Abstract
The specific targeting of critical signaling molecules may provide efficient therapies for T-cell acute lymphoblastic leukemia (T-ALL). However, target identification and drug development are limited by insufficient numbers of primary T-ALL cells and by their high rate of spontaneous apoptosis. We established a human interleukin-7 (IL-7)–dependent T-ALL cell line, TAIL7, that maintains several biologic and signaling properties of its parental leukemia cells. TAIL7 cells are pre–T-ALL cells that proliferate in response to IL-7 and IL-4. IL-7 stimulation of TAIL7 cells prevents spontaneous in vitro apoptosis and induces cell activation and cell cycle progression. The signaling events triggered by IL-7 include down-regulation of p27kip1 and hyperphosphorylation of retinoblastoma protein (Rb). Stimulation of TAIL7 cells by IL-7 leads to phosphorylation of Janus kinase 3 (JAK3), signal transducer and activator of transcription 5 (STAT5), Akt/PKB (protein kinase B), and extracellular-regulated kinase 1 and 2 (Erk1/2). Importantly, specific blockade of JAK3 by its inhibitor WHI-P131 abrogates the IL-7–mediated proliferation and survival of TAIL7 cells, suggesting that activation of JAK3 is critical for IL-7 responsiveness by these cells. Because TAIL7 cells seem to be a biologic surrogate for primary leukemia T cells, this cell line constitutes a valuable tool for the study of the signaling pathways implicated in T-ALL. Exploitation of this cell line should allow the identification of molecular targets and promote the rational design and validation of antileukemia signaling inhibitors.
Introduction
Despite the considerable success of current therapies for childhood T-cell acute lymphoblastic leukemia (T-ALL), induction failure, disease relapse, and treatment-related complications represent considerable challenges. More effective and less toxic therapies are necessary to improve outcome and the therapeutic index. In the postgenome era, a feasible strategy is to target molecules and pathways that play critical roles in the biology of the tumor, a strategy validated by the success of STI-571 in chronic myelogenous leukemia and by the promising results of clinical trials with other signaling inhibitors.1-3 An inherent obstacle is the availability of malignant T cells in numbers sufficient to perform the signaling experiments necessary for target identification, drug screening, and preclinical validation.
We have previously shown that interleukin-7 (IL-7) promotes the survival and proliferation of T-ALL cells by modulating B-cell lymphoma 2 (Bcl-2) expression and cell cycle progression through down-regulation of the cdk inhibitor p27Kip1.4 IL-7 has been implicated in normal T-cell survival and proliferation, and, in immature T cells, plays a nonredundant antiapoptotic role by up-regulating Bcl-2 expression.5 Mice deficient in IL-7 or lacking the IL-7 receptor are lymphopenic because of a defect in T-cell expansion at an early stage of differentiation, and the few mature T cells that develop are functionally impaired.5-7 Importantly, IL-7 has been implicated in leukemogenesis, because IL-7 transgenic mice develop lymphoid malignancies.8 Malignant T-ALL cells express functional IL-7 receptors,9 and IL-7 stimulates blast colony formation and DNA synthesis, suggesting that it may play an important regulatory role in the pathophysiology of T-ALL.9,10 Finally, IL-7 is produced in the bone marrow (BM) and thymic stroma11 and, therefore, is present in the microenvironments where malignant T cells develop.
In an effort to develop better tools for antileukemia target validation in T-ALL, we have established a T-cell Acute-Leukemia IL-7–dependent cell line (TAIL7) derived from the leukemia cells of a 7-year-old male patient. After more than 12 months in continuous culture, TAIL7 cells maintain several features also found in the patient's primary malignant cells, including (1) the monoclonal T-cell receptor γ (TCR-γ) rearrangement; (2) the immunophenotype of T-cell blasts arrested at an immature thymocyte stage of maturation, pre–T-ALL; (3) the proliferative response to IL-7 and IL-4; (4) the engraftment and establishment of leukemia in nonobese diabetic severe combined immune deficiency (NOD/SCID) mice; and, importantly, (5) the signaling events triggered by IL-7 stimulation, namely down-regulation of p27kip1, and hyperphosphorylation of the retinoblastoma protein (Rb). Finally, using a Janus kinase 3 (JAK3)–specific inhibitor, we demonstrate that blockade of signaling pathways triggered by IL-7 significantly abrogates the proliferation and survival of TAIL7 cells. Our results indicate that TAIL7 may constitute a biologically relevant tool for the identification of molecular targets and the development of novel therapeutic agents for T-ALL.
Materials and methods
T-cell ALL specimen
Primary cells were obtained from the peripheral blood of a 7-year-old male patient with relapse T-ALL. Informed consent and approval by the Dana-Farber Cancer Institute Institutional Review Board were obtained according to federal guidelines. The specimen contained more than 95% of leukemia T cells with an immature CD4/CD8 double-positive phenotype: CD1– CD2+ CD5+ CD7+ CD3– CD4+ CD8+.
Establishment and maintenance of TAIL7 cell line
Primary cells were thawed, separated by density centrifugation, and cultured in 24-well plates at 2 × 106 cells/mL in RPMI-1640 supplemented with 10% fetal bovine serum (FBS; RPMI-10) and 10 ng/mL recombinant IL-7 (Endogen, Woburn, MA). After 9 days in culture at 37°C, 5% CO2, dead cells were removed using the magnetically activated cell sorter (MACS) Dead-cell Removal kit (Miltenyi Biotec, Auburn, CA), and viable cells were recultured under similar conditions. This strategy was used for the initial 3 cycles of leukemia T-cell culture and IL-7–mediated expansion. For further passages, every 7 to 9 days, viable leukemia cells were separated by density centrifugation and replated in IL-7. TAIL7 cells have been maintained on IL-7 continuous culture for more than a year. To confirm IL-7 responsiveness and dependency, viability and proliferation assays were performed periodically. When necessary, viable TAIL7 cells were starved in either RPMI-10 for 5 to 10 days or RPMI without FBS for 2 to 3 days prior use. For signaling experiments, cultured cells were washed twice with phosphate-buffered saline (PBS) and incubated for 15 minutes at 37°C with prewarmed PBS alone or with IL-7. The reaction was stopped by placing samples on ice and adding ice-cold PBS. Cells were washed twice with cold PBS, and whole cell lysates were prepared.
Immunophenotype
Primary leukemia cells and TAIL7 cells were phenotyped by using standard methodology.12 The fluorochrome-conjugated monoclonal antibodies (mAbs) (Table 1) were obtained from BD Pharmingen (San Diego, CA) and Beckman-Coulter (Miami, FL). Samples were analyzed in a Coulter Elite or XL flow cytometer (Beckman-Coulter), and data were acquired as listmode files. At least 5000 positive events were acquired for each sample.
Immunophenotype of parental leukemia T-cells and the TAIL7 cell line
. | Primary cells . | TAIL7 . |
|---|---|---|
| CD1a | - | - |
| CD2 | +++ | +++ |
| CD3 | - | - |
| CD4 | ++ | ++ |
| CD8 | ++ | +++ |
| CD5 | +++ | +++ |
| CD7 | +++ | +++ |
| TCRαβ | - | - |
| TCRγδ | - | - |
| MHC I | +++ | +++ |
| MHC II | - | - |
| CD25 | - | - |
| CD44 | ++ | ++ |
| CD45RA | - | - |
| CD45RO | +++ | +++ |
| CD34 | - | - |
| CD38 | +++ | +++ |
| CD13 | - | - |
| CD19 | - | - |
| CD33 | - | - |
| CD56 | - | - |
. | Primary cells . | TAIL7 . |
|---|---|---|
| CD1a | - | - |
| CD2 | +++ | +++ |
| CD3 | - | - |
| CD4 | ++ | ++ |
| CD8 | ++ | +++ |
| CD5 | +++ | +++ |
| CD7 | +++ | +++ |
| TCRαβ | - | - |
| TCRγδ | - | - |
| MHC I | +++ | +++ |
| MHC II | - | - |
| CD25 | - | - |
| CD44 | ++ | ++ |
| CD45RA | - | - |
| CD45RO | +++ | +++ |
| CD34 | - | - |
| CD38 | +++ | +++ |
| CD13 | - | - |
| CD19 | - | - |
| CD33 | - | - |
| CD56 | - | - |
Expression of the indicated surface molecules was classified according to the percentage of positive cells as compared with the appropriate isotype control: -, less than 20%; +, 20% to 40%; ++, 40% to 70%; and +++, more than 70%.
Clonality assessment and cytogenetics analysis
The characterization of immunoglobulin heavy chain (IgH), T-cell receptor (TCR), and TAL1 gene rearrangements on the primary leukemia and TAIL7 cells was performed by using previously described primers and protocols.13,14 Sequencing was performed by using the BigDye Terminators Cycle Sequencing kit (Applied Biosystems, Foster City, CA). V, D, and J segments were identified by using the ImMunoGeneTics (IMGT) database.15 Cytogenetic analysis was performed by using a standard G-banding technique. The consensus karyotypes were derived by analysis of more than 15 metaphase cells.
Engraftment into NOD/SCID mice
To assess tumorigenicity, viable TAIL7 cells (5 × 106 cells) were transplanted intravenously into irradiated NOD/SCID mice. Animals were killed when moribund. Blood was collected via retro-orbital puncture, and bone marrow was extracted by flushing of long bones. Femurs, spleen, liver, kidneys, and, when possible, thymus and lymph nodes were collected for histologic analysis.
Proliferation assays
TAIL7 cells were cultured in flat-bottom 96-well plates at 4 × 105 cells/well, in standard RPMI-10 or in the presence of IL-7 (10 ng/mL), IL-2 (100 U/mL), IL-4 (10 ng/mL), IL-9 (50 U/mL), IL-15 (20 ng/mL), or phytohemagglutinin (PHA; 1 μg/mL) + phorbol 12-myristate 13-acetate (PMA; 1 ng/mL). Cultures were performed in triplicates for the indicated time points. To assess DNA synthesis, 3H-Thymidine (1 μCi/well [0.037 MBq/well]) was added for 16 hours prior to cell harvest and measured by using a liquid scintillation counter. Proliferation index (PIndex) was calculated as PIndex = (mean cpm for each experimental condition)/(mean cpm for medium alone).
Cell viability and activation
Quantitative determination of cell viability was performed by using an Annexin V–based apoptosis assay (R&D Systems, Flanders, NJ). Briefly, cells were suspended in the appropriate binding buffer, stained with fluorescein isothiocyanate (FITC)–conjugated Annexin V and propidium iodide (PI) at room temperature for 15 minutes, and subsequently analyzed by flow cytometry. Because activated cells have an increased size and light refraction, activation status was assessed by measuring changes in these physical parameters in viable TAIL7 cells (Annexin V– and PI-negative).
Cell cycle analysis
Cell cycle distribution was performed by assessment of DNA content using a PI staining. Cells were resuspended in PBS and fixed with ice-cold 80% ethanol for at least 30 minutes. PI (2.5 mg/mL) and ribonuclease A (50 mg/mL) were added, and samples were incubated in the dark, for 30 minutes at 37°C. Flow cytometric analysis was performed by using Lysis II (Becton Dickinson, Mountain View, CA) or Coulter's XL2 software, and analysis of cell cycle histograms was carried out by using ModFit LT (Verity, Topsham, ME) or WinCycle DNAAnalysis software (Phoenix Flow Systems, San Diego, CA).
Immunoprecipitation and immunoblotting
Equal amounts of protein (50 μg/sample) were analyzed by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) or 6% SDS-PAGE (for Rb phosphorylation), transferred onto nitrocellulose membranes, and immunoblotted with the indicated mouse mAbs or rabbit antiserum: p27kip1 mAb (BD Transduction Laboratories, Lexington, KY), Phospho-Erk (extracellular-related kinase) mAb (Santa Cruz Biotechnology, Santa Cruz, CA), Rb mAb (BD Pharmingen), ZAP-70 (zeta-associated protein 70) mAb and phospho-STAT5 (signal transducer and activator of transcription 5) mAb (Upstate Biotechnology, Lake Placid, NY), and P-Akt antiserum (Cell Signaling Technology). Immunodetection was performed by incubation with horseradish peroxidase (HRP)–conjugated antimouse (1:5000) or antirabbit IgG (1:10 000) (Promega, Madison, WI) and developed by chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ). Stripping and reprobing of the immunoblots were done as described.16 Tyrosine phosphorylation of JAK3 was assessed by immunoprecipitation, using equal amounts of protein (250 μg/sample) with anti-JAK3–specific antiserum agarose conjugate (Santa Cruz Biotechnology), followed by immunoblotting with the antiphosphotyrosine mAb 4G10 (Upstate Biotechnology) and anti-JAK3 antiserum (Santa Cruz Biotechnology).
Evaluation of signaling-specific inhibitors and chemotherapeutic drugs
The effects of the JAK3 inhibitor WHI-P131 (Calbiochem, San Diego, CA) on TAIL7 cells were evaluated by using proliferation and viability studies, as described in “Proliferation assays” and “Cell viability and activation.” WHI-P131 was used at final concentrations of 50, 100, and 200 μM. WHI-P131 is a dimethoxyquinazoline compound designed to bind to a key residue in the catalytic site of JAK3.17 It does not inhibit JAK1, JAK2, or other kinases and, at the doses tested, is not toxic to JAK3-negative cells.17 The effects of the mammalian target of rapamycin (mTOR)–specific inhibitor rapamycin (Calbiochem) and the chemotherapeutic drugs dexamethasone and doxorubicin (Sigma-Aldrich, Sintra, Portugal) were evaluated in TAIL7 viability studies.
Real-time quantitative PCR
Real-time quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) analysis was performed as previously described.18 Briefly, TAL1, TAL2, HOX11, HOX11L2, LYL1, LMO1, LMO2, BHLHB1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels were analyzed in parallel experiments by using 100 ng RNA extracted from the cell lines analyzed. Real-time RT-PCR reactions were performed in an ABI PRISM 7700 Sequence Detection System instrument (Applied Biosystems). RNA copy numbers were calculated by using standard curves consisting of serial 10-fold dilutions from 107 to 102 copies of in vitro–transcribed RNA. Samples containing less than 106 copies of GAPDH message per 100 ng RNA were considered to have poor quality and were not analyzed for oncogene expression. PCR analysis of the Tal1d variant, which results from a 90-kb deletion in chromosome band 1p32 adjacent to the TAL1 locus, was performed in genomic DNA extracted from the cell lines analyzed.19
TAIL7 fingerprint
For DNA profiling of TAIL7 cells, analysis of several loci of variable number of tandem repeats (VNTRs) and short-tandem repeats (STRs) was performed. For VNTR profiling, TAIL7 genomic DNA was digested with HaeIII, electrophoresed in a 1% agarose gel, transferred to Biodyne A membrane (Pall Life Sciences, Ann Arbor, MI), and sequentially hybridized with 1 of 7 different alkaline phosphatase–conjugated oligonucleotide probes: MS1 (D1S7), PH30 (D4S139), LH1 (D5S110), CEB42 (D8S358), D17S79 (Invitrogen Life Technologies, Carlsbad, CA), D2S44, or D10S28 (LifeCodes, Stamford, CT), plus the ACES 2.0 Marker Probe Plus (Invitrogen) according to manufacturer's instructions. DNA from the well-characterized K562 cell line was included as control. The size of the “alleles” was calculated by using the Kodak 1D software (Eastman Kodak Company, Rochester, NY). For STR profiling, 18 loci fragments were amplified in multiplexed PCR reactions using fluorescently labeled primers, and the length of the individual-specific “alleles” was determined in an ABI 377 DNA Sequencer (Applied Biosystems).
Results
Establishment of an IL-7–dependent leukemia T-cell line
The very high rate of spontaneous apoptosis and lack of significant in vitro expansion of leukemia T cells represent considerable obstacles to the study of the biology of T-ALL and the evaluation of novel therapeutic agents. To circumvent these limitations, we established a leukemia T-cell line that could be expanded by in vitro culture while maintaining the biologic and signaling properties of primary leukemia cells. We selected IL-7 as the stimulus for the leukemia cells, based on its in vitro effects on T-ALL cells,4,9,10,20 its presence in the leukemia microenvironment, and its putative role in leukemogenesis. An IL-7–responsive T-cell line, TAIL7, was derived from leukemia cells obtained from the peripheral blood of a 7-year-old boy at the time of relapse, when his peripheral blood contained more than 95% of T-cell blasts. Primary cells were cultured in the presence of 10 ng/mL recombinant IL-7 for periods of 7 to 9 days, as described in “Materials and methods.” TAIL7 cells have been in continuous culture in IL-7 for more than 12 months. The average 7-day expansion rate of TAIL7 cells is 3.1-fold ± 0.5-fold (n = 17 determinations). Viability and proliferation assays were performed routinely and showed that TAIL7 cells maintain their responsiveness to IL-7 and progressively undergo apoptotic cell death in its absence (data not shown).
TAIL7 cells are immature T-ALL cells sharing the phenotype and clonality of their parental leukemia cells
To determine whether TAIL7 cells replicate the patient's primary leukemia T cells, immunophenotypic and clonality analyses were performed in the uncultured primary leukemia cells and TAIL7 cells after various periods of time in continuous cell culture (1, 2, 4, and 6 months of IL-7 culture). Primary leukemia and TAIL7 cells share an identical phenotype, CD7+ CD2+ CD5+ CD3– CD1a–, and lack B-cell, natural killer (NK) cell, and myeloid cell markers (Table 1), which is consistent with an immature thymocyte21 or pre–T-ALL lymphoblast22 phenotype. TAIL7 cells also express CD4 and CD8 and, consistent with their lack of CD3, do not express either TCRαβ or TCRγδ on the cell surface. Further evidence indicating that they are arrested in development as pre–T-ALL lymphoblasts is their lack of expression of CD34 and CD45RA and positivity for CD45RO.23,24 Finally, analysis of TAIL7 cells at different culture time points showed that no significant phenotypic changes occurred, thus suggesting that continuous exposure to IL-7 did not induce cell differentiation.
The presence of TCR and IgH rearrangements was investigated to ascertain the clonality of TAIL7 cells and their parental T-ALL cells. An extensive series of putative TCRδ, TCRγ, TAL1, and IgH junctional rearrangement regions were PCR-amplified followed by heteroduplex analysis. A single monoallelic TCRγ rearrangement (VγI-Jγ1.3/3.2) was observed on both the TAIL 7 cells and the primary leukemia cells (Table 2), thus excluding the occurrence of subclone derivation or ongoing recombination on TAIL7 cells.
Rearrangement of TCRγ (Vγ1/Jγ1.3/3.2) identified on primary leukemia cells (day 0) and TAIL7 cells
Day . | TCR γV8*01 . | N region . | TCR γJ2*01 . |
|---|---|---|---|
| 0 | 5′.. .TATTACTGTGCCACCTGGGA---- | CACCTC | GAATTATTATAAGAAACTCTTTGGCA... 3′ |
| 26 | 5′.. .TATTACTGTGCCACCTGGGA---- | CACCTC | GAATTATTATAAGAAACTCTTTGGCA... 3′ |
| More than 70 | 5′.. .TATTACTGTGCCACCTGGGA---- | CACCTC | GAATTATTATAAGAAACTCTTTGGCA... 3′ |
Day . | TCR γV8*01 . | N region . | TCR γJ2*01 . |
|---|---|---|---|
| 0 | 5′.. .TATTACTGTGCCACCTGGGA---- | CACCTC | GAATTATTATAAGAAACTCTTTGGCA... 3′ |
| 26 | 5′.. .TATTACTGTGCCACCTGGGA---- | CACCTC | GAATTATTATAAGAAACTCTTTGGCA... 3′ |
| More than 70 | 5′.. .TATTACTGTGCCACCTGGGA---- | CACCTC | GAATTATTATAAGAAACTCTTTGGCA... 3′ |
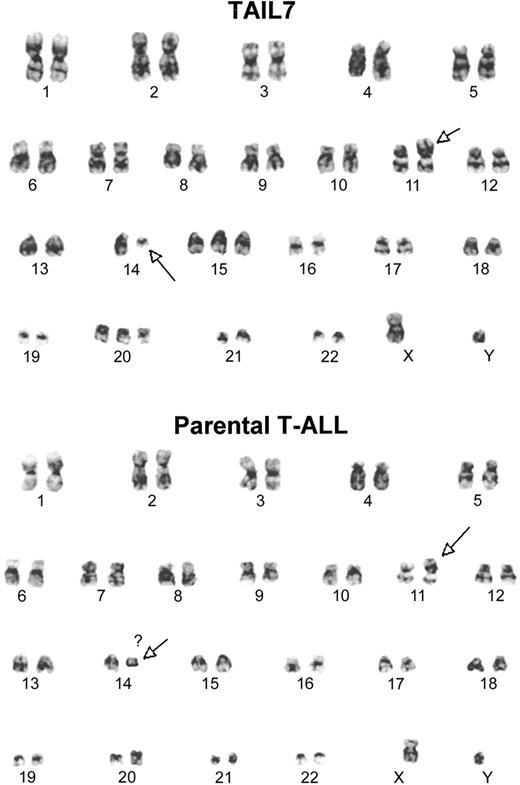
Cytogenetic analysis of TAIL7 cells and their parental primary leukemia cells revealed that they share identical karyotypes, which are consistent with 46 XY t(11;14)(p13;q11) (Figure 1). Although several metaphases in both TAIL7 and parental leukemia cells showed other chromosomal changes (gains, loses, and some breaks), this was the only clonal abnormality consistently observed in all metaphases.
TAIL7 cells and their parental primary leukemia cells share a common karyotype. Cytogenetic analysis was performed by using a standard G-banding technique. The t(11;14)(p13;q11) was the only clonal chromosomal abnormality consistently detected in all metaphases.
TAIL7 cells and their parental primary leukemia cells share a common karyotype. Cytogenetic analysis was performed by using a standard G-banding technique. The t(11;14)(p13;q11) was the only clonal chromosomal abnormality consistently detected in all metaphases.
IL-7 induces TAIL7 cell proliferation, activation, survival, and cell cycle progression
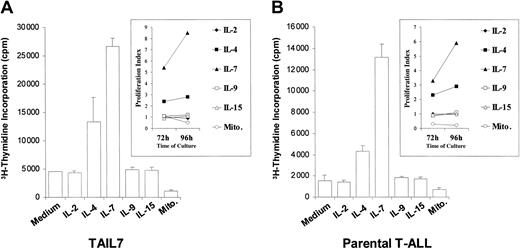
We next evaluated whether TAIL7 cells maintain the pattern of responsiveness to IL-7 and other γc-signaling cytokines observed for their parental primary leukemia cells. TAIL7 cells showed a robust proliferative response to IL-7 and a weaker response to IL-4 (Figure 2A). No proliferation was observed in response to IL-2, IL-9, or IL-15 (Figure 2A). Also, TAIL7 cells failed to proliferate when cultured with PHA plus PMA, which instead resulted in lower levels of 3H-thymidine incorporation (Figure 2A). Importantly, the response of TAIL7 cells to these cytokines and mitogens was in complete concordance with the pattern observed for the parental leukemia cells from which this cell line was derived (Figure 2B). As expected, culture of TAIL7 cells with IL-7 resulted in an increased percentage of cells in S and G2/M phases of the cell cycle (Figure 3A) and in increased cell size and complexity, as illustrated by the physical changes detected by flow cytometry (Figure 3B; FSC versus SSC signals). Finally, stimulation of TAIL7 cells with IL-7 promoted cell viability, which was more pronounced after more stringent preculture starvation conditions (Figure 3C). Again, these findings are consistent with observations using the patient's primary leukemia cells, although the effect of IL-7 on survival of the parental leukemia cells was even more pronounced than that observed for the cell line (average 30% increase on viability at 96 hours versus 17% for the TAIL7 cells). Strikingly, these functional responses triggered by the IL-7 stimulation of TAIL7 cells are equivalent not only to those observed for the parental leukemia cells but also to those observed in all the IL-7–responsive primary leukemia samples previously tested (70% of the patients analyzed4 ).
TAIL7 cells and their parental leukemia T cells proliferate in response to IL-7 and IL-4. TAIL7 cells (A) and patient's peripheral blood blasts (B) were cultured for 96 hours in medium alone or the indicated cytokine or mitogens (Mito.), and proliferation was measured by 3H-thymidine incorporation. Insets: Proliferation index (described in “Materials and methods”) for each condition at 72 and 96 hours. Results are expressed as mean ± SD of triplicates and are representative of 2 independent experiments for the primary leukemia cells and 3 independent experiments for the TAIL7 cell line.
TAIL7 cells and their parental leukemia T cells proliferate in response to IL-7 and IL-4. TAIL7 cells (A) and patient's peripheral blood blasts (B) were cultured for 96 hours in medium alone or the indicated cytokine or mitogens (Mito.), and proliferation was measured by 3H-thymidine incorporation. Insets: Proliferation index (described in “Materials and methods”) for each condition at 72 and 96 hours. Results are expressed as mean ± SD of triplicates and are representative of 2 independent experiments for the primary leukemia cells and 3 independent experiments for the TAIL7 cell line.
IL-7 activates TAIL7 cells and promotes cell cycle progression and increased viability. (A) TAIL7 cells were cultured for 72 hours with or without IL-7, and the cell cycle phase distribution was examined by PI staining and flow cytometry. Results are representative of 3 independent experiments. (B) At 72 hours, activation was assessed by flow cytometry, by gating on the live cell population, and by measuring the changes in cell size (forward light scatter [FSC]) and complexity (sideward light scatter [SSC]). The “percentage of activated cells” shown was defined relative to the distribution of the control population (medium alone). Similar results were obtained in 7 independent experiments. (C) At 96 hours, cells were stained with Annexin V–FITC/PI, and the percentage of viable cells was assessed by flow cytometry. Results are expressed as mean and standard deviation of 3 samples for each condition and are representative of 5 independent experiments.
IL-7 activates TAIL7 cells and promotes cell cycle progression and increased viability. (A) TAIL7 cells were cultured for 72 hours with or without IL-7, and the cell cycle phase distribution was examined by PI staining and flow cytometry. Results are representative of 3 independent experiments. (B) At 72 hours, activation was assessed by flow cytometry, by gating on the live cell population, and by measuring the changes in cell size (forward light scatter [FSC]) and complexity (sideward light scatter [SSC]). The “percentage of activated cells” shown was defined relative to the distribution of the control population (medium alone). Similar results were obtained in 7 independent experiments. (C) At 96 hours, cells were stained with Annexin V–FITC/PI, and the percentage of viable cells was assessed by flow cytometry. Results are expressed as mean and standard deviation of 3 samples for each condition and are representative of 5 independent experiments.
TAIL7 cells are tumorigenic
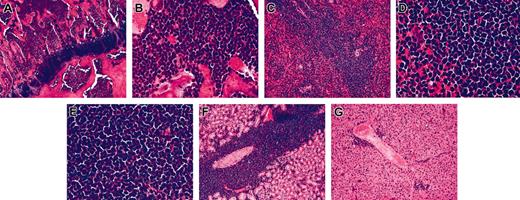
To determine tumorigenicity, TAIL7 cells were transplanted intravenously into irradiated NOD/SCID mice (5 × 106 viable cells/mouse). In all animals tested (n = 6), TAIL7 cells engrafted and developed into a leukemia involving multiple organs. Animals were moribund and killed at 64.3 ± 2.3 days after cell injection (range, 63-67 days). Histologic analyses showed, in all animals, that the BM was packed with leukemia cells (Figure 4A-B). Splenomegaly and enlarged lymph nodes were a common occurrence in the animals that received transplants with large numbers of leukemia cells present in the spleen and lymph nodes (Figure 3C-E). Leukemia cells were also observed invading the liver and kidneys (Figure 4F-G). No evidence of thymus enlargement or mediastinal mass was observed. Importantly, the time-interval of TAIL7 growth and leukemia development in NOD/SCID mice was similar to that observed for primary leukemia cells, which is longer than that observed for established, stimuli-independent leukemia cell lines (data not shown).
TAIL7 cells engraft NOD/SCID mice and are tumorigenic. Viable TAIL7 cells were transplanted into NOD/SCID mice (5 × 106 cells/animal). Mice were killed when moribund, and the indicated organs were collected for histological analysis. Bone marrow; original magnification × 10 (A) and × 40 (B). Spleen; original magnification × 10 (C) and × 40 (D). (E) Lymph node; original magnification, × 40. (F) Kidney; original magnification, × 10. (G) Liver; original magnification, × 10.
TAIL7 cells engraft NOD/SCID mice and are tumorigenic. Viable TAIL7 cells were transplanted into NOD/SCID mice (5 × 106 cells/animal). Mice were killed when moribund, and the indicated organs were collected for histological analysis. Bone marrow; original magnification × 10 (A) and × 40 (B). Spleen; original magnification × 10 (C) and × 40 (D). (E) Lymph node; original magnification, × 40. (F) Kidney; original magnification, × 10. (G) Liver; original magnification, × 10.
IL-7–triggered signaling events on TAIL7 cells
To further investigate whether the TAIL7 cell line is a reliable substitute for primary leukemia cells, we have compared the signaling events triggered by IL-7 stimulation in TAIL7 cells with those of their parental leukemia cells. This is particularly important as TAIL7 cells are to be used as surrogate “primary” leukemia T-ALL cells for the identification of signaling molecules that may constitute therapeutic targets.
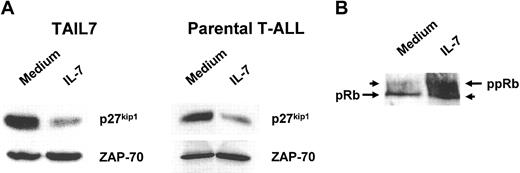
We evaluated the effect of IL-7 on the expression of the cell cycle regulator p27kip1, because we have previously demonstrated that IL-7 induces cell cycle progression in T-ALL cells through down-modulation of the activity of this cdk inhibitor.4 As shown in Figure 5A, we observed a marked down-regulation of p27kip1 in TAIL7 cells following IL-7 stimulation. IL-7 also induced up-regulation of the hyperphosphorylated form of Rb, a critical substrate of cyclin/cdk activity, indicating that cells are committed to cell cycle progression (Figure 5B). An identical pattern of IL-7–mediated p27kip1 down-regulation (Figure 5C) and hyperphosphorylation of Rb (data not shown) was found in parental primary leukemia cells.
IL-7 down-regulates p27kip1 expression and induces hyperphosphorylation of Rb in TAIL7 cells. (A) Lysates of TAIL7 cells cultured for 96 hours were resolved by 10% SDS-PAGE and immunoblotted with a p27kip1-specific antibody. The membrane was stripped and reprobed for ZAP-70 to demonstrate equal protein loading. (B) Lysates were analyzed by 6% SDS-PAGE and immunoblotted for Rb. The antibody recognizes both hypophosphorylated (pRb) and hyperphosphorylated (ppRb) forms of Rb. Representative results of 3 independent experiments are shown.
IL-7 down-regulates p27kip1 expression and induces hyperphosphorylation of Rb in TAIL7 cells. (A) Lysates of TAIL7 cells cultured for 96 hours were resolved by 10% SDS-PAGE and immunoblotted with a p27kip1-specific antibody. The membrane was stripped and reprobed for ZAP-70 to demonstrate equal protein loading. (B) Lysates were analyzed by 6% SDS-PAGE and immunoblotted for Rb. The antibody recognizes both hypophosphorylated (pRb) and hyperphosphorylated (ppRb) forms of Rb. Representative results of 3 independent experiments are shown.
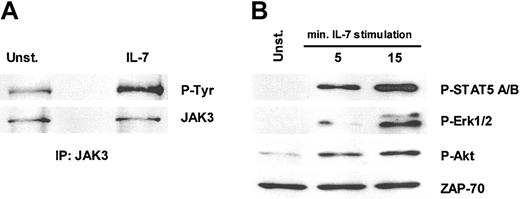
One of the earliest events in IL-7 signaling is the activation of the γc-associated protein tyrosine kinase JAK3.25 As shown in Figure 6A, IL-7 mediates tyrosine phosphorylation of JAK3 in TAIL7 cells, thus confirming that a functional IL-7 receptor complex exists in these cells. We further assessed the capacity of IL-7 to trigger the JAK/STAT, MAP (mitogen-activated protein)/ERK kinase (MEK)/Erk and phosphatidyl inositol-3 kinase (PI3K)/Akt pathways, which are common mediators of cytokine signaling. As shown in Figure 6B, there is a striking up-regulation of phospho-STAT5, phospho-Erk1/2, and phospho-Akt on IL-7 stimulation of TAIL7 cells, thus indicating that all 3 pathways are activated by IL-7.
IL-7 stimulation induces the activation of JAK/STAT, ras/MAPK, and PI3K/Akt pathways in TAIL7 cells. TAIL7 cells were stimulated with 50 ng/mL IL-7 at 37°C for 15 minutes, or the indicated periods, or left unstimulated (Unst.). (A) Lysates were immunoprecipitated with anti-JAK3 agarose conjugate antiserum, resolved by 10% SDS-PAGE, and immunoblotted with an antiphosphotyrosine mAb (P-Tyr), stripped, and reprobed with anti-JAK3 antiserum. (B) Lysates were resolved by 10% SDS-PAGE and immunoblotted. Phosphorylated STAT5, Erk, and Akt were probed with phospho-specific antibodies. ZAP-70 antibody was used as a loading control. Blot was stripped and sequentially reprobed. P-STAT5 recognizes phosphorylated Y694 and Y699 of STAT5a and STAT5b. P-Erk recognizes phosphorylated Y204 of Erk1 and Erk2. P-Akt recognizes phosphorylated S473 of Akt. Results are representative of 3 independent experiments.
IL-7 stimulation induces the activation of JAK/STAT, ras/MAPK, and PI3K/Akt pathways in TAIL7 cells. TAIL7 cells were stimulated with 50 ng/mL IL-7 at 37°C for 15 minutes, or the indicated periods, or left unstimulated (Unst.). (A) Lysates were immunoprecipitated with anti-JAK3 agarose conjugate antiserum, resolved by 10% SDS-PAGE, and immunoblotted with an antiphosphotyrosine mAb (P-Tyr), stripped, and reprobed with anti-JAK3 antiserum. (B) Lysates were resolved by 10% SDS-PAGE and immunoblotted. Phosphorylated STAT5, Erk, and Akt were probed with phospho-specific antibodies. ZAP-70 antibody was used as a loading control. Blot was stripped and sequentially reprobed. P-STAT5 recognizes phosphorylated Y694 and Y699 of STAT5a and STAT5b. P-Erk recognizes phosphorylated Y204 of Erk1 and Erk2. P-Akt recognizes phosphorylated S473 of Akt. Results are representative of 3 independent experiments.
Significantly, the cell cycle–related events triggered by IL-7 stimulation of TAIL7 cells are consistent with those observed by using not only the parental leukemia cells but also any of the IL-7–responsive T-ALL specimens previously tested.4 Moreover, the signaling pathways and the functional responses activated by IL-7 stimulation of TAIL7 cells and primary leukemia cells are equivalent (J.T.B. et al, manuscript in preparation). These findings suggest that the TAIL7 cells are representative of a significant number of T-ALL specimens and may serve as a valid tool for the study of the signaling pathways that are implicated in the pathophysiology of T-ALL as well as for the rational design of antileukemia signaling inhibitors.
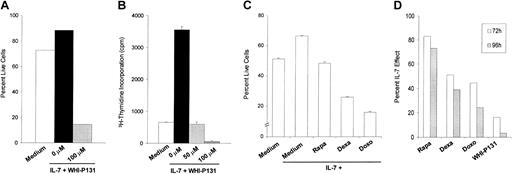
JAK3-specific inhibitor WHI-P131 abolishes the IL-7–promoted proliferation and survival of TAIL7 cells
Because the JAK/STAT pathway is activated by IL-7 stimulation of TAIL7 cells (Figure 6), we evaluated the effect of the JAK3-specific inhibitor WHI-P131 on TAIL7 cell survival and proliferation. WHI-P131 is a dimethoxyquinazoline derivative that specifically inactivates JAK3 but not other JAK family members, by interacting with its catalytic site.17 It has been shown previously that WHI-P131 induces the apoptosis of JAK3-positive B-lineage leukemia cell lines (optimal dose, 100 μM) but not JAK3-negative cells.17 WHI-P131 completely abrogates the stimulatory effect of IL-7 on TAIL7 cells, inhibiting both cell proliferation (Figure 7A) and survival (Figure 7B). This inhibitory effect was dose dependent (Figure 7A and data not shown) and demonstrated that the specific blockade of JAK3 effectively abrogates the stimulatory effect of IL-7. As shown in Figure 7, at 100 μM, the WHI-P131-mediated inhibition of TAIL7 cells surpassed that observed in the absence of IL-7, suggesting that some degree of constitutive activation of JAK3 exists in TAIL7 cells. This finding is in agreement with the low basal level of JAK3 tyrosine phosphorylation observed in unstimulated TAIL7 cells, which was up-regulated by IL-7 (Figure 6A). Despite the apparent constitutive activation of JAK3, we did not find any evidence of constitutive phosphorylation of its direct target STAT5 (Figure 6B). Notwithstanding, we observed that in TAIL7 cells, WHI-P131 markedly reduced the IL-7–mediated phosphorylation of STAT5 (data not shown).
JAK3 inhibitor WHI-P131 abrogates IL-7–induced proliferation and viability in TAIL7 cells. TAIL7 cells were cultured for 72 hours in medium alone, in the presence of IL-7, or with IL-7 plus 100 μM WHI-P131 (J3In.). (A) Viability was assessed by Annexin V–FITC/PI staining and FACS analysis. (B) Proliferation was examined by 3H-Thymidine incorporation and is expressed as mean ± SD of triplicates. (C) Effects of rapamycin (Rapa; 10 nM), dexamethasone (Dexa; 100 nM), and doxorubicin (Doxo; 100 nM) on IL-7–mediated viability of TAIL7 cells. Cultures were performed in duplicates for 96 hours and results are expressed as mean ± SD. (D) For each condition, TAIL7 cell viability was normalized, comparing to the IL-7–mediated effect (100%). Results at 72 hours and 96 hours of drug treatment are shown for a representative experiment.
JAK3 inhibitor WHI-P131 abrogates IL-7–induced proliferation and viability in TAIL7 cells. TAIL7 cells were cultured for 72 hours in medium alone, in the presence of IL-7, or with IL-7 plus 100 μM WHI-P131 (J3In.). (A) Viability was assessed by Annexin V–FITC/PI staining and FACS analysis. (B) Proliferation was examined by 3H-Thymidine incorporation and is expressed as mean ± SD of triplicates. (C) Effects of rapamycin (Rapa; 10 nM), dexamethasone (Dexa; 100 nM), and doxorubicin (Doxo; 100 nM) on IL-7–mediated viability of TAIL7 cells. Cultures were performed in duplicates for 96 hours and results are expressed as mean ± SD. (D) For each condition, TAIL7 cell viability was normalized, comparing to the IL-7–mediated effect (100%). Results at 72 hours and 96 hours of drug treatment are shown for a representative experiment.
We have previously demonstrated that the IL-7–mediated stimulation of primary T-ALL cells is inhibited by the mTOR-specific inhibitor rapamycin.4 On TAIL7 cells, we observed that rapamycin also abrogates IL-7–promoted survival (Figure 7C), mimicking our findings on primary leukemia T cells. Finally, we evaluated the effects on TAIL7 cell survival of 2 chemotherapeutic drugs known for their potent cytotoxic activity on T-ALL cells: dexamethasone and doxorubicin. At concentrations cytotoxic to primary leukemia T cells,26,27 both drugs promote significant apoptosis of TAIL7 cells (Figures 7C). Strikingly, the effect of the JAK3-specific inhibitor WHI-P131 on TAIL7 is superior to that caused by the chemotherapeutic drugs (Figure 7D) and mimics its inhibitory activity on IL-7–promoted proliferation and survival of primary T-ALL cells (data not shown). These findings indicate that TAIL7 cells retain their sensitivity to conventional drugs and are useful surrogates of primary leukemia T cells for the evaluation of novel drugs with therapeutic potential.
T-cell oncogene expression and DNA fingerprinting of TAIL7 cells
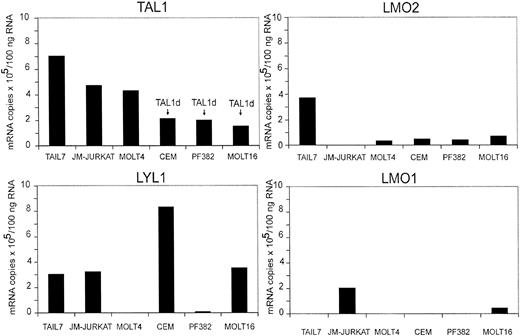
To further characterize TAIL7 cells, we have studied the expression of the T-cell oncogenes TAL1, TAL2, LYL1, LMO1, LMO2, HOX11, HOX11L2, and BHLHB1, which have been shown to be aberrantly expressed in T-cell leukemia.18,28 TAIL7 cells express high levels of TAL1, which are similar to those observed in JM-Jurkat cells and significantly higher than in most cell lines expressing oncogenic levels of TAL1 resulting from the presence of Tal1d rearrangement (Figure 8). TAIL7 cells also express high levels of LMO2 and intermediate levels of LYL1. We have not observed detectable expression of mRNAs for LMO1 (Figure 8), TAL2, HOX11, HOX11L2, or BHLHB1 (data not shown) in TAIL7 cells.
TAIL7 cells express the oncogenes TAL1, LMO2, and LYL1. Expression levels of the T-cell oncogenes TAL1, TAL2, LYL1, LMO1, LMO2, HOX11, HOX11L2, and BHLHB1 were determined by using quantitative RT-PCR analysis of TAIL7 cells and 5 immortalized T-cell leukemia lines. DNA PCR analysis to detect the Tal1d variant was performed as described previously.19
TAIL7 cells express the oncogenes TAL1, LMO2, and LYL1. Expression levels of the T-cell oncogenes TAL1, TAL2, LYL1, LMO1, LMO2, HOX11, HOX11L2, and BHLHB1 were determined by using quantitative RT-PCR analysis of TAIL7 cells and 5 immortalized T-cell leukemia lines. DNA PCR analysis to detect the Tal1d variant was performed as described previously.19
Finally, to establish a genetic “identity” of the TAIL7 cell line, a DNA profile was performed by using validated fingerprint techniques of different single locus variable numbers of tandem repeats (VNTRs) and short tandem repeats (STRs). The results are presented in Table 3. This DNA fingerprint may be useful for future genetic authentication of this cell line and to resolve potential problems of cell line misidentification and contamination, which have occurred with other cell lines and hampered the usefulness and accuracy of several studies.29
DNA fingerprint of TAIL7 cells
. | Fragment size . | . | |
|---|---|---|---|
. | Allele 1, bp . | Allele 2, bp . | |
| VNTR locus | |||
| D1S7 | 12 872 | 4147 | |
| D2S44 | 2775 | 1967 | |
| D4S139 | 6691 | 5597 | |
| D5S110 | 2594 | 1967 | |
| D8S358 | 4034 | 2023 | |
| D10S28 | 1850 | 1634 | |
| D17S79 | 1727 | 1494 | |
| STR locus* | |||
| D16S539 | 150 (09) | 154 (10) | |
| D18S51 | 284 (12) | 300 (16) | |
| D19S253 | 209 (07) | 229 (12) | |
| D21S11 | 219 (28) | 237 (32.2) | |
| D3S1358 | 131 (16) | 135 (17) | |
| D5S818 | 150 (11) | 154 (12) | |
| HUM CSF1PO | 308 (10) | 320 (13) | |
| HUM F13A01 | 187 (05) | 187 (05) | |
| HUM F13B | 182 (09) | 187 (10) | |
| HUM TPOX | 114 (08) | 118 (09) | |
| D10S1237 | 215 (16) | 227 (19) | |
| D7S820 | 244 (11) | 248 (12) | |
| D8S1179 | 104 (13) | 104 (13) | |
| HUM FES/FPS | 148 (10) | 152 (11) | |
| HUM FIBRA/FGA | 283 (21) | 283 (21) | |
| HUM TH01 | 193 (08) | 200 (09.3) | |
| HUM vWA | 147 (16) | 151 (17) | |
| Penta E | 421 (13) | 446 (18) | |
| Sex | |||
| Amelogenine† | 107 (X) | 113 (Y) | |
. | Fragment size . | . | |
|---|---|---|---|
. | Allele 1, bp . | Allele 2, bp . | |
| VNTR locus | |||
| D1S7 | 12 872 | 4147 | |
| D2S44 | 2775 | 1967 | |
| D4S139 | 6691 | 5597 | |
| D5S110 | 2594 | 1967 | |
| D8S358 | 4034 | 2023 | |
| D10S28 | 1850 | 1634 | |
| D17S79 | 1727 | 1494 | |
| STR locus* | |||
| D16S539 | 150 (09) | 154 (10) | |
| D18S51 | 284 (12) | 300 (16) | |
| D19S253 | 209 (07) | 229 (12) | |
| D21S11 | 219 (28) | 237 (32.2) | |
| D3S1358 | 131 (16) | 135 (17) | |
| D5S818 | 150 (11) | 154 (12) | |
| HUM CSF1PO | 308 (10) | 320 (13) | |
| HUM F13A01 | 187 (05) | 187 (05) | |
| HUM F13B | 182 (09) | 187 (10) | |
| HUM TPOX | 114 (08) | 118 (09) | |
| D10S1237 | 215 (16) | 227 (19) | |
| D7S820 | 244 (11) | 248 (12) | |
| D8S1179 | 104 (13) | 104 (13) | |
| HUM FES/FPS | 148 (10) | 152 (11) | |
| HUM FIBRA/FGA | 283 (21) | 283 (21) | |
| HUM TH01 | 193 (08) | 200 (09.3) | |
| HUM vWA | 147 (16) | 151 (17) | |
| Penta E | 421 (13) | 446 (18) | |
| Sex | |||
| Amelogenine† | 107 (X) | 113 (Y) | |
Variable number of tandem repeats (VNTR) and short tandem repeats (STR) were determined as described.
For STR, the allele nomenclature is shown in parentheses.
For amelogenine, the sex chromosome is indicated in parentheses.
Discussion
The development of effective therapies that specifically target critical components of the tumor signaling machinery requires the availability of significant numbers of malignant cells that accurately reflect the biology of the native cancer. This is particularly pertinent for the high-throughput screening of the increasing number of potentially active compounds made accessible by combinatorial chemistry, computational chemistry, and phage display. To expedite the development of signaling inhibitors active for T-cell leukemia and to circumvent the limitations because of an insufficient number of primary leukemia cells and the poor ex vivo survival and viability of these cells, we have established an IL-7–dependent cell line, TAIL7, from a pediatric patient. TAIL7 cells share the immunophenotype and clonality of their parental leukemia cells, as well as their responsiveness to IL-7 and IL-4. Stimulation of TAIL7 cells with IL-7 triggers signaling events, including the activation of the JAK/STAT pathway, hyperphosphorylation of Rb, and down-regulation of p27kip1. Importantly, specific blockade of JAK3 abrogates the proliferative and survival effects of IL-7 on TAIL7 cells. These results validate the use of this cell line for the screening of anti–T-ALL specific agents.
Tumor cell lines have been widely used for several decades to test and develop anticancer drugs. However, significant problems with established cell lines have been reported, including multiple transformations and derivation, misidentification and cross-contamination with other cell line(s).29 The screening and validation of agents that inhibit signaling molecules or pathways in a specific manner particularly requires cell lines that preserve the patterns of responsiveness to microenvironmental stimuli and the signaling pathways engaged by such stimuli. Most available cell lines derived from patients with T-ALL have accumulated genetic lesions that may alter their phenotype, responsiveness to particular stimuli, and, importantly, the integrity of the signaling pathways that are critical for their survival and growth. For example, “classical” leukemia T-cell lines, such as Jurkat, HPB-ALL, CEM, MOLT4, are immortalized, growth factor independent, and often unresponsive to stimuli that affect primary leukemia cells.29-31 Thus, they are fundamentally different from primary leukemia cells, which undergo spontaneous apoptosis ex vivo. In accordance, we tested Jurkat and SupT1, 2 commonly used T-ALL lines, for IL-7 responsiveness and observed that IL-7 does not increase their high viability or proliferation (data not shown). Interestingly, the chemotherapeutic drugs dexamethasone and doxorubicin are cytotoxic to TAIL7 cells at concentrations that promote apoptosis of primary leukemia T cells26,27 but not of Jurkat cells.32,33 Notably, the fact that the rate of apoptosis of primary T-ALL cells can be reduced by cytokines20,34 or stromal cells35 suggests that in vivo microenvironmental signals are continuously sustaining the leukemia cells,36 a physiologic dependence that the available T-ALL cell lines no longer require. Therefore, malignant T-cell lines that maintain the biologic and molecular signatures of their parental malignant cells are necessary for the in-depth study of the biology of T-ALL, the identification of therapeutic targets, and the design and development of more specific antileukemia drugs.
Because the possibility existed that long-term cultured TAIL7 cells could acquire growth-factor independence via constitutive activation of proliferation and survival pathways, we routinely assessed the levels of phosphorylation of STAT5, Erk1/2, and Akt in unstimulated TAIL7 cells. No constitutive phosphorylation of STAT5 or Erk1/2 was observed (Figure 6B), as well as STAT1 or STAT3 (data not shown). Low levels of Akt phosphorylation were detected (as in Figure 6B), which were negligible in comparison to the levels found in cell lines that show constitutive activation of PI3K (such as Jurkat). Importantly, no changes in intensity or pattern of responsiveness to IL-7 were observed in the periodic evaluation of the signaling properties of TAIL7 cells (data not shown). We have also confirmed that TAIL7 cells do not produce IL-7, thus excluding the possibility of an autocrine loop. These observations are in agreement with the slow proliferation rate of TAIL7 cells and with the blockade of proliferation and induction of cell death on IL-7 withdrawal, as periodically assessed.
Several lines of evidence implicate IL-7 in the pathophysiology of lymphoblastic leukemia, particularly that of T-cell ALL. First, leukemic T cells express receptors for IL-7 (IL7-Rα and γc) and are responsive to IL-7, which promotes their survival and proliferation.4,9,10,20,37 Second, IL-7 is present in the microenvironments where T-cell leukemia develops. IL-7 is produced by BM “stromal” cells38 and by BM endothelial cells and mesenchymal stem cells from both healthy donors and leukemia patients (J.A.Y. and A.A.C. et al, unpublished observations, December 2002). Third, studies with IL-7 transgenic mice have shown that the deregulated expression of IL-7 perturbs thymic T-cell development and promotes malignant transformation of T and B lymphocytes.8 Fourth, different studies have shown that IL-7, although not essential on its own, can cooperate in malignant transformation.39-41 Finally, IL-7 is involved in the development of normal thymocytes, which represent the normal counterparts of malignant T cells. It has been shown that IL-7 stimulates the growth of immature double-negative and mature single-positive thymocytes in thymic organ cultures.5,11,42,43 In addition, stimulation of the IL-7R plays a critical role in the maintenance of the peripheral T-cell pool,6,44 and defective IL-7R expression results in T–B+NK+ severe combined immunodeficiency.45,46 On the basis of these lines of evidence, we have selected IL-7 as the cytokine for the establishment of growth factor–dependent TAIL7 cells. We have attempted to generate IL-7–dependent cell lines from other patients with IL-7–responsive T-ALL (n = 8). Cultures of IL-7–supported leukemia cells could be maintained for periods of 2 to 4 months, but no long-term cell lines could be established, with the exception of a BM endothelium-dependent, IL-7-responsive T-ALL line (TLP-3; characterization in progress).
It is our contention that the dissection of the mechanisms involved in leukemic T-cell growth and survival in model cell lines that recapitulate the pathobiology of the disease will lead to the identification of optimal targets for intervention and subsequent development of more specific antileukemia drugs. An approach successfully used in other tumors is to define the signaling pathways that mediate the critical stimuli implicated in tumor cell survival or growth.1,2,47,48 We observed that in both TAIL7 cells and their parental leukemia T cells, stimulation by IL-7 triggers the phosphorylation of various substrates, and the activation of major cytokine-induced pathways, such as JAK/STAT, ras/MAPK, and PI3K/Akt pathways. Activation of the JAK/STAT pathway has been shown in primary T-ALL cells and TEL-JAK2 transgenic mice develop T-cell leukemia.49-51 The magnitude of the inhibitory effect of the JAK3 inhibitor WHI-P131 on TAIL7 cell survival and proliferation suggests that some degree of “constitutive” activation of JAK3 occurs in TAIL7 cells, in addition to that mediated by the stimulation with IL-7. We have also found that WHI-P131 inhibits phosphorylation of STAT5, a critical downstream target of JAK3 and that WHI-P131 is also an effective inhibitor of primary T-ALL cells (J.T.B. et al, manuscript in preparation). Activation of MAPK pathway has been implicated in thymic T-cell selection,52 and mutations that render p21ras constitutively active have been found in a wide variety of human tumors.47 Likewise, PI3K/Akt pathway activation has been shown to play a major role in the progression of many different cancers.53,54 Consistent with our studies on primary leukemia T-cells,4 IL-7 stimulation of TAIL7 cells also modulates the expression and activation of cell cycle regulators, as demonstrated by the down-regulation of p27kip1 and hyperphosphorylation of Rb. Moreover, the mTOR-specific inhibitor rapamycin, which up-regulates the endogenous levels of p27kip1, promotes apoptosis of both IL-7–stimulated TAIL7 and primary T-ALL cells. In contrast, rapamycin does not affect the viability of healthy T cells.32,55 The TAIL7 cell line can thereby be used to screen for small molecules that specifically inhibit critical steps during cell cycle progression and to evaluate the signal transduction pathways that are affected. Finally, because TAIL7 cells express a defined pattern of T-cell oncogenes (high levels of TAL1 and LMO2, and intermediate levels of LYL1) they are a valuable tool for the analysis of transformation pathways in T-ALL, specifically to define the role of misexpression of these transcription factors in promoting tumor cell survival and proliferation.
The development of T-cell leukemia in children with severe combined immunodeficiency who were treated with gene therapy using a retroviral vector driving the constitutive expression of the γc gene56 has highlighted the potential role for cytokine-mediated signaling in human leukemogenesis. In these unfortunate cases, the integration of the retroviral vector in the vicinity of the LMO2 oncogene resulted in the activation of expression of this leukemogenic transcription factor, which may cooperate with the constitutive expression of the γc chain, and consequent activation of JAK3 and STAT5 pathway, to promote the occurrence of leukemia.
In conclusion, we established a cell line, TAIL7, derived from a patient with T-ALL that is dependent on IL-7 and that retains the IL-7–triggered engagement of defined functional and signaling events observed not only in their parental leukemia cells but also in IL-7–responsive primary leukemias, which account for most T-ALL cases.4 This novel cell line is presently being used as a model system for the dissection of the signaling pathways implicated in T-ALL survival and growth, for the identification of therapeutic targets, and, most important, for the screening of putative antileukemia agents. Because TAIL7 reflects the cellular and molecular properties of primary T-cell leukemia, it represents a valuable research tool.
Prepublished online as Blood First Edition Paper, November 13, 2003; DOI 10.1182/blood-2002-12-3861.
Supported by grants from the National Institutes of Health (P01-CA68484) and Fundação para a Ciência e a Tecnologia (FCT-Portugal; PRAXIS-13240 and POCTI-34914). J.T.B. was supported by the Program PRAXIS XXI, FCT, Portugal. J.P.V. was also funded by FCT-Portugal. J.A.Y. was partially funded by the Children's Leukemia Research Association. A.A.F. is a fellow of the Leukemia/Lymphoma Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
J.T.B. was a student of the GABBA program of the University of Porto, Portugal.
We thank Dr Marcela Araújo and Dr António F. Ferreira Neto for the assistance on the clonality and DNA fingerprinting studies, respectively. We thank Dr W. Nicholas Haining and Thomas Keenan for the critical reading of the manuscript. We thank Joana Brandão and Ana Silva for technical assistance.

![Figure 3. IL-7 activates TAIL7 cells and promotes cell cycle progression and increased viability. (A) TAIL7 cells were cultured for 72 hours with or without IL-7, and the cell cycle phase distribution was examined by PI staining and flow cytometry. Results are representative of 3 independent experiments. (B) At 72 hours, activation was assessed by flow cytometry, by gating on the live cell population, and by measuring the changes in cell size (forward light scatter [FSC]) and complexity (sideward light scatter [SSC]). The “percentage of activated cells” shown was defined relative to the distribution of the control population (medium alone). Similar results were obtained in 7 independent experiments. (C) At 96 hours, cells were stained with Annexin V–FITC/PI, and the percentage of viable cells was assessed by flow cytometry. Results are expressed as mean and standard deviation of 3 samples for each condition and are representative of 5 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/5/10.1182_blood-2002-12-3861/6/m_zh80050457290003.jpeg?Expires=1769111093&Signature=pA6ayo9fxf2scdBKcADNjA7uZMS1ga7YNRqDLfNSsZtj7girBRfatx6oJiJzgAfcH4GSODoErnc22ewzYUiVpcDkktxpo7rrWjJzHb9iaDu-l760qH0skBmLGSz2YnTq9ZTznxVo3QAzNyyzYqKK7HKzLY49BPaCsuXHsXb3PMqNQFO6Ryp7pnIC5sOkL~-MdAUWLuo8ilxDePcFcDdheoogYdUwE3g~3Bxtm272SCnAdtrZXEHSwu0vtW93cjucts2RzKxcs2xaHrg3oGyOSNtDT4wHeSm7Ytsk7hwSe7kU69DI5OZf0XDFTuVDYhJwxnT~I4qhpsz5uJnMblaYbA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal