Abstract
The PTPN11 gene encodes SHP-2 (Src homology 2 domain–containing protein tyrosine Phosphatase), a nonreceptor tyrosine protein tyrosine phosphatase (PTPase) that relays signals from activated growth factor receptors to p21Ras (Ras) and other signaling molecules. Mutations in PTPN11 cause Noonan syndrome (NS), a developmental disorder characterized by cardiac and skeletal defects. NS is also associated with a spectrum of hematologic disorders, including juvenile myelomonocytic leukemia (JMML). To test the hypothesis that PTPN11 mutations might contribute to myeloid leukemogenesis, we screened the entire coding region for mutations in 51 JMML specimens and in selected exons from 60 patients with other myeloid malignancies. Missense mutations in PTPN11 were detected in 16 of 49 JMML specimens from patients without NS, but they were less common in other myeloid malignancies. RAS, NF1, and PTPN11 mutations are largely mutually exclusive in JMML, which suggests that mutant SHP-2 proteins deregulate myeloid growth through Ras. However, although Ba/F3 cells engineered to express leukemia-associated SHP-2 proteins cells showed enhanced growth factor–independent survival, biochemical analysis failed to demonstrate hyperactivation of the Ras effectors extracellular-regulated kinase (ERK) or Akt. We conclude that SHP-2 is an important cellular PTPase that is mutated in myeloid malignancies. Further investigation is required to clarify how these mutant proteins interact with Ras and other effectors to deregulate myeloid growth.
Introduction
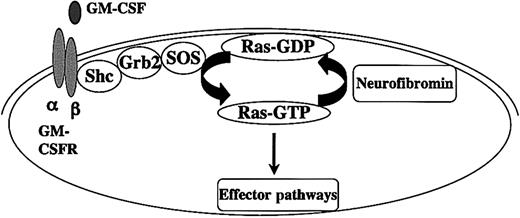
Juvenile myelomonocytic leukemia (JMML) is a relentless myeloproliferative disorder of young children characterized by overproduction of myeloid cells that infiltrate hematopoietic and nonhematopoietic tissues.1,2 A hallmark of JMML is a hypersensitive pattern of granulocyte-macrophage colony-forming unit (CFU-GM) progenitor growth in cultures stimulated with granulocyte-macrophage colony-stimulating factor (GM-CSF).3 Studies of human samples and mouse models strongly implicate hyperactive Ras in the pathogenesis of JMML. Approximately 25% of JMML bone marrow specimens show oncogenic RAS mutations.4,5 In addition, the incidence of JMML is increased by more than 200-fold in children with neurofibromatosis type 1 (NF1).6,7 The NF1 gene encodes neurofibromin, a guanosine triphosphatase (GTPase)–activating protein (GAP) that negatively regulates Ras output by accelerating GTP hydrolysis.8,9 Genetic and biochemical analysis of JMML samples have shown that NF1 functions as a tumor suppressor by negatively regulating Ras.10-12 Taken together, oncogenic RAS mutations or inactivation of NF1 are found in approximately 50% of JMML samples and appear to be mutually exclusive5,12,13 (Figure 1).
Interaction between Ras and neurofibromin in healthy and JMML cells. GM-CSF binding to its surface receptor leads to dimerization and recruits Janus kinase 2 (JAK2), which creates docking sites for adapter molecules by phosphorylating tyrosine residues on the β chain. The guanine nucleotide dissociation factor SOS induces guanosine diphosphate (GDP) dissociation from Ras at the plasma membrane. Ras is then free to bind to GTP, which interacts with effectors such as Raf-1 to activate kinase signaling cascades. Neurofibromin negatively regulates growth by accelerating hydrolysis of Ras-GTP to inactive Ras-GDP. Oncogenic point RAS mutations and loss of NF1 contribute to aberrant proliferation and survival in JMML by elevating Ras-GTP levels. JMML cells are hypersensitive to GM-CSF in vitro, and studies in stains of mutant mice suggest that GM-CSF plays a central role in the aberrant growth of Nf1-deficient cells in vivo.
Interaction between Ras and neurofibromin in healthy and JMML cells. GM-CSF binding to its surface receptor leads to dimerization and recruits Janus kinase 2 (JAK2), which creates docking sites for adapter molecules by phosphorylating tyrosine residues on the β chain. The guanine nucleotide dissociation factor SOS induces guanosine diphosphate (GDP) dissociation from Ras at the plasma membrane. Ras is then free to bind to GTP, which interacts with effectors such as Raf-1 to activate kinase signaling cascades. Neurofibromin negatively regulates growth by accelerating hydrolysis of Ras-GTP to inactive Ras-GDP. Oncogenic point RAS mutations and loss of NF1 contribute to aberrant proliferation and survival in JMML by elevating Ras-GTP levels. JMML cells are hypersensitive to GM-CSF in vitro, and studies in stains of mutant mice suggest that GM-CSF plays a central role in the aberrant growth of Nf1-deficient cells in vivo.
Missense mutations in PTPN11 were shown to cause Noonan syndrome (NS), a developmental disorder characterized by cardiac defects, facial dysmorphism, and skeletal malformations.14,15 SHP-2 (Src Homology 2 domain–containing protein tyrosine Phosphatase-2), the nonreceptor tyrosine protein tyrosine phosphatase (PTPase) encoded by PTPN11, contains 2 src homology 2 (SH2) domains and a catalytic PTPase domain. The SHP-2 PTPase is activated by binding to phosphotyrosyl peptides through its N-SH2 domain.16,17 The SHP-2 crystal structure predicts that these interactions induce a conformational shift that relieves inhibition of the PTPase by the N-SH2 domain.18 Most of the mutations reported in NS kindreds are found in exons 3 and 8, which encode segments of the N-SH2 and PTPase domains, respectively. Molecular modeling implies that almost all of these exon 3 mutations activate phosphatase activity by altering N-SH2 amino acids that interact with the PTPase domain.14,15
SHP-2 participates in signal transduction downstream of growth factor receptors to regulate multiple responses, including proliferation, differentiation, and migration.19,20 The protein is expressed at high levels in hematopoietic cells and undergoes rapid tyrosine phosphorylation on activation of the c-KIT, interleukin 3 (IL-3), GM-CSF, and erythropoietin receptors.19,21,22 SHP-2 most often plays a positive role in transducing signals, which is mediated, at least in part, through the Ras/Raf/ERK (extracellular-regulated kinase) cascade in hematopoietic and nonhematopoietic cells.19,20,23 Loss of Ptpn11 function has profound effects on the developing hematopoietic system.24-27 Ptpn11-deficient yolk sacs contain markedly reduced numbers of hematopoietic colony-forming cells, and mutant embryonic stem cells do not contribute to hematopoiesis in chimeras.25 These and other data implicate SHP-2 as a crucial effector of hematopoietic cell fates that modulates signaling from activated growth factor receptors.
Children with NS show a spectrum of hematologic abnormalities, including isolated monocytosis, myeloid disorders with features of chronic myelomonocytic leukemia (CMML) that remit spontaneously and, rarely, JMML.28-30 Bone marrow cells from NS patients with JMML show characteristic GM-CSF hypersensitivity in methylcellulose cultures.29 These observations, data implicating hyperactive Ras in the pathogenesis of JMML, and the role of SHP-2 in relaying signals from hematopoietic growth factor receptors to Ras identify PTPN11 as an excellent candidate gene that might be mutated in cases of JMML without abnormalities in RAS or NF1. Indeed, a recent study reported somatic PTPN11 mutations in approximately 35% of JMML samples.31 We screened a well-characterized panel of JMML specimens for PTPN11 mutations and confirm that somatic mutations in PTPN11 are common in this disorder. Genetic evidence implicates these mutations as conferring a growth advantage by deregulating Ras. However, although Ba/F3 cells engineered to express mutant SHP-2 proteins consistently show persistent survival on growth factor withdrawal, biochemical analysis did not reveal hyperphosphorylation of the Ras effectors ERK and Akt. In addition, given the known association of oncogenic RAS mutations in other disorders of myelopoiesis, we screened a number of other patients with myeloid disorders for mutations in PTPN11.
Patients, materials, and methods
Leukemia samples
Archived bone marrow or peripheral blood specimens from patients with hematologic malignancies were collected by our laboratory or accrued by the Hematopoietic Tissue Cell Bank of the University of California–San Francisco (UCSF) Comprehensive Cancer Center. Fifty-one patients with JMML; 60 patients with other disorders of myelopoiesis, including acute myelogenous leukemia (AML), chronic myelomonocytic leukemia (CMML), chronic myelogenous leukemia (CML), and myelodysplasia (MDS); and 20 healthy control subjects were screened for mutations in PTPN11. An additional 95 DNA samples were analyzed from a national pediatric AML trial conducted by the Children's Cancer Group between 1996 and 2002. Approval for these studies was obtained from the UCSF Committee on Human Research.
Mutation detection
Denaturing high-performance liquid chromatography (DHPLC) facilitates accurate, high-throughput screening of amplified genomic DNA for point mutations with a sensitivity of approximately 95%.32 Polymerase chain reaction (PCR) using previously published primers for exons 1 to 6 and 8 to 15 were performed according to previously published methods.15 Exon 7 was amplified using forward primer 5′GAAGTAATGCTGATCCAGGC3′, reverse primer 5′AAGAGCACACGACCCTGAGG3′, and Accuprime Taq (Invitrogen, Carlsbad, CA). DNA (50-100 ng) was used for each 50 μL reaction. PCR products were visualized on agarose gels prior to DHPLC analysis. Concentrations of primers (Integrated DNA Technologies, Coralville, IA), dNTPs (Roche, Indianapolis, IN), Mg++ (Applied Biosystems, Foster City, CA), and Amplitaq Gold (Applied Biosystems), as well as PCR conditions, were optimized. DHPLC was conducted on a Helix HPLC (Varian, Palo Alto, CA) using a DNA Sep column (Transgenomics, Omaha, NE) and analyzed according to previously published methods.14,15 Abnormal spectrographs were enzyme purified using 1 U alkaline shrimp phosphatase (Roche) and 1 U exonuclease I (USB, Cleveland, OH), incubated for 1 hour at 37°C, and heat inactivated for 15 minutes at 95°C. Purified products were subsequently sequenced by way of a Prism 3700 Sequencer (ABI, Foster City, CA). The procedures for analyzing RAS and NF1 for mutations have been described.5,10,12
SHP-2 expression constructs
Oligonucleotide primers containing attB sites for use with Gateway cloning technology (Life Technologies, Rockville, MD) as well as murine Ptpn11 gene-specific sequences were used to amplify cDNA sequences from the ATG start codon to nucleotide 1862. Kozak sequences were incorporated into the primer sequence to allow efficient translation. Then, 30 cycles of amplification using Elongase polymerase (Invitrogen) were used to generate a PCR product for use in the BP reaction to generate an entry clone. Next, using the LR enzyme mix from Gateway technology, Ptpn11 was cloned into a mouse stem cell virus (MSCV)–based retroviral vector 33 containing a puromycin resistance cassette that we modified for use with the Gateway system. The QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) was used to generate point mutations into the Ptpn11 expression constructs, which were confirmed by sequencing.
Analysis of Ba/F3 cells
MSCV-puro plasmids engineered to express wild-type SHP-2, the D61Y, or the E76K mutant proteins were cotransfected with plasmids encoding retroviral gag-pol and env proteins into Phoenix cells (a generous gift of Gary Nolan, Stanford University) using Lipofectamine2000 (Invitrogen). Supernatants from transfected cells were used to transduce Ba/F3 cells. Transduced cells were selected for growth in 2.5 μg/mL puromycin for 4 days, and expression of wild-type and mutant SHP-2 was confirmed by Western blot. Ba/F3 parental cells, cells transduced with a control MSCV-puro vector, and lines expressing wild-type or mutant SHP-2 proteins were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and 1 ng/mL recombinant murine IL-3 (Peprotech, Rocky Hill, NJ). After puromycin selection, transduced cells were washed twice in RPMI/10% FBS and grown in IL-3–free media. A total of 1 × 106 cells were seeded into 10-cm dishes, and viable cells were counted by trypan blue exclusion for 2 to 3 weeks. To assess signal transduction, Ba/F3 cells were deprived of IL-3 for 6 hours and then stimulated with 10 ng/mL IL-3 for 10 minutes. Cell lysis and immunoblotting for ERK and Akt were performed as previously described.9 A monoclonal antibody from Transduction Laboratories, Becton Dickinson Biosciences (catalog no. 610621; La Jolla, CA) was used to assess SHP-2 expression.
Results
Missense PTPN11 mutations in JMML specimens
On the basis of case reports of JMML in patients with NS, we first investigated 51 JMML specimens for PTPN11 mutations, including 2 from children with a clinical diagnosis of NS. These studies uncovered missense mutations in 16 of 49 JMML samples from patients without NS, which are summarized in Table 1. PTPN11 mutations were also identified in both patients with NS. Representative DHPLC and sequencing data are shown in Figure 2A-B. Fifteen of 16 PTPN11 mutations detected in sporadic cases of JMML occurred in exon 3, which encodes a segment of the N-SH2 domain (Table 1). The only exception was an exon 4 mutation that we identified in a specimen from a 2-month-old infant. This mutation has also been reported in NS.15 Although the nature of this PTPN11 mutation and the early age of diagnosis are suggestive of NS, detailed clinical information was not available in this case. Both of the JMML specimens from children known to have NS demonstrated different PTPN11 mutations from the patients without NS, including one substitution in exon 13 (Table 1).
PTPN11 mutations detected in JMML
Nucleotide . | Substitution . | Amino acid . | No. cases . |
|---|---|---|---|
| 178 | G>C | Gly60Arg | 1 |
| 181 | G>T | Asp61Tyr | 1 |
| G>A | Asp61Asn | 1 | |
| 205 | G>A | Glu69Lys | 1 |
| 214 | G>A | Ala72Thr | 2 |
| 215 | C>T | Ala72Val | 2 |
| 218 | C>T | Thr73Ile | 1 (JMML/NS) |
| 226 | G>A | Glu76Lys | 3 |
| G>C | Glu76Gln | 1 | |
| 227 | A>G | Glu76Gly | 3 |
| 417 | G>C | Glu139Asp | 1 |
| 1517 | A>C | Gly506Pro | 1 (JMML/NS) |
Nucleotide . | Substitution . | Amino acid . | No. cases . |
|---|---|---|---|
| 178 | G>C | Gly60Arg | 1 |
| 181 | G>T | Asp61Tyr | 1 |
| G>A | Asp61Asn | 1 | |
| 205 | G>A | Glu69Lys | 1 |
| 214 | G>A | Ala72Thr | 2 |
| 215 | C>T | Ala72Val | 2 |
| 218 | C>T | Thr73Ile | 1 (JMML/NS) |
| 226 | G>A | Glu76Lys | 3 |
| G>C | Glu76Gln | 1 | |
| 227 | A>G | Glu76Gly | 3 |
| 417 | G>C | Glu139Asp | 1 |
| 1517 | A>C | Gly506Pro | 1 (JMML/NS) |
PTPN11 mutations in JMML specimens. (A) DNA samples from 3 patients with JMML were amplified by using exon 3–specific primers, and the products were analyzed on DHPLC. (B) PTPN11 mutations corresponding to the abnormal DHPLC spectrographs shown in panel A. Abnormal nucleotides are marked with arrows. (Top) Mutation at nucleotide 181 (G>T); (middle) mutation at nucleotide 182 (A>T); (bottom) mutation at nucleotide 215 (C>T). (C) Absence of the normal PTPN11 allele in a JMML sample; (lane 1) 1-kb ladder, (lane 2) uncut (u) amplified DNA from healthy bone marrow, (lanes 3-5) DNA amplified from a healthy specimen (WT, lane 3), a patient with a heterozygous mutation (A, lane 4), and the patient with the homozygous mutation (B, lane 5). Note the presence of an abnormal (uncut) band (white asterisk) in both patients, with absence of the normal digested bands in patient B. (Lanes 6-8) The same 3 specimens were cut with MaeIII, which cleaves at a novel site within the mutant but not the normal allele (lane 6). (Lanes 7-8) An abnormal band (white asterisk) is visible in both patients A and B, and loss of the upper normal band is seen in patient B (lane 8).
PTPN11 mutations in JMML specimens. (A) DNA samples from 3 patients with JMML were amplified by using exon 3–specific primers, and the products were analyzed on DHPLC. (B) PTPN11 mutations corresponding to the abnormal DHPLC spectrographs shown in panel A. Abnormal nucleotides are marked with arrows. (Top) Mutation at nucleotide 181 (G>T); (middle) mutation at nucleotide 182 (A>T); (bottom) mutation at nucleotide 215 (C>T). (C) Absence of the normal PTPN11 allele in a JMML sample; (lane 1) 1-kb ladder, (lane 2) uncut (u) amplified DNA from healthy bone marrow, (lanes 3-5) DNA amplified from a healthy specimen (WT, lane 3), a patient with a heterozygous mutation (A, lane 4), and the patient with the homozygous mutation (B, lane 5). Note the presence of an abnormal (uncut) band (white asterisk) in both patients, with absence of the normal digested bands in patient B. (Lanes 6-8) The same 3 specimens were cut with MaeIII, which cleaves at a novel site within the mutant but not the normal allele (lane 6). (Lanes 7-8) An abnormal band (white asterisk) is visible in both patients A and B, and loss of the upper normal band is seen in patient B (lane 8).
We noticed absence of the normal PTPN11 allele on the DHPLC tracing of a JMML specimen with a mutation at nucleotide 215. This C>T mutation ablates a BglI cleavage site in the normal sequence and creates a new MaeIII site. Digestion of PCR products amplified from exon 3 confirmed that the normal allele was absent in this case (Figure 2C). To address whether loss of the normal allele exists in other cases of JMML with PTPN11 mutations, we performed PCR amplification followed by allele-specific cleavage of 15 additional cases. All of these leukemias retained the normal PTPN11 allele (data not shown).
Distribution of PTPN11, RAS, and NF1 mutations in JMML
If SHP-2 functions in a growth control signaling pathway that includes the GM-CSF receptor, Ras, and neurofibromin in myeloid cells, PTPN11 mutations might be restricted to leukemia samples without RAS or NF1 mutations. This hypothesis is based on the idea that mutating any component would deregulate the entire cascade and that another mutation would confer little, in any, additional selective advantage. Indeed, previous studies that included most of the JMML specimens investigated in this report revealed RAS and NF1 mutations in mutually exclusive subsets.5,10,12,13 Similarly, BRAF mutations are largely restricted to melanomas without RAS mutations.34 The 49 JMML specimens from patients without NS were divided into 3 groups: (1) samples from patients with a clinical diagnosis of NF1 or an NF1 mutation, (2) samples with RAS (KRAS or NRAS mutations), or (3) all other JMML samples. When these groups were compared, there was a statistically significant difference in the frequency of PTPN11 mutations in group 3 versus groups 1 and 2 (Table 2). These data provide genetic evidence that mutant SHP-2 proteins contribute to leukemogenesis through a Ras-dependent mechanism. Two specimens that were assigned to the NF1 group showed PTPN11 mutations. In one patient with clinical evidence of NF1, extensive molecular analysis did not disclose either loss of the normal NF1 allele or a truncating mutation in the coding region in the leukemic clone. Because some patients with NS show clinical features of NF1,35,36 it is possible that this child was misdiagnosed. The other sample was from a 3-year-old patient without clinical evidence of NF1 who was included in a series of JMML cases studied for NF1 mutations.13 Molecular analysis of the bone marrow demonstrated a nonsense mutation (G4614A) in exon 27a of NF1, but both NF1 alleles were retained. It is formally possible that this mutant allele retains some GAP activity, or that this represents an instance in which leukemic outgrowth in an NF1 patient resulted from a somatic PTPN11 mutation rather than loss of the normal NF1 allele.
Incidence of somatic PTPN11 mutations in subsets of patients with JMML (n = 49)
Patient group . | No. cases . | No. with mutations . | P* . |
|---|---|---|---|
| JMML only | 20 | 14 | NA |
| RAS mutation | 7 | 0 | < .001 |
| NF1 or NF1 mutation | 22 | 2 | < .001 |
Patient group . | No. cases . | No. with mutations . | P* . |
|---|---|---|---|
| JMML only | 20 | 14 | NA |
| RAS mutation | 7 | 0 | < .001 |
| NF1 or NF1 mutation | 22 | 2 | < .001 |
Refers to difference between JMML only and other groups (by chi-square test). NA indicates not applicable
PTPN11 mutations in other hematopoietic malignancies
The prevalence of PTPN11 mutations in JMML suggested that somatic mutations might also exist in other myeloid malignancies. To address this question, we analyzed specimens from children and adults with CMML, CML, AML, MDS, and therapy-related MDS/AML. PTPN11 mutations were detected in 7 of these samples (11%), including 1 of 4 from patients with CMML, 0 of 11 with CML, 3 of 28 with AML, 2 of 7 with MDS, and in 1 of 10 with therapy-related MDS/AML (t-MDS/AML). Each mutation was in exon 3 (Table 3), and many of the same mutations were also found in patients with JMML. On the basis of the results of these studies, we performed a focused analysis of exons 3, 4, 5, 8, and 13 in a cohort of 95 pediatric AML specimens and detected 2 mutations in exon 3. The absence of PTPN11 mutations in CML is consistent with genetic and biochemical data, implicating Ras as a downstream target of Bcr-Abl in myeloid leukemogenesis.37,38 Five of the 9 samples with PTPN11 mutations also had chromosome 7 abnormalities (monosomy 7). This finding is of interest, because monosomy 7 has previously been reported in myeloid malignancies with RAS or NF1 mutations, including JMML.39-43
PTPN11 mutations detected in myeloid malignancies
Nucleotide . | Substitution . | Amino acid . | Diagnosis . |
|---|---|---|---|
| 179 | G>C | Gly60Ala | AML |
| 181 | G>T | Asp61Tyr | AML |
| 182 | A>T | Asp61Val | MDS |
| 188 | A>G | Tyr63Cys | CMML |
| 211 | T>C | Phe71Lys | t-MDS |
| 215 | C>T | Ala72Val | AML(1) MDS (1) |
| 226 | G>A | Glu76Lys | AML |
| 227 | A>G | Glu76Gly | AML |
Nucleotide . | Substitution . | Amino acid . | Diagnosis . |
|---|---|---|---|
| 179 | G>C | Gly60Ala | AML |
| 181 | G>T | Asp61Tyr | AML |
| 182 | A>T | Asp61Val | MDS |
| 188 | A>G | Tyr63Cys | CMML |
| 211 | T>C | Phe71Lys | t-MDS |
| 215 | C>T | Ala72Val | AML(1) MDS (1) |
| 226 | G>A | Glu76Lys | AML |
| 227 | A>G | Glu76Gly | AML |
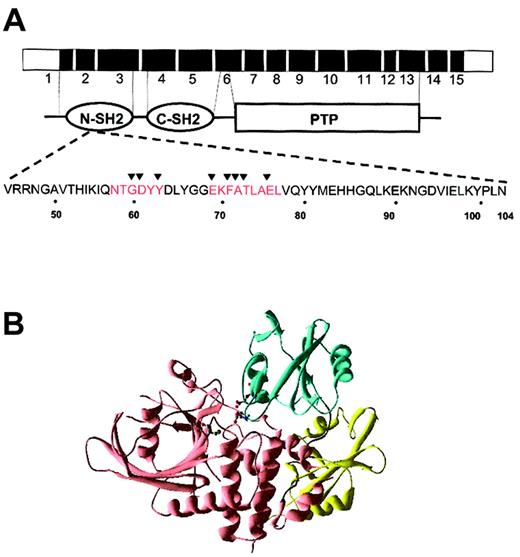
Overall, 25 of the 27 PTPN11 mutations identified in JMML and other myeloid malignancies change amino acids within the N-SH2 domain (exon 3) with codons 61, 72, and 76 affected in 4, 6, and 9 cases, respectively (Tables 1,3). By contrast, although exon 3 is also commonly involved in NS, the overall spectrum is much broader with mutations also seen in exons 2, 4, 7, 8, and 13.14,15 Moreover, only 2 of the leukemia-associated amino acid substitutions identified within exon 3 have been reported in NS.14,15 The JMML specimens analyzed by Tartaglia et al31 displayed a similar pattern of PTPN11 mutations. On the basis of the SHP-2 crystal structure, each PTPN11 mutation is predicted to disrupt the inhibitory interaction of the N-SH2 domain with the PTPase domain (Figure 3A-B).
Sites of exon 3 mutations and predicted effects on SHP-2 structure. (A) Schematic of the PTPN11 gene with functional domains. The amino acid sequence of the N-SH2 domain is highlighted below. The interaction sites between the N-SH2 and PTP domains are indicated in red. The sites of the exon 3 mutations reported here are indicated by the arrowheads. (B) The catalytic cysteine, Cys459, is shown (green dots), as are 2 of the residues mutated in leukemia samples, D61 and E76 (red dots). These residues make critical contacts with the catalytic domain, and the residues' mutation is predicted to disrupt the inhibition of the catalytic domain by the amino-terminal SH2 domain. The N-terminal SH2 domain is shown in blue, the C-terminal SH2 domain in yellow, and the catalytic domain in pink. The figure was generated using Swiss-PdbViewer.
Sites of exon 3 mutations and predicted effects on SHP-2 structure. (A) Schematic of the PTPN11 gene with functional domains. The amino acid sequence of the N-SH2 domain is highlighted below. The interaction sites between the N-SH2 and PTP domains are indicated in red. The sites of the exon 3 mutations reported here are indicated by the arrowheads. (B) The catalytic cysteine, Cys459, is shown (green dots), as are 2 of the residues mutated in leukemia samples, D61 and E76 (red dots). These residues make critical contacts with the catalytic domain, and the residues' mutation is predicted to disrupt the inhibition of the catalytic domain by the amino-terminal SH2 domain. The N-terminal SH2 domain is shown in blue, the C-terminal SH2 domain in yellow, and the catalytic domain in pink. The figure was generated using Swiss-PdbViewer.
Functional and biochemical studies in Ba/F3 cells
Ba/F3 cells have been used to investigate SHP-2 activation in hematopoietic cells44-46 and to interrogate the functional consequences of leukemia-associated mutant proteins.47-51 We, therefore, expressed the mutant D61Y and E76K SHP-2 proteins identified in samples from patients with JMML in this pro-B cell line, which is dependent on IL-3 for survival and proliferation. Ba/F3 cells were infected with retroviral vectors engineered to coexpress wild-type or mutant SHP-2 with a puromycin resistance gene, then cultured with IL-3 and puromycin. This infection/selection procedure was performed on 3 independent occasions to ensure that the biologic effects of expressing mutant SHP-2 proteins were reproducible. After 4 days, the transduced Ba/F3 cells were transferred to medium without IL-3 to assess the effects of expressing wild-type or mutant SHP-2 on survival and proliferation. At this time, cells were also collected to measure SHP-2 expression and to interrogate Ras effector cascades. Wild-type and mutant SHP-2 proteins were expressed at similar levels that were higher than in parental Ba/F3 cells (Figure 4A, bottom panel). We investigated ERK and Akt phosphorylation in cells that were deprived of serum and IL-3 for 6 hours, then stimulated with IL-3. Surprisingly, we did not observe increased levels of phosphorylated ERK or Akt in resting or IL-3–stimulated Ba/F3 cells that expressed either mutant SHP-2 protein (Figure 4A). Signal transduction experiments were performed under a variety of experimental conditions that included varying the concentration of IL-3 and the time course with similar results (data not shown). We also assessed the effects of wild-type and mutant SHP-2 proteins on the survival and growth of transduced Ba/F3 cells after IL-3 withdrawal. The cells were cultured in triplicate at 1 × 106 cells per plate and were counted every other day beginning on day 8. Under these conditions, expression of the E76K mutant consistently enhanced the survival of Ba/F3 cells (Figure 4B). We also observed subtle, but reproducible, effects of the D61Y SHP-2 protein (Figure 4B). Importantly, Ba/F3 cells transduced with mutant SHP-2 proteins did not expand during the 2- to 3-week culture period, but sustained higher numbers of viable cells. However, Ba/F3 cells that expressed either mutant SHP-2 protein frequently demonstrated growth factor–independent proliferation after prolonged time in culture. This was never observed in cells that had been transduced with either empty vector or with the wild-type SHP-2 virus.
Effects of expressing wild-type and mutant SHP-2 proteins on ERK and Akt activation and survival in Ba/F3 cells. (A) Duplicate aliquots of Ba/F3 cells transduced with retroviruses encoding various SHP-2 constructs (wild-type [WT], D61Y, or E76) were collected after 4 days of growth and selection in medium containing IL-3 and puromycin. Parental Ba/F3 cells are labeled P. The cells were starved for 5 hours and lysed without stimulation (–) or after exposure to 10 ng/mL IL-3 for 10 minutes (+). The bottom panel shows SHP-2 expression, which was equivalent in cells transduced with the WT, D61Y, or E76K vectors and elevated above the levels in parental Ba/F3 cells or in cells infected with the empty vector (not shown). Ba/F3 cells expressing all of the SHP-2 constructs showed low levels of phosphorylated ERK (p-ERK) and Akt (p-Akt) after starvation, with robust and equivalent activation in response to IL-3. Parental and transduced Ba/F3 expressed total levels of ERK2 and Akt. (B) Ba/F3 cell counts after IL-3 withdrawal. Ba/F3 cells infected with the MSCV-puro vector (▪), and cells expressing either WT SHP-2 (♦), the D61Y SHP-2 mutant protein (▵), or the E76K SHP-2 mutant protein (○) were plated in triplicate at 1 × 106 cells/plate in the absence of IL-3. Cells were counted starting on day 8.
Effects of expressing wild-type and mutant SHP-2 proteins on ERK and Akt activation and survival in Ba/F3 cells. (A) Duplicate aliquots of Ba/F3 cells transduced with retroviruses encoding various SHP-2 constructs (wild-type [WT], D61Y, or E76) were collected after 4 days of growth and selection in medium containing IL-3 and puromycin. Parental Ba/F3 cells are labeled P. The cells were starved for 5 hours and lysed without stimulation (–) or after exposure to 10 ng/mL IL-3 for 10 minutes (+). The bottom panel shows SHP-2 expression, which was equivalent in cells transduced with the WT, D61Y, or E76K vectors and elevated above the levels in parental Ba/F3 cells or in cells infected with the empty vector (not shown). Ba/F3 cells expressing all of the SHP-2 constructs showed low levels of phosphorylated ERK (p-ERK) and Akt (p-Akt) after starvation, with robust and equivalent activation in response to IL-3. Parental and transduced Ba/F3 expressed total levels of ERK2 and Akt. (B) Ba/F3 cell counts after IL-3 withdrawal. Ba/F3 cells infected with the MSCV-puro vector (▪), and cells expressing either WT SHP-2 (♦), the D61Y SHP-2 mutant protein (▵), or the E76K SHP-2 mutant protein (○) were plated in triplicate at 1 × 106 cells/plate in the absence of IL-3. Cells were counted starting on day 8.
Discussion
We find that missense mutations in PTPN11 are common in JMML and exist in other myeloid malignancies. On the basis of a number of considerations, these amino acid substitutions are almost certain to represent pathologic mutations. First, we did not detect any of these leukemia-associated mutations in 22 healthy bone marrow specimens, and they were not identified in more than 100 control subjects screened by Tartaglia et al.14,15 Second, as would be expected if PTPN11 mutations result in a gain of function, we have not identified deletions, insertions, or substitutions leading to premature termination of protein translation. Third, the data shown in Table 2 argue strongly that these alterations are not random but are functionally equivalent to oncogenic mutations in RAS or inactivation of NF1. Fourth, on the basis of the crystal structure of SHP-2, each of these mutations is predicted to disrupt the inhibitory interaction of the N-SH2 domain with the PTPase domain (Figure 3A-B). Indeed, in an elegant series of experiments in the Xenopus animal cap assay, O'Reilly et al52 generated alanine substitutions corresponding to D61 and E76. These investigators showed that mutant SHP-2 proteins exhibited elevated PTPase activity and conferred a gain-of-function elongation phenotype which is known to involve activation of Ras/ERK signaling downstream of the fibroblast growth factor (FGF) receptor.52 Similarly, our data in the Ba/F3 cell line demonstrate that these mutations have phenotypic consequences in hematopoietic cells. Finally, Tartaglia et al31 independently found a similar incidence and spectrum of somatic PTPN11 mutations in a different series of JMML samples.
Although many of the somatic PTPN11 mutations identified in leukemia specimens alter the same codons as in NS, the spectrum is distinct with respect to the pattern of amino acid substitutions and specificity for exon 3. Furthermore, the 2 mutations that we detected in JMML specimens from children with NS are uncommon, with only one being previously reported. The distribution of PTPN11 mutations found in JMML suggests that these alleles might be deleterious in embryonic life. Consistent with this idea, the D61Y and E76K mutant proteins show higher phosphatase activities than the most common substitution found in children with NS (N308D).31
Expressing the D61Y and E76K mutations enhanced the survival of transduced Ba/F3 cells that were deprived of IL-3. Interestingly, the E76K mutation, which shows higher PTPase activity,31 is more potent in this assay. Although mutant SHP-2 proteins did not acutely induce IL-3–independent proliferation, there was a lower rate of attrition than in control cells expressing either empty vector or wild-type SHP-2. This is reminiscent of the effects of the E2A-HLF fusion protein in Ba/F3 cells.47 Despite genetic evidence that the PTPN11 mutations found in JMML deregulate growth through a Ras-dependent mechanism, we did not detect aberrant activation of ERK or Akt in transduced Ba/F3 cells. Substitutions at the D61 and E76 positions of SHP-2 perturb Ras signaling in other systems.31,52 The discrepancies between these studies and our data in Ba/F3 cells might be due to differences in the expression levels of mutant proteins and/or the cellular context. It is interesting that Tartaglia et al31 reported relatively modest levels of ERK activation in COS-7 cells with basal and serum-stimulated kinase levels that were equivalent to wild-type but prolonged activation in cells expressing mutant SHP-2 proteins. Furthermore, although those researchers observed increased proliferation in COS-7 cells, our data in hematopoietic cells support the idea that the predominant effect of mutant SHP-2 proteins is to reduce the requirement for growth factors in cell survival. This model is consistent with data from cultured Nf1-deficient myeloid cells, which also survive in the absence of exogenous growth factors.53
The absence of hyperphosphorylation of ERK and Akt in transduced Ba/F3 cells is not inconsistent with the idea that mutant SHP-2 proteins deregulate Ras signaling in primary myeloid cells. However, these data raise the possibility that leukemia-associated mutant alleles undermine myeloid growth control through a Ras-independent mechanism. Indeed, although data from Drosophila, Xenopus, and mammalian cells place SHP-2 upstream of Ras in a variety of cell types, other observations support a more complex role that includes functions either parallel to or downstream of Ras.19,52,54,55 Importantly, SHP-2 phosphatase activity is essential for all of its known biologic functions.17 Src kinases, which are activated by dephosphorylation, are attractive potential direct or indirect targets of SHP-2 that regulate the growth of many cell types. Biochemical analysis in myeloid lineage cells may uncover how mutant SHP-2 proteins perturb growth control through effects on Ras and other signaling molecules.
Our data raise the possibility that PTPN11 mutations cooperate with other genetic lesions to induce JMML. This idea is consistent with the clinical observation that hematologic abnormalities, including some JMML-like myeloid disorders, may remit spontaneously in children with NS. Along these lines, it is interesting that we detected loss of the normal PTPN11 allele in one JMML and a second sample showed both heterozygous inactivation of NF1 and a PTPN11 mutation. Our studies to date also suggest that PTPN11 mutations are relatively common in myeloid malignancies with monosomy 7. Expressing leukemia-associated PTPN11 alleles in primary murine hematopoietic cells will help to elucidate the cellular and phenotypic consequences of these mutations and the requirement for cooperating events to induce myeloid disease in vivo.
The high prevalence of PTPN11 mutations in JMML is intriguing and may indicate a specific role for SHP-2 in regulating GM-CSF signaling, perhaps in the context of fetal and neonatal hematopoiesis. Phosphorylation of Tyr-577 on the β common chain of the activated GM-CSF receptor provides a docking site for Shc, which recruits Grb2, Gab2, SHP-2, and the p85 subunit of phosphatidylinositol-3 kinase (PI3K), and induces downstream activation of Akt.21,22,45 Interestingly, SHP-2 also interacts directly with β common at Tyr612 and at Tyr695. Although the functional importance of these sites is uncertain, Tyr612 can induce Gab-2 phosphorylation independent of Tyr577.45 The general idea that mutant SHP-2 molecules might deregulate Ras by aberrantly amplifying signals from activated growth factor receptors is consistent with data from Xenopus.52 In this system, mutant SHP-2 proteins showed elevated PTPase activities and promoted elongation in the absence of FGF. Interestingly, ectopic expression of the D61A and E76A mutants was insufficient to induce mesoderm induction but reduced the requirement for FGF to complete this process.52 By analogy, the PTPN11 mutations found in JMML might contribute to leukemogenesis by hyperactivating Ras signaling at physiologic levels of GM-CSF. This idea is consistent with the hypersensitive pattern of CFU-GM colony growth observed when JMML bone marrows or Nf1 mutant hematopoietic cells are cultured in methylcellulose and with the profound attenuation of the murine JMML-like myeloproliferative disease with Gmcsf ablation.3,11,56,57 Elucidating how mutant SHP-2 proteins interact biochemically with the GM-CSF receptor, with adapter molecules, and with other phosphotyrosyl substrates in primary myeloid cells will extend our knowledge of normal and leukemic signal transduction. The D61A and E76A mutants created by O'Reilly et al52 retain the capacity to bind phosphotyrosyl substrates, and it will be important to confirm that this is also true of leukemia-associated mutant SHP-2 molecules.
In 1994, Sawyers and Denny37 pointed out that Ras signaling is perturbed in myeloid malignancies by distinct genetic mechanisms such as oncogenic RAS point mutations, the BCR-ABL translocation, and NF1 inactivation. Since then, the FLT3 and c-KIT receptors have joined the list of mutant proteins that appear to contribute to myeloid leukemogenesis, at least in part, through hyperactive Ras. SHP-2 represents the first tyrosine phosphatase that functions as an oncogene in human cancer and genetic data support the hypothesis that these mutations might deregulate myeloid growth through a Ras-dependent mechanism. However, the biochemical data presented here do not exclude the possibility that other pathways affected by mutant SHP-2 may contribute to myeloid neoplasia. Fully characterizing how SHP-2 relays signals from activated growth factor receptors to Ras and other molecules and how aberrant signals contribute to tumorigenesis may uncover novel therapeutic targets.
Prepublished online as Blood First Edition Paper, November 26, 2003; DOI 10.1182/blood-2003-09-3287.
Supported by grants from the National Institutes of Health (grants K23 CA80915 and CHRC HD28825 [M.L.L.], P01 CA40046 [M.M.L. and K.M.S.], CA80916 [P.D.E.], and HL04409 [J.G.]); by Cancer Center Core (grant P30 CA82103, University of California at San Francisco [UCSF] Tissue Cell Bank); by an award from the U.S. Army Chronic Myelogenous Leukemia Program (Project CM020058); and by the Frank Campini Foundation (M.L.L. and K.M.S.). K.H.L. is the Hammond Research Fellow of the National Childhood Cancer Foundation.
Coded specimens from patients with pediatric acute myelogenous leukemia (AML) were supplied by the National Childhood Cancer Foundation.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Gary Gilliland for providing Ba/F3 cells and for helpful comments. We thank Dr Robert Hawley for the MSCV vector and Dr Gary Nolan for Phoenix packaging cells. We also acknowledge all of the families and physicians who generously provided blood or bone marrow for analysis.


![Figure 4. Effects of expressing wild-type and mutant SHP-2 proteins on ERK and Akt activation and survival in Ba/F3 cells. (A) Duplicate aliquots of Ba/F3 cells transduced with retroviruses encoding various SHP-2 constructs (wild-type [WT], D61Y, or E76) were collected after 4 days of growth and selection in medium containing IL-3 and puromycin. Parental Ba/F3 cells are labeled P. The cells were starved for 5 hours and lysed without stimulation (–) or after exposure to 10 ng/mL IL-3 for 10 minutes (+). The bottom panel shows SHP-2 expression, which was equivalent in cells transduced with the WT, D61Y, or E76K vectors and elevated above the levels in parental Ba/F3 cells or in cells infected with the empty vector (not shown). Ba/F3 cells expressing all of the SHP-2 constructs showed low levels of phosphorylated ERK (p-ERK) and Akt (p-Akt) after starvation, with robust and equivalent activation in response to IL-3. Parental and transduced Ba/F3 expressed total levels of ERK2 and Akt. (B) Ba/F3 cell counts after IL-3 withdrawal. Ba/F3 cells infected with the MSCV-puro vector (▪), and cells expressing either WT SHP-2 (♦), the D61Y SHP-2 mutant protein (▵), or the E76K SHP-2 mutant protein (○) were plated in triplicate at 1 × 106 cells/plate in the absence of IL-3. Cells were counted starting on day 8.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/6/10.1182_blood-2003-09-3287/6/m_zh80060458500004.jpeg?Expires=1770427445&Signature=VfuLW6CXoidHhOfM5V5q-9fwnzDp~UTEHziP508PrImj3edHy4sn1mQeSPXdagAjyDV8OhYlhTvm0poHCaY6MHdV~VCaKm1udmswU495bkDkHaC2l~IrF1peLDIhHFew4X3dcf-5hPMx05EGrSV~x~VHzIrie4evqRzYupwzpH~3OtXmdmvhCbGIJMwKir~BBLKsuvbv7SYa-uzjZ0QpiwTPh9mYtYo3muhyajc7YhBl5jbYlEs~mYxCAwzkyKyCBB5A1zgd0AjbHkpY~d1EGXnSzNehKuyvOPKtaf~dY-FGpPZ5xK7jxnKPvLLcjAXWZGZKts637ojnLorOU2Sc5A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal