Abstract
Ahi-1/AHI-1 (Abelson helper integration site-1) encodes a family of protein isoforms containing one Src homology 3 (SH3) domain and multiple tryptophan-aspartic acid 40 (WD40)–repeat domains. The function of these proteins is unknown, but involvement in leukemogenesis has been suggested by the high frequency of Ahi-1 mutations seen in certain virus-induced murine leukemias. Here we show that in both mice and humans, Ahi-1/AHI-1 expression is highest in the most primitive hematopoietic cells with specific patterns of down-regulation in different lineages. Cells from patients with chronic myeloid leukemia (CML; n = 28) show elevated AHI-1 transcripts in all disease phases and, in chronic phase, in the leukemic cells at all stages of differentiation, including quiescent (G0) CD34+ cells as well as terminally differentiating cells. In the most primitive lin–CD34+CD38– CML cells, transcripts for the 2 shorter isoforms of AHI-1 are also increased. Although 15 of 16 human lymphoid and myeloid leukemic cell lines showed aberrant control of AHI-1 expression, this was not seen in blasts obtained directly from patients with acute Philadelphia chromosome–negative (Ph–) leukemia (n = 15). Taken together, our results suggest that down-regulation of AHI-1 expression is an important conserved step in primitive normal hematopoietic cell differentiation and that perturbations in AHI-1 expression may contribute to the development of specific types of human leukemia.
Introduction
Ahi-1 is a recently identified gene that is commonly activated by proviral insertional mutations in v-abl– or myc-induced murine leukemias and lymphomas.1 The Ahi-1 locus is also involved in the development of the acute myeloid leukemia (AML) that arises in NF-1 heterozygous mice2 and in Moloney murine leukemia virus (Mo-MuLV)–induced rat T-cell lymphomas.3 Although the function of the Ahi-1 protein has not yet been determined, it is known to have multiple Src homology 3 (SH3) binding sites, an SH3 domain and multiple tryptophan-aspartic acid 40 (WD40)–repeat domains, suggesting that it has signaling functions. In mice and rats, the Ahi-1 gene encodes 2 major RNA species (5 kilobase [kb] and 4.2 kb) and several shorter splice variants that can be found in several different organs. Murine pre–B- and T-cell leukemic cells with insertional mutations in Ahi-1 show both increased expression of Ahi-1 and expression of truncated Ahi-1/viral fused transcripts, including splice variants in which the SH3 domain is deleted.1 A search of GenBank databases using mouse and rat Ahi-1 cDNA sequences identified 3 human cDNA isoforms of very high homology. Notably, the shorter human isoform II lacks the SH3 domain, and isoform III, although also shorter than isoform I, contains additional coding sequences not present in either isoform I or II. The Ahi-1/AHI-1 gene is thus also subject to alternative splicing in normal cells.
The repeated finding of Ahi-1 mutations in different types of experimental hematopoietic malignancies whose generation could be associated with genes of limited leukemogenic activity (eg, v-abl, c-myc, and NF-1)2,4-7 is consistent with the concept that mutant forms of Ahi-1 can serve as cooperating oncogenes. The fact that ABL, MYC, and NF-1 have been implicated in the genesis of human leukemias2,8-12 is intriguing and further underscores the likelihood that alterations in human AHI-1 are involved in the development of human hematopoietic malignancies.
To investigate this possibility, we first examined the changes in expression of Ahi-1/AHI-1 that occur during normal hematopoiesis and then looked for potential changes in AHI-1 expression in various types of human leukemic cells. Ahi-1/AHI-1 transcript levels were found to be highest in the most primitive types of normal hematopoietic cells and were then down-regulated during their early differentiation. Marked deregulation of AHI-1 expression was seen in a broad spectrum of human leukemic cell lines and in cells obtained directly from patients with Philadelphia chromosome–positive (Ph+) but not Ph– leukemias. These findings reinforce the concept that alterations in expression of this gene may be important in the development of Ph+ leukemias.
Patients, materials, and methods
Hematopoietic cells
Mouse bone marrow (BM) cells were harvested from 8- to 10-week-old C57BL/6 mice and then washed, and low-density cells were isolated as described.13 Heparin-treated blood and BM cells were obtained with informed consent from newly diagnosed patients, including 28 chronic myeloid leukemia (CML) patients with high white blood cell (WBC) counts (Table 1), 12 patients with pre-B–acute lymphocytic leukemia (ALL; Table 2), and 15 patients with acute Ph– myeloid leukemia (AML; Table 3), as part of routine clinical diagnostic or follow-up procedures. Samples of normal adult human BM cells were also obtained from harvests of local allogeneic transplants or from cadaveric donors (Northwest Tissue Center, Seattle, WA). In all cases, informed consent was obtained, and the procedures used including the Helsinki Protocol were approved by the Research Ethics Board of the University of British Columbia. Low-density cells were isolated from human samples by centrifugation on Ficoll-Hypaque (Amersham Pharmacia, Piscataway, NJ) and then cryopreserved in dimethyl sulfoxide and fetal calf serum (FCS) until required. In some instances, samples obtained from CML patients were first enriched for CD34+ cells by immunomagnetic removal on columns (StemCell Technologies, Vancouver, BC, Canada) of cells expressing the following lineage markers (lin+ cells): CD2, CD3, CD14, CD16, CD19, CD24, CD56, CD66b, and glycophorin A, as described.16
Clinical and progenitor data for CML samples
Clinical status . | No. of samples studied . | Patient age, y . | WBC count, ×109/L . | CFCs/103 CD34+ cells . | LTC-ICs/104 CD34+ cells . | Ph+ LTC-IC samples/total* . |
|---|---|---|---|---|---|---|
| Chronic phase | 18 | 23-56 | 75-640 | 42-120 | 17-1000 | 8/18 |
| Accelerated phase | 8 | 45-71 | 35-460 | 30-160 | 42-480 | 1/8 |
| Blast crisis, myeloid | 2 | 46-60 | 60-110 | 15-50 | ND | ND |
Clinical status . | No. of samples studied . | Patient age, y . | WBC count, ×109/L . | CFCs/103 CD34+ cells . | LTC-ICs/104 CD34+ cells . | Ph+ LTC-IC samples/total* . |
|---|---|---|---|---|---|---|
| Chronic phase | 18 | 23-56 | 75-640 | 42-120 | 17-1000 | 8/18 |
| Accelerated phase | 8 | 45-71 | 35-460 | 30-160 | 42-480 | 1/8 |
| Blast crisis, myeloid | 2 | 46-60 | 60-110 | 15-50 | ND | ND |
CFC and LTC-IC assays were performed as previously described.14 Values for LTC-ICs are the LTC-IC—derived CFC numbers.
ND indicates not done.
LTC-IC was genotyped as Ph- and Ph+ by cytogenetic analysis of Giemsa-banded metaphases from individually processed colonies generated from LTC-IC—derived CFCs.15
Clinical and progenitor data for pre-B—ALL samples
Ph status . | No. of samples studied . | Patient age range, y . | WBC count, ×109/L (% blasts range) . | CFC range, ×106/L . |
|---|---|---|---|---|
| Ph+ | 8 | 36-61 | 3-31 (66-98) | 10-20 |
| Ph- | 4 | 19-33 | 5-10 (88-96) | 8-16 |
Ph status . | No. of samples studied . | Patient age range, y . | WBC count, ×109/L (% blasts range) . | CFC range, ×106/L . |
|---|---|---|---|---|
| Ph+ | 8 | 36-61 | 3-31 (66-98) | 10-20 |
| Ph- | 4 | 19-33 | 5-10 (88-96) | 8-16 |
Clinical data for AML samples
FAB subtype . | No. of samples studied . | Patient age, y . | WBC count, × 109/L (% blasts) . | Cytogenetics . |
|---|---|---|---|---|
| M1 | 2 | 50-66 | 6-80 (35-100) | t(8;20) |
| M2 | 4 | 25-52 | 11-78 (38-50) | t(7;11), Inv(16), t(7;16)(q32,q23), del17(q21.1) |
| M4 + M4eos | 5 | 29-66 | 23-305 (55-90) | Inv(16), +14, +22, del15(q12,q23) |
| M5 + M5a | 3 | 20-47 | 47-168 (72-92) | Inv(16) |
| MDS/AML | 1 | 70 | 2 (50) | Inv(16) |
FAB subtype . | No. of samples studied . | Patient age, y . | WBC count, × 109/L (% blasts) . | Cytogenetics . |
|---|---|---|---|---|
| M1 | 2 | 50-66 | 6-80 (35-100) | t(8;20) |
| M2 | 4 | 25-52 | 11-78 (38-50) | t(7;11), Inv(16), t(7;16)(q32,q23), del17(q21.1) |
| M4 + M4eos | 5 | 29-66 | 23-305 (55-90) | Inv(16), +14, +22, del15(q12,q23) |
| M5 + M5a | 3 | 20-47 | 47-168 (72-92) | Inv(16) |
| MDS/AML | 1 | 70 | 2 (50) | Inv(16) |
FAB indicates French-American-British; MDS, myelodysplastic syndrome.
Cell purification
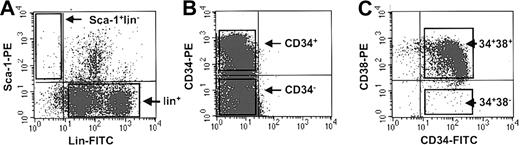
Mouse BM cells were suspended in Hanks solution with 2% FCS at 5 × 106 to 10 × 106 cells/mL and then incubated with 6 μg/mL of 2.4G2 (anti-Fc receptor antibody),17 then with a mixture of biotinylated lineage-specific rat monoclonal antibodies (anti-B220 [RA3-6B2], anti–Gr-1 [RB6-8C5], anti–Mac-1 [M1/70], anti–Ly-1 [53-7.3], Ter119 [anti[en]Ly-76]), and finally with phycoerythrin (PE)–labeled anti–Sca-1 (anti–Ly-6A/E; E13-161.7) for 30 minutes at 4° C as described.18 Antibody-labeled cells were then washed once, incubated with allophycocyanin (APC)–labeled streptavidin (SA-APC) for 30 minutes at 4° C, washed twice again, and finally resuspended in 1 μg/mL propidium iodide (PI; Sigma, St Louis, MO). A FACStarPlus or FACSVantage (Becton Dickinson Immunocytometry Systems, San Jose, CA) was used to isolate populations of greater than 99% pure, viable Sca-1+lin– cells and lin+ cells (Figure 1A). Later subsets of terminally differentiating cells were further purified based on surface expression Gr-1, Mac-1, Ly-1, B220, and Ter119 using directly conjugated PE- or APC-conjugated monoclonal antibodies against these antigens to stain mouse BM cells as described at the beginning of this section.
FACS profiles of primitive murine and human BM cells. (A) Representative dot plots showing the gates used to isolate Sca-1+lin– and lin+ populations of murine BM cells. (B) Representative dot plot showing the gates used to isolate CD34+ and CD34– populations from low-density normal human BM cells. (C) Representative dot plot showing the gates used to isolate the CD38– and CD38+ subsets of CD34+ cells from lin– normal human BM cells.
FACS profiles of primitive murine and human BM cells. (A) Representative dot plots showing the gates used to isolate Sca-1+lin– and lin+ populations of murine BM cells. (B) Representative dot plot showing the gates used to isolate CD34+ and CD34– populations from low-density normal human BM cells. (C) Representative dot plot showing the gates used to isolate the CD38– and CD38+ subsets of CD34+ cells from lin– normal human BM cells.
Lin–CD34+CD38+, lin–CD34+CD38–, and lin+CD34– cells were purified by fluorescent-activated cell sorting (FACS) of thawed low-density or lin– normal human BM or CML patients' cells after staining with antihuman CD34–fluorescein isothiocyanate (FITC; 8G12) and antihuman CD38-PE (Becton Dickinson Immunocytometry Systems) for 30 minutes at 4° C as described19 (Figure 1B). To isolate subsets of CD34+ cells in G0 or G1/S/G2/M, lin– normal and CML cells were first incubated for 45 minutes at 37° C in 10 μM Hoechst 33342 (Hst; Molecular Probes, Eugene, OR). Pyronin Y (Py; 5 μg/mL; Sigma) was then added and the incubation continued at 37° C for another 45 minutes. Cells were finally labeled with anti-CD34–FITC for an additional 20 minutes and washed in the continuing presence of 10 μM Hst and 2.5 μg/mL Py plus 1 μg/mL PI. Gates were set to collect HstloPylo (G0) and all remaining (G1/S/G2/M) cells as separate fractions within the PI–CD34+ population as described in detail previously.16,20
Culture of leukemic cell lines
Leukemic cell lines were grown at 37° C in a humidified atmosphere of air containing 5% CO2. Growth factor–independent cell lines were cultured in RPMI 1640 medium plus 10% FCS and 10–4 M 2-mercaptoethanol (Sigma). Mo7E and TF-1 cells lines were cultured in RPMI plus 10% FCS and 10 ng/mL human interleukin 3 (IL-3; Novartis, Basel, Switzerland).
Conventional reverse transcriptase–polymerase chain reaction (RT-PCR) analyses and cDNA cloning
Total RNA was extracted from aliquots of cells using the Absolutely RNA Microprep kit (Stratagene, La Jolla, CA) or TRIzol (Invitrogen, Burlington, ON, Canada). Poly(A) (polyadenylated) RNA was isolated from total RNA using a MicroPoly(A)Pure Kit (Ambion, Austin, TX). The RT reaction was performed in 20-μL volumes with superscript II reverse transcriptase (Invitrogen) using random hexamer oligonucleotides (Amersham Pharmacia) as described.13 PCR was then performed on the cDNAs obtained in 50 μL of 1 × PCR buffer (Invitrogen) containing 20 mM Tris-HCL(pH 8.4), 50 mM KCl, 2 mM MgCl2, 200 μM of each deoxynucleoside triphosphate (dNTP; Invitrogen), 1 unit of Taq polymerase, and 10 pM of specific primers for amplification of the AHI-1 isoform III (5′-GCCATCTTACCGCTCTATGATG-3′ and 5′-GAAGGAGGTGTCTCTGTGAGTC-3′). Thirty-five cycles of amplification (94°C for 30 seconds, 62°C for 60 seconds, 72°C for 60 seconds) were then performed. The PCR product obtained was purified on agarose gels and cloned in a pCR2.1-TOPO vector with a T7 promoter (Invitrogen). A cDNA fragment containing AHI-1 isoform II was digested with XhoI from clone COL01816 (obtained from Dr Sumio Sugano of Human Genome Center, Tokyo, Japan) and cloned into the pCR2.1-TOPO vector. The plasmids were isolated using the Maxi Prep Kit (Qiagen, Hilden, Germany) and each plasmid was further verified by restriction enzyme digestion and sequencing.
In vitro transcription of AHI-1 isoform cDNAs
Vectors containing cDNAs for human AHI-1 isoform I (AL136797 obtained from RZPD, Berlin, Germany) and isoforms II and III constructed as described in the previous section were used for in vitro transcription, respectively. These were linearized by restriction enzyme digestion and transcribed to generate sense RNA transcripts using the T7 MEGAScript Transcription Kit (Ambion). The RNA was treated with RNAse-free DNAse and recovered by LiCl precipitation as suggested by the manufacturer. RNA concentrations were determined by a Ribogreen assay (Molecular Probes) and absorbance measures at optical density 260 (OD260) using a Genequant II spectrophotometer (Pharmacia, Orangeville, ON, Canada). Two micrograms of transcribed RNA were separated on a 1.2% agarose gel to ensure that each standard RNA template was a single, pure species free of DNA contamination. Based on the length of each transcript, the molar mass was calculated and converted into a number of transcripts per microgram of RNA: transcript number/μg RNA = No./(transcript length × MW), where molecular weight (MW) = 340 × 106 μg/mol, and No. = Avogadro number.
Absolute standard curves and real-time RT-PCR analyses
One or 0.1 μg of the RNA templates prepared from the different AHI-1 isoforms were reverse transcribed with random hexamers in a 20-μL reaction volume using SuperScript II as described in “Conventional reverse transcriptase–polymerase chain reaction (RT-PCR) analyses and cDNA cloning.” The cDNA mixture generated was then serially diluted in steps of 10 or 100 down to the equivalent of a single transcript copy by performing 3 independent serial dilutions and assaying each dilution in duplicate. Real-time PCR was performed using 25 μL of 2 × SYBY Green PCR Master Mix (Applied Biosystems, Foster City, CA), 1 μL of 20 pM of specific primers, 1 to 2 μL cDNA, and water to a final volume of 50 μL as described previously.21 Specific forward and reverse primers to produce approximately 100–base pair (bp) amplicons for optimal amplification in real-time PCR of reverse transcribed cDNA for AHI-1 isoform I to detect all 3 isoforms were 5′-CTGTCACAGAGGTGATACGTTC-3′ and 5′-GACTGTTGTGAGGAAACTGCTG-3′. Primers for the specific detection of AHI-1 isoform I were 5′-GCTCCTCAAAAGCAATCAATCAA-3′ and 5′-CTCATTTCAGAATGTGTCATAGAT-3′, for AHI-1 isoform II were 5′-CATGCTGACCGCTCAAGAGATT-3′ and 5′-GTGTTGAATTCAGCAAAGTGACT-3′, for AHI-1 isoform III were 5′-CCTGAGATAAAGGAGAGATCCC-3′ and 5′-GAAGGAGGTGTCTCTGTGAGCT-3′, and for mouse Ahi-1 were 5′-GCCAGTGCCCACTAAGCCTGA-3′ and 5′-GCTGAGAGTCCTCAGGGGTAC-3′. Optimal reaction conditions for the amplification of human and murine AHI-1/Ahi-1 were as follows: 40 cycles of 3-step PCR (94°C for 15 seconds, 60°C for 20 seconds, 72°C for 30 seconds with a single fluorescence measurement) after initial denaturation (94°C for 5 minutes). Real-time PCR was also subjected to melt curve analysis for verification of true target amplicons and absence of primer dimers. Real-time PCR and data analysis were performed on an iCycler iQ system, using iCycler iQ Real-time Detection Software (BIO-RAD, Hercules, CA). An absolute standard curve was generated by plotting the threshold cycle (Ct) values with 95% confidence intervals against the logarithm of the initial transcript numbers. The transcript numbers of experimental RNAs were then calculated after real-time amplification from the linear regression of the standard curve.
Northern blot analysis
Twenty micrograms of total RNA or 5 μg of Poly(A) RNA isolated from various leukemic cell lines was separated on 1.2% ready-to-use seakem gold agarose gels (Cambrex Bio Science, Rockland, ME), transferred onto nylon membranes, and hybridized with a human 1.4-kb AHI-1 cDNA fragment. The membranes were then washed and rehybridized with an actin probe. The intensity for each specific signal was measured using a phosphorimager STORM 860 and the ImageQuant software from Molecular Dynamics (Sunnyvale, CA). The expression of AHI-1 was then normalized according to the level of expression of actin in the same sample.
Results
Ahi-1/AHI-1 is expressed at higher levels in the more primitive subsets of normal murine and human BM cells
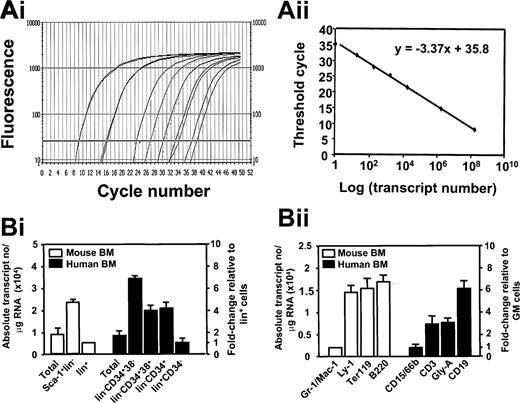
Because initial real-time RT-PCR measurements showed that both glyceraldehyde phosphate dehydrogenase (GAPDH) and actin transcript levels vary in different subpopulations of normal and leukemic cells, as previously reported,22,23 we quantified absolute Ahi-1/AHI-1 transcript levels by comparison of the test material to a standard curve generated from the relevant cloned AHI-1 isoforms as described in “Patients, materials, and methods” and illustrated for isoform I in Figure 2A.
Measurement of absolute numbers of Ahi/AHI-1 transcripts in subpopulations of normal murine and human BM cells. (Ai) Real-time RT-PCR SYBR Green1 fluorescence versus cycle number of 10- or 100-fold serial dilutions of cDNA prepared from an in vitro–transcribed sense RNA transcript of a vector containing the full-length AHI-1 cDNA. (Aii) Linear regression generated by plotting the Ct values, with 95% confidence intervals, against the logarithm of the initial transcript numbers from a series of measurements of independently diluted vector transcript. (Bi) Ahi-1 transcript numbers per microgram of RNA calculated as described “Patients, materials, and methods” from real-time RT-PCR analyses of RNA extracts obtained from each FACS-purified population shown (murine BM subpopulations, n = 3; normal human BM subpopulations, n = 4). (Bii) A similar analysis performed on RNA isolated from specific subsets of lin+ murine (n = 3) and human BM cells (n = 4). The number of Ahi-1/AHI-1 transcripts from each population was calculated by an absolute standard curve. Values shown are the mean ± SEM.
Measurement of absolute numbers of Ahi/AHI-1 transcripts in subpopulations of normal murine and human BM cells. (Ai) Real-time RT-PCR SYBR Green1 fluorescence versus cycle number of 10- or 100-fold serial dilutions of cDNA prepared from an in vitro–transcribed sense RNA transcript of a vector containing the full-length AHI-1 cDNA. (Aii) Linear regression generated by plotting the Ct values, with 95% confidence intervals, against the logarithm of the initial transcript numbers from a series of measurements of independently diluted vector transcript. (Bi) Ahi-1 transcript numbers per microgram of RNA calculated as described “Patients, materials, and methods” from real-time RT-PCR analyses of RNA extracts obtained from each FACS-purified population shown (murine BM subpopulations, n = 3; normal human BM subpopulations, n = 4). (Bii) A similar analysis performed on RNA isolated from specific subsets of lin+ murine (n = 3) and human BM cells (n = 4). The number of Ahi-1/AHI-1 transcripts from each population was calculated by an absolute standard curve. Values shown are the mean ± SEM.
Using this approach we found that Sca-1+lin– cells purified from normal adult mouse BM (Figure 1A) contained 5-fold more total Ahi-1 transcripts than the lin+ cells (25 × 103 vs 5 × 103 transcripts/μg RNA; Figure 2Bi) and, within the different subsets of lin+ cells, Ahi-1 transcript levels were 6- to 7-fold lower in the granulocyte/macrophage lineage (Gr-1+/Mac-1+ cells) compared with the T-lymphoid (Ly-1+), erythroid (Ter119+), and B-lymphoid (B220+) lineages (Figure 2Bii).
Analysis of FACS-purified subpopulations of normal adult human BM cells (Figure 1B-C) showed a similar pattern of down-regulated AHI-1 expression during normal human hematopoietic cell differentiation, with an overall 6-fold decrease from the most primitive lin–CD34+CD38– subset to the most mature lin+CD34– cells (34 × 103 vs 5 × 103 transcripts/μg RNA; Figure 2B). In addition, human lin+ BM cells expressing markers of the granulopoietic lineage (CD15/66b) showed significantly reduced expression of AHI-1 compared with levels measured in T (CD3+), erythroid (glycophorin A+), and B (CD19+) cells (Figure 2Bii). This conservation of the pattern of changes in Ahi-1/AHI-1 expression between mice and humans during the multi-step differentiation of hematopoietic cells suggests that the products of this gene may play important roles in the regulation of these differentiation events.
AHI-1 expression is deregulated in several human leukemic cell lines
As a preliminary screen for evidence of AHI-1 involvement in human leukemia, we next analyzed the levels of AHI-1 transcripts in 16 established human leukemic cell lines. These included lines with myeloid (n = 8), B-cell (n = 3), and T-cell (n = 5) phenotypes (Figure 3A). The results of this survey showed AHI-1 transcript levels to be significantly higher (up to 40-fold) in 15 of these cell lines by comparison to normal human bone marrow lin+CD34– cells. The lines showing the highest increases in AHI-1 transcript levels included an AML line (THP-1), a line derived from a CML patient in blast crisis (K562), and 3 T-cell leukemia virus (HTLV-1)–associated adult T-cell leukemia/lymphoma (ATLL) cell lines (MT-4, Hut102, and Hut78). Northern blot analysis using a probe derived from the 5′ end of human AHI-1 cDNA demonstrated full-length (6.5 kb) transcripts in most of the cell lines tested at increased levels by comparison to normal (unfractionated) BM cells (Figure 3B; Table 4). Elevated levels of shorter AHI-1 transcripts (4.2, 2.0, and/or 1.2 kb) were also evident in 4 of the lines (K562, Raji, Hut78, and Hut102). These results were confirmed and extended by Northern blot analysis of poly(A) RNA (Table 5).
Highly deregulated expression of AHI-1 in human leukemic cell lines. (A) The panel shows absolute AHI-1 transcript levels calculated as described in “Patients, materials, and methods” from real-time RT-PCR analyses of RNA isolated from 16 human leukemic cell lines. Data for total normal human BM cells (data from Figure 2) are shown for comparison. Values shown are the mean ± SEM of data from 3 independent RNA isolates from each cell line. The horizontal dotted line indicates the highest increases in AHI-1 transcript levels found in 5 human cell lines. Below is a schematic diagram of the full-length human AHI-1 isoform I with arrows indicating the position of the specific primers at the 5′ end of the gene used to amplify all transcript isoforms concomitantly (see also Figure 4A). *Stop codons. (B) Detection of different species of AHI-1 transcripts by Northern blot analysis of total RNA from the cell lines analyzed in panel A. Lanes were loaded with 20 μg of total RNA and the blots were probed using a 1.4-kb AHI-1 cDNA fragment.
Highly deregulated expression of AHI-1 in human leukemic cell lines. (A) The panel shows absolute AHI-1 transcript levels calculated as described in “Patients, materials, and methods” from real-time RT-PCR analyses of RNA isolated from 16 human leukemic cell lines. Data for total normal human BM cells (data from Figure 2) are shown for comparison. Values shown are the mean ± SEM of data from 3 independent RNA isolates from each cell line. The horizontal dotted line indicates the highest increases in AHI-1 transcript levels found in 5 human cell lines. Below is a schematic diagram of the full-length human AHI-1 isoform I with arrows indicating the position of the specific primers at the 5′ end of the gene used to amplify all transcript isoforms concomitantly (see also Figure 4A). *Stop codons. (B) Detection of different species of AHI-1 transcripts by Northern blot analysis of total RNA from the cell lines analyzed in panel A. Lanes were loaded with 20 μg of total RNA and the blots were probed using a 1.4-kb AHI-1 cDNA fragment.
Quantitation of altered AHI-1 isoform expression in human leukemic cell lines from Northern blot analysis of total RNA
. | Relative transcript levels for different AHI-1 mRNA species . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| Cells . | 6.5 kb . | 5.0 kb . | 4.2 kb . | 2.0 kb . | 1.2 kb . | ||||
| NBM | 1 | 1 | 1 | 1 | 1 | ||||
| U937 | 6 | — | 7 | — | — | ||||
| HL60 | 14 | — | 12 | — | — | ||||
| K562 | 18 | 21 | 35 | 19 | — | ||||
| THP-1 | 14 | — | 16 | — | — | ||||
| IM9 | 3 | — | 3 | — | — | ||||
| Daudi | 4 | — | 3 | — | — | ||||
| Raji | 12 | — | 25 | — | — | ||||
| Jurkat | 6 | — | 5 | — | — | ||||
| CCL119 | 12 | — | 14 | — | — | ||||
| MT-4 | 14 | — | 12 | — | — | ||||
| Hut102 | 46 | 34 | 40 | — | 15 | ||||
| Hut 78 | 49 | — | 52 | — | 16 | ||||
. | Relative transcript levels for different AHI-1 mRNA species . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| Cells . | 6.5 kb . | 5.0 kb . | 4.2 kb . | 2.0 kb . | 1.2 kb . | ||||
| NBM | 1 | 1 | 1 | 1 | 1 | ||||
| U937 | 6 | — | 7 | — | — | ||||
| HL60 | 14 | — | 12 | — | — | ||||
| K562 | 18 | 21 | 35 | 19 | — | ||||
| THP-1 | 14 | — | 16 | — | — | ||||
| IM9 | 3 | — | 3 | — | — | ||||
| Daudi | 4 | — | 3 | — | — | ||||
| Raji | 12 | — | 25 | — | — | ||||
| Jurkat | 6 | — | 5 | — | — | ||||
| CCL119 | 12 | — | 14 | — | — | ||||
| MT-4 | 14 | — | 12 | — | — | ||||
| Hut102 | 46 | 34 | 40 | — | 15 | ||||
| Hut 78 | 49 | — | 52 | — | 16 | ||||
The levels of each type of AHI-1 transcript were quantified and normalized to actin transcript levels as described in “Patients, materials, and methods” and then the ratio normalized again to the corresponding value obtained for unfractionated normal bone marrow (NBM) cells.
— indicates no signal to be detected.
Quantitation of altered AHI-1 isoform expression in human leukemic cell lines from Northern blot analysis of poly(A) RNA
. | Relative transcript levels for different AHI-1 mRNA species . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| Cells . | 6.5 kb . | 5.0 kb . | 4.2 kb . | 2.0 kb . | 1.2 kb . | ||||
| NBM | 1 | 1 | 1 | 1 | 1 | ||||
| KG1 | 10 | 10 | 12 | — | — | ||||
| TF1 | 12 | 14 | 15 | — | 12 | ||||
| U937 | 14 | — | 15 | — | — | ||||
| HL60 | 16 | — | 10 | — | — | ||||
| K562 | 25 | — | 50 | 30 | — | ||||
| Raji | 14 | — | 32 | — | — | ||||
| Jurkat | 8 | 7 | 8 | — | 6 | ||||
| CCL119 | 13 | 13 | 14 | — | 5 | ||||
| MT-4 | 18 | 14 | 10 | — | 5 | ||||
| Hut102 | 40 | 34 | 42 | — | 18 | ||||
| Hut 78 | 57 | 62 | 65 | — | 22 | ||||
. | Relative transcript levels for different AHI-1 mRNA species . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| Cells . | 6.5 kb . | 5.0 kb . | 4.2 kb . | 2.0 kb . | 1.2 kb . | ||||
| NBM | 1 | 1 | 1 | 1 | 1 | ||||
| KG1 | 10 | 10 | 12 | — | — | ||||
| TF1 | 12 | 14 | 15 | — | 12 | ||||
| U937 | 14 | — | 15 | — | — | ||||
| HL60 | 16 | — | 10 | — | — | ||||
| K562 | 25 | — | 50 | 30 | — | ||||
| Raji | 14 | — | 32 | — | — | ||||
| Jurkat | 8 | 7 | 8 | — | 6 | ||||
| CCL119 | 13 | 13 | 14 | — | 5 | ||||
| MT-4 | 18 | 14 | 10 | — | 5 | ||||
| Hut102 | 40 | 34 | 42 | — | 18 | ||||
| Hut 78 | 57 | 62 | 65 | — | 22 | ||||
The levels of each type of AHI-1 transcript were quantified and normalized to actin transcript levels as described in “Patients, materials, and methods” and then the ratio normalized again to the corresponding value obtained for unfractionated normal bone marrow (NBM) cells.
— indicates no signal to be detected.
To quantify more precisely the levels of expression of different AHI-1 isoforms in different cell populations, real-time RT-PCR analyses were undertaken using specific primers to detect the 3′ end of each isoform (Figure 4A) and absolute levels of each isoform were calculated as described in “Patients, materials, and methods” using a standard curve for the corresponding cloned isoform. In normal human BM cells, transcripts for isoform III were the most prevalent, with slightly fewer transcripts for isoform I and approximately 10-fold fewer transcripts for isoform II, which lacks the SH3 domain (Figure 4B). In most of the 15 leukemic cell lines that showed an overall up-regulated expression of AHI-1 relative to normal BM, transcripts for isoforms I and II were consistently elevated, with the greatest increases (up to 35-fold) affecting isoform II. In contrast, isoform III, which contains additional coding sequences not present in isoforms I and II, showed increased expression in only half of the lines and, in those where this isoform was up-regulated, the increase relative to normal BM cells was small (maximum increase of 4-fold).
Expression of AHI-1 isoforms are variably altered in human leukemic cell lines. (A) Schematic diagram of human AHI-1 isoforms (i-iii). The numbers refer to the positions of the nucleotides. Open reading frames are indicated by thick bars, boxes, and triangles. Untranslated sequences are indicated by thin lines. In-frame stop codons are indicated by asterisks. The position of the specific primers used to detect each isoform are indicated by the arrows. (B) Absolute AHI-1 isoform transcript levels were calculated as described in “Patients, materials, and methods” from real-time RT-PCR analyses of a single set of the same RNA samples used to generate the data shown in Figure 3A but, in this case, specific primers to detect each isoform separately (A) and the corresponding standard curves were used. Lane 1 indicates unfractionated normal BM cells; lanes 2-9, various myeloid cell lines known as Mo7E, AML5, KG1, TF1, U937, HL60, K562, THP1; lanes 10-17, various lymphoid cell lines known as Im9, Daudi, Raji, Jurkat, CC119, MT4, Hut102, and Hut78.
Expression of AHI-1 isoforms are variably altered in human leukemic cell lines. (A) Schematic diagram of human AHI-1 isoforms (i-iii). The numbers refer to the positions of the nucleotides. Open reading frames are indicated by thick bars, boxes, and triangles. Untranslated sequences are indicated by thin lines. In-frame stop codons are indicated by asterisks. The position of the specific primers used to detect each isoform are indicated by the arrows. (B) Absolute AHI-1 isoform transcript levels were calculated as described in “Patients, materials, and methods” from real-time RT-PCR analyses of a single set of the same RNA samples used to generate the data shown in Figure 3A but, in this case, specific primers to detect each isoform separately (A) and the corresponding standard curves were used. Lane 1 indicates unfractionated normal BM cells; lanes 2-9, various myeloid cell lines known as Mo7E, AML5, KG1, TF1, U937, HL60, K562, THP1; lanes 10-17, various lymphoid cell lines known as Im9, Daudi, Raji, Jurkat, CC119, MT4, Hut102, and Hut78.
AHI-1 expression is deregulated in CML at all stages of leukemic cell differentiation
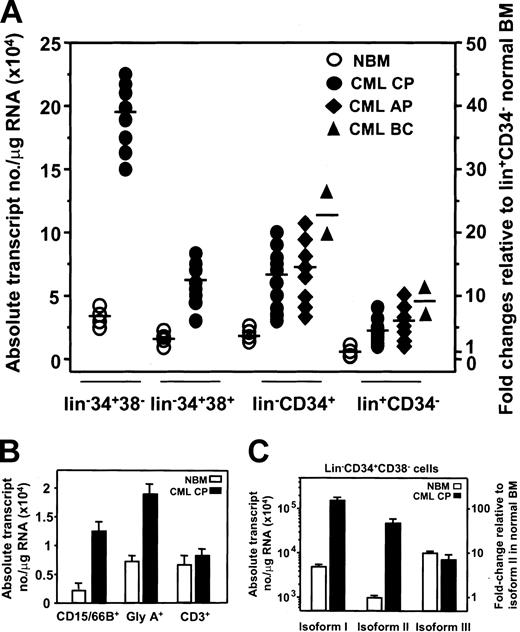
Based on the results of the cell line survey, we next examined cells obtained directly from a total of 28 patients with CML, including samples representative of all stages of this disease (Table 1). Eight of the chronic-phase samples had been previously shown to contain predominantly Ph+ cells in all compartments, including the most primitive but rare cells detectable, such as long-term culture-initiating cells (LTC-ICs),24 to the most mature cells that make up the bulk of the cells in the BM of CML patients. These samples were included in the present analysis to ensure that AHI-1 gene expression could be examined in the most primitive types of CML cells, as, in most CML patients, the LTC-IC compartment is made up of predominantly normal cells.25 As shown in Figure 5A, relative to the corresponding populations in normal human BM (n = 4), AHI-1 transcript levels were elevated at all stages of CML cell differentiation in chronic-phase samples but particularly so in the lin–CD34+CD38– compartment where the increase averaged 7-fold. AHI-1 transcripts were also consistently elevated in the lin–CD34+ and lin+CD34– cells from CML patients with accelerated-phase disease.
Enhanced expression of AHI-1 in primary CML cells. (A) Absolute AHI-1 transcript levels were calculated as described in “Patients, materials, and methods” from real-time RT-PCR analyses of RNA isolated from FACS-purified lin–CD34+CD38–, lin–CD34+CD38+, lin–CD34+, and lin+CD34– normal BM (n = 4) and CML cells in chronic phase (n = 18), accelerated phase (n = 8), and myeloid blast crisis (n = 2). The mean levels of AHI-1 expression in each population are indicated by the horizontal bars. (B) A similar analysis performed on RNA isolated from later subsets of lin+ normal human BM cells (n = 4) and chronic-phase CML cells (n = 6). (C) Real-time RT-PCR analyses of RNA isolated from FACS-purified lin–CD34+CD38– normal BM (n = 4) and chronic-phase CML cells (n = 8) using the same isoform-specific primers shown in Figure 4A and the corresponding standard curves. Values shown are the mean ± SEM.
Enhanced expression of AHI-1 in primary CML cells. (A) Absolute AHI-1 transcript levels were calculated as described in “Patients, materials, and methods” from real-time RT-PCR analyses of RNA isolated from FACS-purified lin–CD34+CD38–, lin–CD34+CD38+, lin–CD34+, and lin+CD34– normal BM (n = 4) and CML cells in chronic phase (n = 18), accelerated phase (n = 8), and myeloid blast crisis (n = 2). The mean levels of AHI-1 expression in each population are indicated by the horizontal bars. (B) A similar analysis performed on RNA isolated from later subsets of lin+ normal human BM cells (n = 4) and chronic-phase CML cells (n = 6). (C) Real-time RT-PCR analyses of RNA isolated from FACS-purified lin–CD34+CD38– normal BM (n = 4) and chronic-phase CML cells (n = 8) using the same isoform-specific primers shown in Figure 4A and the corresponding standard curves. Values shown are the mean ± SEM.
The majority of the more primitive CD34+ leukemic cells are cycling in contrast to the CD34+ cells in normal BM.20 Nevertheless, the leukemic CD34+ population does contain a subset of quiescent (G0) cells20 and in vitro these are relatively insensitive to imatinib mesylate, an inhibitor of the BCR-ABL kinase.26 It was therefore important to determine whether the up-regulation of AHI-1 expression seen in the total CD34+ leukemic population was also a feature of these quiescent CML progenitors. The method of dual Hst/Py labeling of CD34+ cells was used to isolate as separate populations the G0 (HstloPylo) and the G1+S+G2+M (Hstlo/+ Py+) CD34+ cells (Figure 6A) from 6 chronic-phase CML patients' samples, in which the LTC-ICs were predominantly Ph+, and from 3 normal BM samples as controls. AHI-1 transcripts were consistently higher in the G0 fraction of CD34+ cells from both normal BM and CML samples. However, when the levels in each fraction were compared between normal and CML samples, there were more AHI-1 transcripts in both the G0 and the G1/S/G2/M subpopulations of CD34+ CML cells, with the greater increase being seen in the G0 subpopulation (∼3-fold; Figure 6B).
Increased expression of AHI-1 in quiescent versus cycling CD34+ CML cells. (A) Representative examples of Hst and Py staining of viable (PI–) CD34+ cells from normal human BM (left) and from a CML sample (right). (B) Absolute AHI-1 transcript levels calculated as described in “Patients, materials, and methods” from real-time RT-PCR analyses of RNA isolated from G0 and G1/S/G2/M fractions of normal BM CD34+ cells (n = 3) and CD34+ cells from chronic-phase CML samples (n = 6). Values shown are the mean ± SEM.
Increased expression of AHI-1 in quiescent versus cycling CD34+ CML cells. (A) Representative examples of Hst and Py staining of viable (PI–) CD34+ cells from normal human BM (left) and from a CML sample (right). (B) Absolute AHI-1 transcript levels calculated as described in “Patients, materials, and methods” from real-time RT-PCR analyses of RNA isolated from G0 and G1/S/G2/M fractions of normal BM CD34+ cells (n = 3) and CD34+ cells from chronic-phase CML samples (n = 6). Values shown are the mean ± SEM.
Because specific patterns of Ahi-1/AHI-1 expression were found in different lineages of normally maturing hematopoietic cells (Figure 2) and the output of different (leukemic) blood cell types are known to be differentially affected in chronic-phase CML patients (increased myelopoiesis and decreased erythropoiesis), it was of interest to examine AHI-1 expression in specific subsets of CML lin+ cells. As shown in Figure 5B, by comparison to the same phenotype of cells from normal adult BM (n = 3), AHI-1 transcript levels were higher in terminally differentiating granulopoietic (CD15/66b+) and erythroid (glycophorin A+) cells, both of which are part of the Ph+ clone but not from the T cells (CD3+) (which are not part of the Ph+ clone) from all chronic-phase CML samples examined (n = 6).
Aberrant splicing of AHI-1 mRNA in CML cells
To investigate whether the altered expression of AHI-1 characteristic of primary leukemic CML AHI-1 might extend to an alteration in their output of different AHI-1 isoforms, additional real-time RT-PCR analyses of RNA from various normal BM and CML subpopulations were undertaken. As summarized in Table 6 and Figure 5C, the distribution of transcripts for AHI-1 isoforms I, II, and III remained relatively constant with increasing differentiation from the lin–CD34+CD38– compartment to the majority lin+CD34– population, with transcripts for isoform III being the most prevalent and for isoform II the least abundant. By comparison to normal BM, transcripts for all 3 isoforms were elevated (∼3- to 5-fold) in the predominant lin+CD34– cells (Table 6). In the more primitive subpopulations of CML cells, the pattern seen for the total AHI-1 transcripts, with the greatest increase being seen in the lin–CD34+CD38– subset (Figure 5A), was mirrored by the changes exhibited for isoforms I and II (Table 6; Figure 5C). In contrast, although transcripts for isoform III were elevated to a similar extent as isoforms I and II in the total CD34+ cells, there was no further increase in isoform III transcripts in the lin–CD34+CD38– subset. A similar finding was obtained from comparative real-time RT-PCR analyses of the levels of transcripts for the different AHI-1 isoforms in the G0 and the G1/S/G2/M subsets of CD34+ cells from normal BM and CML samples (Table 7). These showed higher transcript levels of all isoforms in the G0 vs G1/S/G2/M fractions of CD34+ cells from both normal BM and CML patients. The levels of transcripts for isoforms I and II were also approximately 3- to 5-fold higher in the CML cells by comparison to their normal BM counterparts, but there was no difference in the levels of transcripts for isoform III in the G0 subset of CD34+ from the CML samples and normal BM.
Absolute levels of transcripts for different AHI-1 isoforms in subpopulations of cells in normal BM and chronic-phase CML samples
Phenotype and origin of cells . | No. of transcripts/μg RNA × 104 (fold change vs NBM) . | . | . | ||
|---|---|---|---|---|---|
| . | Isoform I . | Isoform II . | Isoform III . | ||
| Lin-CD34+CD38- | |||||
| NBM | 0.5 ± 0.06 | 0.15 ± 0.03 | 1.05 ± 0.02 | ||
| CML | 15.6 ± 0.3 (31) | 4.8 ± 0.6 (32) | 0.85 ± 0.15 (0.8) | ||
| Lin-CD34+CD38+ | |||||
| NBM | 0.27 ± 0.05 | 0.05 ± 0.01 | 0.50 ± 0.08 | ||
| CML | 1.48 ± 0.12 (5.4) | 0.16 ± 0.03 (3.2) | 1.00 ± 0.12 (2) | ||
| Lin-CD34+ | |||||
| NBM | 0.35 ± 0.09 | 0.06 ± 0.01 | 0.65 ± 0.10 | ||
| CML | 1.6 ± 0.25 (4.5) | 0.19 ± 0.03 (3.2) | 1.60 ± 0.30 (2.5) | ||
| Lin+CD34- | |||||
| NBM | 0.1 ± 0.03 | 0.01 ± 0.00 | 0.20 ± 0.02 | ||
| CML | 0.45 ± 0.08 (4.5) | 0.05 ± 0.01 (5) | 0.65 ± 0.08 (3.3) | ||
Phenotype and origin of cells . | No. of transcripts/μg RNA × 104 (fold change vs NBM) . | . | . | ||
|---|---|---|---|---|---|
| . | Isoform I . | Isoform II . | Isoform III . | ||
| Lin-CD34+CD38- | |||||
| NBM | 0.5 ± 0.06 | 0.15 ± 0.03 | 1.05 ± 0.02 | ||
| CML | 15.6 ± 0.3 (31) | 4.8 ± 0.6 (32) | 0.85 ± 0.15 (0.8) | ||
| Lin-CD34+CD38+ | |||||
| NBM | 0.27 ± 0.05 | 0.05 ± 0.01 | 0.50 ± 0.08 | ||
| CML | 1.48 ± 0.12 (5.4) | 0.16 ± 0.03 (3.2) | 1.00 ± 0.12 (2) | ||
| Lin-CD34+ | |||||
| NBM | 0.35 ± 0.09 | 0.06 ± 0.01 | 0.65 ± 0.10 | ||
| CML | 1.6 ± 0.25 (4.5) | 0.19 ± 0.03 (3.2) | 1.60 ± 0.30 (2.5) | ||
| Lin+CD34- | |||||
| NBM | 0.1 ± 0.03 | 0.01 ± 0.00 | 0.20 ± 0.02 | ||
| CML | 0.45 ± 0.08 (4.5) | 0.05 ± 0.01 (5) | 0.65 ± 0.08 (3.3) | ||
Comparison of transcript levels of AHI-1 isoforms in G0 and G1/S/G2/M CML cells
Phenotype and origin of cells . | No. of transcripts/μg RNA × 104 (fold change vs NBM) . | . | . | ||
|---|---|---|---|---|---|
| . | Isoform I . | Isoform II . | Isoform III . | ||
| G0 | |||||
| NBM | 0.38 ± 0.03 | 0.06 ± 0.005 | 0.96 ± 0.05 | ||
| CML | 1.35 ± 0.04 (3.6) | 0.19 ± 0.02 (3.1) | 1.10 ± 0.07 (1.1) | ||
| G1/S/G2/M | |||||
| NBM | 0.22 ± 0.02 | 0.02 ± 0.005 | 0.26 ± 0.04 | ||
| CML | 0.90 ± 0.03 (4.1) | 0.11 ± 0.02 (5.5) | 0.72 ± 0.05 (2.7) | ||
Phenotype and origin of cells . | No. of transcripts/μg RNA × 104 (fold change vs NBM) . | . | . | ||
|---|---|---|---|---|---|
| . | Isoform I . | Isoform II . | Isoform III . | ||
| G0 | |||||
| NBM | 0.38 ± 0.03 | 0.06 ± 0.005 | 0.96 ± 0.05 | ||
| CML | 1.35 ± 0.04 (3.6) | 0.19 ± 0.02 (3.1) | 1.10 ± 0.07 (1.1) | ||
| G1/S/G2/M | |||||
| NBM | 0.22 ± 0.02 | 0.02 ± 0.005 | 0.26 ± 0.04 | ||
| CML | 0.90 ± 0.03 (4.1) | 0.11 ± 0.02 (5.5) | 0.72 ± 0.05 (2.7) | ||
Expression of AHI-1 in blasts from patients with acute Ph+ or Ph– leukemia
AHI-1 transcript levels were then analyzed in cells from patients with various types of acute leukemia. These included FACS-purified subpopulations of CD34+ and CD34– cells from patients with CML in myeloid blast crisis (n = 2; Table 1), pre-B–ALL (Ph+/BCR-ABL+, n = 8; and Ph–/BCR-ABL–, n = 4; Table 2), and Ph– AML (> 35% blasts, n = 15; Table 3). AHI-1 transcript levels were found to be elevated in both the lin–CD34+ and lin+CD34– cells present in the 2 CML patients with blast-phase disease (Figure 5A). The results for the pre-B–ALL analyses are shown in Figure 7A. It can be seen that AHI-1 transcript levels were slightly elevated in the CD34+CD19+ cells obtained from 4 (3 Ph+ and 1 Ph–) of the 12 ALL samples studied and not in the CD34+CD19– (presumably normal) cells from any of these samples. The 3 Ph+ ALL samples showing the highest levels of AHI-1 transcripts in the CD34+CD19+ cells were those in which the CD34+CD19+ population contained the highest proportion of Ph+ blasts (94%-100%), as shown by fluorescence in situ hybridization (FISH) analysis (data not shown). However, overall the AHI-1 transcript levels in CD34+CD19+ cells obtained from Ph+ and Ph– ALL samples compared with normal controls were not significantly different (P = .25). Additional studies with much larger groups of ALL patients will be required to determine whether AHI-1 expression is differentially affected in the leukemic blasts of patients with Ph+ disease. The results for the AML analyses are shown in Figure 7B. These revealed no obvious deviations from the pattern of AHI-1 expression characteristic of the corresponding populations of normal BM cells. AHI-1 isoforms were also found to be expressed at similar levels in the same subpopulations of AML cells as in their counterparts in normal BM (data not shown).
Comparison of AHI-1 transcript levels in different subpopulations of primary AML and ALL cells. Absolute AHI-1 transcript levels calculated as described in “Patients, materials, and methods” from real-time RT-PCR analyses of RNA isolated from the populations indicated. (A) Comparison of CD19– and CD19+ subpopulations of normal human BM CD34+ cells (open = 4) and CD34+ ALL cells with Ph+ (n = 8) or Ph– (n = 4) genotypes. (B) Comparison of different subpopulations of normal human BM cells (n = 4) and different FAB subtypes of AML (n = 15). The mean levels of AHI-1 expression in each population are indicated by the horizontal bars.
Comparison of AHI-1 transcript levels in different subpopulations of primary AML and ALL cells. Absolute AHI-1 transcript levels calculated as described in “Patients, materials, and methods” from real-time RT-PCR analyses of RNA isolated from the populations indicated. (A) Comparison of CD19– and CD19+ subpopulations of normal human BM CD34+ cells (open = 4) and CD34+ ALL cells with Ph+ (n = 8) or Ph– (n = 4) genotypes. (B) Comparison of different subpopulations of normal human BM cells (n = 4) and different FAB subtypes of AML (n = 15). The mean levels of AHI-1 expression in each population are indicated by the horizontal bars.
Discussion
Ahi-1/AHI-1 is a recently identified and widely expressed gene that encodes a family of proteins of as yet undetermined function(s).1 However, the fact that these proteins contain multiple SH3-binding sites, an SH3 domain and multiple WD40-repeat domains, all of which are known to be important mediators of protein-protein interactions, suggests Ahi-1/AHI-1 proteins are involved in cell-signaling activities. In the present study, we first demonstrated that the expression of Ahi-1/AHI-1 is regulated at multiple stages of hematopoiesis in a fashion that is conserved between mouse and humans. Ahi-1/AHI-1 is expressed at the highest level in the most primitive hematopoietic cells and is then rapidly down-regulated as the cells begin to differentiate. Comparison of AHI-1 expression in quiescent (G0) versus proliferating (G1/S/G2/M) cells in the CD34+ fraction of normal human BM cells showed AHI-1 transcript levels to be approximately 2- to 3-fold higher in the G0 cells. However, because this strategy also selects for the LTC-IC–containing CD38– subset of CD34+ cells, it is difficult from these studies to distinguish between the operation of regulatory mechanisms associated with cell cycle control rather than differentiation. Interestingly, the same pattern of down-regulated AHI-1 expression was found to apply to the 3 most prevalent transcript isoforms expressed in normal hematopoietic cells, even though absolute transcript levels for these 3 AHI-1 isoforms varied over an approximate 10-fold range at each stage of differentiation studied. Thus down-regulation of Ahi-1/AHI-1 expression may be critical to normal hematopoietic cell differentiation, as has now been reported for a number of genes.27-30 In addition, our study suggests that regulation of Ahi-1/AHI-1 gene expression continues during the terminal maturation of hematopoietic cells in a lineage-specific fashion, suggesting a potential involvement of AHI-1 proteins in the correct execution of specific differentiation programs.
A survey of AHI-1 transcript levels in a variety of human leukemic cell lines provided initial evidence for the likely involvement of deregulated AHI-1 in human leukemia. In most of these lines, including some with myeloid, T-, or B-lineage phenotypes, AHI-1 transcripts were significantly increased above the level found in any normal human BM cell. These findings are similar to those recently reported for murine leukemias and lymphomas harboring insertional Ahi-1 mutations.1 AHI-1 transcripts were found to be present at the highest levels in 3 lines established from patients with HTLV-1–induced ATLL (Hut102, Hut78, and MT-4). ATLL is a T-cell malignancy that appears in 3% to 5% of HTLV-1–infected individuals depending on as yet unknown cofactors.31 One of these may be Tax, an HTLV-1–encoded oncoprotein. Tax triggers T-cell proliferation and interferes with many cellular pathways.32-35 It has also been implicated in the initiation of a preleukemic state36 and the control of telomerase reverse transcriptase expression, suggesting a role in genome instability.37 The present studies suggest a possible involvement of AHI-1 as another “cofactor” in the genesis of ATLL.
The observation of increased AHI-1 transcript levels in K562 cells, particularly the shorter versions, prompted us to examine primary sources of Ph+ leukemic cells for evidence of aberrant AHI-1 expression. A series of studies revealed that AHI-1 transcript levels were elevated not only in the blasts from the acute phase of CML but also in the leukemic blasts from a subset of patients with Ph+ pre-B–ALL. In contrast, parallel analyses of AHI-1 transcripts in the blast population obtained from 15 patients with Ph– AML revealed no example of a significant perturbation of AHI-1 expression.
Not only were Ahi-1 transcript numbers increased in the leukemic blasts from patients with acute Ph+ leukemia, but AHI-1 transcripts were also elevated in the neoplastic cells present during the accelerated and chronic phases of CML when the differentiation of mature CML blood cell types is not qualitatively perturbed. A comparison could therefore be made of phenotypically identical normal BM and CML cells at different stages of differentiation. These included the most primitive compartment of lin–CD34+CD38– cells that contain the CML stem cells, an intermediate population of lin–CD34+CD38+ cells that contain most of the cells with short-term in vitro colony-forming cell (CFC) ability, and, finally, the lin+CD34– cells that comprise the majority of cells that are undergoing terminal myeloid and erythroid differentiation. At each stage, real-time RT-PCR measurements of absolute transcript levels showed AHI-1 expression to be significantly enhanced in the CML cells.
In the 2 more mature compartments, the majority of the cells are known to be dominated by the neoplastic Ph+ cells in patients with chronic-phase disease. However, in the most primitive compartment, residual normal cells are often predominant and cannot be selectively removed.25,26 Therefore to allow an analysis of AHI-1 expression in the most primitive types of chronic-phase CML cells, we selected samples from patients in whom these were known to outnumber their normal counterparts several-fold.19 Comparison of the transcript levels measured in these cells with their normal BM counterparts showed that the magnitude of the elevation in AHI-1 expression in CML cells was not uniform during their differentiation but was most marked (∼40-fold) in the lin–CD34+CD38– subset. Subsequently this difference was reduced to approximately 15-fold in the remaining CD34+ cells and approximately 5-fold in the terminally differentiating lin+CD34– cells. The increased expression of AHI-1 seen in CML CD34+ cells relative to in normal BM CD34+ cells was also found to be more marked in the quiescent (G0) subset. However, these cells would also have been selectively enriched in their content of lin–CD34+CD38– cells,16 which could explain the increased AHI-1 transcripts detected. In a separate study, we have found the levels of BCR-ABL transcripts to also be highest in the lin–CD34+CD38– CML cells with progressive decreases in their absolute levels as the cells differentiate.38 Together, these findings suggest a strong biologic connection between BCR-ABL expression and deregulation of AHI-1 expression.
The present studies also provide evidence of splicing perturbations affecting AHI-1 expression in human leukemic cells. Variation of the splicing process has been previously associated with tumorigenesis.39 In 4 of the cell lines analyzed (K562, Raji, Hut102, and Hut78 cells), increases in the production of shorter transcripts (4.2, 2.0, and/or 1.2 kb) were documented. In primary CML cells, a general pattern of up-regulated expression of the AHI-1 transcript isoforms I and II was seen in all populations analyzed, and a lack of up-regulated expression specific to isoform III was noted in the most primitive chronic-phase CML cells. The isoform II of AHI-1 lacks the SH3 domain and would thus be expected to encode truncated proteins with distinct properties, possibly including gain-of-function activities. The presence of elevated levels of transcripts for such an entity could thus be envisaged to have significant biologic consequences. Isoform III contains additional sequences not shared by the other 2 isoforms. This would suggest a possible dominant-negative function. Although it not clear at present how this would be mediated, the unique sequences possessed by inform III could be envisaged to confer novel interactive abilities with other proteins.
In summary, Ahi-1/AHI appears to encode a family of signaling proteins whose levels are normally markedly down-regulated during early hematopoietic cell differentiation. This may, in fact, be important to the loss of stem cell self-renewal capacity and the activation and subsequent execution of normal hematopoietic differentiation programs. Accordingly, mechanisms that impede the early down-regulation of expression of this gene in primitive hematopoietic cells or that inappropriately stimulate its expression in their immediate progeny might be predicted to have leukemogenic sequelae. The complexity of isoforms encoded by AHI-1 and their potential to be differentially perturbed by variable effects on transcript production suggests a multiplicity of outcomes from even a single mutation affecting AHI-1 expression. Identification of the targets of AHI-1 proteins and tests of the biologic effects of overexpression or knocked out expression of specific AHI-1 isoforms in normal and leukemic cells should offer important approaches to the further elucidation of the role of AHI-1 in human leukemia.
Prepublished online as Blood First Edition Paper, January 29, 2004; DOI 10.1182/blood-2003-11-4026.
Supported by the Cancer Research Society of Canada (X.J.) with additional support from the National Cancer Institute of Canada (NCIC; C.E. and A.E.) with funds from the Canadian Cancer Society and the Terry Fox Run and Genome BC. Y.Z. was a recipient of a Stem Cell Network Fellowship, W.-Y. C. was a recipient of a BC Cancer Summer Studentship, E.P. was a recipient of a University of British Columbia Summer Studentship, and S.K. was a recipient of a Canadian National Science and Engineering Research Council Studentship.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank G. Thornbury, R. Zapf, and C. Smith for FACS operation; K. Saw, L. Zhou, and G. Shaw for excellent technical assistance; the staff of the Stem Cell Assay Laboratory for initial processing, cryopreservation, and isolation of lin– cells from adult normal and leukemic BM and blood samples; and R. Premji for assistance in preparing the manuscript. We also thank Sumio Sugano (Human Genome Center, Tokyo, Japan) for the AHI-1 cDNA clone COLO1816.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal