Abstract
The ATM/p53-dependent DNA damage response pathway plays an important role in the progression of lymphoid tumors. Inactivation of the ATM or TP53 gene is frequent in B-cell lymphocytic leukemia (B-CLL) and leads to aggressive disease. Although the ATM and p53 pathways overlap, they are not congruent, and it is unclear how the mechanism of tumor progression differs between ATM- and p53-deficient tumors. Using microarray analysis of ATM-mutant, TP53-mutant, and ATM/TP53 wild-type B-CLLs, we show that after exposure to DNA damage transcriptional responses are entirely dependent on ATM function. The p53 proapoptotic responses comprise only a part of ATM-regulated transcription; additionally, ATM regulates prosurvival responses independently of p53. Consequently, the greater severity of the TP53-mutant B-CLLs compared with ATM-mutant B-CLLs is consistent with the additive effect of defective apoptotic and elevated survival responses after DNA damage in these tumors. We also show that transcription expression profiles of ATM-deficient, TP53-deficient, and wild-type B-CLLs are indistinguishable before irradiation. Therefore, damage-induced transcriptional fingerprinting can be used to stratify tumors according to their biologic differences and simultaneously identify potential targets for treating refractory tumors.
Introduction
Chronic B-cell lymphocytic leukemia (B-CLL) is the most common leukemia in western countries and is characterized by the proliferation of mature B cells.1 Considerable clinical heterogeneity exists among patients with B-CLL. Some patients have stable disease for many years, whereas others have rapidly progressing disease and short life expectancy. Recently, a number of genetic markers have been identified that directly influence the progression of disease. The mutational status of the immunoglobulin heavy chain (VH) gene distinguishes biologically distinct B-CLL subtypes,2,3 and B-CLL tumors with unmutated VH genes are generally associated with rapid clinical progression, whereas those that have accumulated somatic mutations in the VH gene characteristically have a more indolent outcome. However, it is now clear that this simple subdivision does not always adequately predict clinical behavior, and the mutational state of genes associated with the DNA damage response pathway has been shown to provide more powerful predictive information.4
The DNA damage response pathway plays a crucial role in the etiology and clinical behavior of malignant cells. Inducing tumor cell death by ionizing radiation (IR) and many cytotoxic drugs causes DNA double-strand breaks (DSBs),5 and the inactivation of genes that regulate the response to DSBs is commonly found in human cancer. The ATM protein is the principal integrator of the various cellular responses to DSBs.6 Critically, ATM is responsible for activating the p53 tumor–suppressor protein, leading to the up-regulation of p53-responsive genes that promote cell-cycle arrest and apoptosis. Outcomes of p53 activation, however, depend on the cellular context and the severity of the induced DNA damage, implying that in a given cell other cellular factors influence whether the outcome of the response will be survival or death. The roles of ATM and p53 do not completely overlap because biallelic loss of ATM results in unusual sensitivity to IR, whereas p53 deficiency confers radioresistance.7
Approximately one third of B-CLL tumors have inactive ATM or TP538-11 and exhibit defects in the p53 damage response and in IR-induced apoptosis.4,11 Tumors that have developed TP53 or ATM mutations are associated with poor clinical outcome even when associated with mutated VH genes. There are, however, differences that distinguish ATM-mutant tumors from TP53-mutant tumors at the clinical and cellular levels. Compared with ATM-mutant tumors, TP53-mutant B-CLL is more clinically aggressive and exhibits a complete absence of DNA damage–induced apoptosis in vitro. ATM-mutant tumors show a more prolonged clinical course and retain a capacity for apoptosis after DNA damage induced in vitro, though at a reduced level.10,11
Microarray analysis has shown that B-CLL tumors have a transcriptional profile similar to that of memory B cells, and a number of genes have been identified that can be used to predict VH mutation status of the tumor12,13 or resistance to DNA damage.14 However, the expression signatures that define tumors with mutations in the ATM or TP53 genes have not been determined.
Here we have used microarray technology to analyze the expression of 12 627 transcripts before and after IR in ATM-mutant, TP53-mutant, and ATM/TP53 wild-type B-CLL tumors. Expression profiles of the 3 subsets were indistinguishable before IR, but after IR they allowed grouping of the tumors according to their ATM or TP53 gene status. Furthermore, damage-induced expression profiles suggested a cooperative role for ATM and p53 in inducing proapoptotic responses after DNA damage but an opposing role in inducing survival responses. Therefore, expression profiling of the DNA damage response in B-CLL not only improves the stratification of DNA damage–resistant B-CLL tumors, it identifies potential therapeutic targets in leukemias with a specific type of DNA damage response defect.
Patients, materials, and methods
B-CLL patients
Diagnostic samples from 16 patients with B-CLL (6 ATM-mutant, 5 TP53-mutant, and 5 ATM/TP53 wild-type)11 who had not received previous treatment were included in the study. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque centrifugation, as previously described.11 All samples were frozen in liquid nitrogen as viable cells in 90% fetal calf serum/10% dimethyl sulfoxide (DMSO). Samples were thawed at 37°C, washed in saline buffer, and cultured overnight in RPMI 1640 medium supplemented with 10% fetal calf serum and glutamine. The viability of each sample was then assessed by trypan blue dye exclusion and was found to be greater than 95% in all patients. Samples from B-CLL patients with white blood cell (WBC) counts greater than 100 × 109/L were used for microarray analysis. Double CD19/CD5 labeling followed by fluorescence-activated cell sorter (FACS) analysis was performed in 5 representative B-CLL samples, revealing 90% to 95% cells positive for both markers. In 4 additional B-CLL samples, single CD5 labeling showed a range of 95% to 99% positive cells.
cRNA synthesis and microarray hybridization
We analyzed each of 16 B-CLL tumors before and 10 hours after exposure to 5 Gy IR. B-CLL tumor cells were exposed to 5 Gy Co 60 γ-rays at a dose rate of 1.25 Gy/min. Doses of IR and lengths of postirradiation time were chosen on the basis of our previous experience, suggesting that in B-CLL cells the peak up-regulation of p53-responding proteins occurs at 10 hours.11 Total RNA was obtained using Trizol reagent (Invitrogen, Carlsbad, CA) and was purified with RNeasy columns (Qiagen, Valencia, CA). Five micrograms RNA was used to generate double-stranded cDNA using an oligo dT-primer containing the T7 RNA polymerase promoter site and the SuperScript Choice System kit (Invitrogen). cDNA was purified by phenol/chloroform-isoamyl alcohol extraction, and biotinylated cRNA was synthesized by in vitro transcription using the BioArray High Yield RNA transcription kit (Affymetrix, Santa Clara, CA). Fifteen micrograms purified cRNA (RNeasy columns; Qiagen) was fragmented and hybridized to human U95Av2 Genechips (Affymetrix) for 16 hours at 45°C following the Affymetrix protocol. Gene chips were then washed and labeled with streptavidinphycoerythrin (strep-PE), and the signal was amplified using biotinylated antistreptavidin followed by another round of staining with strep-PE. Washing and staining were performed on the Affymetrix fluidics station according to the recommended fluidics protocol. Labeled gene chips were scanned using a confocal argon ion laser (Agilent Technologies, Palo Alto, CA).
Data collection and normalization
Expression values were obtained for all 32 hybridizations using Affymetrix Microarray Suite 5.0 software. Data quality was assessed using MAS 5.0 report files and GeneSpring 4.2.1 (Silicon Genetics, San Carlos, CA).15 Arrays were included in the analysis only if they met minimum requirements for data distribution—3′ to 5′ ratio (less than 3), background (less than 100), noise, and percentage call. To allow comparisons between samples using Affymetrix DMT 1.0, global scaling was applied to a target intensity of 100. For analysis with GeneSpring 4.2.1 software, raw data were exported from MAS 5.0, and values were normalized to the median signal value for each array. For comparisons among unirradiated tumors, expression was normalized to the median signal value per gene across all arrays. For comparisons between tumors on the basis of irradiation-responsive genes, normalizations were performed to the mean level of gene expression in the unirradiated control.
Data filtering and analysis
The U95Av2 gene chip contains 12 627 transcripts, including control bacterial genes. To devise a list of informative genes, we used Genespring 4.2.1 and generated experimental interpretations to compare tumors according to immunoglobulin VH status, ATM and TP53 mutation, or exposure to IR. In all cases, variance was determined using a global error model based on the appropriate replicates. We excluded genes whose signal strengths did not significantly exceed background values (control signal filter was set at a minimum of 37) and genes whose expression did not reach a threshold value for reliable detection (based on the Affymetrix MAS 5.0 probability of detection [P ≤ .1]) in at least 3 of 5 or 6 replicate samples. Finally, genes whose levels of expression did not vary between response to IR or tumor type by more than 1.5-fold were also excluded. The remaining genes were considered informative and were subjected to parametric analysis of variance (ANOVA) if the number for comparison was greater than 2 or to t test between 2 conditions using a global error model with the variance statistic derived from replicates. Finally, to reduce false-differential gene expression, a Benjamini-Hochberg or a Bonferroni16 multiple testing correction filter was applied. All microarray data are available as MAGE-ML files (http://xml.coverpages.org/mageML.html) and will be submitted in MIAME-compliant format to the Array Express database held at the European Bioinformatics Institute (http://www.ebi.ac.uk/arrayexpress/).
Gene signatures that distinguish DNA damage response between the 3 tumor types were identified from informative genes using hierarchical average pairwise linkage clustering (GeneSpring 4.2.1 and CLUSTER (http://www.microarrays.org/software.html).17 To identify the groups of genes with similar patterns of behavior in response to DNA damage, we also evaluated a number of nonhierarchical methods including SOM, K-means, K mediods, and KenTau.18 Results were compared using weighted kappa scores18,19 and showed that the K-means approach provided reliable clusters (data not shown). Similarity was measured in this case using Pearson correlation by which 100 iterations were performed with 4 repeats to find 4 optimal clusters.
Protein analysis
Eleven samples used for microarray analysis and another 24 B-CLL samples that were not subjected to microarray analysis were used to validate microarray results at the protein level. The ATM and TP53 status of tumors not analyzed by microarray chips had previously been determined.4,10,11
Western blotting of unirradiated and irradiated B-CLL samples was performed as described previously.11 The following antibodies were used: rabbit anti-Fas (Calbiochem, San Diego, CA), rabbit anti-Bbc3 (Abcam, Cambridge, United Kingdom), mouse anti-PCNA (Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-p21 (Calbiochem), mouse anti-NFκB (Upstate Biotechnology, Lake Placid, NY), and mouse anti-Mdm2 (generously donated by Dr R. Grand, CR-UK Institute for Cancer Studies, University of Birmingham, United Kingdom). To verify that equivalent amounts of each sample were loaded, the filters were additionally probed with anti–actin-IgG (AC74; Sigma, St Louis, MO). Densitometry was performed routinely on each of the tested proteins and on actin bands.
Results
Clinical and biologic features of B-CLL subsets
Sixteen tumors representative of 3 B-CLL genetic subtypes were selected for microarray experiments (Tables 1, 2, 3). Five tumors (ATM1, ATM2, ATM4, ATM5, ATM6) had biallelic mutations or loss of heterozygosity (LOH) combined with mutation in the remaining allele of ATM (Tables 1, 2, 3).11 One tumor (ATM3) had only a single ATM mutation.11 Five tumors (TP53-1, TP53-2, TP53-3, TP53-4, TP53-5) had single TP53 mutations.11 Three TP53-mutant tumors (TP53-1, TP53-2, TP53-5)11 had lost the second TP53 allele, and in 5 B-CLL tumors (WT 1-5) both ATM and TP53 genes were wild type. All wild-type tumors were associated with somatic mutations within the VH gene, whereas all ATM-mutant tumors and 4 of 5 TP53-mutant tumors had unmutated VH genes.11 One tumor with a TP53 mutation (TP53-5) was associated with somatic mutations in the VH gene.11 All wild-type tumors had normal p53 up-regulation as measured by protein expression after IR, and all ATM-mutant and TP53-mutant tumors exhibited defects in the up-regulation of p53 (Tables 1, 2, 3).11 Clinical observation demonstrated a mild clinical course in patients with ATM and TP53 wild-type tumors, whereas patients with ATM or TP53 mutations had aggressive disease that was particularly marked in TP53-mutant tumors (Tables 1, 2, 3).11
Clinical and cellular features of ATM/TP53 wild-type B-CLL tumors analyzed in microarray experiments
Tumor . | ATM/TP53 status . | p53 induction . | VH status . | Clinical course . |
|---|---|---|---|---|
| WT1 | wt | N | Mutated | S |
| WT2 | wt | N | Mutated | S |
| WT3 | wt | N | Mutated | S |
| WT4 | wt | N | Mutated | S |
| WT5 | wt | N | Mutated | S |
Tumor . | ATM/TP53 status . | p53 induction . | VH status . | Clinical course . |
|---|---|---|---|---|
| WT1 | wt | N | Mutated | S |
| WT2 | wt | N | Mutated | S |
| WT3 | wt | N | Mutated | S |
| WT4 | wt | N | Mutated | S |
| WT5 | wt | N | Mutated | S |
WT and wt indicate wild type; N, normal; S, stable.
Clinical and cellular features of ATM-mutant B-CLL tumors analyzed in microarray experiments
Tumor . | ATM mutation . | p53 induction . | VH status . | Clinical course . |
|---|---|---|---|---|
| ATM1 | 8600G > A (2867G > E) and LOH across ATM | D | Unmutated | P |
| ATM2 | 1058delGT and 5464G > A (1822E > Q) | D | Unmutated | P |
| ATM3 | 8266A > T (2756K > stop) | D | Unmutated | P |
| ATM4 | 2144insA and 4393insA | D | Unmutated | P |
| ATM5 | 8084G > C (2695G > A) and 7351G > A (2451A > T) | D | Unmutated | P |
| ATM6 | 8839A > C (2947T > S) and LOH across ATM | D | Unmutated | P |
Tumor . | ATM mutation . | p53 induction . | VH status . | Clinical course . |
|---|---|---|---|---|
| ATM1 | 8600G > A (2867G > E) and LOH across ATM | D | Unmutated | P |
| ATM2 | 1058delGT and 5464G > A (1822E > Q) | D | Unmutated | P |
| ATM3 | 8266A > T (2756K > stop) | D | Unmutated | P |
| ATM4 | 2144insA and 4393insA | D | Unmutated | P |
| ATM5 | 8084G > C (2695G > A) and 7351G > A (2451A > T) | D | Unmutated | P |
| ATM6 | 8839A > C (2947T > S) and LOH across ATM | D | Unmutated | P |
D indicates defective; P, progressive.
Clinical and cellular features of TP53-mutant B-CLL tumors analyzed in microarray experiments
Tumor . | TP53 mutation . | p53 induction . | VH status . | Clinical course . |
|---|---|---|---|---|
| TP53-1 | 472C > G (158R > G) and LOH across TP53 | D | Unmutated | P |
| TP53-2 | 844C > T (282R > W) and LOH across TP53 | D | Unmutated | P |
| TP53-3 | 1207ins133bp | D | Unmutated | P |
| TP53-4 | 210ins2bp | D | Unmutated | P |
| TP53-5 | 701A > G (234Y > C) and LOH across TP53 | D | Mutated | P |
Tumor . | TP53 mutation . | p53 induction . | VH status . | Clinical course . |
|---|---|---|---|---|
| TP53-1 | 472C > G (158R > G) and LOH across TP53 | D | Unmutated | P |
| TP53-2 | 844C > T (282R > W) and LOH across TP53 | D | Unmutated | P |
| TP53-3 | 1207ins133bp | D | Unmutated | P |
| TP53-4 | 210ins2bp | D | Unmutated | P |
| TP53-5 | 701A > G (234Y > C) and LOH across TP53 | D | Mutated | P |
D indicates defective; P, progressive.
Previous studies documented the apoptotic responses of freshly isolated tumor cells to ionizing radiation.10,11 Exposure to ionizing radiation led to substantial apoptosis in ATM and TP53 wild-type tumor populations within 4 days, whereas ATM- and TP53-mutant tumors exhibited clear defects in IR-induced apoptosis. TP53-mutant B-CLLs showed a virtual absence of apoptosis, whereas ATM-mutant tumors revealed a level of apoptosis that was intermediate between that of wild-type tumors and TP53-mutant tumors.10,11
We reasoned that the distinctive clinical and biologic features of ATM- and TP53-mutant B-CLL could reflect a unique pathogenesis of these subsets or a difference in their response to treatment. Therefore, we performed transcriptional profiling on tumor cells before and after ionizing radiation.
Comparing expression profiles in B-CLL subsets before irradiation
We performed microarray analysis on unpurified B-CLL samples containing an excess of tumor cells (90%-99% of population). Recent microarray studies used magnetic bead separation to obtain a pure population of B-CLL tumor cells because it was suggested that contamination with the normal cells might affect gene expression profiles.12,13 Less than 10% of nontumor cells might have been present in our viable samples and might have led to a small reduction in sensitivity of the analysis. With respect to magnetic bead separation, it is unclear how the process itself affected gene expression, and our aim was to avoid further manipulation, which might also have resulted in a change of expression profiles.
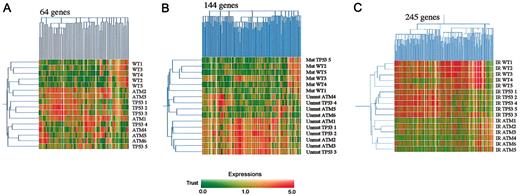
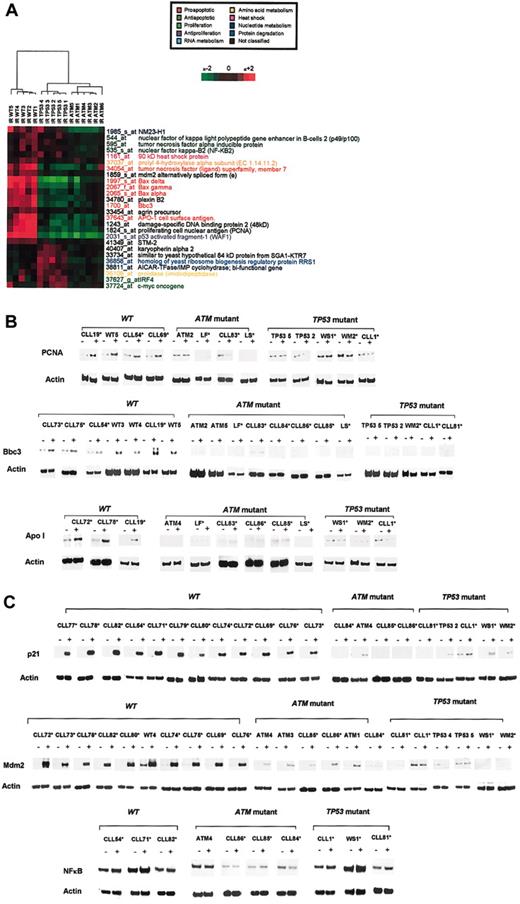
Affymetrix U95Av2 oligonucleotide microarrays consisting of 12 627 transcripts were used to analyze baseline gene expression in the 16 B-CLL samples. Filtering for reliability and detection (“Materials and methods”) left 6596 genes. An initial analysis was performed to determine whether mutations in ATM or TP53 led to distinct baseline transcriptional fingerprints. ANOVA identified 64 genes whose expression differed significantly (range, 0.0000001-0.0499; P < .05) between B-CLL tumors based on ATM and TP53 mutational status. Given that multiple testing of large sample sets may result in false detection, we applied the Benjamini-Hochberg correction to recalculate the significance. Using this stringent technique, no significant differences were seen between tumors. Furthermore, 2-dimensional hierarchical clustering of the 64 genes using average linkage cluster analysis with standard correlation showed no evidence of distinct expression profiles for tumor types based on the presence of ATM inactivation or TP53 mutations (Figure 1A).
Expression profiles inATM-mutant,TP53-mutant, andATM/TP53wild-type B-CLLs. (A) Hierarchical clustering of ATM-mutant, TP53-mutant, and ATM/TP53 wild-type (wt) B-CLLs based on the expression of 64 genes in unirradiated tumors. (B) Hierarchical clustering of VH-unmutated and VH-mutated B-CLLs based on the expression of 144 genes in unirradiated tumors. (C) Hierarchical clustering of ATM-mutant, TP53-mutant, and ATM/TP53 wt B-CLLs based on the change in gene expression of 245 genes after IR. The horizontal axis of the color bar (below) represents normalized levels of gene expression on a continuous scale. Red indicates overexpression and green indicates underexpression relative to the mean. The vertical axis of the color bar indicates reliability of the data. Dark or unsaturated colors represent low trust, and bright, saturated colors represent high trust. Signal values per gene were normalized to the median value across all experiments.
Expression profiles inATM-mutant,TP53-mutant, andATM/TP53wild-type B-CLLs. (A) Hierarchical clustering of ATM-mutant, TP53-mutant, and ATM/TP53 wild-type (wt) B-CLLs based on the expression of 64 genes in unirradiated tumors. (B) Hierarchical clustering of VH-unmutated and VH-mutated B-CLLs based on the expression of 144 genes in unirradiated tumors. (C) Hierarchical clustering of ATM-mutant, TP53-mutant, and ATM/TP53 wt B-CLLs based on the change in gene expression of 245 genes after IR. The horizontal axis of the color bar (below) represents normalized levels of gene expression on a continuous scale. Red indicates overexpression and green indicates underexpression relative to the mean. The vertical axis of the color bar indicates reliability of the data. Dark or unsaturated colors represent low trust, and bright, saturated colors represent high trust. Signal values per gene were normalized to the median value across all experiments.
In contrast, immunoglobulin heavy chain gene VH mutation status was correlated with global gene expression patterns, in agreement with previous reports.12,13 Expression of 144 genes differed significantly (range 0.0000001-0.0499; P < .05) between VH mutated and VH unmutated tumors, but after multiple testing correction by Benjamini-Hochberg only 35 genes remained significantly different (P < .05). Using the stringent set of 35 genes and the list of 144 genes without multiple testing correction in hierarchical 2-dimensional average cluster linkage analysis, we identified gene expression patterns that correlated with mutational status of the VH locus (Figure 1B). This observation was reinforced by analysis in one tumor that carried a TP53 mutation but also had a somatically mutated VH gene, a genetic combination rarely observed whose transcriptional profile clustered with the mutated VH group (Figure 1B).
Klein et al12 and Rosenwald et al13 previously used a supervised learning-based algorithm to generate a predictive gene list for VH mutation status. Five of these predictive genes (lipoprotein lipase, septin, tropomyosin, Pak 1, and Zapp 70), which were down-regulated in VH-mutated B-CLL compared with VH-unmutated tumors, also appeared on our list of 144 differentially expressed genes. However, 15 of the VH status “predictive” genes were excluded from our analysis because of their low or unreliable expression. Our results, therefore, are consistent with previous studies12,13 suggesting a common global gene expression pattern among B-CLLs. However, we found a small subset of genes that apparently differentiated VH-mutated from VH-unmutated tumors.12,13 This is not surprising bearing in mind that VH-mutated and VH-unmutated tumors in our present study were also selected to be either ATM- or TP53-mutant (mostly VH-unmutated tumors) or ATM/TP53 wild-type (all VH-mutated). It is possible that some of the observed differences between VH-mutated and -unmutated tumors in this study were a reflection of the common pathogenesis of ATM- and TP53-mutant tumors rather than of VH mutation status. Further studies are required to elucidate differences between B-CLLs with other combinations of ATM, TP53, and VH status.
Gene expression profiling allows discrimination of ATM-mutant, TP53-mutant, and ATM/TP53 wild-type tumors in response to IR
The poor clinical outcomes of B-CLL tumors bearing ATM or TP53 mutations might result from differences in their responses to DNA damage, induced during tumor progression or as a consequence of treatment. This part of the study was focused solely on the effect of irradiation treatment by normalizing the response of each gene in the 16 irradiated tumors to the level of that gene in the corresponding tumors before irradiation (Figure 1C). Five hundred thirteen of the 1385 genes that remained after initial filtering were found to be significantly different among ATM-mutant, TP53-mutant, and ATM/TP53 wild-type tumors (P < .05), and hierarchical average linkage clustering based on these genes clearly separated the tumors into 3 subgroups. Of the 513 genes, 245 were found by Benjamini-Hochberg multiple testing correction to be highly significantly differentially expressed between ATM-mutant, TP53-mutant, and ATM/TP53 wild-type B-CLLs (P < .05) (Figure 1C).
The normalization procedure was specifically designed to detect irradiation-responsive genes. However, some genes may be up- or down-regulated in wild-type tumors to levels similar to those seen before irradiation in either ATM- or TP53-mutant tumors. In these circumstances it would be unlikely that the regulation is of biologic significance in the irradiated state. We examined this possibility by comparing the list of 245 genes that differentiate between tumors after irradiation with the 64 genes detected as different among unirradiated tumors. We found that only 3 genes (37037_at, 37174_at, and 41349_at) fell reliably into both categories, indicating that such genes do not significantly contribute to postradiation transcription patterns.
To identify clusters of genes with similar responses to IR in 3 B-CLL subtypes, we applied a nonhierarchical, heuristic, partitional clustering algorithm, K-means clustering, to 245 differentially expressed genes. Variability was described most effectively by 13 K-means clusters, but these could be broadly simplified into 4 major clusters of gene expression (Figures 2, 3, 4, 5). Clusters 1 and 2 represented genes normally up-regulated in response to IR in the presence of functionally active ATM and p53, whereas clusters 3 and 4 represented genes whose transcription was not markedly changed under the same circumstances but became up-regulated after IR in the presence of functionally inactive p53 (cluster 3) or ATM (cluster 4), respectively. Despite the fact that these 4 clusters represented diverse cellular signals, 3 were indicative of a particular type of cellular response.
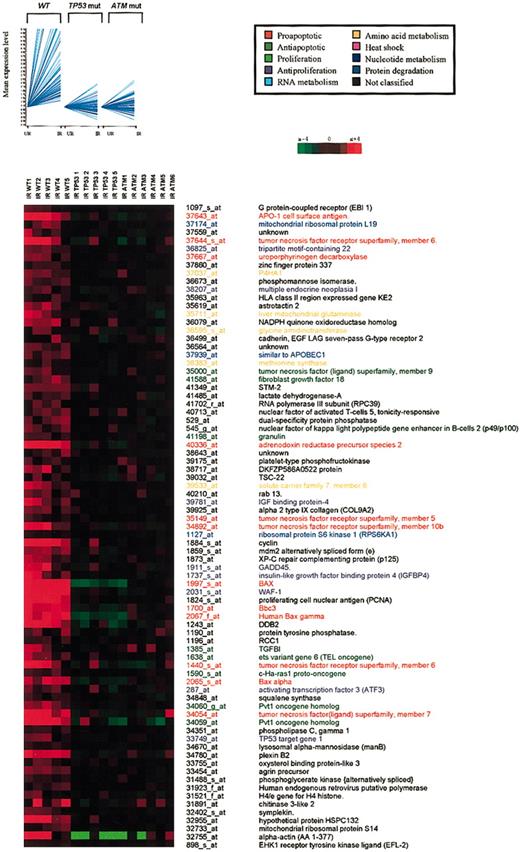
Genes selectively up-regulated inwttumors. Cluster 1. (Top) Seventynine genes were obtained by K-means clustering with Pearson correlation (GeneSpring 4.2.1) of 245 differentially expressed genes identified by parametric t test with Benjamini-Hochberg multiple testing correction. UR indicates unirradiated; IR, irradiated. Signal values per gene were normalized to the mean of the unirradiated controls for each sample. (Bottom) The cluster was visualized using TREEVIEW (http://www.microarrays.org/software.html). Columns represent individual B-CLL samples, and rows correspond to genes. Color changes within a row indicate expression levels relative to the average of the same population. Functional classification of the genes is indicated by different colors (key).
Genes selectively up-regulated inwttumors. Cluster 1. (Top) Seventynine genes were obtained by K-means clustering with Pearson correlation (GeneSpring 4.2.1) of 245 differentially expressed genes identified by parametric t test with Benjamini-Hochberg multiple testing correction. UR indicates unirradiated; IR, irradiated. Signal values per gene were normalized to the mean of the unirradiated controls for each sample. (Bottom) The cluster was visualized using TREEVIEW (http://www.microarrays.org/software.html). Columns represent individual B-CLL samples, and rows correspond to genes. Color changes within a row indicate expression levels relative to the average of the same population. Functional classification of the genes is indicated by different colors (key).
Genes up-regulated inwtandTP53-mutant tumors. Cluster 2. (Top and bottom) Cluster 2, composed of 61 genes, is represented as described for cluster 1.
Genes up-regulated inwtandTP53-mutant tumors. Cluster 2. (Top and bottom) Cluster 2, composed of 61 genes, is represented as described for cluster 1.
Genes up-regulated inTP53-mutant B-CLLs. Cluster 3. (Top and bottom) Cluster 3 contains 86 genes selectively up-regulated in B-CLL cells in TP53-mutant tumors, represented as described for cluster 1.
Genes up-regulated inTP53-mutant B-CLLs. Cluster 3. (Top and bottom) Cluster 3 contains 86 genes selectively up-regulated in B-CLL cells in TP53-mutant tumors, represented as described for cluster 1.
Genes up-regulated inATM-mutant tumors (cluster 4). Top and bottom: Cluster 4 contains 19 genes selectively up-regulated in ATM-mutant tumors and is represented as described for cluster 1.
Genes up-regulated inATM-mutant tumors (cluster 4). Top and bottom: Cluster 4 contains 19 genes selectively up-regulated in ATM-mutant tumors and is represented as described for cluster 1.
Virtually the whole transcriptional response to ionizing radiation is dependent on ATM function
Comparison of transcriptional profiles before and after exposure to ionizing radiation showed that only 7 of 1385 IR-responsive genes were similarly regulated in all 3 tumor types—ATM/TP53 wild-type, ATM-mutant, and TP53-mutant B-CLLs. Assuming ATM is required for p53-dependent transcription, this observation implies that virtually all significant gene transcription after IR is directly or indirectly controlled by ATM and that the DNA damage response is, therefore, almost completely dependent on ATM function in B-CLL cells. ATM and p53 activation also suppressed transcriptional activity of some genes whose differential expression after IR was only revealed in ATM- and TP53-mutant tumors, respectively (Figures 4-5). Strikingly, no p53 responses were regulated independently of ATM.
Responses defective in ATM- and TP53-mutant tumors after IR include genes concerned with initiation of apoptosis and cell-cycle regulation
Seventy-nine genes (cluster 1) were up-regulated in ATM/TP53 wild-type tumors after IR but not in TP53-mutant or ATM-mutant tumors (Figure 2). Prominent in this cluster were proapoptotic genes (Bax, PUMA/Bbc3, adrenodoxin reductase, the tumor necrosis factor [TNF] family of receptors and ligands, including ApoI/Fas, Trail receptor 2 (DR5), 4-1BB-L, and CD70) and cell-cycle regulators (p21WAF-1, RCC1, Gadd45).20-24 In this cluster, we also identified several genes involved in the regulation of transcription (TSC-22, NFAT, RNA III polymerase),25 DNA repair (DDB2, xeroderma pigmentosum C, PCNA),26 and cellular metabolism (uroporphyrinogen decarboxylase, NADPH oxoreductase, methionine synthase, phosphomannose isomerase, mitochondrial glutaminase). The presence of genes involved in a range of extracellular processes, such as cell junction formation, cell signaling, and cell migration also indicated a role in the communicating cell status to the surrounding tissues. Overall, the defective up-regulation of major proapoptotic and growth-inhibitory genes in TP53- and ATM-mutant tumors was consistent with the defect in IR-induced apoptosis.10,11
Genes that show impaired transcriptional activation only in ATM-mutant tumors are predominantly concerned with cell survival
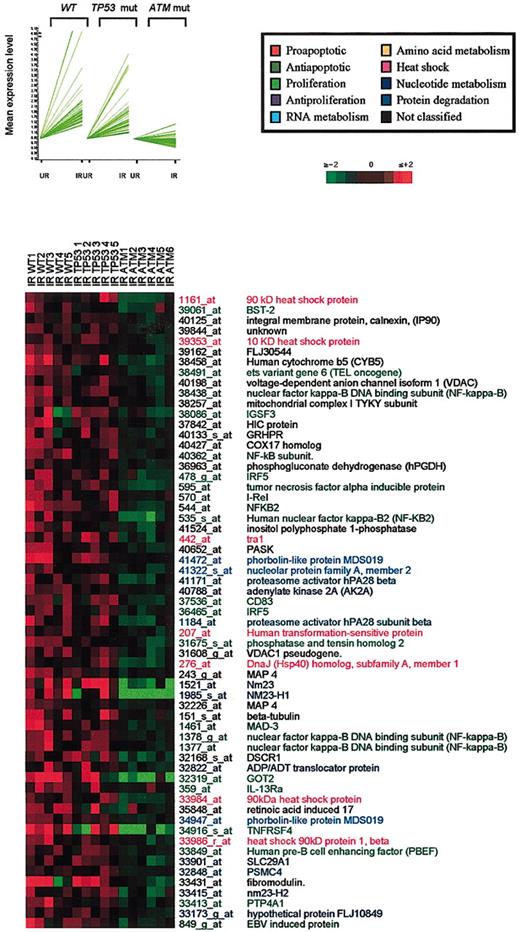
Cluster 2 formed a group of 61 genes transcriptionally up-regulated in response to IR in wild-type cells and TP53-mutant, but not in ATM-mutant B-CLL tumors (Figure 3). In contrast to cluster 1, most genes in this cluster displayed functions supportive of cell survival and growth. Prominent prosurvival genes in this category included several members of the NFκB family27 and NFκB regulators such as Mad3,28 a group of heat-shock proteins (90 kDa, 70 kDa, 10 kDa, and 40 kDa),29,30 nucleotide and RNA metabolism genes, and the proteosome genes hPA28 and PSMC4. Genes supporting B-cell proliferation (IGSF3, IRF5, CD83, PTP4A1, PBEF, OX40, OX40 ligand, interleukin-13 receptor [IL-13R], BST-2, TEL, and nucleoside diphosphate kinase [NM23], identified as one of the genes discriminating between B-CLL and memory B cells)12,31,32 were also identified in this cluster.
Transcription of many ATM-dependent genes associated with cell survival is suppressed by p53 function
A third cluster containing 86 genes up-regulated in ATM/TP53 wild-type tumors but not in ATM mutants showed particularly high levels of up-regulation in TP53-mutant tumors (Figure 4). Interestingly, clusters 3 and 2 showed functional similarities. It appears, therefore, that in response to IR, ATM activation leads to the induction of a spectrum of genes associated with cell survival and that transcription of most of these is actively suppressed by p53. Cluster 3 included genes involved in the regulation of transcription (DEAD box RNA helicases, topoisomerase II, EZH2, LIM, IRF-4),33-37 protein synthesis (threonyl, isoleucine, glycyl and valyl tRNA synthetase), ribosome biogenesis (BOP1, RRS1), innate immunity (Toll-like receptor I),38 cellular proliferation (Fuse-binding protein, c-Myc, Ikaros, fibroblast growth factor [FGF], and prolactin),39,40 antiapoptotic activity (cFLIP, plekstrin, PAK2, TR3 orphan receptor),41 and transmembrane trafficking (transferrin receptor, exportin, and ABC1).42
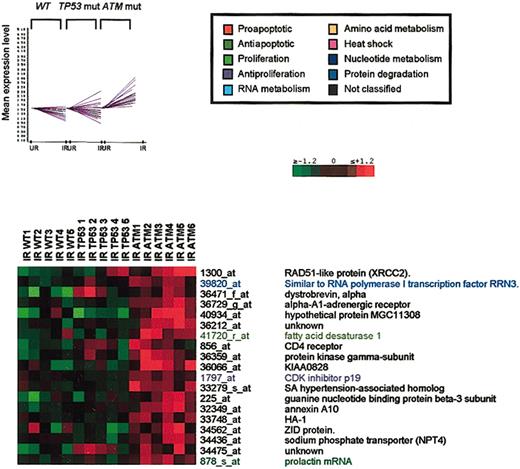
Transcriptional activation of many genes is revealed only in ATM-mutant B-CLLs
A fourth cluster of 19 genes showed transcriptional activation only in ATM-mutant tumors (Figure 5). This observation must reflect an active suppression of transcriptional activation after ATM activation that is released during absent ATM function. This set of genes included those with a range of diverse cellular functions involved in DNA repair (Rad51 homolog), transcriptional regulation (RRN3),43 transmembrane signaling (dystrobrevin), hDM2 regulation (p19ARF),44,45 calcium and sodium metabolism (NPT4), and extracellular matrix assembly (annexin). Cluster 4 also involved the fatty acid desaturase gene with antiapoptotic properties.46
Signature of 26 IR-responsive genes provides high discrimination among ATM/TP53WT, ATM-mutant, and TP53-mutant B-CLL tumors
To identify a subset of genes that would accurately discriminate between the 3 B-CLL subtypes, we applied highly stringent Bonferroni multiple correction testing to the 1385 IR-responsive genes. The resultant set of 26 transcripts clearly distinguished wild-type B-CLLs from TP53- and ATM-mutant tumors, whereas TP53- and ATM-mutant tumors were distinguishable at a second level of branching (Figure 6A). Prominent among this subset were many proapoptotic and cell-cycle–regulating genes, including Bax, PUMA (Bbc3), PCNA, APO-1, Mdm2, and WAF-1. These genes were normally up-regulated in wild-type tumors but not in ATM-mutant or TP53-mutant B-CLLs.
Transcripts discriminative betweenATM-mutant,TP53-mutant, andwtB-CLLs. (A) Twenty-six differentially expressed transcripts were identified after Bonferroni multiple testing discriminated among TP53/ATM wt, TP53-mutant, and ATM-mutant tumors. (B-C) Expression at the protein level, before and after irradiation, of a set of 6 selected genes (in panel B, PCNA, Bbc3, Apo1; in panel C, p21, Mdm2, NFκB) in ATM-mutant, TP53-mutant, and ATM/TP53 wt B-CLL tumors. Asterisks indicate independent ATM-mutant, TP53-mutant, and ATM/TP53 wt B-CLL tumors not used in microarray experiments. Actin expression was used as a loading control.
Transcripts discriminative betweenATM-mutant,TP53-mutant, andwtB-CLLs. (A) Twenty-six differentially expressed transcripts were identified after Bonferroni multiple testing discriminated among TP53/ATM wt, TP53-mutant, and ATM-mutant tumors. (B-C) Expression at the protein level, before and after irradiation, of a set of 6 selected genes (in panel B, PCNA, Bbc3, Apo1; in panel C, p21, Mdm2, NFκB) in ATM-mutant, TP53-mutant, and ATM/TP53 wt B-CLL tumors. Asterisks indicate independent ATM-mutant, TP53-mutant, and ATM/TP53 wt B-CLL tumors not used in microarray experiments. Actin expression was used as a loading control.
Next, we asked whether selected p53-dependent damage-responsive genes from the set of 26 transcripts were also upregulated at the protein level in a manner discriminating between genotypically distinct B-CLLs. To provide “proof of principal” and to determine whether the genes most relevant biologically could be used as indicators among ATM-mutant, TP53-mutant, and wild-type B-CLLs, we used Western blotting to analyze the proteins of 6 representative genes crucial for inducing the p53 pathway or for tumor killing. We analyzed 11 samples that were used in the microarray experiments and 24 additional, independent B-CLL samples that were not subjected to microarray analysis. ATM and TP53 status of these independent B-CLL tumors was previously determined.10,11 Western blots showed that the expression of proteins Bbc3, ApoI, PCNA, MDM2, and p21 was up-regulated after IR in wild-type tumors but not in ATM- or TP53-mutant B-CLLs (Figure 6B-C). In contrast, and consistent with our microarray data, the expression of NFκB was up-regulated in representative wild-type and TP53-mutated but not in ATM-mutated tumors. Consequently, we conclude that the change in discriminating transcripts after DNA damage could also be readily seen at the protein level and could be used to distinguish ATM-mutant, TP53-mutant, and wild-type B-CLL tumors.
Furthermore, the discriminating set of 26 transcripts is consistent with a biologic difference between clinically progressive ATM- or TP53-mutant tumors and nonprogressive B-CLLs, in which neither ATM nor TP53 was mutated, and suggests that contamination with normal cells was not a significant problem in our experiments.
Discussion
Microarray analysis of cellular transcription has already demonstrated great potential in the diagnosis and risk stratification of hematopoietic malignancies.47,48 However, it is likely that the tumor cell response to treatment may also affect the clinical outcomes of many cancers. The purpose of this study was to identify genes that are expressed differentially between aggressive ATM- or TP53-mutant tumors and more benign wild-type B-CLLs. This meant making use of a form of damage, by ionizing radiation, to which both ATM and p53 responded. We show here that global gene expression profiles of biologically and clinically distinct ATM-mutant, TP53-mutant, and ATM/TP53 wild-type B-CLL tumors are similar before exposure to DNA-damaging agents but become clearly distinguishable after DNA damage. These results suggest that the more severe clinical outcomes associated with ATM- and TP53-mutant B-CLL tumors may depend on their reduced ability to respond appropriately to DNA damage, induced either by medical treatment or by endogenous causes. There is, therefore, a requirement to recognize the mechanisms underlying the poor treatment response in ATM- and TP53-mutant tumors and to develop novel therapeutic targets for tumors that have acquired these mutations.
Our results provide an important insight into the relationship between p53 and ATM in the DNA damage response. Interestingly, ATM transcriptionally activated fewer than one third from the subset of 245 radiation-responsive genes (cluster 1) through the p53 pathway (Figure 7). Functionally, these genes reflected biologically distinct cellular responses to DNA damage, including activating apoptosis and regulating the cell cycle as previously reported.24 Notably, the list included many genes active in the death receptor and the mitochondrial pathways of apoptosis,49 consistent with p53-dependent transcriptional activation as a potent inducer of apoptosis in B-CLL cells with the ATM/TP53 wild-type sequence. Overall, however, p53 responses represented only a subset of ATM-dependent genes.
ATM/p53 regulation network inTP53-mutant,ATM-mutant, and wild-type tumors based on their differential global expression profiles. Regulation of IR-induced transcriptional responses by ATM and p53. All transcriptional responses to IR are under ATM control, whereby a subset of responses involves p53 and either supports apoptosis (cluster 1) or inhibits IR-induced survival (cluster 3). Responses independent of p53 support damage-induced survival (cluster 2). A small subset of genes involved in DNA repair and transcription regulation is suppressed by ATM after IR (cluster 4).
ATM/p53 regulation network inTP53-mutant,ATM-mutant, and wild-type tumors based on their differential global expression profiles. Regulation of IR-induced transcriptional responses by ATM and p53. All transcriptional responses to IR are under ATM control, whereby a subset of responses involves p53 and either supports apoptosis (cluster 1) or inhibits IR-induced survival (cluster 3). Responses independent of p53 support damage-induced survival (cluster 2). A small subset of genes involved in DNA repair and transcription regulation is suppressed by ATM after IR (cluster 4).
Notably, more than half of the transcripts from the subset of 245 IR-responsive genes (147 transcripts) were up-regulated in a p53-independent manner (clusters 2 and 3) (Figure 7). In contrast to the proapoptotic p53 response, the function of these genes was broadly consistent with promoting cell survival. In addition to genes with direct antiapoptotic function, this group included genes involved in energy production, nucleotide metabolism, and proteosome function. Eleven genes had direct links to the heat-shock response, including the regulator NM23.29 The effect of inducing these genes is likely to counteract the induction of the proapoptotic ATM/p53 pathway after DNA damage. We observed that p53 suppressed the up-regulation of 86 of the prosurvival class of genes (cluster 3) because their up-regulation was only revealed in p53-deficient cells. Consequently, B-CLL cells in which p53 is nonfunctional but in which ATM is still active are not only deficient in activating apoptosis, they also receive additional survival signals through ATM activation. This is reflected in the extreme resistance of p53-mutant B-CLL cells to apoptosis in response to ionizing radiation compared with ATM-mutant tumors.11 Interestingly, our observations are in contrast to those of previous reports suggesting that the greater resistance of TP53-mutant cells to apoptosis after IR compared with ATM-mutant cells is caused by a p53 pathway that is independent of ATM activation.50 We find no evidence of such a pathway in B-CLL, where virtually all radiation-induced transcriptional responses in B-CLL, including those with opposing cellular consequences, required ATM.
Transcription factors that mediate the p53-independent ATM response are unknown, but the radiation-responsive genes NFκB and AP-1 are potential candidates. In resting memory B cells, the physiological counterpart of B-CLL, NFκB, has antiapoptotic activity and supports cell survival.27 We observed defective radiation-induced up-regulation of NFκB itself and of several NFκB-responsive genes in ATM-mutant tumors, including the B-cell survival supporting genes A20 and CD83. Similarly, several genes that have antiapoptotic function and contain a binding motif for the AP-1 transcription factor, such as BST-2, MAD3, FLJ1084, CYB5, and PSMC4, were also up-regulated in tumors with the ATM wild-type sequence but not in ATM-mutant tumors.
In principle, it is possible that differences in gene expression among the 3 genotypes in part reflected differences of distribution in the cell cycle. This appeared not to be the case in the present study. We have previously undertaken fluorescence-activated cell sorter (FACS) analysis of cells from the same tumors. Before irradiation, most cells of the 3 B-CLL genotypes—ATM and TP53 mutated and wild type—were in the G1 phase. This was likely a reflection of the slow cycling times of the mass of tumor cells in vivo. Ten hours after irradiation, there was no change in cell-cycle distribution either within or between genotypes, and most cells were still in G1.
It is also possible that the loss of ATM or p53 results only in a degree of shift in the kinetics of expression of some ATM-dependent genes, especially if there is some redundancy with, for instance, ATM. In a range of other genes, however, for which there is no such redundancy, the loss of ATM results in an alteration of expression.
Paradoxically, apart from a defect in IR-induced apoptosis, ATM–/– cells also show increased sensitivity to the killing effects of ionizing radiation, a feature observed in A-T patients and in cultured A-T cells and not shared by p53–/– mice or p53–/– cells in culture.50,51 Based on these observations, radiation toxicity was attributed to an ATM pathway that does not involve p53.50 It is conceivable that the unusual radiosensitivity of ATM–/– cells may, at least in part, be a consequence of the loss of survival responses we have demonstrated here. Consistent with our findings, defects in NFκB, NM23, and some heat-shock proteins had been already associated with increased radiosensitivity.52,53 It is important to stress, however, that ATM–/– cells can lack a repair function, a transcription-independent mechanism, that can also cause radiosensitivity.5 Furthermore, the outcome—either a defect in damage-induced apoptosis or an increased radiosensitivity—will also almost certainly depend on the cellular context, and cells from different tissues may be differentially prone to ATM-dependent apoptosis or survival.
Our results suggest that after exposure to ionizing radiation the cell undergoes a period during which biochemical processes that may either kill the cell or allow its survival are simultaneously at play. This provides a mechanism for regulating the threshold for apoptotic death in B-CLL cells contingent on the strength of input signals and the biochemical makeup of the cell. Such a mechanism may allow cells that correctly repair DNA damage and are not at risk for mutagenesis to survive, whereas those in which the damage is too great are killed. Confirmation of our hypothesis requires an understanding of the temporal relationship between proapoptotic and survival responses in B-CLL cells after IR and an understanding of the dose effects of IR. However, identifying a class of genes that appeared to be transcriptionally up-regulated by ATM but were repressed by p53 (cluster 3) in our study provides strong evidence for a feedback link between p53-dependent and p53-independent branches of the ATM pathway. Repressed genes could include those that may be required for triggering apoptosis at an appropriate level of damage. Consistent with this theory, many genes that promote growth or are antiapoptotic, such as cFLIP/CLARP2, fall into this category.
In conclusion, we propose a model in which the ATM-dependent response to ionizing radiation involves a complex regulatory network that includes positive and negative regulators of apoptosis (Figure 7). These findings have implications for therapeutic strategies that target ATM function in the management of cancer. Current approaches are largely based on the use of antisense therapy to inhibit ATM function to radiosensitize tumor tissue54 and the additional possibility of using small-molecule antagonists. Our findings suggest that in lymphoid tumors, inhibiting ATM function will be more successful if targeted toward the prosurvival arm of the ATM response. In contrast, in TP53-mutant tumors, the additional inactivation of ATM may prove valuable by reducing the survival signals induced by DNA damage. Ideally, strategies to improve DNA damage–induced killing of TP53-mutant tumors should combine the inhibition of ATM prosurvival functions with the restoration of proapoptotic ATM/p53 function. Our results highlight the complexity of the cellular responses regulated by ATM and TP53 and demonstrate the value of expression profiling in a dynamic, responsive transcriptional context.
Prepublished online as Blood First Edition Paper, September 4, 2003; DOI 10.1182/blood-2003-04-1161.
Supported by the Leukaemia Research Fund UK, Cancer Research UK, the Kay Kendall Leukaemia Fund of the UK, and theAtaxia-Telangiectasia Society of the UK.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Paul Biggs for technical assistance and Dr A. R. Pettitt (University of Liverpool) for generously allowing us to use his ATM- and TP53-mutant samples in Western blots to validate our microarray findings.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal