD38- and MPB Lin-CD34+CD38- were used as the starting material in reverse transcriptase–PCR (RT-PCR) for the amplification of β-actin, as one of the highly expressed housekeeping genes, and β-glucoronidase, as one of the genes present as a single transcript per cell.
Purification of HSPC populations from FB and MPB. (A) Lineage-depleted population from several fetal blood (n = 10) and MPB (n = 6) samples were pooled separately and stained with CD34 antibody conjugated to APC and CD38 antibody conjugated to PE. Using a stringent sorting gate (R1), the CD34+CD38- population in FB (i) and MPB (iii) were sorted and reanalysis of the sorted populations ensured the high purity of FB Lin-CD34+CD38- (ii) and MPB Lin-CD34+CD38- (iv) cells. (B) Amplification of β-actin and β-glucoronidase to verify the integrity and maintenance of the abundant of primary transcripts after cDNA amplification in FB Lin-CD34+CD38- and MPB Lin-CD34+CD38-. Different concentrations (7 ng, 0.7 ng, and 0.07 ng) of amplified cDNA from FB Lin-CD34+CD38- and MPB Lin-CD34+CD38- were used as the starting material in reverse transcriptase–PCR (RT-PCR) for the amplification of β-actin, as one of the highly expressed housekeeping genes, and β-glucoronidase, as one of the genes present as a single transcript per cell.
Verification of SSH reactions between FB-HSPCs and MPB-HSPCs
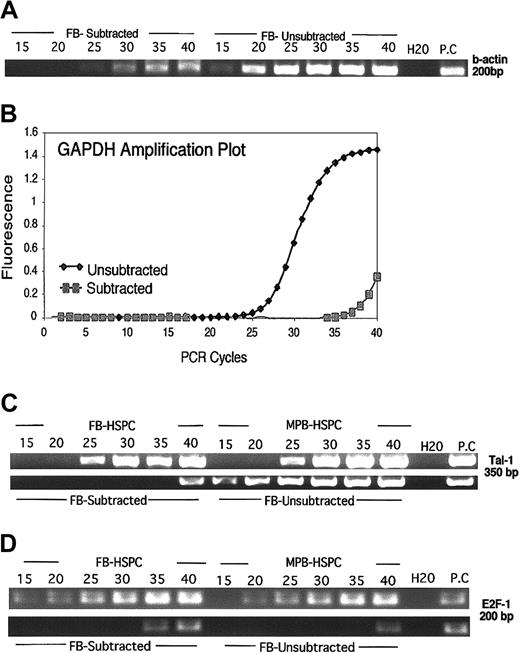
In order to verify the efficiency of SSH reactions, the abundance of equally expressed transcripts in starting populations of FB-HSPCs and MPB-HSPCs from unsubtracted control reactions was compared with subtracted (SSH) reactions. Ideally, an efficient SSH reaction would allow the majority of common transcripts to be removed and would, therefore, require a higher number of PCR cycles to amplify and detect residual common transcripts within subtracted cDNA compared with the unsubtracted cDNA. Using semiquantitative PCR, amplified products from subtracted and unsubtracted samples were first analyzed for the abundant housekeeping transcript β-actin from 15 cycles PCR up to 40 cycles in 5-cycle intervals. As illustrated in Figure 3A, the presence of β-actin is observed after 15 cycles in the unsubtracted sample, whereas it is only clearly detectable in the subtracted template at 30 cycles. This difference in detectability indicated that the SSH was extremely efficient to remove equally expressed genes in both populations.
Efficient SSH between FB-HSPCs versus MPB-HSPCs. The amplified cDNA of FB-HSPCs was subtracted from excess amount of MPB-HSPCs in a 2-step hybridization at 68°C to remove equally expressed genes. The differentially regulated genes in subtracted and unsubtracted libraries were amplified in suppression and nested PCRs. (A) In order to evaluate the quality of subtracted library, 50 ng of FB-subtracted and FB-unsubtracted DNA was used as a template to assess β-actin amplification in a semiquantitative PCR. An aliquot (7 mL) of amplified product was taken at 15 cycles and after every 5 cycles to 40 cycles followed by 2% agarose gel fractionation to estimate the abundance of β-actin at different cycles. (B) Q-PCR analysis of GAPDH, as one of the other housekeeping genes, verified the quality of FB-subtracted by using 10 ng of DNA from FB-subtracted (Ct = 36; ▦) and FB-unsubtracted (Ct = 23; ♦) libraries. (C) Amplification of Tal-1 as one of the key transcription factors involved in the occurrence of hematopoiesis. Semiquantitative PCR, the same methodology explained in Figure 2A, using 20 ng of amplified cDNA from FB-HSPCs and MPB-HSPCs, indicated equal expression of Tal-1 in both populations (Figure 2C top). In the FB-subtracted library, however, using 50 ng of DNA from FB-subtracted and FB-unsubtracted libraries, Tal-1 amplicon is detectable after 40 cycles in the FB-subtracted library as opposed to its appearance only after 15 cycles in the FB-unsubtracted library. (D) Amplification of E2F1 as one of the critical transcription factors involved in the cycling cells indicated a moderately higher expression in FB-HSPCs than MPB-HSPCs (Figure 2D top). In a semiquantitative approach, using 50 ng DNA as template, E2F1 is detectable in FB-HSPCs after 15 cycles but is present in MPB-HSPCs after 20 cycles. However, in an efficient SSH (Figure 2D bottom), using 100 ng DNA of subtracted and unsubtracted libraries, E2F1 is detected after 35 cycles in FB-subtracted but is observed at 40 cycles in the FB-unsubtracted library, indicating efficient SSH to enrich differentially regulated genes in FB-HSPCs.
Efficient SSH between FB-HSPCs versus MPB-HSPCs. The amplified cDNA of FB-HSPCs was subtracted from excess amount of MPB-HSPCs in a 2-step hybridization at 68°C to remove equally expressed genes. The differentially regulated genes in subtracted and unsubtracted libraries were amplified in suppression and nested PCRs. (A) In order to evaluate the quality of subtracted library, 50 ng of FB-subtracted and FB-unsubtracted DNA was used as a template to assess β-actin amplification in a semiquantitative PCR. An aliquot (7 mL) of amplified product was taken at 15 cycles and after every 5 cycles to 40 cycles followed by 2% agarose gel fractionation to estimate the abundance of β-actin at different cycles. (B) Q-PCR analysis of GAPDH, as one of the other housekeeping genes, verified the quality of FB-subtracted by using 10 ng of DNA from FB-subtracted (Ct = 36; ▦) and FB-unsubtracted (Ct = 23; ♦) libraries. (C) Amplification of Tal-1 as one of the key transcription factors involved in the occurrence of hematopoiesis. Semiquantitative PCR, the same methodology explained in Figure 2A, using 20 ng of amplified cDNA from FB-HSPCs and MPB-HSPCs, indicated equal expression of Tal-1 in both populations (Figure 2C top). In the FB-subtracted library, however, using 50 ng of DNA from FB-subtracted and FB-unsubtracted libraries, Tal-1 amplicon is detectable after 40 cycles in the FB-subtracted library as opposed to its appearance only after 15 cycles in the FB-unsubtracted library. (D) Amplification of E2F1 as one of the critical transcription factors involved in the cycling cells indicated a moderately higher expression in FB-HSPCs than MPB-HSPCs (Figure 2D top). In a semiquantitative approach, using 50 ng DNA as template, E2F1 is detectable in FB-HSPCs after 15 cycles but is present in MPB-HSPCs after 20 cycles. However, in an efficient SSH (Figure 2D bottom), using 100 ng DNA of subtracted and unsubtracted libraries, E2F1 is detected after 35 cycles in FB-subtracted but is observed at 40 cycles in the FB-unsubtracted library, indicating efficient SSH to enrich differentially regulated genes in FB-HSPCs.
To complement this analysis, Q-PCR was used to compare the expression level of another highly abundant housekeeping gene, glycerol aldehyde phosphate dehydrogenase (GAPDH),24 that is equally expressed in both FB-HSPCs and MPB-HSPCs (Figures 4B,5C), in subtracted and unsubtracted template cDNA (Figure 3B). The cycle threshold (Ct) for GAPDH in the unsubtracted sample was observed to be 26 cycles as opposed to 37 cycles in the subtracted sample, thereby providing additional evidence for the efficiency of SSH of MPB-HSPCs from FB-HSPCs. Similar observations were made by comparing the relative abundance of hematopoietic-specific transcripts for lower expressed genes, such as the transcription factor Tal-126 involved in definitive hematopoiesis.27 Tal-1 was shown to be equally expressed in starting populations of Lin-CD34+CD38- cells derived from FB and MPB (Figure 3C top) but could only be detected after 40 cycles of PCR in the subtracted cDNA (Figure 3C bottom). Conversely, efficient SSH would allow for more abundantly expressed genes from tester FB-HSPCs to be detected by using lower numbers of PCR cycles. E2F1 is a transcription factor shown to be more highly expressed in BM CD34+ than MPB CD34+.28 The significance of E2F1 in cycling status of human hematopoietic cells has also been demonstrated.29,30 Since FB-HSPCs differ from MPB-HSPCs based on cell cycle status, we examined expression of E2F1 in FB-HSPCs versus MPB-HSPCs. E2F1 was found to be more highly expressed in FB-HSPCs than MPB-HSPCs (Figure 3D top) using semiquantitative PCR. Using DNA template from FB-HSPC–subtracted libraries, E2F1 is detectable only after 35 cycles compared with 40 cycles in unsubtracted sample (Figure 3D bottom), indicating the enrichment of E2F1 in FB-HSPCs. Analysis of Tal-1 and E2F1 from subtracted libraries further confirms that the resulting FB-HSPC–subtracted library is enriched in differentially expressed genes. Taken together, these results indicate that the resulting subtracted cDNA is highly enriched for transcripts expressed differentially in purified FB-HSPCs compared with its adult counterpart derived from MPB-HSPCs, since the hematopoietic transcription factors common to both adult and fetal human HSPCs have been efficiently removed. SSH was repeated 2 more times to ensure the quality and reproducibility of the subtracted library through amplification profile of β-actin and E2F1 (data not shown).
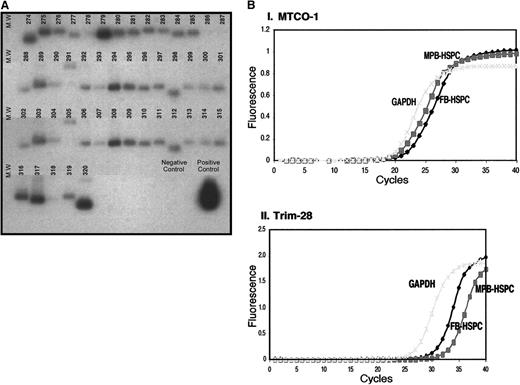
Screening of colonies in the FB-subtracted library using southern hybridization. (A) The blot of FB274 to FB320 is shown as a representative to explain the screening strategy to isolate colonies of FB-subtracted library for further analysis. The nested PCR products of FB-subtracted library were ligated to TA vector and transformed into DH5a cells. Bacterial plasmids were extracted, restriction digested with EcoRI, and gel fractionated on 1.5% agarose gel. Plasmid DNA were then transferred to nylon membrane and hybridized with MPB-HSCs amplified cDNA that was subsequently digested with RsaI and used as a probe. Colonies with the least signal or no hybridization signal were selected for further analysis. (B) Verification of the screening strategy. One of the colonies, MTCO-1, that showed a high hybridization signal with the cDNA of MPB-HSPCs was selected to further analyze its expression in FB-HSPCs and MPB-HSPCs in a Q-PCR approach. (Bi) Using 400 pg of amplified cDNA of FB-HSPCs and MPB-HSPCs, it is shown that there is not dramatic difference between the expression profile of MTCO-1 in both populations (FB-HSPCs Ct = 21.0 and MPB-HSPCs Ct = 20.5). (Bii) However, Trim-28 as one of the colonies with low hybridization signal with cDNA of MPB-HSPCs turned out to be expressed more in FB-HSPCs (Ct = 26.5) than MPB-HSPCs (Ct = 29.0).
Screening of colonies in the FB-subtracted library using southern hybridization. (A) The blot of FB274 to FB320 is shown as a representative to explain the screening strategy to isolate colonies of FB-subtracted library for further analysis. The nested PCR products of FB-subtracted library were ligated to TA vector and transformed into DH5a cells. Bacterial plasmids were extracted, restriction digested with EcoRI, and gel fractionated on 1.5% agarose gel. Plasmid DNA were then transferred to nylon membrane and hybridized with MPB-HSCs amplified cDNA that was subsequently digested with RsaI and used as a probe. Colonies with the least signal or no hybridization signal were selected for further analysis. (B) Verification of the screening strategy. One of the colonies, MTCO-1, that showed a high hybridization signal with the cDNA of MPB-HSPCs was selected to further analyze its expression in FB-HSPCs and MPB-HSPCs in a Q-PCR approach. (Bi) Using 400 pg of amplified cDNA of FB-HSPCs and MPB-HSPCs, it is shown that there is not dramatic difference between the expression profile of MTCO-1 in both populations (FB-HSPCs Ct = 21.0 and MPB-HSPCs Ct = 20.5). (Bii) However, Trim-28 as one of the colonies with low hybridization signal with cDNA of MPB-HSPCs turned out to be expressed more in FB-HSPCs (Ct = 26.5) than MPB-HSPCs (Ct = 29.0).
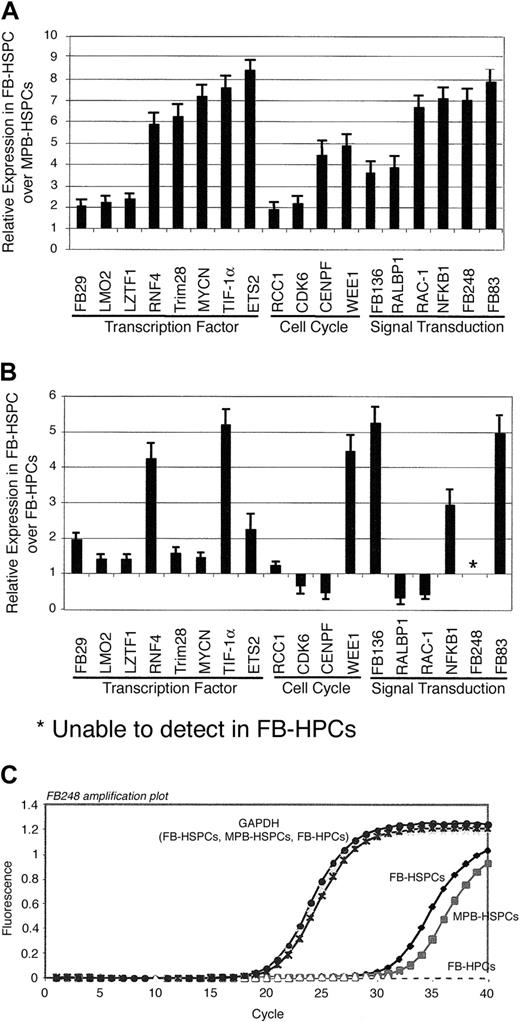
Quantitative measurement of selected differentially regulated genes in FB-HSPCs versus MPB-HSPCs and FB-HPCs. (A) The cDNA of pooled FB-HSPCs and MPB-HSPCs was used in Q-PCR to evaluate the expression level of some of the differentially regulated genes in the group of transcription factors, cell cycle regulators, and signal transduction genes. Q-PCR reactions were performed in duplicate and GAPDH was used as an internal control in Q-PCR reactions, and the relative expression of these genes in FB-HSPCs versus MPB-HSPCs was calculated by 2-ΔΔCt equation (“Materials and methods”). Error bars indicate standard errors. (B) The same methodology was used to calculate the relative expression of the selected transcripts in FB-HSPCs versus FB-HPCs. *Lack of expression in FB-HPCs. (C) Amplification plot of FB248 as the transcript exclusively expressed in HSPC populations but not in FB-HPC. GAPDH amplification is shown as an internal control for Q-PCR reaction. However, FB248 amplification demonstrates the presence of amplicon in FB-HSPCs (Ct = 28.5) and MPB-HSPCs (Ct = 31.0) but not in FB-HPCs.
Quantitative measurement of selected differentially regulated genes in FB-HSPCs versus MPB-HSPCs and FB-HPCs. (A) The cDNA of pooled FB-HSPCs and MPB-HSPCs was used in Q-PCR to evaluate the expression level of some of the differentially regulated genes in the group of transcription factors, cell cycle regulators, and signal transduction genes. Q-PCR reactions were performed in duplicate and GAPDH was used as an internal control in Q-PCR reactions, and the relative expression of these genes in FB-HSPCs versus MPB-HSPCs was calculated by 2-ΔΔCt equation (“Materials and methods”). Error bars indicate standard errors. (B) The same methodology was used to calculate the relative expression of the selected transcripts in FB-HSPCs versus FB-HPCs. *Lack of expression in FB-HPCs. (C) Amplification plot of FB248 as the transcript exclusively expressed in HSPC populations but not in FB-HPC. GAPDH amplification is shown as an internal control for Q-PCR reaction. However, FB248 amplification demonstrates the presence of amplicon in FB-HSPCs (Ct = 28.5) and MPB-HSPCs (Ct = 31.0) but not in FB-HPCs.
Sequence analysis and annotation of differentially expressed transcripts derived from SSH of FB-HSPCs from MPB-HSPCs
To characterize individual transcripts differentially expressed in FB-HSPCs, amplified cDNA of MPB-HSPCs (RsaI digested) was used to probe colonies generated by subcloning FB-HSPC–subtracted transcripts into bacterial clones. Plasmid DNA was prepared from individual colonies containing FB-HSPC–inserted transcripts, electrophoresed, and transferred to membranes for hybridization against labeled cDNA generated from purified adult MPB-HSPCs. A representative example of 47 clones from 320 screened is shown in Figure 4A. Inserts with low hybridization signal were selected for further sequence analysis based on their relative low expression in MPB-HSPCs, suggesting they were unique to FB-HSPCs.
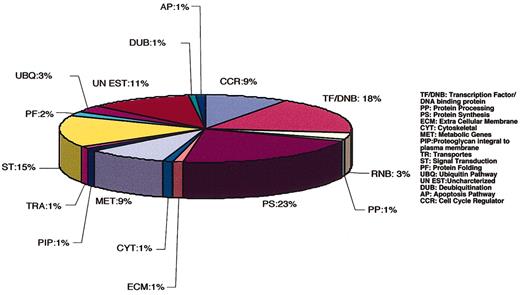
To demonstrate the efficiency of this screening and selection method to identify clones containing genes highly enriched in FB-HSPC versus MPB-HSPC, Q-PCR was used on representative clones given high (cytochrome c oxidase-I, MTCO-1) and low Trim28 signal after hybridization with labeled cDNA from MPB-HSPCs. Q-PCR results indicated that there is no difference in expression of MTCO-1 in freshly purified FB-HSPC versus MPB-HSPC (Figure 4Bi), whereas expression of Trim28 was higher in FB-HSPCs (Figure 4Bii), thereby validating our clone selection criteria. Using this strategy, 97 clones from 320 indicated low signal upon hybridization with labeled MPB-HSPCs (Table 2) and were subsequently selected and sequenced. The BLAST (http://www.ncbi.nlm.nih.gov/genome/seq/page.cgi?F=HsBlast.html&&ORG=Hs) analysis identified the closest transcript (gene) in human genome, and the SMART program (http://www.ncbi.nlm.nih.gov/BLAST) showed the presence of domain structure(s), if any, in the amino acid sequences of the selected transcripts/genes. Using Gene Ontology Consortium (http://www.Geneontology.org/) and search of the available literature we categorized identified genes that led to the annotation of differentially expressed transcripts into 16 groups, including DNA binding, cell cycle regulator, protein synthesis, and transporters (Figure 6).
Annotation of sequenced clones identified from the FB-subtracted library
Clone . | Accession no. . | Name of gene . | Annotation . | Domain(s) . |
|---|---|---|---|---|
| FB11 | BC034041 | Homo sapiens, LIM domain only 2 | TF/DNB | LIM |
| FB271 | BC042483 | Homo sapiens, leucine zipper transcription factor-like 1 | TF/DNB | LIM |
| FB70, 305 | NM_005378 | Homo sapiens MYCN mRNA | TF/DNB | HLH, PFAM |
| FB83, 197, 245 | BC028689 | Homo sapiens transcriptional intermediary factor 1 (TIF 1), mRNA | TF/DNB | BROMO, PHD, BBC, B-BOX, RING |
| FB63 | NM_005762 | Homo sapiens tripartite motif-containing 28 | TF/DNB | BROMO, PHD, BBC, B-BOX, RING |
| FB109, 194 | J04102 | Human ETS2 mRNA | TF/DNB | ETS |
| FB170 | NM_002938 | Homo sapiens ring finger protein 4 (RNF4), mRNA | TF/DNB | PFAM |
| FB291 | NM_007265 | Homo sapiens suppressor of S cerevisiae gcr2 (HSGT1), mRNA | TF/DNB | — |
| FB18 | AF060511 | Homo sapiens clone 016b10 My016 protein mRNA | TF/DNB | PFAM BoIA |
| FB183, 184 | AL832296 | EST | TF/DNB | HOX |
| FB29 | NM_015339 | Activity-dependent neuroprotective protein | TF/DNB | HOX, ZNF |
| FB225 | NM_014311 | Homo sapiens SMUG1 mRNA | TF/DNB | PFAM:UDG |
| FB2 | NM_001259 | Homo sapiens cyclin-dependent kinase 6 (CDK6) | CCR | S-TKc |
| FB91 | NM_003390 | Homo sapiens WEE1 mRNA for protein kinase | CCR | STYKc |
| FB111 | D00591 | Homo sapiens RCC1 gene | CCR | PFAM:RCC1 |
| FB25 | AL162078.1 | Homo sapiens mRNA; cDNA DKFZp761H229 | CCR | TPR |
| FB65 | AF189722 | Homo sapiens PDZ-binding kinase mRNA | CCR | STYKc |
| FB288 | NM_017943 | Homo sapiens F-box only protein 34 (FBXO34), mRNA | CCR | FBOX |
| FB251 | NM_016343 | Homo sapiens centromere protein F, 350/400ka (mitosin) (CENPF) | DNR | — |
| FB300 | AF007128 | 1-Homo sapiens clone 23870 mRNA | CCR | LRR, FBOX |
| FB128 | NM_138609 | Homo sapiens H2A histone family, member Y (H2AFY), transcript variant 1, mRNA | CRR | A1pp, H2A |
| FB100 | AF273028 | Homo sapiens NUDT10 mRNA | CRR | PFAM NUDIX |
| FB124, 240 | NM_018449 | Homo sapiens ubiquitin associated protein 2 (UBAP2), mRNA | UBQ | UBA |
| FB278 | BC019111 | Homo sapiens, sequestosome | UBQ | Contain UBA, Znf_zz and PB1 domains |
| FB145 | NM_005153 | Homo sapiens ubiquitin-specific protease 10 | DUB | PFAM |
| FB4, 33 | NM_004607 | Homo sapiens tubulin-specific chaperone a (TBCA) | PF | — |
| FB301 | NM_006788 | Homo sapiens raIA binding protein 1 (RALBP1), mRNA | ST | RhoGAP |
| FB205, 212 | AF498964 | Homo sapiens rac1 gene | ST | RHO |
| FB179, 250 | AF213884 | Homo sapiens nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (NFKB1) gene | ST | Death, ANK, PFAM:RHD, IPT |
| FB269 | AF111162 | Homo sapiens guanine nucleotide exchange factor mRNA | ST | Sec7 |
| FB48 | Y09160 | H sapiens Sub 1.5 mRNA | ST | RhoGEF, PH |
| FB114 | AF267853 | Homo sapiens HTPHLP mRNA | ST | Phosducin |
| FB44 | U50648 | Human interferon inducible RNA-dependent protein kinase | ST | DSRM, STYKc |
| FB26 | AF139768 | Homo sapiens type II transmembrane protein MDL-1 | ST | TM, CLECT |
| FB90, 96, 129, 136 | BC033731 | Homo sapiens, DIP13 beta | ST | PHD, PTB |
| FB248 | AL832326 | EST | ST | L27, GuKc, PDZ, SH3 |
| FB98 | BC030545 | EST | ST | PHD |
| FB5 | AK097395.1 | Highly similar to SUPEROXIDE DISMUTASE Homo sapiens, clone MGC | MET | PFAM |
| FB35 | AF339086 | Homo sapiens NADH dehydrogenase | MET | PFAM:oxidored |
| FB172, 181 | AY063361 | Homo sapiens isolate EICh03-98 NADH dehydrogenase subunit 4 gene | MET | PFAM:oxidored |
| FB184 | BC008906 | Homo sapiens, enoyl Coenzyme A hydratase | MET | PFAM:ECH |
| FB196 | NM_005918 | Homo sapiens malate dehydrogenase 2, NAD (mitochondrial) (MDH2) | MET | — |
| FB293 | NM_006708 | Homo sapiens glyoxalase I (GLO1), mRNA | MET | — |
| FB144 | NM_004595 | Homo sapiens spermine synthase | MET | PFAM:Spermine_synth |
| FB23, 38, 40, 226 | BC019669 | Homo sapiens, eukaryotic translation elongation factor 1 alpha 1 | PS | PFAM, GTP, EFTU, D2, D3 |
| FB171 | NM_021121 | Homo sapiens eukaryotic translation elongation factor 1 beta 2 (EEF1B2), transcript variant 2 | PS | PFAM:EF1BD |
| FB102 | NM_006397 | Homo sapiens ribonuclease H2, large subunit (RNASEH2A), mRNA | PS | — |
| FB22, 123 | NM_000990 | Homo sapiens ribosomal protein L27a | PS | — |
| FB6, 224 | BC027620 | Homo sapiens, ribosomal protein S6 | PS | PFAM: Ribosomal_S6e |
| FB139, 140 | BC029732 | Homo sapiens, ribosomal protein S15 | PS | PFAM:Ribosomal_S15 |
| FB265 | NM_000972 | Homo sapiens ribosomal protein L7a | PS | PFAM:Ribosomal_L7Ae |
| FB125 | NM_000971 | Homo sapiens ribosomal protein L7 (RPL7), mRNA | PS | Ribosomal_L30 |
| FB42 | NM_000996 | Homo sapiens ribosomal protein L35a (RPL35A), mRNA | PS | PFAM:Ribosomal_L35Ae |
| FB28 | NM_000981 | Homo sapiens ribosomal protein L19 | PS | PFAM:Ribosomal_L19e |
| FB81 | NM_007104 | Homo sapiens ribosomal protein L10a | PS | PFAM:Ribosomal_L1 |
| FB220 | BC008003 | Homo sapiens, ribosomal protein L3 | PS | PFAM:Ribosomal_L3 |
| FB203 | BC040610 | Homo sapiens, Similar to ribosomal protein L4 | PS | — |
| FB262 | X06705 | Human PLA-X mRNA | PS | PFAM:Ribosomal_L7Ae |
| FB286 | BC007755 | Homo sapiens, glycyl-tRNA synthetase | PS | PFAM |
| FB49 | NM_001109 | Homo sapiens a disintegrin and metalloproteinase domain 8 (ADAM8) mRNA | PIP | PFAM Reprolysin, Pfam:Pep 1 DISIN, AGR, EGF, TM |
| FB30 | NM_005200 | Homo sapiens cell matrix adhesion regulator | ECM | — |
| FB20 | AL121895 | Human DNA sequence contains EPB41L1 gene encoding the erythrocyte membrane protein band | CYT | B41 |
| FB313 | BC009357 | Homo sapiens, transgelin 2 | PP | CH, PFAM:calponin |
| FB259 | NM_003883 | Histon deacetylation3 (HDAC3) | AP | PFAM-Histon Deacetyl |
| FB290 | NM_001007 | Homo sapiens ribosomal protein S4, X-linked (RPS4X), mRNA | RNB | S4, PFAM:KOW |
| FB229 | NM_005839 | Homo sapiens serine/arginine repetitive matrix 1 (SRRM1), mRNA | RNB | PW1 |
| FB243 | NM_018834 | Homo sapiens matrin 3 (MATR3), mRNA | RNB | ZnFU1, RRM |
| FB126 | NM_014252 | Homo sapiens solute carrier family 25 (mitochondrial carrier; ornithine transporter) mRNA | TRA | PFAM:mito_carr |
| FB287 | AF151028 | Homo sapiens HSPC194 mRNA | Others | PFAM 0136 |
| FB307 | NM_016424 | Homo sapiens LUC7A protein | Others | PFAM DUF 259 |
| FB200 | XM_084664 | Eythroid differentiation-related factor | Others | TM |
| FB314 | AF210651 | Homo sapiens NAG18 (NAG18) mRNA | Others | — |
| FB209 | XM_058792 | Homo sapiens similar to nuclear pore complexinteracting protein | Others | — |
| FB302 | BC031346 | Homo sapiens, clone IMAGE mRNA | Others | — |
| FB304 | AL832781 | Homo sapiens mRNA | Others | — |
| FB318 | AF225417 | Homo sapiens 88.8 kDa protein mRNA | Others | — |
| FB299 | XM_058813 | Homo sapiens similar to RIKEN cDNA | Others | — |
| FB131 | BC014508 | Homo sapiens similar to signal peptidase complex | Others | PFAM:Peptidase_S26 |
Clone . | Accession no. . | Name of gene . | Annotation . | Domain(s) . |
|---|---|---|---|---|
| FB11 | BC034041 | Homo sapiens, LIM domain only 2 | TF/DNB | LIM |
| FB271 | BC042483 | Homo sapiens, leucine zipper transcription factor-like 1 | TF/DNB | LIM |
| FB70, 305 | NM_005378 | Homo sapiens MYCN mRNA | TF/DNB | HLH, PFAM |
| FB83, 197, 245 | BC028689 | Homo sapiens transcriptional intermediary factor 1 (TIF 1), mRNA | TF/DNB | BROMO, PHD, BBC, B-BOX, RING |
| FB63 | NM_005762 | Homo sapiens tripartite motif-containing 28 | TF/DNB | BROMO, PHD, BBC, B-BOX, RING |
| FB109, 194 | J04102 | Human ETS2 mRNA | TF/DNB | ETS |
| FB170 | NM_002938 | Homo sapiens ring finger protein 4 (RNF4), mRNA | TF/DNB | PFAM |
| FB291 | NM_007265 | Homo sapiens suppressor of S cerevisiae gcr2 (HSGT1), mRNA | TF/DNB | — |
| FB18 | AF060511 | Homo sapiens clone 016b10 My016 protein mRNA | TF/DNB | PFAM BoIA |
| FB183, 184 | AL832296 | EST | TF/DNB | HOX |
| FB29 | NM_015339 | Activity-dependent neuroprotective protein | TF/DNB | HOX, ZNF |
| FB225 | NM_014311 | Homo sapiens SMUG1 mRNA | TF/DNB | PFAM:UDG |
| FB2 | NM_001259 | Homo sapiens cyclin-dependent kinase 6 (CDK6) | CCR | S-TKc |
| FB91 | NM_003390 | Homo sapiens WEE1 mRNA for protein kinase | CCR | STYKc |
| FB111 | D00591 | Homo sapiens RCC1 gene | CCR | PFAM:RCC1 |
| FB25 | AL162078.1 | Homo sapiens mRNA; cDNA DKFZp761H229 | CCR | TPR |
| FB65 | AF189722 | Homo sapiens PDZ-binding kinase mRNA | CCR | STYKc |
| FB288 | NM_017943 | Homo sapiens F-box only protein 34 (FBXO34), mRNA | CCR | FBOX |
| FB251 | NM_016343 | Homo sapiens centromere protein F, 350/400ka (mitosin) (CENPF) | DNR | — |
| FB300 | AF007128 | 1-Homo sapiens clone 23870 mRNA | CCR | LRR, FBOX |
| FB128 | NM_138609 | Homo sapiens H2A histone family, member Y (H2AFY), transcript variant 1, mRNA | CRR | A1pp, H2A |
| FB100 | AF273028 | Homo sapiens NUDT10 mRNA | CRR | PFAM NUDIX |
| FB124, 240 | NM_018449 | Homo sapiens ubiquitin associated protein 2 (UBAP2), mRNA | UBQ | UBA |
| FB278 | BC019111 | Homo sapiens, sequestosome | UBQ | Contain UBA, Znf_zz and PB1 domains |
| FB145 | NM_005153 | Homo sapiens ubiquitin-specific protease 10 | DUB | PFAM |
| FB4, 33 | NM_004607 | Homo sapiens tubulin-specific chaperone a (TBCA) | PF | — |
| FB301 | NM_006788 | Homo sapiens raIA binding protein 1 (RALBP1), mRNA | ST | RhoGAP |
| FB205, 212 | AF498964 | Homo sapiens rac1 gene | ST | RHO |
| FB179, 250 | AF213884 | Homo sapiens nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (NFKB1) gene | ST | Death, ANK, PFAM:RHD, IPT |
| FB269 | AF111162 | Homo sapiens guanine nucleotide exchange factor mRNA | ST | Sec7 |
| FB48 | Y09160 | H sapiens Sub 1.5 mRNA | ST | RhoGEF, PH |
| FB114 | AF267853 | Homo sapiens HTPHLP mRNA | ST | Phosducin |
| FB44 | U50648 | Human interferon inducible RNA-dependent protein kinase | ST | DSRM, STYKc |
| FB26 | AF139768 | Homo sapiens type II transmembrane protein MDL-1 | ST | TM, CLECT |
| FB90, 96, 129, 136 | BC033731 | Homo sapiens, DIP13 beta | ST | PHD, PTB |
| FB248 | AL832326 | EST | ST | L27, GuKc, PDZ, SH3 |
| FB98 | BC030545 | EST | ST | PHD |
| FB5 | AK097395.1 | Highly similar to SUPEROXIDE DISMUTASE Homo sapiens, clone MGC | MET | PFAM |
| FB35 | AF339086 | Homo sapiens NADH dehydrogenase | MET | PFAM:oxidored |
| FB172, 181 | AY063361 | Homo sapiens isolate EICh03-98 NADH dehydrogenase subunit 4 gene | MET | PFAM:oxidored |
| FB184 | BC008906 | Homo sapiens, enoyl Coenzyme A hydratase | MET | PFAM:ECH |
| FB196 | NM_005918 | Homo sapiens malate dehydrogenase 2, NAD (mitochondrial) (MDH2) | MET | — |
| FB293 | NM_006708 | Homo sapiens glyoxalase I (GLO1), mRNA | MET | — |
| FB144 | NM_004595 | Homo sapiens spermine synthase | MET | PFAM:Spermine_synth |
| FB23, 38, 40, 226 | BC019669 | Homo sapiens, eukaryotic translation elongation factor 1 alpha 1 | PS | PFAM, GTP, EFTU, D2, D3 |
| FB171 | NM_021121 | Homo sapiens eukaryotic translation elongation factor 1 beta 2 (EEF1B2), transcript variant 2 | PS | PFAM:EF1BD |
| FB102 | NM_006397 | Homo sapiens ribonuclease H2, large subunit (RNASEH2A), mRNA | PS | — |
| FB22, 123 | NM_000990 | Homo sapiens ribosomal protein L27a | PS | — |
| FB6, 224 | BC027620 | Homo sapiens, ribosomal protein S6 | PS | PFAM: Ribosomal_S6e |
| FB139, 140 | BC029732 | Homo sapiens, ribosomal protein S15 | PS | PFAM:Ribosomal_S15 |
| FB265 | NM_000972 | Homo sapiens ribosomal protein L7a | PS | PFAM:Ribosomal_L7Ae |
| FB125 | NM_000971 | Homo sapiens ribosomal protein L7 (RPL7), mRNA | PS | Ribosomal_L30 |
| FB42 | NM_000996 | Homo sapiens ribosomal protein L35a (RPL35A), mRNA | PS | PFAM:Ribosomal_L35Ae |
| FB28 | NM_000981 | Homo sapiens ribosomal protein L19 | PS | PFAM:Ribosomal_L19e |
| FB81 | NM_007104 | Homo sapiens ribosomal protein L10a | PS | PFAM:Ribosomal_L1 |
| FB220 | BC008003 | Homo sapiens, ribosomal protein L3 | PS | PFAM:Ribosomal_L3 |
| FB203 | BC040610 | Homo sapiens, Similar to ribosomal protein L4 | PS | — |
| FB262 | X06705 | Human PLA-X mRNA | PS | PFAM:Ribosomal_L7Ae |
| FB286 | BC007755 | Homo sapiens, glycyl-tRNA synthetase | PS | PFAM |
| FB49 | NM_001109 | Homo sapiens a disintegrin and metalloproteinase domain 8 (ADAM8) mRNA | PIP | PFAM Reprolysin, Pfam:Pep 1 DISIN, AGR, EGF, TM |
| FB30 | NM_005200 | Homo sapiens cell matrix adhesion regulator | ECM | — |
| FB20 | AL121895 | Human DNA sequence contains EPB41L1 gene encoding the erythrocyte membrane protein band | CYT | B41 |
| FB313 | BC009357 | Homo sapiens, transgelin 2 | PP | CH, PFAM:calponin |
| FB259 | NM_003883 | Histon deacetylation3 (HDAC3) | AP | PFAM-Histon Deacetyl |
| FB290 | NM_001007 | Homo sapiens ribosomal protein S4, X-linked (RPS4X), mRNA | RNB | S4, PFAM:KOW |
| FB229 | NM_005839 | Homo sapiens serine/arginine repetitive matrix 1 (SRRM1), mRNA | RNB | PW1 |
| FB243 | NM_018834 | Homo sapiens matrin 3 (MATR3), mRNA | RNB | ZnFU1, RRM |
| FB126 | NM_014252 | Homo sapiens solute carrier family 25 (mitochondrial carrier; ornithine transporter) mRNA | TRA | PFAM:mito_carr |
| FB287 | AF151028 | Homo sapiens HSPC194 mRNA | Others | PFAM 0136 |
| FB307 | NM_016424 | Homo sapiens LUC7A protein | Others | PFAM DUF 259 |
| FB200 | XM_084664 | Eythroid differentiation-related factor | Others | TM |
| FB314 | AF210651 | Homo sapiens NAG18 (NAG18) mRNA | Others | — |
| FB209 | XM_058792 | Homo sapiens similar to nuclear pore complexinteracting protein | Others | — |
| FB302 | BC031346 | Homo sapiens, clone IMAGE mRNA | Others | — |
| FB304 | AL832781 | Homo sapiens mRNA | Others | — |
| FB318 | AF225417 | Homo sapiens 88.8 kDa protein mRNA | Others | — |
| FB299 | XM_058813 | Homo sapiens similar to RIKEN cDNA | Others | — |
| FB131 | BC014508 | Homo sapiens similar to signal peptidase complex | Others | PFAM:Peptidase_S26 |
The National Center for Biotechnology Information (NCBI) was used to identify the closest gene/transcript to the query. The SMART program was used to identify the presence of domain structure in some of the transcripts and queries. The data were annotated based on the information gained from NCBI, SMART, Gene consortium ontology, and literature search.
LIM indicates Zinc-binding domain present in Lin 11, IsI-1, and Mec-3; TF/DNB, transcription factor/DNA binding protein; HLH, helix-loop-helix; PFAM, a comprehensive database of protein domain families based on seed alignments; BROMO, bromo domain; PHD, pleckstrin homology domain; BBC, B-Box C-terminal domain; B-BOX, B-Box–type zinc finger; RING, Ring finger domain; ETS, erythroblast transformation–specific domain, http://smart.embl-heidelberg.de; BoIA, a receptor for ubiquitination targets; ZNF, zinc finger protein; UDG, uracil DNA glycosylase; CRR, chromatin regulator; S-KTc, serine threonine protein kinase; STYKc, protein kinase, unclassified specificity; TPR, tetratricopeptide repeats; F-BOX, a receptor for ubiquitination targets; DNR, DNA repair; LRR, leucine-rich repeat; A1pp,Appr-1′-p processing enzyme; H2A, histone 2A; NUDIX, http://smart.embl-heidelberg.de; UBQ, ubiquitin pathway; UBA, ubiquitin-associated; Znf_zz, zinc-binding domain present in dystrophin CREB-binding protein; PB1, PB1 domain; DUB, deubiquitination; PF, protein folding; ST, signal transduction; RhoGAP, GTPase-activator protein for Rho-like GTPases; RHO, Rho (Ras homology) subfamily of Ras-like small GTPases; ANK, ankyrin; RHD, see http://smart.embl-heidelberg.de; IPT, Ig-like, plexins, transcription factors; Sec7, Sec7 domain; RhoGEF, guanine nucleotide exchange factor for Rho/Rac/Cdc42-motif; DSRM, double-stranded RNA binding motif; TM, transmembrane domain; CLECT, C-type lectin (CTL) or carbohydrate-recognition domain (CRD); MET, metabolic genes; ECH, see http://smart.embl-heidelberg.de; PS, protein synthesis; EETU, see http://smart.embl-heidelberg.de; PIP, proteoglycan integral to plasma membrane; DISIN, homologs of snake disintegrins; AGR, see http://smart.embl-heidelberg.de; EGF, epidermal growth factor-like domain; ECM, cell adhesion and extracellular matrix; CYT, cytoskeletal; PP, protein processing; CH, calponin homology domain; AP, apoptosis; S4, S4 RNA binding domain; KOW, Kyprides Ouzonis Woese motif; PW1, PW1 domain in splicing factors; ZnFU1, U1-like zinc finger; RRM, RNA recognition motif; TRA, transporter; and –, no known domain associated with clone.
Illustration of annotated genes identified in FB-HSPC–subtracted library. Blast analysis (http://www.ncbi.nlm.nih.gov/genome/seq/page.cgi?F=HsBlast.html&&ORG=Hs) was used to identify the closest transcript in the gene bank, and SMART program (http://www.ncbi.nlm.nih.gov/BLAST) identified the presence of domain(s) in the amino acid sequences of the selected transcripts. Gene Ontology Consortium (http://www.Geneontology.org/) and literature search were used to annotate selected transcripts differentially expressed in FB-HSPCs into 16 groups shown in a pie diagram. CCR indicates cell cycle regulator; TF/DNB, transcription factor/DNA binding protein; RNB, RNA binding; PP, protein processing; PS, protein synthesis; ECM, extracellular membrane; CYT, cytoskeletal; MET, metabolic genes; PIP, proteoglycan integral to plasma membrane; TRA, transports; ST, signal transduction; PF, protein folding; UBQ, ubiquitin pathway; UN EST, uncharacterized; DUB, deubiquitination; and AP, apoptosis pathway.
Illustration of annotated genes identified in FB-HSPC–subtracted library. Blast analysis (http://www.ncbi.nlm.nih.gov/genome/seq/page.cgi?F=HsBlast.html&&ORG=Hs) was used to identify the closest transcript in the gene bank, and SMART program (http://www.ncbi.nlm.nih.gov/BLAST) identified the presence of domain(s) in the amino acid sequences of the selected transcripts. Gene Ontology Consortium (http://www.Geneontology.org/) and literature search were used to annotate selected transcripts differentially expressed in FB-HSPCs into 16 groups shown in a pie diagram. CCR indicates cell cycle regulator; TF/DNB, transcription factor/DNA binding protein; RNB, RNA binding; PP, protein processing; PS, protein synthesis; ECM, extracellular membrane; CYT, cytoskeletal; MET, metabolic genes; PIP, proteoglycan integral to plasma membrane; TRA, transports; ST, signal transduction; PF, protein folding; UBQ, ubiquitin pathway; UN EST, uncharacterized; DUB, deubiquitination; and AP, apoptosis pathway.
Quantitative analysis of differentially expressed genes in FB-HSPCs using Q-PCR
On the basis of the biologic differences of purified FB-HSPCs to other HSPCs derived from adult sources that possess identical phenotype, we concentrated our analysis on transcription factors, cell cycle regulators, and signal transduction that we hypothesized were more likely to account for these functional differences. In order to verify the authenticity and quantify the expression of these category of genes identified from FB-HSPC–subtracted library, we performed Q-PCR24,31-33 on newly isolated (Figure 1B) FB-HSPCs and MPB-HSPCs and extended our study to more mature FB hematopoietic cells lacking repopulating cells but enriched in hematopoietic progenitor cells, termed FB-HPCs, previously shown to be enriched in the Lin-CD34+CD38+ subpopulation.5 Required primers were designed from an area of the gene of interest that was not present in the clone originating from SSH to verify the expression of the gene and without limiting the analysis to the specific sequenced region represented in the subclone containing the insert.
Using 2-ΔΔCt equation, explained in “Materials and methods,”18 the relative expression of the candidate genes was calculated in FB-HSPCs compared with MPB-HSPCs and FB-HPCs. This approach provided a quantitative measure of genes in the population of cells enriched for HSPCs from in utero versus adult hematopoietic development (Figure 5A). All of the genes selected for this analysis were found to be more highly expressed in FB-HSPCs than MPB-HSPCs (Figure 5A), confirming the quality of the FB-HSPC–subtracted library. We also compared the relative expression of candidate genes in FB-HSPCs versus FB-HPCs (Figure 5B) to examine the potential role of these genes at 2 different stages of the fetal hematopoietic hierarchy. Q-PCR data analysis in FB-HSPCs versus FB-HPCs indicated that most differential expression of genes from FB-HSPCs versus MPB-HSPCs was also differentially expressed in FB progenitors devoid of repopulating activity. Interestingly, some of the candidate genes, such as CDK6, RALBP-1, and Rac1, are more highly expressed in FB-HPCs than FB-HSPCs, indicating that some of these genes are not exclusively highly expressed in the stem cell compartment of hematopoietic cells and may play some roles in later stages of in utero hematopoietic development. However, one of the transcripts (FB248) differentially expressed in FB-HSPCs compared with MPB-HSPCs (Figure 5A) is not detectable in FB-HPCs (Figure 5B-C), indicating its potential significance in FB-HSPCs. Analysis of amino acid sequences of FB248 indicated the presence of Src homology-3 (SH3) domains, suggesting its role in signal transduction of FB-HSPCs. Further analysis of this transcript is being pursued in our laboratory.
Discussion
Previous studies from our laboratory identified a novel source of HSPCs from fetal circulation.8 Unlike hematopoietic stem progenitors derived from later stages of human ontogeny, such as adult or full-term cord blood, FB-HSPCs were less refractory to retroviral gene transfer,9 retained repopulating activity in NOD/SCID mice at a higher frequency,8,9 and demonstrated the presence of human repopulating stem cells actively traversing the cell cycle.9 Surprisingly, the phenotype of purified FB-HSPCs is identical to other sources of human HSPCs at later developmental stages and is almost exclusively in the Lin-CD34+CD38- subpopulation. The molecular basis of the functional differences between FB and adult HSPCs had yet to be examined. Despite circulating characteristics of FB-HSPCs and MPB-HSPCs, distinct mechanisms most likely govern the origin and nature of these FB-HSPCs in the circulation. MPB-HSPCs are mobilized by G-CSF,34,35 whereas unknown cellular and molecular mechanisms are responsible for circulating HSPCs in fetal blood. To better define the molecular basis for the distinct functional proliferative and differentiative characteristics of FB-HSPCs, here we use SSH to identify differentially expressed genes in FB-HSPCs that were minimally expressing in adult HSPC counterparts that lack active cycling status and are refractory to retroviral gene transfer. Our approach incorporates repeated SSH reactions between FB-HSPCs and MPB-HSPCs to provide a subset of candidate genes thought to be differentially expressed in FB-HSPCs and quantitatively verifies the differential expression of candidates using real-time Q-PCR from freshly isolated samples of FB-HSPCs and MPB-HSPCs. The study approach represents the first to compare gene expression of 2 human hematopoietic stem cells populations found at different stages of human ontogeny and to apply the results to functional characteristics of these subsets. Due to the nature of the differences between FB-HSPCs and adult HSPCs, our investigation focused on those genes most likely to control intrinsic factors of in utero–developing FB-HSPCs, including transcription factors, cell cycle regulators, and signal transduction genes. We have annotated differentially expressed FB-HSPC genes to 3 specific characteristics distinct to FB-HSPCs that were not demonstrated in functional assays to adult counterparts (Table 3).
Annotation and association of functional unique properties of FB-HSPCs to candidate regulatory genes differentially expressed in FB-HSPCs versus adult HSPCs
. | Cellular properties of FB-HSPCs . | Possible regulatory gene(s) . |
|---|---|---|
| 1 | Higher efficiency of gene transfer due to higher cycling rate.9 | Cell cycle inducer: MYCN,36 CDK,37 ETS2,38 and E2F129,30 |
| Cell cycle regulator: WEE1,39,40 RCC1,41 CENPF42 | ||
| 2 | Higher number of myeloid progenitor.8 | TRIM2843 |
| 3 | Higher number of erythroid progenitors.8 | LMO2,44 RNF4,45,46 and NF-KB147 |
. | Cellular properties of FB-HSPCs . | Possible regulatory gene(s) . |
|---|---|---|
| 1 | Higher efficiency of gene transfer due to higher cycling rate.9 | Cell cycle inducer: MYCN,36 CDK,37 ETS2,38 and E2F129,30 |
| Cell cycle regulator: WEE1,39,40 RCC1,41 CENPF42 | ||
| 2 | Higher number of myeloid progenitor.8 | TRIM2843 |
| 3 | Higher number of erythroid progenitors.8 | LMO2,44 RNF4,45,46 and NF-KB147 |
LMO2 indicates Lim only domain-2.
In the group of transcription factors, previous reports have shown the expression pattern and the role of ring finger proteins (RNF) in embryonic period and its role on erythropoiesis.45,46 However, it is yet to be determined that the RNF4 identified in our library possess any role in the hematopoietic system. MYC oncogenes have been shown to be important in cell proliferation and differentiation.48,49 N-myc protooncogene (MYCN) has a key role in cell cycling through cooperation with cyclin D1 resulting in the generation of pre–B- and B-cell lymphoma in double transgenic mice36 and has been associated with a positive feedback loop between E2F1,50,51 thereby confirming our findings since both E2F1 and MYCN are found to be up-regulated in FB-HSPCs (Table 3). The ETS family of transcription factor contains an evolutionarily conserved DNA domain that regulates the expression of a wide range of genes through interaction with other transcription factors and cofactors.52,53 Ets2 has been shown to be involved in the regulation of cell cycle through interaction with cdc2 and cyclin D1 at the G2/M phase of the cell cycle.38 In addition to known transcription factors, our study identifies several expressed sequence tags (ESTs) with unknown function in FB-HSPCs. Amino acid sequence analysis, using SMART program, identified domain motifs in some of the ESTs, such as homeobox domain (HOX) in FB29, similar to domains in HOX genes known to induce self-renewal in mouse HSPCs.
In the group of cell cycle regulators, cyclin-dependent kinase 6 (CDK6) is an inducer of the G1 phase of the cell cycle.37 Regulator of chromatin condensation-1 (RCC1) plays a key role in mitotic spindle assembly and is predominantly localized in the chromosomes of mitotic cells.54 Previous studies have shown that the generation of Ras-related nuclear protein (Ran-GTP) in the vicinity of chromosomes by RCC1 is important for the fidelity of mitotic assembly in human,41 supporting the higher frequency of cycling status of FB-HSPCs. The presence of CENPF, which accumulates during the G2 and rapidly degrades after mitosis,42 may be an indication of active cycling status of FB-HSPCs. Human WEE1 has been shown to play a key role in the negative regulation of entry to the mitotic phase of the cell cycle through induction of the inhibitory role of CDC2/cyclin B kinase.39,40 The presence of WEE1 in our differentially expressed FB-HSPC library also indicates the active regulation of cell cycling activity of FB-HSPCs that is not abnormally disrupted.
In the group of signal transduction genes, our identification of genes involved in the Ras signaling pathway suggests that the Ras pathway is significant to cycling status of FB-HSPCs, consistent with the role of Ras in the progression of G0 to S phase during activation of quiescent cells.55-57 Ral binding protein 1 (RALBP-1),58 downstream molecule of Ral signaling pathway, plays a key role in the activation of GTPase activity of Rac1 and CDC42.58,59 Interestingly, Rac1 was also identified as one of the transcripts differentially expressed in our library and found to be more expressed in FB-HSPCs than MPB-HSPCs. Very recent investigation has identified the significance of Rac1 in repopulating ability of HSPCs where Rac1-deficient mice fail to engraft in the bone marrow of irradiated mice.60 Our library also identifies homologs of mouse Lsc oncogene (human Sub1.5) as one of the activators of Rho family GTPases,61 which is consistent with previous studies indicating the higher abundance of Sub1.5 in human CD34+ progenitor cells.62 Recent investigation has shown the role of NF-KB1 in cell cycling63 and normal erythropoiesis,47 and several studies64-66 also demonstrated the activation of NF-KB1 by Rho, CDC42, and Rac1 proteins indicating their interaction in FB-HSPCs. We also identified a guanine nucleotide exchange factor that is considered to be one of the activators of Ras signaling pathways through activation of GTP-bound state of Ras superfamily proteins.65 Interestingly, this study also identified an EST transcript (FB248) with domain in receptor targeting proteins Lin-2 and Lin-7 (L27), guanylate kinase homologs (GuKc), domain present in PSD-95, Dlg, and ZO-1/2 (PDZ), SH3 domains, which is exclusively expressed in FB-HSPCs and MPB-HSPCs but not in FB-HPCs (Figure 5B-C). The presence of SH3 domain67 signifies the potential role of this transcript in signal transduction of FB-HSPCs.
Using the most widely used purified population of human HSPCs (Lin-CD34+CD38-), our study identifies differentially expressed genes in HSPCs during early stages of in utero human hematopoietic development that are not expressed in adult HSPCs. Creation and validation of this novel library will facilitate future work to define the functional effect and causal relationship of these genes in human HSPCs to be read out using in vitro and in vivo assays in misexpression approaches that include overexpression using retroviral transduction of MPB-HSPCs, or loss of function, through RNA interference. Since it appears that FB248 expression is restricted to HSPCs and not more differentiated progenitors, our laboratory is pursuing the potential function and role of this specific transcript in the context of human HSPC signal transduction.
Prepublished online as Blood First Edition Paper, December 4, 2003; DOI 10.1182/blood-2003-09-3209.
Supported by the National Cancer Institute of Canada (NCIC), and a Canada Research Chair in Stem Cell Biology and Regenerative Medicine (M.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal