Abstract
Constitutively activating mutations of FMS-like tyrosine kinase 3 (FLT3) occur in approximately one third of patients with acute myeloid leukemia (AML) and are associated with poor prognosis. Altered FLT3 signaling leads to antiapoptotic and proliferative signaling pathways. We recently showed that these mutations can also contribute to the differentiation arrest that characterizes leukemia. In this report we investigated the mechanism by which internal tandem duplication (ITD) mutation of FLT3 signaling blocks differentiation. Normally, myeloid differentiation requires the induction of CCAAT/enhancer-binding protein α (C/EBPα) and PU.1 expression. Expression of both genes was repressed by FLT3/ITD signaling in 32Dcl3 (32D) cells and this repression was overcome by treatment with a FLT3 inhibitor, allowing differentiation to proceed. We also observed increased expression of C/EBPα and PU.1 accompanied by signs of differentiation in 2 of 3 primary AML samples from patients with FLT3/ITD mutations receiving a FLT3 inhibitor, CEP-701, as part of a clinical trial. Forced expression of C/EBPα was also able to overcome FLT3/ITD-mediated differentiation block, further proving the importance of C/EBPα in this process.
Introduction
FMS-like tyrosine kinase 3 (FLT3) is a member of the class III receptor tyrosine kinase family that also includes c-kit receptor tyrosine kinase (KIT) and FMS, 2 other receptors with important roles in hematopoiesis. FLT3 is preferentially expressed on hematopoietic stem/progenitor cells and plays a role in both differentiation and proliferation.1 Somatic mutations of FLT3 involving internal tandem duplications (ITDs) of the juxtamembrane domain or D835 point mutations in the activation loop have been identified in approximately 17% to 34% and 7% of acute myeloid leukemia (AML) patients, respectively.2-6 The ITD mutations appear to activate the tyrosine kinase domain of the FLT3 receptor through constitutive dimerization and result in autophosphorylation of the receptor.7 Constitutively activated FLT3 contributes to leukemic transformation and portends an especially poor prognosis for patients with this mutation.8-10
Uncontrolled proliferation, antiapoptotic advantages, and a block in differentiation characterize acute leukemia. The role of FLT3/ITDs in giving a proliferative and antiapoptotic advantage to cells has been reported.11,12 Recently, we demonstrated that FLT3/ITD expression also contributes to a blockage of granulocyte colony-stimulating factor (G-CSF)–mediated differentiation in 32Dcl3 (32D) cells.13 However, the mechanism by which this block occurs is not known.
The development of mature granulocytes from hematopoietic progenitor cells is regulated by a complex network of transcription factors.14 The family of CCAAT/enhancer-binding proteins (C/EBPs) plays a key role in myeloid differentiation. One member of the C/EBP family, C/EBPα, is prominently expressed in early myeloid progenitor cells, and its expression decreases as these progenitors further differentiate into mature granulocytes.15 Absence of neutrophil development and G-CSF signaling is observed in C/EBPα-deficient mice.16 Overexpression of C/EBPα in the HL-60 and U937 human leukemia cell lines or murine 32D cells leads to the development of neutrophils.17,18 Recently, mutations in the C/EBPα gene have been identified in AML. The mutant C/EBPα blocks wild-type C/EBPα DNA binding and transactivation of granulocyte target genes in a dominant-negative manner, with a resultant failure in granulocytic differentiation.19
PU.1 is a member of the Ezb transformation–specific sequence (Ets) family of transcription factors and is expressed in granulocytic, monocytic, and B-lymphoid cells.20,21 PU.1 expression levels increase during the differentiation of granulocytes.21-23 PU.1-deficient mice exhibit defects in the development of neutrophils, macrophages, and B cells.24,25 Mutant alleles of the PU.1 gene have been described in 9 of 126 AML patients in one study.26 DNA binding and transactivation of the macrophage-CSF (M-CSF) receptor promoter, a direct PU.1 target gene, were deficient in most PU.1 mutants, which affected the DNA-binding domain. Additionally, these mutations impaired the ability of PU.1 to synergize with PU.1-interacting proteins such as AML1 or c-Jun in the activation of PU.1 target genes, suggesting disruption of PU.1 function contributes to the block in differentiation found in AML patients. Both C/EBPα and PU.1 are needed for the full commitment and differentiation of the granulocytic lineage.14 During terminal granulocytopoiesis, C/EBPα induces PU.1 gene expression.18,27 A recent report using microarray data showed that FLT3/ITD can suppress C/EBPα and PU.1 expression in comparison with wild-type FLT3.28 Whether suppression of C/EBPα plays a role in the blockage of differentiation mediated by FLT3/ITD is unknown.
To investigate the possible mechanism of FLT3/ITD-mediated block in differentiation of 32D cells, the effect of FLT3/ITD expression on C/EBPα and its downstream target, PU.1, was examined in this study. Induction of C/EBPα and PU.1 expression in response to G-CSF was suppressed in FLT3/ITD-expressing 32D cells. This suppression could be overcome through inhibition of FLT3 tyrosine kinase activity. Similar results were also observed in primary blast samples from some FLT3/ITD-positive AML patients in response to therapy with a FLT3 kinase inhibitor. Forced expression of C/EBPα was able to induce PU.1 expression and overcome the FLT3/ITD-mediated block in granulocytic differentiation of 32D cells. This supports the FLT3/ITD-mediated suppression of C/EBPα expression as the major mechanism by which FLT3/ITD signaling contributes to blocked differentiation.
Materials and methods
Reagents
Recombinant murine interleukin 3 (IL-3) was purchased from Pepro Tech (Rocky Hill, NJ). Rabbit antihuman FLT3, rabbit antirat C/EBPα, rabbit antimouse PU.1 antibody, and mouse antihuman heat shock protein 90 (HSP 90) antibody were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse antihuman CD13, CD33, CD117, CD34, CD38, human leukocyte antigen–DR (HLA-DR), CD15, and CD11b monoclonal antibodies were purchased from Becton Dickinson (San Jose, CA). Monoclonal mouse antiphosphotyrosine antibody (4G10) and recombinant protein A–agarose were purchased from Upstate Biotechnology (Lake Placid, NY). Horseradish peroxidase–conjugated secondary antibody and enhanced chemiluminescence (ECL) detection system were from Amersham (Arlington Heights, IL). CEP-701 was graciously provided by Cephalon (West Chester, PA).
Cells
The 32D cells were cultured in RPMI 1640, supplemented with 1 ng/mL recombinant mouse IL-3, 10% heat-inactivated fetal calf serum (FCS), and antibiotics (penicillin and streptomycin) at 37°C with 5% CO2. For the differentiation assay, 32D cells were washed twice with phosphate-buffered saline (PBS) and then transferred to medium containing 20 ng/mL G-CSF (Amgen, Thousand Oaks, CA).
The 32D cells expressing the C/EBPα-estrogen receptor ligand-binding domain fusion gene, 32D/α-ER, have been previously described.18 The 32D/α-ER cells were cultured in IL-3 containing Iscove modified Dulbecco medium (IMDM) supplemented with 2 μg/mL puromycin. To induce activation of C/EBPα-ER, estradiol at a final concentration of 1 μM was added to the medium.
Primary AML blast cells from bone marrow were collected after obtaining patient consent under a Johns Hopkins University Institutional Review Board–approved protocol and purified by Ficoll-Hypaque (Amersham, Piscataway, NJ) density centrifugation.
DNA constructs and retroviral transduction
Constructs of pBabePuro retroviral vectors containing FLT3/ITD or wild-type FLT3 and transduction of 32D cells with these vectors have been described previously.13 Similarly, pBabe-Neo retroviral vectors containing FLT3/ITD or wild-type FLT3 were made and used to transduce 32D/α-ER cells. Stable transfectants were then selected by limiting dilution in 96-well plates with 1 mg/mL G418.
Northern blot analysis
Total RNA was extracted from 1 × 107 cells (RNeasy Mini Kit; Qiagen, Valencia, CA). The RNA samples were separated on 1% formaldehyde-denaturing agarose gels and transferred to nylon membranes (NEN, Boston, MA). The cDNAs of C/EBPα, PU.1, myeloperoxidase (MPO), lysozyme, lactoferrin, and actin were used as probes. All probes were labeled with 32P–deoxycytidine triphosphosphate (32P-dCTP) using a random primer labeling kit (Stratagene, Cedar Creek, TX). Probes were hybridized to blots in 6 × SSC (saline sodium citrate), 50% formamide, 0.1% SDS (sodium dodecyl sulfate), 2 × Denhardt reagent, and 0.125 mg/mL salmon sperm DNA at 42°C for 16 hours. The membranes were washed twice with 2 × SSC/0.1% SDS for 30 minutes at 42°C and then exposed to XAR film (Kodak, Rochester, NY) at –80°C for 2 to 24 hours.
Immunoprecipitation and Western blot analysis
Cells were washed twice with ice-cold PBS and lysed for 30 minutes in ice-cold nonidet P-40 (NP-40) lysis buffer (20 mM Tris-HCl, pH 7.4; 150 mM NaCl; 100 mM NaF; 10% glycerol; 1% NP-40; and 10 mM EDTA [ethylenediaminetetraacetic acid]) containing protease and phosphatase inhibitors (2 mM sodium orthovanadate, 50 μg/mL antipain, 5 μg/mL aprotinin, 1 μg/mL leupeptin, and 10 μg/mL phenylmethylsulfony fluoride; Sigma, St Louis, MO). To detect FLT3, clarified lysates (500 μg) were incubated with antibody against human FLT3 and with protein A–agarose at 4°C. The immunoprecipitates were washed 3 times with ice-cold TBS-T (10 mM Tris-HCl, pH 7.4; 100 mM NaCl; 0.1% Tween-20), resuspended in SDS sample buffer, heated, and separated by 8% SDS–polyacrylamide gel electrophoresis (PAGE) gel. Gels were blotted onto polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA) and stained with the indicated antibody. Antibody binding was detected by incubation with a horseradish peroxidase–conjugated secondary antibody, followed by chemiluminescence detection. For detection of PU.1, 50 μg of protein lysate was resuspended in SDS sample buffer, electrophoresed in 10% SDS-PAGE gels, and then transferred to PVDF membrane, and blotted with anti-PU.1 antibody. Protein lysate used for detection of C/EBPα was prepared from 1 × 106 cells that were lysed in 2 × Sample buffer (100 mM Tris-HCl, pH 6.8; 4% SDS; 20% glycerol; 200 mM dithiothreitol; 4% β-Mercaptoethanol). The lysates were subjected to Western blotting with antibody against C/EBPα.
Cellular differentiation and proliferation assay
Cellular differentiation was assessed either morphologically or immunophenotypically. Wright-Giemsa stains of cytospun 32D cells were examined by × 40 microscopy. Antigen expression profiles of primary AML blasts were determined by multiparameter flow cytometric analysis using a FACSCalibur automated flow cytometry system (Becton Dickinson). Cellular proliferation was determined by counting the viable cell numbers on the basis of trypan blue exclusion.
Cell cycle analysis
Cells (1 × 106) were washed twice with PBS, fixed with 70% methanol, treated with 20 μg/mL RNase, and stained with 50 μg/mL propidium iodide. Cells were subjected to flow cytometric analysis of DNA content using a flow cytometer (FACSort; Becton Dickinson). The percentages of cell cycle distribution were calculated by Cell Quest software (Becton Dickinson).
Real-time reverse transcription–polymerase chain reaction (RT-PCR)
Total RNA was isolated from patients' blast cells using Rneasy columns (Qiagen). The cDNA was synthesized from 2 μg RNA from each sample using SuperScript First-Strand Synthesis System (Invitrogen, Carlsbad, CA) in a 40-μL reaction system. Two microliters of cDNA was amplified using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) in the iCycler iQ Real-Time Detection System (Bio-Rad). PCR were performed at 50°C for 2 minutes, at 95°C for 3 minutes, followed by 40 cycles of denaturation at 95°C for 15 seconds, and annealing and extension at 60°C for 1 minute. The primer pairs were as follows: C/EBPα: 5′ TGGACAAGAACAGCAACGAG 3′, 5′ TTGTCACTGGTCAGCTCCAG 3′; PU.1: 5′ GTGCCCTATGACACGGATCT 3′, 5′ GAAGCTCTCGAACTCGCTGT 3′; actin: 5′ TGCGTGACATTAAGGAGAAG 3′, 5′ GCTCGTAGCTCTTCTCCA 3′.
Relative gene expression levels were calculated using standard curve generated by serial dilution of plasmids containing human C/EBPα, PU.1, or actin cDNA. The measured amount of C/EBPα and PU.1 expression in each sample was normalized by actin expression.
Results
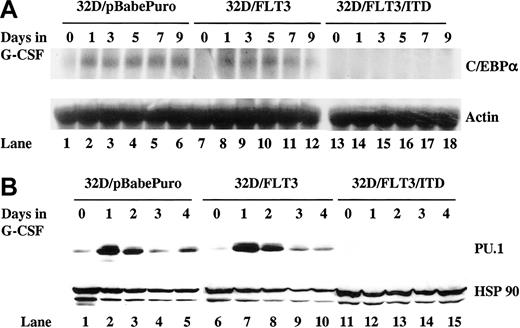
Induction of C/EBPα and PU.1 by G-CSF is inhibited in 32D cells expressing FLT3/ITD
Normally, induction of C/EBPα and PU.1 expression is observed during G-CSF–mediated differentiation of 32D cells. In order to investigate whether FLT3/ITD interferes with induction of C/EBPα and PU.1 expression, stable transfectants of 32D cells expressing FLT3/ITD (32D/FLT3/ITD), wild-type FLT3 (32D/FLT3), or empty vector (32D/pBabePuro) were established. Clones expressing comparable levels of FLT3/ITD or FLT3 were selected to allow a direct comparison of the effects of the different constructs as previously described.13 The 32D/FLT3/ITD are unable to differentiate to granulocytes in response to G-CSF, both morphologically and functionally, in contrast to 32D/FLT3 and 32D/pBabePuro cells.13 In this study, transfected 32D cells were exposed to G-CSF (20 ng/mL) for 9 days and the expression of C/EBPα and PU.1 was analyzed in 3 independent clones (representative data are shown). G-CSF was unable to induce C/EBPα and PU.1 expression in 32D/FLT3/ITD cells (Figure 1A-B). The basal level of PU.1 expression was also suppressed (Figure 1B lane 11 vs 1 and 6). In contrast, induction of C/EBPα and PU.1 was observed by day 1 in 32D/FLT3 and 32D/pbabePuro cells in response to G-CSF (Figure 1A-B). FLT3 ligand (FL) stimulation of 32D/FLT3 cells did not suppress induction of C/EBPα or PU.1 (data not shown). Thus, induction of C/EBPα and PU.1 expression is inhibited in 32D cells due to the expression of FLT3/ITD.
Induction of C/EBPα and PU.1 by G-CSF is inhibited in 32D cells expressing FLT3/ITD. The 32D/pBabePuro, 32D/FLT3, and 32D/FLT3/ITD cells were washed and transferred from medium containing IL-3 (1 ng/mL) to medium containing G-CSF (20 ng/mL) for 9 days. Every other day, fresh G-CSF–containing medium was added to cells to replace the old medium. (A) Total cellular RNA from 32D/pBabe (lanes 1-6), 32D/FLT3 (lanes 7-12), and 32D/FLT3/ITD (lanes 13-18) cells was prepared from 1 × 107 cells after 0, 1, 3, 5, 7, and 9 days in G-CSF. RNA (10 μg) from each time point was then subjected to Northern blotting with a 1.1-kb NcoI-NcoI fragment of rat C/EBPα (top). The same membrane was stripped and then reprobed with actin cDNA probe (bottom). (B) Total protein lysates from 32D/pBabe (lanes 1-5), 32D/FLT3 (lanes 6-10), and 32D/FLT3/ITD (lanes 11-15) cells were prepared from 1 × 107 cells after 0, 1, 2, 3, and 4 days in G-CSF. Protein lysates (50 μg) from each time point were then subjected to Western blotting with antibody against PU.1 (top). The same membrane was stripped and then incubated with antibody against HSP 90 (bottom).
Induction of C/EBPα and PU.1 by G-CSF is inhibited in 32D cells expressing FLT3/ITD. The 32D/pBabePuro, 32D/FLT3, and 32D/FLT3/ITD cells were washed and transferred from medium containing IL-3 (1 ng/mL) to medium containing G-CSF (20 ng/mL) for 9 days. Every other day, fresh G-CSF–containing medium was added to cells to replace the old medium. (A) Total cellular RNA from 32D/pBabe (lanes 1-6), 32D/FLT3 (lanes 7-12), and 32D/FLT3/ITD (lanes 13-18) cells was prepared from 1 × 107 cells after 0, 1, 3, 5, 7, and 9 days in G-CSF. RNA (10 μg) from each time point was then subjected to Northern blotting with a 1.1-kb NcoI-NcoI fragment of rat C/EBPα (top). The same membrane was stripped and then reprobed with actin cDNA probe (bottom). (B) Total protein lysates from 32D/pBabe (lanes 1-5), 32D/FLT3 (lanes 6-10), and 32D/FLT3/ITD (lanes 11-15) cells were prepared from 1 × 107 cells after 0, 1, 2, 3, and 4 days in G-CSF. Protein lysates (50 μg) from each time point were then subjected to Western blotting with antibody against PU.1 (top). The same membrane was stripped and then incubated with antibody against HSP 90 (bottom).
Inhibition of FLT3 tyrosine kinase activity restores G-CSF–mediated C/EBPα and PU.1 induction in 32D/FLT3/ITD cells
To further confirm that FLT3/ITD-mediated constitutive activation of FLT3 receptor leads to the suppression of C/EBPα and PU.1 expression in 32D/FLT3/ITD cells, CEP-701 was used to inhibit FLT3 tyrosine kinase activity. CEP-701 is an indolocarbazole derivative that is very potent (50% inhibitory concentration [IC50]: 2 nM) for inhibiting FLT3 in BaF3/FLT3/ITD and 32D/FLT3/ITD cell lines, as well as primary AML samples expressing FLT3/ITDs.13,29 In the presence of 5 nM CEP-701 and 20 ng/mL G-CSF, induction of C/EBPα and PU.1 in 32D/FLT3/ITD cells was detected by day 3 (Figure 2A lanes 10-14; Figure 2B lanes 8-10; Figure 2C lanes 10-12). This restoration of C/EBPα and PU.1 induction was also observed in 2 additional clones (data not shown) and was accompanied by morphologic differentiation and up-regulation of myeloid-specific differentiation markers including MPO and lysozyme.13 Thus, the tyrosine kinase activity of FLT3/ITD is required for the observed inhibition of C/EBPα and PU.1 expression.
CEP-701 restores G-CSF–mediated induction of C/EBPα and PU.1 expression in 32D/FLT3/ITD cells. (A) 32D/FLT3/ITD cells were treated without (lanes 1-7) or with (lanes 8-14) CEP-701 (5 nM) in the presence of G-CSF (20 ng/mL) with replacement of the medium every other day. Total cellular RNA was prepared from 1 × 107 32D/FLT3/ITD cells on days 0, 1, 3, 5, 7, 9, and 11. RNA (10 μg from each sample) was then subjected to Northern blotting with C/EBPα and actin cDNA probes. (B) Total protein lysates were prepared from 1 × 106 32D/FLT3/ITD cells on days 0, 1, 3, 7, and 9 treated without (lanes 1-5) or with (lanes 6-10) 5 nM CEP-701 in the presence of G-CSF. The whole protein lysates from each time point were then subjected to Western blotting with antibody against C/EBPα and HSP 90. Protein lysates from parental 32D cells (lane 11) were used as a positive control. (C) Total protein lysates were prepared from 1 × 107 32D/FLT3/ITD cells on days 0, 1, 2, 3, 4, and 5 treated without (lanes 1-6) or with (lane 7-12) 5 nM CEP-701 in the presence of G-CSF. Protein lysates (50 μg) from each time point were then subjected to Western blotting with antibody against PU.1 and HSP 90.
CEP-701 restores G-CSF–mediated induction of C/EBPα and PU.1 expression in 32D/FLT3/ITD cells. (A) 32D/FLT3/ITD cells were treated without (lanes 1-7) or with (lanes 8-14) CEP-701 (5 nM) in the presence of G-CSF (20 ng/mL) with replacement of the medium every other day. Total cellular RNA was prepared from 1 × 107 32D/FLT3/ITD cells on days 0, 1, 3, 5, 7, 9, and 11. RNA (10 μg from each sample) was then subjected to Northern blotting with C/EBPα and actin cDNA probes. (B) Total protein lysates were prepared from 1 × 106 32D/FLT3/ITD cells on days 0, 1, 3, 7, and 9 treated without (lanes 1-5) or with (lanes 6-10) 5 nM CEP-701 in the presence of G-CSF. The whole protein lysates from each time point were then subjected to Western blotting with antibody against C/EBPα and HSP 90. Protein lysates from parental 32D cells (lane 11) were used as a positive control. (C) Total protein lysates were prepared from 1 × 107 32D/FLT3/ITD cells on days 0, 1, 2, 3, 4, and 5 treated without (lanes 1-6) or with (lane 7-12) 5 nM CEP-701 in the presence of G-CSF. Protein lysates (50 μg) from each time point were then subjected to Western blotting with antibody against PU.1 and HSP 90.
Inhibition of FLT3 signaling up-regulates C/EBPα and PU.1 expression and leads to increased expression of differentiation markers in blasts of 2 of 3 FLT3/ITD-positive patients receiving an oral FLT3 inhibitor
Expression of C/EBPα is required for the differentiation of myeloid lineage cells. If C/EBPα expression is inhibited due to FLT3/ITD signaling, an up-regulation of C/EBPα expression might follow the inhibition of FLT3 tyrosine kinase activity. Several FLT3/ITD-positive AML patients have participated in a clinical trial of oral CEP-701 at our institute. Sufficient bone marrow blasts for analysis of C/EBPα expression by Northern blotting were available from one patient and were examined prior to and after 4 weeks of therapy with CEP-701. This bone marrow sample consisted of more than 90% blasts by visual inspection before and after 4 weeks of therapy. After treatment with CEP-701 for 4 weeks, an increase of C/EBPα expression was observed (Figure 3A lane 2 vs 1). Real-time RT-PCR was also used to examine expression of C/EBPα and PU.1 in the blasts of 3 patients before and after treatment with a FLT3 tyrosine kinase inhibitor. More than a 2-fold increase of C/EBPα and PU.1 expression was observed in patient no. 2 and no. 3, while little change was observed in patient no. 1 (Figure 3B). Patient no. 2 is the same patient whose Northern blots appear in Figure 3A. To study whether or not any signs of differentiation occurred due to the increased level of C/EBPα and PU.1 expression, these blast samples were analyzed by flow cytometric analysis. In patient no. 2, the blasts prior to therapy were positive for CD38 and HLA-DR, as well as the myeloid-associated markers CD13, CD33, and CD117. The blast population showed little expression of CD15 and CD11b, markers associated with myeloid differentiation.30,31 After 4 weeks of FLT3 inhibitor therapy, the bone marrow still consisted of more than 90% blasts by morphologic examination. When analyzed by flow cytometric analysis, the blasts now showed signs of differentiation with an increase of CD15 expression from 4.0% to 51.5% (Figure 3C). The population of leukemic cells from patient no. 3 showed an immunophenotypic profile similar to that noted in patient no. 2 prior to CEP-701 therapy. After therapy, the blasts showed evidence of differentiation, with complete loss of CD34, HLA-DR, and CD117 expression, and increased CD11b expression from 11.8% to 70.0% (Figure 3D). In contrast, the posttherapy marrow specimen from patient no. 1 showed little in the way of immunophenotypic changes in the blast population. These data suggest that inhibition of FLT3 tyrosine kinase activity may reverse the suppression of C/EBPα and PU.1 expression and induce some signs of differentiation in at least some cases of AML.
Inhibition of FLT3 kinase activity leads to an up-regulation of C/EBPα and PU.1 and an increase in the expression of differentiation markers in blasts of 2 of 3 FLT3/ITD-positive AML patients. (A) AML bone marrow blasts were collected before and after 4 weeks of treatment with CEP-701 from a FLT3/ITD-positive AML patient who was enrolled in the CEP-701 clinical trial and whose bone marrow blasts did not decrease with therapy. Total cellular RNA was prepared and 7 μg from each sample was subjected to sequential Northern blotting with a 0.7-kb EcoRI-HindIII fragment of human C/EBPα cDNA17 and actin cDNA probe. (B) Bone marrow blasts were obtained before and after 2 or 4 weeks of treatment with CEP-701 from 3 FLT3/ITD-positive AML patients whose bone marrow did not show a decrease of blasts in response to therapy. C/EBPα and PU.1 mRNA expression were analyzed using real-time RT-PCR analysis. Levels of C/EBPα and PU.1 were expressed relative to actin. Error bars indicate SD. (C-D) Bone marrow blasts were collected prior to and after 2 or 4 weeks of treatment with CEP-701 from 2 FLT3/ITD-positive AML patients. Antigen expression profiles of these blasts were determined by multiparameter flow cytometric analysis. AML blasts, characterized by low side-scattering value and expression of CD45, CD117, and CD33 (data not shown), were specifically gated (left). Expression of CD15 or CD11b by these blasts was then analyzed (right).
Inhibition of FLT3 kinase activity leads to an up-regulation of C/EBPα and PU.1 and an increase in the expression of differentiation markers in blasts of 2 of 3 FLT3/ITD-positive AML patients. (A) AML bone marrow blasts were collected before and after 4 weeks of treatment with CEP-701 from a FLT3/ITD-positive AML patient who was enrolled in the CEP-701 clinical trial and whose bone marrow blasts did not decrease with therapy. Total cellular RNA was prepared and 7 μg from each sample was subjected to sequential Northern blotting with a 0.7-kb EcoRI-HindIII fragment of human C/EBPα cDNA17 and actin cDNA probe. (B) Bone marrow blasts were obtained before and after 2 or 4 weeks of treatment with CEP-701 from 3 FLT3/ITD-positive AML patients whose bone marrow did not show a decrease of blasts in response to therapy. C/EBPα and PU.1 mRNA expression were analyzed using real-time RT-PCR analysis. Levels of C/EBPα and PU.1 were expressed relative to actin. Error bars indicate SD. (C-D) Bone marrow blasts were collected prior to and after 2 or 4 weeks of treatment with CEP-701 from 2 FLT3/ITD-positive AML patients. Antigen expression profiles of these blasts were determined by multiparameter flow cytometric analysis. AML blasts, characterized by low side-scattering value and expression of CD45, CD117, and CD33 (data not shown), were specifically gated (left). Expression of CD15 or CD11b by these blasts was then analyzed (right).
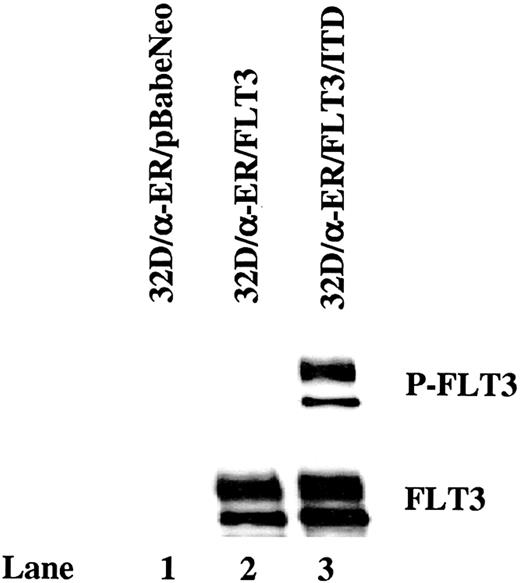
FLT3/ITD is constitutively autophosphorylated in 32D cells expressing C/EBPα-ER and transforms these cells to IL-3–independent growth
If the FLT3/ITD-mediated block of differentiation in 32D cells is caused by its suppression of C/EBPα expression, forced expression of C/EBPα might be expected to overcome the blockage of differentiation. To test this hypothesis, FLT3/ITD, wild-type FLT3, and pBabeNeo vector were introduced into 32D/α-ER cells via retroviral transduction. The 32D/α-ER cells are 32D cells expressing a fusion protein (C/EBPα-ER) containing the entire C/EBPα polypeptide and the ligand-binding domain of the estrogen receptor under control of the long terminal repeat (LTR) promoter. C/EBPα-ER is inactive but becomes active in the presence of estradiol and activates transcription from C/EBPα-binding sites.32 Stable expression of FLT3/ITD and FLT3 by subclones was confirmed by Western blotting of cell lines cloned by limiting dilution. Clones expressing comparable levels of FLT3/ITD (32D/α-ER/FLT3/ITD) or FLT3 (32D/α-ER/FLT3) were selected (Figure 4). Wild-type FLT3-transfected 32D/α-ER cells showed little endogenous phosphorylation (Figure 4 lane 2). In contrast, FLT3 was tyrosine phosphorylated in 32D/α-ER/FLT3/ITD cells even in the absence of added FL (Figure 4 lane 3). The 32D/α-ER/FLT3/ITD cells proliferated without the addition of IL-3, whereas wild-type FLT3– and pBabeNeo vector–transfected 32D/α-ER cells required IL-3 for their growth (data not shown). Subsequent experiments with 32D/α-ER/FLT3/ITD cells were performed in the absence of IL-3 and G-CSF.
FLT3/ITD is constitutively autophosphorylated in 32D cells expressing C/EBPα-ER. Total cellular protein extracts derived from 1 × 107 32D/α-ER cells transduced with the pBabeNeo vector, pBabeNeo-FLT3, or pBabeNeo-FLT3/ITD were immunoprecipitated with anti-FLT3 antibody. The immunoprecipitates were resolved by 8% SDS-PAGE and subjected to immunoblot analysis with antiphosphotyrosine antibody 4G10 (top). The same membrane was stripped and reprobed with anti-FLT3 antibody (bottom).
FLT3/ITD is constitutively autophosphorylated in 32D cells expressing C/EBPα-ER. Total cellular protein extracts derived from 1 × 107 32D/α-ER cells transduced with the pBabeNeo vector, pBabeNeo-FLT3, or pBabeNeo-FLT3/ITD were immunoprecipitated with anti-FLT3 antibody. The immunoprecipitates were resolved by 8% SDS-PAGE and subjected to immunoblot analysis with antiphosphotyrosine antibody 4G10 (top). The same membrane was stripped and reprobed with anti-FLT3 antibody (bottom).
Activation of C/EBPα induces PU.1 expression, cell cycle arrest, and differentiation in 32D cells expressing FLT3/ITD
To determine whether PU.1 expression is directly suppressed by FLT3/ITD signaling or is instead a result of C/EBPα suppression, we examined its expression in 32D/α-ER/FLT3/ITD cells. When C/EBPα is activated by estradiol, PU.1 expression was readily detected by Northern blotting at the earliest time point examined (Figure 5 lanes 2-4). Induction of PU.1 was also observed in 32D/α-ER/FLT3 and 32D/α-ER/pBabeNeo cells activated by estradiol (data not shown). This data confirms that PU.1 is a downstream target of activated C/EBPα and is suppressed in FLT3/ITD-expressing cells as a result of C/EBPα suppression.
Forced expression of C/EBPα induces PU.1 expression in 32D cells expressing FLT3/ITD. The 32D/α-ER/FLT3/ITD cells were cultured in medium containing 1 μM estradiol for 48 hours. Total cellular RNA from 1 × 107 cells was extracted 0, 8, 24, and 48 hours after incubation with estradiol. Ten micrograms of RNA from each sample was subjected to sequential Northern blotting with PU.1 and actin cDNA probe.
Forced expression of C/EBPα induces PU.1 expression in 32D cells expressing FLT3/ITD. The 32D/α-ER/FLT3/ITD cells were cultured in medium containing 1 μM estradiol for 48 hours. Total cellular RNA from 1 × 107 cells was extracted 0, 8, 24, and 48 hours after incubation with estradiol. Ten micrograms of RNA from each sample was subjected to sequential Northern blotting with PU.1 and actin cDNA probe.
To examine the effect of C/EBPα expression on the proliferation of 32D cells expressing FLT3/ITD, viable cells were counted daily based on trypan blue exclusion. Activation of C/EBPα by estradiol induced a rapid growth arrest compared with the growth curve of these cells without estradiol treatment (Figure 6A). To rule out the possibility that estradiol itself affected the proliferation of these cells, 32D/FLT3/ITD cells were also incubated with estradiol for 3 days. Proliferation of these cells was unaffected (Figure 6A). Similar results were also observed from other clones (data not shown). It was also noted that 32D/α-ER/FLT3/ITD cells grew more slowly when compared with 32D/FLT3/ITD cells. This may be due to the leakiness of C/EBPα-ER activity in the absence of estrogen or activation of C/EBPα-ER by the small amounts of estrogen in the growth medium.
Forced expression of C/EBPα leads to growth arrest and cell cycle arrest in 32D cells expressing FLT3/ITD. (A) A quantity of 1 × 106 32D/α-ER/FLT3/ITD and 32D/FLT3/ITD cells were cultured in the absence or presence of estradiol (1 μM) for 3 days. Viable cells were counted daily on the basis of trypan blue exclusion. Results shown are the means from triplicate assays. Error bars indicate SD. (B) The 32D/α-ER/FLT3/ITD and 32D/FLT3/ITD cells were incubated with estradiol for 24 hours. Cells (1 × 106) were collected before and after exposure to estradiol, treated with 20 μg/mL RNase, and stained with 50 μg/mL propidium iodide. Cells were subjected to flow cytometric analysis of DNA content. Data represented were from 1 of 3 different experiments.
Forced expression of C/EBPα leads to growth arrest and cell cycle arrest in 32D cells expressing FLT3/ITD. (A) A quantity of 1 × 106 32D/α-ER/FLT3/ITD and 32D/FLT3/ITD cells were cultured in the absence or presence of estradiol (1 μM) for 3 days. Viable cells were counted daily on the basis of trypan blue exclusion. Results shown are the means from triplicate assays. Error bars indicate SD. (B) The 32D/α-ER/FLT3/ITD and 32D/FLT3/ITD cells were incubated with estradiol for 24 hours. Cells (1 × 106) were collected before and after exposure to estradiol, treated with 20 μg/mL RNase, and stained with 50 μg/mL propidium iodide. Cells were subjected to flow cytometric analysis of DNA content. Data represented were from 1 of 3 different experiments.
To investigate whether or not activation of C/EBPα blocks the proliferation of 32D/α-ER/FLT3/ITD cells by inducing cell cycle arrest, cell cycle status was analyzed in 3 independent clones (representative data are shown). After 24 hours of estradiol treatment, the proportion of 32D/α-ER/FLT3/ITD cells in G0/G1 phase increased from 57.7% to 72.3% and the percentage of cells in S phase decreased from 32.8% to 14.9% (Figure 6B; Table 1). This indicates a blockage of the G1 to S phase transition in these cells. Estradiol itself did not affect the cell cycle status of 32D/FLT3/ITD cells (Figure 6B; Table 1).
Cell cycle analysis of 32D/α-ER/FLT3/ITD cells exposed to estradiol
Cell lines and treatment . | G0/G1 phase, % . | S phase, % . | G2/M phase, % . | G0/G1-to-S ratio . |
|---|---|---|---|---|
| 32D/α-ER/FLT3/ITD | ||||
| Before estradiol treatment | 57.7 ± 1.9 | 32.8 ± 0.5 | 8.1 ± 1.9 | 1.8 ± 0.1:1 |
| 24 h estradiol treatment | 72.3 ± 1.2 | 14.9 ± 0.5 | 6.5 ± 0.8 | 4.9 ± 0.2:1 |
| 32D/FLT3/ITD | ||||
| Before estradiol treatment | 48.1 ± 0.4 | 35.2 ± 0.5 | 15.8 ± 0.8 | 1.4 ± 0.02:1 |
| 24 h estradiol treatment | 48.1 ± 0.8 | 34.3 ± 0.5 | 16.4 ± 1.2 | 1.4 ± 0.01:1 |
Cell lines and treatment . | G0/G1 phase, % . | S phase, % . | G2/M phase, % . | G0/G1-to-S ratio . |
|---|---|---|---|---|
| 32D/α-ER/FLT3/ITD | ||||
| Before estradiol treatment | 57.7 ± 1.9 | 32.8 ± 0.5 | 8.1 ± 1.9 | 1.8 ± 0.1:1 |
| 24 h estradiol treatment | 72.3 ± 1.2 | 14.9 ± 0.5 | 6.5 ± 0.8 | 4.9 ± 0.2:1 |
| 32D/FLT3/ITD | ||||
| Before estradiol treatment | 48.1 ± 0.4 | 35.2 ± 0.5 | 15.8 ± 0.8 | 1.4 ± 0.02:1 |
| 24 h estradiol treatment | 48.1 ± 0.8 | 34.3 ± 0.5 | 16.4 ± 1.2 | 1.4 ± 0.01:1 |
Flow cytometric analysis of propidium iodide stained 32Dα-ER/FLT3/ITD and 32D/FLT3/ITD cells was performed prior to and after 24 hours of estradiol exposure. Values are the mean percentage or ratio ± SD from 3 different experiments.
We next assessed whether forced expression and activation of C/EBPα would result in the differentiation of 32D/α-ER/FLT3/ITD cells. Activation of C/EBPα by estradiol for 2 days promoted rapid morphologic differentiation to granulocytes (Figure 7A; Table 2). To confirm the findings of morphologic maturation, several markers of functional maturation were also examined by Northern blotting. Induction of MPO, lysozyme, and lactoferrin were all seen in 32D/α-ER/FLT3/ITD cells upon exposure to 1 μM estradiol (Figure 7B). Similar results were also obtained from 2 additional clones of 32D/α-ER/FLT3/ITD cells (data not shown). Neither morphologic (Table 2) nor markers of differentiation (data not shown) were observed in 32D/FLT3/ITD cells treated with estradiol alone. These results, taken together, indicate that induced expression of C/EBPα was able to overcome the blockage of differentiation caused by the expression of FLT3/ITD.
Forced expression of C/EBPα induces differentiation of 32D cells expressing FLT3/ITD. The 32D/α-ER/FLT3/ITD cells were incubated with 1 μM estradiol for 2 days. (A) Cytospins were prepared on days 0 and 2. The morphologic features of the cells were visualized by means of Wright-Giemsa staining followed by light microscopy. Original magnification × 40. (B) Total cellular RNA was prepared from 1 × 107 cells 0, 8, 24, and 48 hours after incubation with estradiol. RNA (10 μg from each sample) was then subjected to sequential Northern blotting with MPO, lysozyme, lactoferrin, and actin cDNA probes.
Forced expression of C/EBPα induces differentiation of 32D cells expressing FLT3/ITD. The 32D/α-ER/FLT3/ITD cells were incubated with 1 μM estradiol for 2 days. (A) Cytospins were prepared on days 0 and 2. The morphologic features of the cells were visualized by means of Wright-Giemsa staining followed by light microscopy. Original magnification × 40. (B) Total cellular RNA was prepared from 1 × 107 cells 0, 8, 24, and 48 hours after incubation with estradiol. RNA (10 μg from each sample) was then subjected to sequential Northern blotting with MPO, lysozyme, lactoferrin, and actin cDNA probes.
Forced expression of C/EBPα induces differentiation of 32D cells expressing FLT3/ITD
Cell lines and treatment . | Myeloblasts, % . | Intermediates, % . | Granulocytes, % . |
|---|---|---|---|
| 32D/a-ER/FLT3/ITD | |||
| Before estradiol treatment | 88.7 ± 1.5 | 11.3 ± 1.5 | 0 |
| 2 d estradiol treatment | 0 | 47.7 ± 3.5 | 52.3 ± 3.5 |
| 32D/FLT3/ITD | |||
| Before estradiol treatment | 89 ± 3.6 | 11 ± 3.6 | 0 |
| 2 d estradiol treatment | 89.7 ± 3.7 | 10.3 ± 3.7 | 0 |
Cell lines and treatment . | Myeloblasts, % . | Intermediates, % . | Granulocytes, % . |
|---|---|---|---|
| 32D/a-ER/FLT3/ITD | |||
| Before estradiol treatment | 88.7 ± 1.5 | 11.3 ± 1.5 | 0 |
| 2 d estradiol treatment | 0 | 47.7 ± 3.5 | 52.3 ± 3.5 |
| 32D/FLT3/ITD | |||
| Before estradiol treatment | 89 ± 3.6 | 11 ± 3.6 | 0 |
| 2 d estradiol treatment | 89.7 ± 3.7 | 10.3 ± 3.7 | 0 |
Differential counts of 32D/α-ER/FLT3/ITD and 32D/FLT3/ITD cell cultures induced by estradiol were performed on day 0 and 2. Values are the mean percentages ± SD of cells from 3 independent counts examined by light microscopy after Wright-Giesma staining of the cytospins. “Intermediates” include promyelocytes, myelocytes, and metamyelocytes.
Discussion
AML is characterized by the uncontrolled proliferation of myeloid cells that accumulate at various stages of development, where their further differentiation is blocked. FLT3-activating mutations are the most common genetic alteration that occurs in AML.2 The presence of FLT3/ITD mutations results in increased leukocytosis and a poor prognosis in patients expressing them.10,33,34 We previously showed that expression of FLT3/ITD mutations in a myeloid precursor cell line causes a block in G-CSF–mediated granulocytic differentiation.13 Thus, FLT3/ITD-transformed hematopoietic cells, such as 32D cells, have a phenotype resembling AML; that is, they show growth factor–independent proliferation and survival, and an altered response to differentiative stimuli.8 Both a murine bone marrow transplant model and transgenic mice expressing constitutively activated FLT3 demonstrate that FLT3 is capable of inducing myeloproliferative disease but is not sufficient by itself to cause AML.35,36 One explanation for the difference of FLT3/ITD-mediated effects on differentiation of primary hematopoietic cells versus 32D cells is that 32D cells are immortalized and thus are already partially transformed. These differences have also been observed in studies on the effects of promyelocytic leukemia/retinoic acid receptor α (PML/RARα), the fusion protein resulting from t(15;17) in M3 AML, on myeloid differentiation. Expression of PML/RARα in the U937 myeloid cell line inhibits the ability of U937 cells to differentiate in response to several stimuli.37 In contrast, expression of PML/RARα in transgenic mice is not sufficient to cause leukemia in any mouse during the first year, implying the necessity of accumulating additional “hits” to fully block differentiation.38 Retroviral transduction of FLT3/ITD into bone marrow cells obtained from PML/RARα transgenic mice results in a short latency acute promyelocytic leukemia–like disease with complete penetrance, suggesting that FLT3 signaling can cooperate with PML/RARα to give the complete differentiation block that characterizes leukemia.39
Granulopoiesis is a complex process governed by a number of transcription factors.14 Among them, C/EBPs are key regulators of granulopoiesis. We observed inhibition of C/EBPϵ induction as a consequence of FLT3/ITD expression in our previous report. However, C/EBPϵ is normally induced relatively late in the process of myeloid differentiation and thus, this block is likely a consequence, rather than a cause, of inhibited differentiation. Since C/EBPα is known to play an important role during early granulopoiesis, it is a major focus of this study. The critical role of C/EBPα in myeloid development was proved when C/EBPα knockout mice were generated and these mice completely lack myeloid development. Induction of C/EBPα function in primary human CD34+ cells leads to granulocytic differentiation.40 Normally, induction of C/EBPα expression is seen as an early event after G-CSF stimulation of 32D cells. We observed C/EBPα induction in parental and wild-type FLT3-expressing 32D cells in response to G-CSF, and this induction was accompanied by granulocytic differentiation. In contrast, G-CSF was unable to induce C/EBPα expression in 32D cells expressing FLT3/ITD, and this was accompanied by a block in differentiation. Thus, suppression of C/EBPα expression might represent the crucial molecular event underlying the blockage of differentiation of 32D cells caused by the expression of FLT3/ITD.
Little information is known about the regulation of C/EBPα expression. It has been shown that the C/EBPα promoter can be autoactivated.41,42 Recently, it has been reported that BCR-ABL regulates C/EBPα expression through induction of heterogeneous ribonucleoprotein E2 (hnRNP E2), which inhibits the translation of C/EBPα mRNA.43 There are no previous reports regarding the regulation of C/EBPα RNA or protein stability, which could also affect the expression level of C/EBPα. Based on the findings in this study, that C/EBPα RNA levels are suppressed in FLT3/ITD cells, it is likely that the mechanism involves C/EBPα transcription or RNA stability. Future investigations will be required to differentiate between these mechanisms.
The ability of the FLT3 tyrosine kinase inhibitor CEP-701 to restore induction of C/EBPα expression in 32D/FLT3/ITD cells in response to G-CSF indicates that the suppression of C/EBPα depends on the tyrosine kinase activity of FLT3/ITD. In 2 of 3 patients with primary AML receiving the FLT3 inhibitor CEP-701, inhibition of FLT3 signaling also resulted in the up-regulation of C/EBPα and expression of several markers of differentiation by the leukemic cells. This implies that FLT3/ITD activates signaling pathways that can lead to suppression of C/EBPα expression in primary AML cells as well as in 32D cells. Forced expression of C/EBPα restores the differentiation of 32D cells expressing FLT3/ITD, reinforcing the hypothesis that suppression of C/EBPα expression may be one of the essential molecular events required for the blockage of differentiation in these cells.
Forced expression of C/EBPα not only promotes a rapid differentiation in 32D cells expressing FLT3/ITD but also leads to growth arrest. This observation is consistent with the findings that C/EBPα is expressed at high levels in terminally differentiated cells, down-regulated during proliferation, and a strong inhibitor of cellular proliferation when overexpressed in cultured cells.32,44-46 In addition, hepatocytes from newborn C/EBPα-deficient mice display increased proliferative activity, and hyperproliferation of alveolar type II cells is also seen in the lungs of these mice.47 More recently, mutations that inactivate C/EBPα have been described in patients with AML.19 Considering this data, suppression of C/EBPα expression may contribute not only to a block in differentiation but also to increased proliferation of transformed cells with the resultant higher blast counts observed in patients with FLT3/ITD-positive AML.
FLT3/ITD signaling contributes to the block in differentiation in the clinical AML cases, but other mutations may also contribute to this effect. Inhibition of the FLT3 signaling, therefore, induces differentiation (as reflected in increased CD11b and CD15 expression in patient no. 2 and no. 3 after CEP-701 treatment) but only to a partial degree. Thus it may explain why no obvious morphologic differentiation was observed in patients no. 2 and 3 after therapy with CEP-701. It is possible that a combination of a FLT3 inhibitor with a differentiating agent would act synergistically to induce clinically significant (ie, morphologic) differentiation. Likewise, C/EBPα is certainly not the only transcription factor regulating differentiation. The other “hits” that occur in acute leukemias often affect other transcription factors that likely also contribute to the observed block in differentiation. In addition, in one patient C/EBPα was expressed at a significant level. This implies that other pathways are also capable of affecting C/EBPα expression and that in this patient's blasts, pathways other than C/EBPα are the dominant mechanism blocking differentiation.
PU.1 is another critical transcription factor during early granulopoiesis. It has been previously reported that C/EBPα-ER induced endogenous PU.1 RNA expression within 8 hours in both 32D and Ba/F3 cells, even in the presence of cycloheximide, indicating that C/EBPα directly activates the PU.1 gene.18,27 Therefore, PU.1 may act downstream of C/EBPα to induce myeloblasts to undergo granulocytic differentiation. However, PU.1 mRNA is also present in fetal liver cells derived from C/EBPα null mice.48 Thus, PU.1 transcription is not wholly dependent on C/EBPα. In this study, PU.1 induction, which normally occurs during G-CSF–mediated differentiation of wild-type FLT3-expressing and parental 32D cells, was not detected in FLT3/ITD-expressing 32D cells. Activation of C/EBPα-ER was able to restore the induction of PU.1 expression in FLT3/ITD-expressing 32D cells. In addition, in both FLT3/ITD-positive AML patients in which FLT3 tyrosine kinase inhibitor therapy increased C/EBPα expression, an increase of PU.1 expression was also observed. Thus, these results strongly suggest that suppression of PU.1 expression due to the expression of FLT3/ITD is a consequence of inhibition of C/EBPα expression.
In summary, the data presented in this report indicates that constitutively activated FLT3/ITD mutants can lead to the suppression of C/EBPα expression. Inhibition of C/EBPα expression is likely the critical molecular event required for the differentiation arrest that was observed in 32D cells expressing FLT3/ITD and may also contribute to the block in differentiation that usually characterizes leukemic cells. Modulation of C/EBPα activity may provide a new approach to the therapy of AML.
Prepublished online as Blood First Edition Paper, October 30, 2003; DOI 10.1182/blood-2003-06-1978.
Supported by grants from National Cancer Institute (NCI; CA90668, CA70970, CA91177), Leukemia and Lymphoma Society, and Children's Cancer Foundation. D.S. is the Douglas Kroll Research Foundation Translational Researcher of the Leukemia and Lymphoma Society. A.D.F. is a Scholar of the Leukemia and Lymphoma Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Kyu-tae Kim for providing actin primers for real-time RT-PCR, Dr Douglas Smith for providing primary AML blast cells from the Leukemia Bank at Johns Hopkins, Dr Daniel G. Tenen and Dr Hanna Radomska for providing the C/EBPα cDNA probe of human origin and for the C/EBPα Western blotting protocol, Shirley Fuller for technical assistance with flow cytometric analysis, and Bruce Ruggeri and Susan Jones-Bolin of Cephalon, Inc for providing CEP-701.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal