Abstract
There are conflicting reports about the involvement of single nucleotide polymorphisms (SNPs) of the ataxia telangiectasia mutated (ATM) gene with cancer, and the consequences of these SNPs for ATM function remain unclear. We therefore sought to identify SNPs of the ATM gene in pediatric Hodgkin disease (HD) and to analyze ATM function in cells from patients with these SNPs. We have identified SNPs of the ATM gene in 5 of 14 children (S1455R, n = 1; H1380Y, n = 1; N1650S, n = 2; and I709I, n = 1). One patient had nonsense-associated altered splicing of the ATM gene. Lymphoblastoid cell lines expressing the S1455R and N1650S exhibited defective ATM-mediated p53 phosphorylation and Chk2 activation; cells expressing the H1380Y exhibited defective c-Abl activation after X-irradiation. Expression of the N1650S in ATM-null fibroblasts conferred only partial hyperradiosensitivity. Furthermore, the introduction of N1650S ATM into U2OS cells, which express wild-type ATM, showed reduced p53-Ser15 phosphorylation, suggesting a dominant-negative effect of the N1650S over the wild-type ATM protein. We conclude that the rare polymorphic variants of the ATM gene that we identified in children with HD encode functionally abnormal proteins, and we discuss the possible genetic risk factors for childhood HD.
Introduction
It is estimated that 0.5% to 1% of the general population are heterozygous carriers of mutations in the ataxia telangiectasia mutated (ATM) gene that is responsible for ataxia telangiectasia (AT), an autosomal recessive, multisystem disorder associated with progressive cerebellar ataxia, bulbar telangiectasia, immunologic deficiency, chromosomal instability, radiation sensitivity, and predisposition to lymphoreticular cancers.1 The heterozygous carriers of the ATM gene, who are clinically normal, have an increased susceptibility to cancer,2-4 particularly breast cancer,4-6 and patients with AT are at high risk of malignancies such as leukemia and malignant lymphoma, including Hodgkin disease (HD).2,7-10 Although most cases of HD occur sporadically, there may be inheritable factors involved in the development of HD,11-15 and a subset of HD is associated with an increased incidence of secondary malignancies, especially in patients who were younger at the time of treatment.16-19 Survivors of HD, like AT carriers, are at increased risk of radiation-induced breast cancer.18,20,21
The 13-kb transcript of the ATM gene encodes a 350-kDa protein kinase whose carboxy-terminal region exhibits homology to the catalytic domain of phosphoinositide-3-kinases22,23 and that plays key roles in intracellular signaling, cell-cycle control, and DNA repair and recombination.24 In response to DNA damage, increased ATM protein kinase activity mediates the activation and stabilization of p53 and results in cell-cycle arrest. p53 is stabilized by ATM-mediated phosphorylation at Ser1525,26 and Ser20. The ATM-dependent phosphorylation of p53 at Ser20 is mediated by the checkpoint kinases Cds1/Chk2 and Chk1.27,28 Phosphorylation of Ser15 affects primarily the activity of p53, whereas phosphorylation of Ser20 of p53 and Ser395 of HDM2, a proto-oncoprotein that promotes the rapid degradation of p53, affects the stability of p53. ATM has been shown to associate physically with c-Abl, but it has never been shown convincingly that full-length ATM phosphorylates c-Abl in vitro or that phosphorylation of Ser465 of c-Abl in vivo is dependent on ATM. c-Abl activation may, however, depend on ATM. After DNA damage in AT cells, c-Abl–mediated phosphorylation of RNA polymerase II carboxy-terminal domain (CTD) is defective,29,30 and irradiation-induced disruption of the BRCA1 and c-Abl complex is dependent upon ATM.31 These results suggest that c-Abl phosphorylation has some role in the DNA damage and repair machinery.
Cells from patients with AT and some ATM heterozygotes have defective multiple cell-cycle checkpoint mechanisms and show increased radiosensitivity.32,33 There is also evidence that ATM haploinsufficiency hypersensitizes mice to sublethal doses of irradiation.34,35 We do not know much about the relation between heterozygous missense mutations or single nucleotide polymorphisms (SNPs) that give rise to functionally defective proteins and AT or cancer susceptibility, but there are a few reports of an association between heterozygous ATM missense mutations and cancer susceptibility. For example, heterozygous carriers of the 7271T>G (V2424G) ATM mutation show increased susceptibility to breast cancer and leukemia,36 and ATM heterozygous mice carrying 7636_7644del show an increased cancer susceptibility.37 Several germ-line SNPs of ATM have been identified in breast cancer,6,36,38-40 colorectal cancer,41 chronic lymphocytic leukemia (CLL),42-44 pediatric leukemia with MLL gene rearrangement,45 and acute lymphoblastic leukemia (ALL) of T-cell lineage.46 A few of these SNPs seem to behave as missense mutations,40 but the meaning or functional consequences of many others is still unclear. The aim of the present study was, therefore, to examine the germ-line ATM gene for the presence of SNPs in children with HD to determine whether there is a link between SNPs of the ATM gene and genetic risk factors for pediatric HD. We screened cord blood DNA (from archived stocks) from healthy neonates for the alleles we found in our patients with HD, to determine the frequencies of those alleles in a healthy population. We also examined the kinase activities of wild-type and variant ATM in Epstein-Barr virus (EBV)–transformed lymphoblastic cell lines (LCLs) from healthy subjects and from patients with HD whose ATM gene contained SNPs. To further clarify the functions of the ATM variants that we found, we examined (1) ATM function in ATM-null cells transfected with wild-type or variant ATM by evaluating clonogenic cell survival after irradiation, (2) in vitro ATM kinase activity of wild-type or variant ATM expressed in human embryonic kidney–derived 293 cells by transfection, and (3) the dominant-negative effect of this variant ATM by transfecting it into ATM-competent osteosarcoma-derived U2OS cells.
Patients, materials, and methods
Patients
For this study, we enrolled 14 Japanese pediatric patients with HD and no symptoms or family history of ataxia telangiectasia who were receiving care at more than 10 independent, unrelated institutions. Mononuclear cells were extracted from peripheral blood samples obtained from these patients and from healthy subjects (unselected healthy volunteers). Blood samples were also obtained from patients' parents, in 2 cases. DNA extracted from neonatal cord blood was obtained from archived stocks of DNA maintained at the Department of Pediatrics, Showa University Fujigaoka Hospital. Written informed consent was obtained from patients or their parents or guardians for the collection and use of all samples, and approval for this study was obtained from the local ethics committee of Tokyo Medical and Dental University.
DNA sequence analysis
To identify SNPs in the ATM coding sequence, we sequenced the open reading frame (ORF) and partial 5′ untranslated region (UTR) of ATM cDNA obtained from fresh peripheral blood mononuclear cells from 14 patients with HD. Messenger RNA was extracted from these mononuclear cells by using the Dynabeads RNA extraction kit (Dynabeads, Oslo, Norway), and reverse transcriptase–polymerase chain reaction (RT-PCR) product was amplified from this mRNA by using the LA RT-PCR kit (TaKaRa, Kyoto, Japan). Amplified PCR products were sequenced by using a cycling sequence method (ABI Big Dye terminator chemistry; PE Applied Biosystems, Foster City, CA) followed by capillary electrophoresis on an ABI 310 automated sequencer. SNPs identified in RT-PCR products were confirmed by sequencing genomic DNA-based PCR products from peripheral blood mononuclear cells.
Estimation of allele frequency
The frequency of ATM mutant alleles that have 4949A>G, 4365T>A, and 4138C>T sequence variations among 100 samples of DNA from normal neonatal cord blood was determined by using (1) the PCR and restriction-fragment length polymorphism (RFLP) method, using the restriction endonuclease TspEI (for allele 4949A>G), or (2) the allele-specific multiplex PCR method (for 4365T>A and 4138C>T).47 The primers and PCR conditions used to examine exons 30, 31, and 35 were as previously reported.48 The primers used to identify alleles 4365A (patient HD1) and 4138T (patient HD4) by SNP-specific multiplex PCR were CTGAAAGATATAAAAAGT (5′-HD1 mutant-specific primer), CCAAGCTCCTCCTAAGCCT (3′-HD1 mutant-specific primer), CCTGCTCCTAATCCACCTT (5′-HD4 mutant-specific primer), and CACATGCGATGGAAAATA (3′-HD4 mutant-specific primer). The PCR conditions were 94°C for 30 seconds, 50°C for 45 seconds, and 72°C for 30 seconds, for each of 35 cycles.
Subcloning of the ATM mutant allele
RNA was obtained from 1 × 106 EBV-transformed LCLs by using the Dynabeads RNA extraction kit. A partial sequence (nucleotides 3880-5907 of ATM) was amplified by using Pyrobest Taq polymerase (TaKaRa) followed by cDNA synthesis using reverse transcriptase XL (TaKaRa). Amplified cDNA was subcloned into the pGEM-T Easy TA cloning system (Promega, Madison, WI). The plasmid, which contained the corresponding SNP of the ATM gene, was digested with KpnI and SwaI, and the insert was subcloned into KpnI and SwaI sites of the ATM expression vector pEBS-YZ5 by using a DNA ligation kit version 2 (TaKaRa).49 We sequenced the resulting expression vector (pEBS-YZ5/4949G) and confirmed that it had a wild-type sequence, except for the single 4949A>G mutation.
Segregation analysis
Segregation analysis was performed in cases HD2 and HD3. Blood samples were obtained, with written informed consent, from both parents in each case, and DNA was extracted from mononuclear cells separated from the blood. The exons corresponding to those that had SNPs in the children's cases were sequenced after genomic PCR amplification. A familial history of illness was obtained from the parents.
Cells, cell lines, and their characteristics
LCLs were established by immortalizing peripheral blood mononuclear cells with EBV (strain B95-8).50 LCLs were thus established from 3 healthy volunteers (controls: LCL-wt1, LCL-wt2, and LCL-wt3) and from 4 patients with HD (LCL-HD1, LCL-HD2, LCL-HD4, and LCL-HD6). LCLs were maintained at approximately 5 × 105 cells/mL in RPMI 1640 medium (Gibco BRL, Gaithersburg, MD) containing 15% fetal calf serum (FCS; Gibco BRL) at 37°C in 5% CO2. The LCL AT65RM,51 which harbors the compound heterozygous ATM mutation 8814_8824del/6527_6528ins7 and results in ATM-null cells, was used as a negative control in studies of ATM protein function; the SV40-transformed AT cell line GM05849C harboring the homozygous ATM mutation 7009_7010delTG, which also results functionally in ATM-null cells (Coriell Cell Repositories, Camden, NJ), was used to examine the function of a variant ATM protein obtained from one of our patients with HD; and human embryonic kidney–derived 293 and osteosarcoma-derived U2OS cells (Human Science Research Resources Bank, Osaka, Japan), which show normal ATM-dependent phosphorylation of p53, were used to investigate the kinase activity of a mutant ATM. These cells were maintained in Dulbecco modified Eagle medium (DMEM; Gibco BRL) containing 10% FCS.
ATM functional assays
Preparation of cell lysates. Cells were X-irradiated, to induce DNA damage, during the logarithmic growth phase. Lysates were prepared, from 1 × 106 cells, by first washing the cells with phosphate-buffered saline (PBS) then incubating them in 150 mM NaCl, 1.0% NP-40, 0.1% sodium dodecyl sulfate (SDS), 0.1% sodium deoxycholate, 5 mM EDTA (ethylenediaminetetraacetic acid), 10 mM Tris-HCl pH 7.4 containing protease inhibitors, for 30 minutes on ice. In all cases, cell lysates were also prepared from unirradiated cells as a control.
Western blot analysis. Protein concentration in the lysates was measured using the DC protein assay (Bio-Rad, Richmond, CA); 30 μg protein samples were denatured in boiling sample buffer, separated by SDS polyacrylamide gel electrophoresis (PAGE), and transferred to polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA). Blots were probed with anti–ATM antibody,51 anti–p53-Ser15 phosphospecific antibody (New England Biolabs, Beverly, MA), anti–p53-Ser20 phosphospecific antibody,28 anti–p53 antibody (clone Do1; Oncogene Research, Boston, MA), anti–Chk2 antibody, anti–c-Abl antibody (24-11; Santa Cruz, Santa Cruz, CA), or anti–α-tubulin antibody (Oncogene Science, Cambridge, MA). Primary antibodies were detected by binding horseradish peroxidase (HRP)–conjugated antirabbit or antimouse second antibody (Amersham Life Science, Buckinghamshire, United Kingdom).
ATM activity in wild-type, HD, and AT cells. Using Western blotting, we examined cell lysates for the presence of p53 phosphorylated at Ser15 (which is generated by direct phosphorylation by ATM after DNA damage) or at Ser20 (which is generated by ATM-dependent Chk2 phosphorylation after DNA damage) to assess the extent of ATM kinase activity occurring in the cells 30 minutes after 5 Gy of X-irradiation.
We also performed in vitro assays, as reported previously, of ATM kinase52 and ATM-dependent Chk253 and c-Abl30,54 kinase activities. Cell lysates were prepared 1 hour after 5 Gy of irradiation for the Chk2 assay or after 10 Gy of X-irradiation for c-Abl assay, and the proteins were immunoprecipitated by using anti–ATM antibody,51 anti–Chk2 antibody, or anti–c-Abl antibody, respectively. The substrate used in the ATM kinase assay was a glutathione S-transferase (GST)–p53 fusion protein (N-terminal 1-100 amino acids); anti–p53-phosphoserine 15 antibody was used to detect phosphorylated Ser15 by Western blotting. The substrates used for Chk2 kinase and c-Abl kinase were recombinant GST-Cdc25C and a GST-CTD fusion protein, respectively; incorporation of phosphate from 32P γATP into the phosphorylated product was detected by autoradiography after SDS-PAGE. GST-p53, GST-Cdc25C, and GST-CTD were visualized by staining the gels with Coomassie brilliant blue. The results were quantified by using NIH image 1.63 software (National Institutes of Health, Bethesda, MD).
N1650S variant ATM activity. We used an in vitro kinase assay, as described above, to compare the in vitro activities of wild-type and N1650S variant ATM in LCLs. To obtain a sufficient quantity of each protein for such analysis, we transfected human embryonic kidney–derived 293 cells with FLAG-tagged wild-type ATM (pEBS-YZ5) and N1650S variant ATM (pEBS-YZ5/4949G) by using Effectene transfection reagent (Qiagen, Hilden, Germany) and prepared cell lysates 24 hours after transfection. The FLAG-tagged proteins were immunoprecipitated with anti–FLAG antibody (M2, Sigma), and Western blotting was used to detect the phosphorylated products, as before. Results were quantified by using NIH image 1.63 software.
We used a clonogenic cell survival assay to assess the effect of the N1650S variant ATM on cultured cells' responses to DNA damage caused by X-irradiation. We subcloned the 4949A>G allele encoding the amino acid change N1650S into ATM-expression vector pEBS-YZ5 (as described above) and stably transfected ATM-null cells (GM05849C cells) with the plasmids pEBS7 (vector alone), pEBS-YZ5 (encoding wild-type ATM), or pEBS-YZ5/4949G (encoding N1650S variant ATM) by using Effectene transfection reagent, as before. Cells were selected by their growth in hygromycin (200 μg/mL). The clonogenic assay was conducted as described previously.49 Briefly, cells were resuspended at a concentration of 1 × 103 cells per 3 mL DMEM containing 10% FCS (Gibco BRL), and the cell suspension was plated into each well of a 6-cm multiwell plate. Cells were X-irradiated with a dose of 0, 0.5, 1, 2, or 5 Gy at a rate of 1 Gy/min, and surviving (Giemsa-stained) colonies were counted after 2 weeks in culture.
To determine whether the 4949A>G mutation had a dominant-negative effect, we transfected U2OS cells with mock and N1650S variant ATM expression vectors (pEBS7 and pEBS-YZ5/4949G, respectively) and examined ATM kinase activities in these cells in culture. Transfection was performed, as described above, when the cells were growing logarithmically, and the cells were X-irradiated (5 Gy) 24 hours after transfection. Cell lysates were prepared 30 minutes after irradiation, and the presence of p53 phospho-Ser15 in the lysates was determined by Western blotting as before. Results were quantified by using NIH image 1.63 software.
Analysis of loss of heterozygosity by laser capture microdissection and pyrosequencing
HD cells were retrieved, from paraffin-wax embedded tumor tissue from patients HD2 and HD4, onto a film on a microtube cap by using an LM200 laser capture microdissection instrument (Arcturus Engineering, Mountain View, CA). The film was incubated in 30 μL digestion buffer (1 mg/mL proteinase K, 10 mM Tris-HCl, pH 8.0, 1 mM EDTA) for 2 hours at 50°C, and pyrosequencing was automatically performed on a PSQ96 system, using enzymes and reagents from a PSQ96 SNP reagent kit (Pyrosequencing AB, Uppsala, Sweden). The sequencing primers used to identify SNPs at position 4138 (5′-TGCTCCTAATCCACCT-3′ forward primer) and position 4949 (5′-TTGGATAACTGCAACAAA-3′ reverse primer) of ATM mRNA were designed such that the terminal residue hybridized to the base immediately adjacent to the SNP (C to T and A to G, respectively).
Results
SNPs in the ATM gene in patients with HD
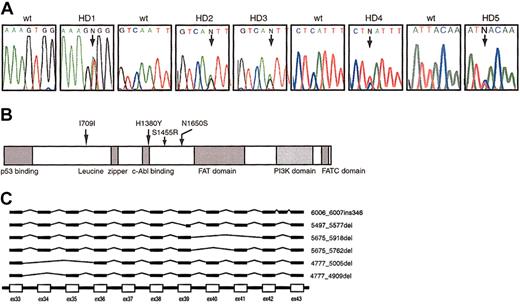
We identified 5 patients with an SNP and 1 patient with nonsense-associated altered splicing (NAS) of the ATM gene (Figure 1).
Polymorphic variations of theATMgene. (A) DNA sequence electrophoretograms with arrows showing the positions of the heterozygous germ line SNPs at 4365T>A in patient HD1, 4949A>G in patients HD2 and HD3, 4138C>T in patient HD4, and 2127T>C in patient HD5. The first codon of the ORF was designated +1. “N” represents indistinguishable nucleotides. (B) Schematic diagram of the ATM protein. The locations of amino acid changes are indicated by arrows in relation to the predicted domains. (C) Schematic diagram of variant species of ATM mRNA found in cells from patient HD6.
Polymorphic variations of theATMgene. (A) DNA sequence electrophoretograms with arrows showing the positions of the heterozygous germ line SNPs at 4365T>A in patient HD1, 4949A>G in patients HD2 and HD3, 4138C>T in patient HD4, and 2127T>C in patient HD5. The first codon of the ORF was designated +1. “N” represents indistinguishable nucleotides. (B) Schematic diagram of the ATM protein. The locations of amino acid changes are indicated by arrows in relation to the predicted domains. (C) Schematic diagram of variant species of ATM mRNA found in cells from patient HD6.
Allele-frequency analysis of SNPs in normal cord blood
We screened 100 samples obtained from an archive stock of DNA from unselected neonatal cord blood for each of the nucleotide changes we found in our patients with HD (Table 1). At the time of submission of this manuscript, none of these SNPs had been catalogued in the AT Mutation Database (http://www.benaroyaresearch.org/bri_investigators/atm.htm; last updated April 2002).55
SNPs detected in patients with HD and the frequency of those SNPs in unselected neonates
Patient . | Nucleotide . | Exon . | Amino acid change . | Allele frequency in neonates, %* . |
|---|---|---|---|---|
| HD1 | 4365T>A | 31 | S1455R | 1 |
| HD2 | 4949A>G | 35 | N1650S | 0 |
| HD3 | 4949A>G | 35 | N1650S | 0 |
| HD4 | 4138C>T | 30 | H1380Y | 3 |
| HD5 | 2127T>C | 16 | I709I | ND |
Patient . | Nucleotide . | Exon . | Amino acid change . | Allele frequency in neonates, %* . |
|---|---|---|---|---|
| HD1 | 4365T>A | 31 | S1455R | 1 |
| HD2 | 4949A>G | 35 | N1650S | 0 |
| HD3 | 4949A>G | 35 | N1650S | 0 |
| HD4 | 4138C>T | 30 | H1380Y | 3 |
| HD5 | 2127T>C | 16 | I709I | ND |
ND indicates not determined.
Archived stocks of 100 samples of DNA from neonatal cord blood.
The SNPs were confirmed by sequencing genomic DNA from fresh peripheral blood mononuclear cells. All of the SNPs found were heterozygous, and 2 unrelated patients (HD2 and HD3) exhibited the same nucleotide change (4949A>G). In both of these cases, 1 parent carried the same heterozygous 4949A>G nucleotide change. The heterozygous 2127T>C mutation seen in patient HD5 was predicted not to involve an amino acid substitution.
NAS of ATM in 1 patient with HD
In patient HD6, we identified wild-type transcripts and 6 aberrant transcripts with NAS (Figure 1C). The ratio of the expression of these mutant transcripts to that of wild-type transcript was approximately 1:1, as determined by sequencing of more than 10 independent clones isolated by TA cloning. Expression of these abnormal transcripts was consistently identified in cells from each of 2 separate fresh blood samples obtained from this patient, confirming that this observation was not an artifact associated with EBV transformation or poor storage conditions.
There was no association between the presence or absence of ATM polymorphisms and the patient's age or histologic or clinical features of HD in the 14 patients included in this study.
Function of variant ATM proteins
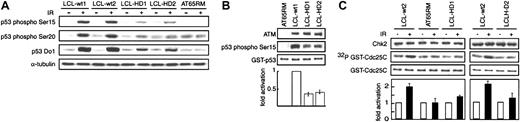
ATM-dependent phosphorylation of p53. We investigated the ATM-dependent response to DNA damage by evaluating the ATM kinase–mediated phosphorylation of p53 at Ser15 and ATM-dependent Chk2-mediated phosphorylation of p53 at Ser20 after a 5-Gy dose of X-irradiation (Figure 2A). We checked p53 phosphorylation after irradiation using several LCLs from healthy subjects, but there was no significant difference between them (not shown). Furthermore, normal p53 phosphorylation after irradiation was shown by cells from 1 patient with HD but no SNPs in the ATM gene (LCL-HD6) and by LCL-HD4 (data not shown). The amount of phosphoserine 15 detected in wild-type cells (LCL-wt1 and LCL-wt2; Figure 2A) 30 minutes after irradiation was far greater than that detected in HD cells (LCL-HD1 and LCL-HD2), and very little phosphoserine 15 was detectable in cells from patients with AT (AT65RM) before or after irradiation. Similarly, the amount of phosphoserine 20 detected in irradiated wild-type cells was far greater than that detected in irradiated HD cells or AT cells. The total amount of p53 in wild-type cells increased after irradiation, as a result of stabilization of p53 by phosphorylation at Ser15 or Ser20; HD cells showed a less substantial increase in total p53 after irradiation. To investigate the activity of ATM in these cells, the immunoprecipitated ATM from AT cells (AT65RM), wild-type cells (LCL-wt1), and HD cells (LCL-HD1 and LCL-HD2) was tested in an in vitro phosphorylation assay using GST-p53 (1-100) as substrate (Figure 2B). Smaller amounts of phosphorylated p53 Ser15 were generated by the ATM from HD cells than by the ATM from wild-type cells, despite the presence of similar amounts of ATM protein, and AT cells contained no detectable ATM or p53 Ser15. Because p53 Ser20 is phosphorylated by Chk2 (another substrate of ATM) in response to DNA damage,56 we performed similar in vitro experiments in which we analyzed the activity of Chk2, which also phosphorylates Cdc25C at Ser216. The results of these experiments showed that ATM-mediated Chk2 kinase activity was impaired in AT65RM, LCL-HD1, and LCL-HD2 cells as well (Figure 2C). These results indicate that the phosphorylating activity of the variant ATM is impaired in comparison with that of the wild-type ATM.
ATM-dependent p53 phosphorylation after DNA damage. (A) Western blot detection of Ser15-phosphorylated p53, Ser20-phosphorylated p53, total p53, and α-tubulin (as a control for the amount of cell lysate loaded) in lysates of immortalized lymphoblastic cells from healthy subjects (LCL-wt1 and LCL-wt2), patients with HD (LCL-HD1 and LCL-HD2), and a patient with AT (AT65RM), before (–) and 30 minutes after (+) X-irradiation (IR; 5 Gy) of the cells in culture. Representative data from 3 independent experiments are shown. (B) Western blot detection of GST-p53 phospho-Ser15 in an in vitro phosphorylation assay with immunoprecipitated ATM from AT cells (AT65RM), wild-type cells (LCL-wt1), and HD cells (LCL-HD1 and LCL-HD2); equivalence of loading of GST-p53 is shown by Coomassie brilliant blue staining (third panel). Extents of phosphorylation of recombinant GST-p53 (1-100) are shown relative to that of LCL-wt1 (= 1) (bottom graph). Data represent mean values from 3 independent experiments. Equal amounts of ATM protein (top panel) were immunoprecipitated from LCL-wt1, LCL-HD1, and LCL-HD2; no ATM protein was immunoprecipitated from AT65RM. (C) Whole lysates of cells from a healthy subject (LCL-wt2), patients HD1 and HD2 (LCL-HD1 and LCL-HD2), and a patient with AT (AT65RM) were immunoprecipitated with anti–Chk2 antibody before (–) and 1 hour after (+) X-irradiation (5 Gy). Immunoprecipitated Chk2 protein (detected by Western blotting with anti–Chk2 antibody; top panel) was used in an in vitro kinase assay with recombinant GST-Cdc25C as the substrate (detected with Coomassie brilliant blue; third panel), phosphorylation of which is shown by 32P incorporation (by autoradiography; second panel). Extents of phosphorylation of recombinant GST-Cdc25C (167-267) after X-irradiation are shown relative to that before X-irradiation (= 1) (bottom graph). Data represent mean values from 3 independent experiments.
ATM-dependent p53 phosphorylation after DNA damage. (A) Western blot detection of Ser15-phosphorylated p53, Ser20-phosphorylated p53, total p53, and α-tubulin (as a control for the amount of cell lysate loaded) in lysates of immortalized lymphoblastic cells from healthy subjects (LCL-wt1 and LCL-wt2), patients with HD (LCL-HD1 and LCL-HD2), and a patient with AT (AT65RM), before (–) and 30 minutes after (+) X-irradiation (IR; 5 Gy) of the cells in culture. Representative data from 3 independent experiments are shown. (B) Western blot detection of GST-p53 phospho-Ser15 in an in vitro phosphorylation assay with immunoprecipitated ATM from AT cells (AT65RM), wild-type cells (LCL-wt1), and HD cells (LCL-HD1 and LCL-HD2); equivalence of loading of GST-p53 is shown by Coomassie brilliant blue staining (third panel). Extents of phosphorylation of recombinant GST-p53 (1-100) are shown relative to that of LCL-wt1 (= 1) (bottom graph). Data represent mean values from 3 independent experiments. Equal amounts of ATM protein (top panel) were immunoprecipitated from LCL-wt1, LCL-HD1, and LCL-HD2; no ATM protein was immunoprecipitated from AT65RM. (C) Whole lysates of cells from a healthy subject (LCL-wt2), patients HD1 and HD2 (LCL-HD1 and LCL-HD2), and a patient with AT (AT65RM) were immunoprecipitated with anti–Chk2 antibody before (–) and 1 hour after (+) X-irradiation (5 Gy). Immunoprecipitated Chk2 protein (detected by Western blotting with anti–Chk2 antibody; top panel) was used in an in vitro kinase assay with recombinant GST-Cdc25C as the substrate (detected with Coomassie brilliant blue; third panel), phosphorylation of which is shown by 32P incorporation (by autoradiography; second panel). Extents of phosphorylation of recombinant GST-Cdc25C (167-267) after X-irradiation are shown relative to that before X-irradiation (= 1) (bottom graph). Data represent mean values from 3 independent experiments.
In summary, the results presented in Figure 2 show that the ATM-dependent DNA damage response is abnormal in cells with single-nucleotide variant ATM from patients with HD. Using an RT-PCR–based RFLP method, we found that the expression levels of the polymorphic and wild-type alleles of ATM in LCL-HD1, LCL-HD2, and LCL-HD4 were equivalent (data not shown).
ATM-mediated activation of c-Abl kinase. Because the SNP identified in cells from patient HD4 was localized within the predicted c-Abl binding domain of ATM,29 we examined the ATM-mediated activation of c-Abl kinase activity in LCL-HD4 by using an in vitro RNA polymerase II CTD phosphorylation assay 1 hour after exposure of the cells to 10 Gy X-irradiation. The results showed that c-Abl kinase activity was reduced in LCL-HD4 as much as it was in AT65RM cells, compared with wild-type cells (Figure 3).
ATM-dependent c-Abl activation in vitro. Whole lysates of cells from a healthy subject (LCL-wt2), patient HD4 (LCL-HD4), and a patient with AT (AT65RM) were immunoprecipitated with anti–c-Abl antibody before (–) and 1 hour after (+) X-irradiation (10 Gy). Immunoprecipitated c-Abl protein (detected by Western blotting with anti–c-Abl antibody; top panel) was used in an in vitro kinase assay with recombinant GST-CTD as the substrate (detected with Coomassie brilliant blue; third panel), phosphorylation of which is shown by 32P incorporation (by autoradiography; second panel). Extents of phosphorylation of recombinant GST-CTD after X-irradiation are shown relative to that before X-irradiation (= 1) (bottom graph). Data represent mean values from 3 independent experiments.
ATM-dependent c-Abl activation in vitro. Whole lysates of cells from a healthy subject (LCL-wt2), patient HD4 (LCL-HD4), and a patient with AT (AT65RM) were immunoprecipitated with anti–c-Abl antibody before (–) and 1 hour after (+) X-irradiation (10 Gy). Immunoprecipitated c-Abl protein (detected by Western blotting with anti–c-Abl antibody; top panel) was used in an in vitro kinase assay with recombinant GST-CTD as the substrate (detected with Coomassie brilliant blue; third panel), phosphorylation of which is shown by 32P incorporation (by autoradiography; second panel). Extents of phosphorylation of recombinant GST-CTD after X-irradiation are shown relative to that before X-irradiation (= 1) (bottom graph). Data represent mean values from 3 independent experiments.
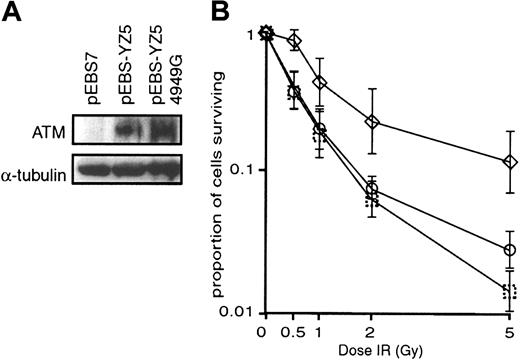
Activity of N1650S variant ATM inATM-null cells and in ATM-competent cells. ATM-null cells (GM05849C) stably transfected with pEBS7 (vector alone) expressed no ATM, but cells transfected with either the wild-type or the variant ATM expression vectors (pEBS-YZ5 or pEBS-YZ5/4949G, respectively) expressed similar amounts of ATM (Figure 4A). We evaluated cell survival by measuring clonogenic activity after irradiation of these cells (Figure 4B).
Clonogenic cell survival ofATM-null cells transfected with wild-type or mutantATM. (A) Western blot analysis showing expression of ATM protein (detected by anti–ATM antibody) in ATM-null (GM05849C) cells stably transfected with pEBS-YZ5 (wild-type ATM expression vector) or pEBS-YZ5/4949G (N1650S variant ATM expression vector). No ATM protein was detectable in mock-transfected (pEBS7) cells, and equal amounts of wild-type and variant ATM were expressed in ATM-null cells. Equivalence of loading of cell lysates is shown by anti–α-tubulin labeling. (B) Cell survival by clonogenic activity 2 weeks after various doses of X-irradiation (IR). The ATM-null cell line GM05849C was transfected with pEBS7 (mock transfectant;  , pEBS-YZ5 (wild-type ATM expression vector; ⋄), or pEBS-YZ5/4949G (N1650S variant ATM expression vector; ○). Data represent mean values from 4 independent experiments.
, pEBS-YZ5 (wild-type ATM expression vector; ⋄), or pEBS-YZ5/4949G (N1650S variant ATM expression vector; ○). Data represent mean values from 4 independent experiments.
Clonogenic cell survival ofATM-null cells transfected with wild-type or mutantATM. (A) Western blot analysis showing expression of ATM protein (detected by anti–ATM antibody) in ATM-null (GM05849C) cells stably transfected with pEBS-YZ5 (wild-type ATM expression vector) or pEBS-YZ5/4949G (N1650S variant ATM expression vector). No ATM protein was detectable in mock-transfected (pEBS7) cells, and equal amounts of wild-type and variant ATM were expressed in ATM-null cells. Equivalence of loading of cell lysates is shown by anti–α-tubulin labeling. (B) Cell survival by clonogenic activity 2 weeks after various doses of X-irradiation (IR). The ATM-null cell line GM05849C was transfected with pEBS7 (mock transfectant;  , pEBS-YZ5 (wild-type ATM expression vector; ⋄), or pEBS-YZ5/4949G (N1650S variant ATM expression vector; ○). Data represent mean values from 4 independent experiments.
, pEBS-YZ5 (wild-type ATM expression vector; ⋄), or pEBS-YZ5/4949G (N1650S variant ATM expression vector; ○). Data represent mean values from 4 independent experiments.
As shown in Figure 4, pEBS-YZ5/4949G-transfected GM05849C cells (ie, those transfected with the single-nucleotide variant ATM) exhibited substantially less clonogenic activity than did wild-type transfectants.
We then analyzed the in vitro kinase activity of the N1650S (4949A>G) ATM variant ectopically expressed in 293 cells and compared the abilities of the immunoprecipitated FLAG-tagged wild-type and N1650S variant ATM proteins to phosphorylate GST-p53 (1-100) at Ser15 in vitro (Figure 5). The N1650S (4949A>G) ATM phosphorylated about 30% less p53 Ser15 than did the wild-type protein.
In vitro phosphorylation of p53 Ser15 by N1650S ATM kinase. FLAG-tagged ATM protein was immunoprecipitated with anti–FLAG antibody from 293 cells transiently transfected with wild-type ATM expression vector pEBS-YZ5, variant ATM expression vector pEBS-YZ5/4949G, or mock transfectant pEBS7. Equal amounts of FLAG-tagged wild-type and variant ATM (detected with anti–FLAG antibody) were immunoprecipitated (top panel). Recombinant GST-p53 (1-100) was subjected to an in vitro phosphorylation assay using the immunoprecipitated, FLAG-tagged wild-type and variant ATM, and GST-p53 phosphorylated at Ser15 was detected by using anti-p53 Ser15 phosphospecific antibody (second panel). Equal amounts of substrate (GST-p53) were present, as shown by Coomassie brilliant blue staining (third panel). The extent of phosphorylation of recombinant GST-p53 (1-100) is shown relative to that of pEBS-YZ5 transfectant (= 1) (bottom graph). Data represent mean values from 3 independent experiments.
In vitro phosphorylation of p53 Ser15 by N1650S ATM kinase. FLAG-tagged ATM protein was immunoprecipitated with anti–FLAG antibody from 293 cells transiently transfected with wild-type ATM expression vector pEBS-YZ5, variant ATM expression vector pEBS-YZ5/4949G, or mock transfectant pEBS7. Equal amounts of FLAG-tagged wild-type and variant ATM (detected with anti–FLAG antibody) were immunoprecipitated (top panel). Recombinant GST-p53 (1-100) was subjected to an in vitro phosphorylation assay using the immunoprecipitated, FLAG-tagged wild-type and variant ATM, and GST-p53 phosphorylated at Ser15 was detected by using anti-p53 Ser15 phosphospecific antibody (second panel). Equal amounts of substrate (GST-p53) were present, as shown by Coomassie brilliant blue staining (third panel). The extent of phosphorylation of recombinant GST-p53 (1-100) is shown relative to that of pEBS-YZ5 transfectant (= 1) (bottom graph). Data represent mean values from 3 independent experiments.
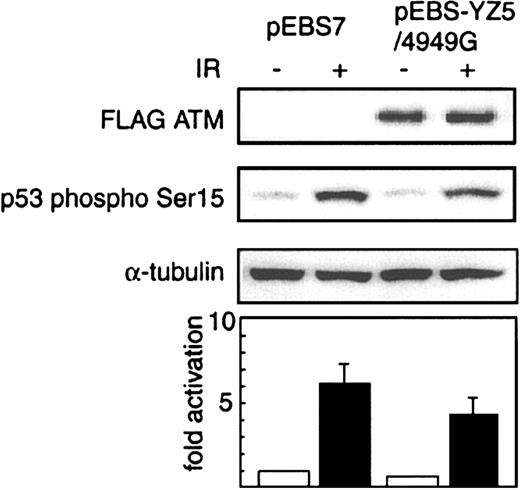
To determine whether transfection with the variant ATM expression vector pEBS-YZ5/4949G had a dominant-negative effect on ATM expression in normal cells, we examined cellular p53-Ser15 phosphorylation in culture, 30 minutes after X-irradiation, in U2OS cells transfected with this variant ATM vector or with vector alone. We analyzed cell lysates for the presence of phosphorylated p53 Ser15 by Western blotting (Figure 6). The pEBS-YZ5/4949G–transfected cells produced 30% less phosphorylated p53 Ser15 than did the mock-transfected cells 30 minutes after irradiation.
Dominant-negative effect of N1650S ATM on p53-Ser15 phosphorylation. Western blot analysis of p53-Ser15 phosphorylation before (–) or 30 minutes after (+) X-irradiation (IR; 5 Gy) in U2OS cells transiently transfected with FLAG-tagged variant ATM expression vector pEBS-YZ5/4949G and expressing FLAG-tagged N1650S ATM, or mock transfectant pEBS7. FLAG-tagged variant ATM was expressed only in pEBS-YZ5/4949G–transfected U2OS cells (detected with anti–FLAG antibody; top panel). Cell lysates were probed with anti–p53-Ser15 phosphospecific antibody (second panel). Anti–α-tubulin antibody (third panel) was used to show equivalence of loading. Extents of phosphorylation of p53 Ser15 after X-irradiation are shown relative to that in pEBS7-transfected cells before X-irradiation (= 1) (bottom graph). Data represent mean values from 3 independent experiments.
Dominant-negative effect of N1650S ATM on p53-Ser15 phosphorylation. Western blot analysis of p53-Ser15 phosphorylation before (–) or 30 minutes after (+) X-irradiation (IR; 5 Gy) in U2OS cells transiently transfected with FLAG-tagged variant ATM expression vector pEBS-YZ5/4949G and expressing FLAG-tagged N1650S ATM, or mock transfectant pEBS7. FLAG-tagged variant ATM was expressed only in pEBS-YZ5/4949G–transfected U2OS cells (detected with anti–FLAG antibody; top panel). Cell lysates were probed with anti–p53-Ser15 phosphospecific antibody (second panel). Anti–α-tubulin antibody (third panel) was used to show equivalence of loading. Extents of phosphorylation of p53 Ser15 after X-irradiation are shown relative to that in pEBS7-transfected cells before X-irradiation (= 1) (bottom graph). Data represent mean values from 3 independent experiments.
These results indicate that this 4949A>G polymorphism, which results in the single amino acid change N1650S, is associated with defective ATM activity and acts in a dominant-negative manner.
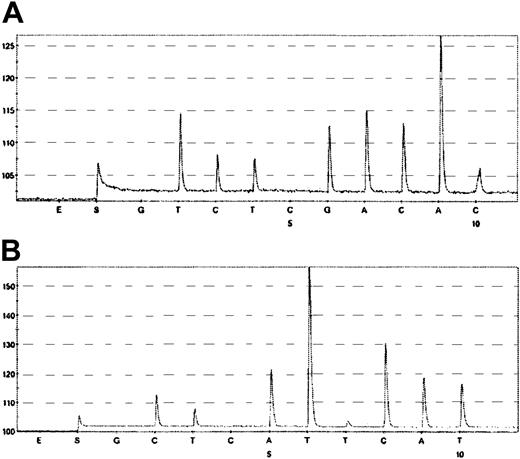
To confirm the allelic status of ATM in HD tumor cells, we looked for loss of heterozygosity by using microcapture, microdissection, and pyrosequencing (Figure 7). HD cells from patients HD2 and HD4 were heterozygous for 4949A>G and wild-type ATM and for 4138C>T and wild-type ATM, respectively, as found in the germ line cells. These results confirm that the ATM gene in HD cells from patients HD2 and HD4 did not undergo loss of heterozygosity.
Loss of heterozygosity analysis by pyrosequencing. (A) Pyrogram of the coding SNP in the ATM gene (codon 4949) of tumor cells retrieved from patient HD2. Data were obtained by using the 5′-T/CTGACAAC-3′ reverse primer (forward sequence GTTGTCAA/G). (B) Pyrogram of the coding SNP in the ATM gene (codon 4138) of tumor cells retrieved from patient HD4. Data were obtained by using the 5′-T/CATTTTCCAT-3′ foward primer. Pyrograms were obtained by sequential addition of the SNP-specific nucleotides shown beneath each trace.
Loss of heterozygosity analysis by pyrosequencing. (A) Pyrogram of the coding SNP in the ATM gene (codon 4949) of tumor cells retrieved from patient HD2. Data were obtained by using the 5′-T/CTGACAAC-3′ reverse primer (forward sequence GTTGTCAA/G). (B) Pyrogram of the coding SNP in the ATM gene (codon 4138) of tumor cells retrieved from patient HD4. Data were obtained by using the 5′-T/CATTTTCCAT-3′ foward primer. Pyrograms were obtained by sequential addition of the SNP-specific nucleotides shown beneath each trace.
Discussion
We looked for polymorphic variations of the germ line ATM gene in childhood HD and identified 4 distinct, rare SNPs in 5 patients and NAS in 1 patient. One of the SNPs was a nucleotide change that did not result in amino acid substitution (2127T>C in HD5). SNPs 4365T>A and 4949A>G, in LCLs from patients with HD, were associated with abnormal responses to DNA damage by the ATM-mediated p53 phosphorylation and Chk2 activation pathways.
Secondary structure prediction by using the GOR IV method57 indicates that the S1455R amino acid substitution (4365T>A nucleotide change in HD1) would result in elongation of an α-helical structure in the ATM protein, but we do not yet know what implication this change might have for the activity of the protein. Our evaluation of the activity of the ATM variant containing the N1650S amino acid change (4949A>G nucleotide change) indicated that this variant provided insufficient complementation of radiosensitivity in the ATM-null cells and that phosphorylation by this variant in ATM-competent cells was inhibited in a dominant-negative manner. These findings indicate that the function of this polymorphic variant is defective under heterozygous conditions, as illustrated by the impaired ATM function in cells from patient HD2.
Segregation analysis for patients HD2 and HD3, who harbored the 4949A>G nucleotide change, showed that one of the parents of each child was heterozygous for this nucleotide change; that is, this was not a de novo nucleotide change in these patients. These parents were free of malignant diseases, including HD, at the time of this study. This finding lends support to the hypothesis that the penetrance of this nucleotide change is not high, such that development of HD requires not only an ATM with the 4949G SNP, but also some environmental factors or additional SNPs of some tumor suppressor gene. Alternatively, our finding favors a recently reported hypothesis12,58 that genetic anticipation is involved in the development of HD (ie, that parents of affected offspring who are carrying the same SNP are likely to have the disease themselves, later in life).
The ATM kinase encoded by ATM containing the SNP 4138C>T (amino acid change H1380Y) in LCL-HD4 exhibited normal p53 phosphorylation activity (data not shown) but defective c-Abl kinase activation (similar to that in cells from patients with AT). This finding is probably a result of a dominant-negative effect of the H1380Y mutation, which is localized within the predicted c-Abl binding domain (amino acids 1372-1383) of ATM, and suggests that the interaction between H1380Y ATM and c-Abl protein is impaired.29,30 These questions would be addressed by further studies of the function of 4138C>T ATM (eg, by subcloning this variant into an expression vector). This rare 4138C>T SNP, which is associated with impaired c-Abl activation, has also been identified in 5 cases of malignant disease (1 case of T-cell ALL, 1 case of chronic myeloid leukemia, 1 case of diffuse large B-cell lymphoma, and 2 cases of breast cancer)39,59-61 and in 1 out of every 88 healthy African individuals.62 We found the H1380Y variant at an allele frequency of 3% in random controls, which corresponds to a carrier frequency of 6% in the Japanese population. This frequency is much higher than those reported for some European populations59,62 and is not consistent with a role for H1380Y in HD. Alternatively, H1380Y could be an allele with a very low penetrance.
We detected 6 types of NAS in patient HD6. Several of these NAS transcripts have been reported previously.55 The NAS 4777_4909del (skipping of exon 34) was reported in 1 patient with AT and in a healthy individual; careful interpretation of this NAS is needed.63 Of the other mutant transcripts, 5675_5918del (skipping of exons 40 and 41) and 6006_6007ins346 are novel, whereas 4777_5005del (skipping of exons 34 and 35), 5497_5577del, and 5675_5762del (skipping of exon 40) have been identified in each of 3 cell lines from patients with AT.63-65 Several splicing variants have been shown to be caused by SNPs in intron sequences, including 5497_5577del, which is caused by the SNP g.29406a>c (IVS 38-2A>C) (ie, a mutation in intron 38 involving a change from A to C at nucleotide –2 of exon 39) in an AT cell line.63 We failed to identify this g.29406a>c (IVS 38-2A>C) mutation in HD6, and further studies are ongoing to identify the genomic alterations that cause this type of unique splicing dysfunction. Complex splicing of ATM transcripts can originate from poor preparations or long blood storage,66 but our NAS transcripts were consistently identified in each of 2 fresh blood samples obtained independently from this patient, which rules out the possibility that our finding was an artifact.
HD is one of the frequently reported hematologic malignancies in patients with AT,7-10 but family members who are heterozygous carriers of ATM mutations have not been demonstrated to have an increased risk of HD. Because 70% to 90% of patients with AT have biallelic germ line mutations of the ATM gene67,68 that result in a truncated protein, most of the carriers are likely to have 1 wild-type allele and 1 truncation mutation of the ATM gene. Previous studies have investigated the possible involvement of germ line ATM mutations with increased risk of secondary malignancies in HD by using a protein truncation test or yeast-based stop codon assay to screen for truncation mutations.19,69 The conclusion drawn from these studies was that the ATM gene is not involved in the pathogenesis of HD. There is, however, one report of 13 ATM variants from 64 individuals with HD (heterozygous or homozygous) in which rare variants of ATM were more frequent in the cohort with HD (27 patients) than among patients with breast cancer following HD (37 patients).70 The authors of this report concluded that ATM truncations or rare variants are not associated with an increased risk of breast cancer after HD, but their observations of multiple germ line variants and homozygotes suggest that rare variants may contribute cancer-susceptibility alleles in a subset of cases. These results, that many more rare ATM variants are found in patients with HD than among the normal population, and our findings (the SNPs we found and the results of our functional analysis of them) support the involvement of polymorphic variations in the pathogenesis of childhood HD.
Several missense mutations have been found in patients with AT,68 but it is still unclear whether all heterozygous carriers of missense mutations will develop classic AT. Missense mutations of the ATM gene have more significant roles in the development of cancer.71 The ATM protein multimerizes and associates with other proteins, forming a functional complex. DNA damage induces intermolecular autophosphorylation of Ser1981 in ATM that causes multimers to dissociate and initiates cellular ATM kinase activity.72,73 It is speculated that SNPs of the kinase domain result in defects in autophosphorylation activity and lead to defective activation of ATM. The role of the SNPs occurring outside of the kinase domain is unclear in this model, but these SNPs are thought to affect the conformational change around Ser1981, the normal multimerization of the ATM protein, ATM's DNA binding capacity, or ATM's interaction with other proteins. It is not surprising, therefore, that some SNPs would have a dominant-negative effect against wild-type ATM protein. Indeed, there is experimental evidence that overexpression of a cDNA encoding an inactive ATM kinase has a dominant interfering effect on ATM kinase activity.74 There are also rare SNPs found in patients with breast cancer (7775C>G encoding the amino acid change S2592C) and in pediatric leukemia with the MLL gene rearrangement (8921C>T encoding the amino acid change P2974L) that alter the function of ATM by dominant interference.45,73 Furthermore, Spring et al37 have reported that mice heterozygous for the ATM mutation 7636_7644del, but not for a truncation mutation, showed an increased susceptibility to tumors. Scott et al73 reported that missense mutations in ATM are associated with defective p53 phosphorylation and that these mutations are incapable of correcting the radiosensitive phenotype of AT cells. However, many rare, missense ATM variants in patients with breast cancer had wild-type properties, and only 1 variant with the missense mutation S2592C had functional relevance.73 The discrepancy between our results and the data of Scott et al may be explained by different localization of the variants studied.
We found no loss of heterozygosity in the ATM gene of HD tumor cells from patients HD2 and HD4. This observation is consistent with N1650S acting in a dominant-negative manner in HD2. Although loss of heterozygosity of ATM has been reported in a patient with diffuse large B-cell lymphoma whose germ line cells were heterozygous for the mutation H1380Y,61 we found no loss of heterozygosity in cells from patient HD4, who harbored this mutation. Our results provide indirect evidence that H1380Y acts in a dominant-negative manner.
In our study of the frequencies of these SNPs in 100 cord blood samples, we found no 4949T>G mutations, whereas 4365T>A and 4138C>T mutations occurred with frequencies of 1% and 3%, respectively. In the presence of genotoxic stresses, individuals who carry such SNPs may be more susceptible to genomic instability, an argument supported by the defects we observed in the ATM-mediated p53 signal transduction pathway and in the ATM-mediated c-Abl kinase activation pathway. These defects may be amplified when additional factors for genomic instability exist and may lead to an accumulation of mutations that contribute to the emergence of neoplastic phenotypes. We conclude that SNPs of the ATM gene may be contributory genetic risk factors for pediatric HD.
Prepublished online as Blood First Edition Paper, September 11, 2003; DOI 10.1182/blood-2003-01-0094.
Supported by Grants-in-Aid for Pediatric Research and for Cancer Research from the Ministry of Health and Welfare, Japan, as part of a comprehensive 10-year strategy for cancer control, and by a Grant-in-Aid from the Ministry of Education, Science and Culture, Japan. D.D. is supported by grants from Telethon (GPO205/01) and The Italian Association for Cancer Research (AIRC).
M.T. and R.T. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Dr Akira Ogawa, Medical and Biological Laboratories, Nagano, Japan, for the kind gift of the GST-p53 (1-100) expression vector, recombinant GST-Cdc25C, and anti–Chk2 antibody. We thank Dr Y. Shiloh for ATM expression vector pEBS-YZ5. The GST-CTD expression vector pGCTD was a kind gift from Dr S. Peterson.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal