Abstract
Retinoid-related molecules (RRMs) are derivatives of retinoic acid and promising antileukemic agents with a mechanism of action different from that of other common chemotherapeutics. Here, we describe a novel chemical series designed against the RRM prototype, CD437. This includes molecules with apoptotic effects in acute promyelocytic leukemia and other myelogenous leukemia cell lines, as well as ST2065, an RRM with antagonistic properties. The most interesting apoptotic agent is ST1926, a compound more powerful than CD437 in vitro and orally active in vivo on severe combined immunodeficiency (SCID) mice that received transplants of NB4 cells. ST1926 has the same mechanism of action of CD437, as indicated by the ability to trans-activate retinoic acid receptor γ, to induce the phosphorylation of p38 and JNK, and to down-regulate the expression of many genes negatively modulated by CD437. ST1926 causes an immediate increase in the cytosolic levels of calcium that are directly related to the apoptotic potential of the RRMs considered. The intracellular calcium elevation is predominantly the result of an inhibition of the mitochondrial calcium uptake. The phenomenon is blocked by the ST2065 antagonist, the intracellular calcium chelator BAPTA (1,2 bis (2-aminophenoxy) ethane-N, N, N′,N′-tetraacetic acid tetrakis (acetoxymethyl ester), and by high concentrations of calcium blockers of the dihydropyridine type, compounds that suppress ST1926-induced apoptosis.
Introduction
Novel compounds inducing apoptosis or programmed cell death (PCD) through mechanisms other than those activated by clinically used chemotherapeutic agents are needed. CD437 is the prototype of a promising class of cytotoxic compounds known as adamantyl-retinoids1-4 or retinoid-related molecules (RRMs).5,6 Throughout this article, RRMs will be used to indicate this class of compounds. CD437 is endowed with antitumor as well as antileukemic activity in various experimental models.7-13 The compound is characterized by a chemical structure similar to that of retinoic acid, and was originally developed as a selective agonist of retinoic acid receptor γ (RARγ).14-16 In cellular models of acute myelogenous leukemia (AML), CD437 induces rapid PCD.2,3,17 In AML and other cell types, the apoptotic process set in motion by CD437 does not require activation of RARγ or any other type of RAR and RXR nuclear retinoic acid receptors.2 Thus, the mechanism of action of the compound is different from that of all-trans-retinoic acid (ATRA) and other classic retinoids. Indeed, the RRM induces apoptosis in cells resistant to ATRA.18,19 In addition, a subline of the acute promyelocytic leukemia (APL)–derived NB4 blast made selectively resistant to CD437 (NB4.437r) maintains sensitivity to ATRA.3
Although still largely obscure, the mechanism of action of CD437 is also different from that of many chemotherapeutics and apoptotic agents.2,3,19,20 Recent evidence suggests that the mitochondrion represents an important target for the RRM.10,19,21 CD437 causes opening of the mitochondrial transition pore, release of cytochrome c into the cytosol, and subsequent activation of the caspase proteolytic cascade.2,3 In myeloid blasts, the process of apoptosis ignited by CD437 does not seem to require gene expression or de novo protein synthesis3 and is associated with the activation of mitogen-activated protein (MAP) kinases, such as p38 and Jun N-terminal kinase (JNK).2,3,11,22
In spite of in vitro and in vivo activity, CD437 has limited clinical potential given the narrow window between therapeutic and toxic doses as well as the relatively unfavorable pharmacokinetic profile. To overcome the associated problems, we sought to design CD437 analogs that may be more powerful, less toxic, and more bioavailable than the lead compound. Here we describe a novel chemical series of CD437 congeners, whose prototype is ST1926. ST1926 is more apoptotic than CD437 in AML cells and is active in vivo in the model of APL represented by the severe combined immunodeficiency (SCID) mouse inoculated with NB4 blasts. With the use of ST1926 and other members of the series, we provide evidence that one of the primary events set in motion by RRMs in the leukemic blast is an increase in cytosolic calcium.
Materials and methods
Chemicals
ATRA, doxorubicin, staurosporine, cyclosporine A, verapamil, antimycin A, oligomycin, pertussis toxin, fenretinide, etoposide, nicardipine, nimodipine, nitrendipine, cyclosporin A, and 1,2 bis (2-aminophenoxy) ethane-N, N, N′, N′-tetraacetic acid tetrakis (acetoxymethyl ester) (BAPTA) were obtained from Sigma, St Louis, MO; carbonylcyanide-4-(trifluoromethoxy)-phenylhydrazone (FCCP) was from Biomol (Plymouth Meeting, PA); U0126, PD169316 and SP600125 were from Calbiochem (La Jolla, CA). CD437 was synthesized by Sigma-Tau Industrie Farmaceutiche Riunite S.p.A. Details on the chemical synthesis of ST1926, ST1898, ST1879, ST2474, ST2306, ST2307, ST2142, ST2064, ST2475, ST1927, ST2016, ST2060, ST2062, ST2065 and ST2067 will be given elsewhere.
Cell culture, treatments, and transfections
NB4,23 NB4.306,2 U937, Kasumi, HL-60, and KG1 (American Type Culture Collection [ATCC], Rockville, MD) cells were cultured in the presence of RPMI-1640 with 10% fetal calf serum (FCS) (20% FCS in the case of Kasumi). COS-7 cells (ATCC) were grown in Dulbecco modified Eagle medium (DMEM) containing 10% FCS. COS-7 cells were transfected with RARα RARβ, and RARγ plasmids in the presence of the DR5-tk-CAT reporter gene and the normalization plasmid pCH110 (containing bacterial β-galactosidase), as described.24-26
Cellular proliferation, viability apoptosis, and caspase-3 activation
Cell number and viability were determined following staining with erythrosin (Sigma).2,3,24 For the determination of the apoptotic index, cells were fixed in methanol and stained with DAPI (4,6 diamidino-2-phenylindole) as described.2,3 The apoptotic index indicates the percentage of cells with morphologic features of nuclear fragmentation following DAPI staining and counting a minimum of 300 nuclei/field under the fluorescence microscope. In some experiments, apoptosis was determined according to the annexin-V assay by flow cytometry (FACSORT system; Becton Dickinson, San Jose, CA) with a commercially available kit (Annexin-V–FLUOS staining kit; Boehringer Mannheim, Mannheim, Germany). Before flow cytometry, cells were counterstained with propidium iodide (PI). Caspase-3 activation was measured fluorometrically with the use of the fluorogenic peptidic substrate DEVD-amc (Ac-Asp-Glu-Val-Asp-AMC; Alexis, Laufelingen, Switzerland), as already described.2,3
MAP kinases and Western blot analysis
Extracts of NB4 cells (from 3 × 106 cells) were subjected to Western blot analysis as reported.2,3 The antibodies directed against cytochrome c, actin, ERK-1, ERK-2, JNK, p38, and the corresponding phosphorylated forms were from Cell Signaling Technology (Beverly, MA). Protein bands were visualized with the enhanced chemiluminescence (ECL) detection kit (Amersham, Little Chalfont, United Kingdom). JNK activity was determined on JNK immunoprecipitates with c-JUN as a substrate, using a commercial kit (Cell Signaling Technology). For the experiments involving cytochrome c determination in the cytosolic and mitochondrial fractions, we followed previously described protocols.2,3
RNA extraction, gene profiling, and Northern blot analysis
RNA was extracted with the TRIzol reagent (Invitrogen S.R.L., S. Giuliano Milanese, Italy) and the polyadenylated fraction selected with the Atlas Pure kit (Clontech, Palo Alto, CA). The 12K human ATLAS plastic filters (Clontech) were used for the microarray experiments. Single-strand 32P-labeled cDNA probes were synthesized from poly(A+) RNA as suggested by the manufacturer. Video imaging was performed with the Storm 860 Phosphoimager (Molecular Dynamics, Sunnyvale, CA). Microarray data were quantitated with ATLAS IMAGE 2.7 analysis software (Clontech). Northern blot analysis was performed as described27 with specific human cDNA probes obtained following reverse transcriptase–polymerase chain reaction (RT-PCR) amplification of the transcripts with specific amplimers (Figure 5).
Effect of ST1926 and CD437 on the mRNA levels of a number of genes in NB4 cells: validation of the microarray results. NB4 cells (150 000/well) were treated with vehicle (control), ST1926, and CD437 at the indicated concentrations for 4 hours. Total RNA was extracted and subjected to Northern blot analysis using the indicated cDNAs as radioactive probes. Probes were amplified by RT-PCR from the total RNA extracted from NB4 cells using the following amplimers: NM_006281 = 5′GACCATGGTGATAAACAGTGAGG 3′ (sense oligonucleotide), 5′TGCATCCATCGCATCCAGAATGG 3′ (antisense oligonucleotide) NM_001416 = 5′CCGAGAAGATGCATGCTCGAGAT 3′ (sense oligonucleotide), 5′GGTCAGCAACATTGAGGGGCATT 3′ (antisense oligonucleotide) NM_002699 = 5′GTTCGCCAAGCAGTTCAAGCAGC 3′ (sense oligonucleotide), 5′CTTGAGAAAGTGGCTCTCGAGCG 3′ (antisense oligonucleotide) NM_000986 = 5′TCCTTTCCAAGAGGAATCCTCGG 3′ (sense oligonucleotide), 5′TTTCCACCAACTCGGGGAGCTGA 3′ (antisense oligonucleotide) NM_021029 = 5′TGGTTAACGTCCCTAAAACCCGC 3′ (sense oligonucleotide), 5′GAACTGGATCACTTGGCCCTTTC 3′ (antisense oligonucleotide) NM_004718 = 5′TAGTGGCTTCACGCAGAAGTTGG 3′ (sense oligonucleotide), 5′GTTTTTGGGCTGCGAAGCCATGT 3′ (antisense oligonucleotide) For each Northern blot, an equivalent amount of RNA was added in each lane as indicated by the ethidium bromide staining of the 18S RNA. The results confirm the data illustrated in Table 2 for a number of selected genes.
Effect of ST1926 and CD437 on the mRNA levels of a number of genes in NB4 cells: validation of the microarray results. NB4 cells (150 000/well) were treated with vehicle (control), ST1926, and CD437 at the indicated concentrations for 4 hours. Total RNA was extracted and subjected to Northern blot analysis using the indicated cDNAs as radioactive probes. Probes were amplified by RT-PCR from the total RNA extracted from NB4 cells using the following amplimers: NM_006281 = 5′GACCATGGTGATAAACAGTGAGG 3′ (sense oligonucleotide), 5′TGCATCCATCGCATCCAGAATGG 3′ (antisense oligonucleotide) NM_001416 = 5′CCGAGAAGATGCATGCTCGAGAT 3′ (sense oligonucleotide), 5′GGTCAGCAACATTGAGGGGCATT 3′ (antisense oligonucleotide) NM_002699 = 5′GTTCGCCAAGCAGTTCAAGCAGC 3′ (sense oligonucleotide), 5′CTTGAGAAAGTGGCTCTCGAGCG 3′ (antisense oligonucleotide) NM_000986 = 5′TCCTTTCCAAGAGGAATCCTCGG 3′ (sense oligonucleotide), 5′TTTCCACCAACTCGGGGAGCTGA 3′ (antisense oligonucleotide) NM_021029 = 5′TGGTTAACGTCCCTAAAACCCGC 3′ (sense oligonucleotide), 5′GAACTGGATCACTTGGCCCTTTC 3′ (antisense oligonucleotide) NM_004718 = 5′TAGTGGCTTCACGCAGAAGTTGG 3′ (sense oligonucleotide), 5′GTTTTTGGGCTGCGAAGCCATGT 3′ (antisense oligonucleotide) For each Northern blot, an equivalent amount of RNA was added in each lane as indicated by the ethidium bromide staining of the 18S RNA. The results confirm the data illustrated in Table 2 for a number of selected genes.
Measurement of intracellular calcium
Intracellular calcium concentrations were measured with the use of the fluorescent probe Fura-2 acetoxymethyl ester (FURA-2).28 Briefly, cells (1 × 107/mL) were loaded with 1 μM FURA-2 (Molecular Probes, Irvine, CA) at 37°C in the dark for 30 minutes, washed twice, resuspended in phosphate-buffered saline (PBS) containing 1.26 mM CaCl2 at 106 cells/mL and then used for the experiments. Dual excitation, alternating at 340 nm and 380 nm, was provided by a spectrophotofluorometer (model LS-50B; Perkin-Elmer, Milano, Italy) equipped with 2 excitation monochromators, and emission was fixed at 480 nm. The temperature was set at 37°C ± 1°C. In some experiments, to eliminate extracellular calcium, cells preloaded with FURA-2 were resuspended in PBS without Ca2+, and 0.5 mM EGTA (ethylene glycol tetraacetic acid) was added to each sample prior to addition of the appropriate stimulus.
In vivo studies
NB4 cells (3 × 106) were intraperitoneally inoculated (as described) in SCID mice (8 mice/group). ST1926 was dissolved in cremophor/ethanol 1:1 solution, and diluted 1:10 in PBS at the concentration of 50 mg/kg; the doses of 30 mg/kg and 40 mg/kg were then prepared by appropriate dilutions in the same vehicle. ATRA was dissolved in the dark in Cremophor EL (Sigma) and kept magnetically stirred; the solution was then diluted 1:10 in PBS at the final concentration of 40 mg/kg. Both compounds were administered intraperitoneally and orally twice per day for 3 weeks starting from the day after cell inoculation, in a volume of 10 mL/kg. During treatments body weight and lethality were registered.
Results
ST1926 is more apoptotic than CD437
Analogs of CD437 were synthesized and tested for apoptotic activity and cytotoxicity in the APL-derived NB4 cell line.3 Apoptotic activity was assessed as the percentage of cells showing signs of nuclear fragmentation (apoptotic index)2,3 following 4 hours of treatment (Table 1). ST1926 is the most effective molecule of the series. ST1898, ST1879, ST2474, ST2306, and ST2307 show intermediate potency and are less effective than CD437. ST2142, ST2064, and ST2475 induce apoptosis only at the highest concentration tested, whereas ST1927, ST2016, ST2060, ST2062, ST2065, and ST2067 are devoid of significant activity. For all active molecules, apoptosis is invariably followed by loss of cell viability, which is evident by 18 to 24 hours (data not shown).
Apoptotic effect of a chemical series of RRMs in NB4 cells
. | Compound . | 1 μM . | 10 μM . | 100 μM . |
|---|---|---|---|---|
| ST1926 |  | 72.0 ± 2.3* | > 95 | > 95 |
| CD437 |  | 35.1 ± 3.0* | 80.0 ± 2.5* | > 95 |
| ST1898 |  | 5.0 ± 2.6 | 25.7 ± 6.5* | 40.1 ± 16.5* |
| ST1879 |  | 2.5 ± 0.8 | 33.3 ± 19.7* | > 95 |
| ST2474 |  | 9.0 ± 4.1 | 58.8 ± 1.8* | 88.2 ± 14.3* |
| ST2306 |  | 5.6 ± 0.1 | 43.9 ± 2.3* | 93.3 ± 1.9* |
| ST2307 |  | 6.2 ± 3.0 | 41.6 ± 4.6* | 46.5 ± 3.8* |
| ST2142 |  | < 1 | 9.9 ± 6.7* | 48.4 ± 6.6* |
| ST2064 |  | < 1 | 2.2 ± 1.7 | 70.4 ± 18.1* |
| ST2475 |  | < 1 | 1.6 ± 1.6 | 26.6 ± 5.9* |
| ST1927 |  | 3.0 ± 1.0 | 2.9 ± 0.8 | 4.1 ± 2.6* |
| ST2016 |  | < 1 | < 1 | Necrosis |
| ST2060 |  | < 1 | < 1 | 1.8 ± 1.3 |
| ST2062 |  | < 1 | < 1 | 1.7 ± 0.8 |
| ST2065 |  | < 1 | < 1 | 1.3 ± 0.3 |
| ST2067 |  | < 1 | 1.1 ± 0.4 | < 1 |
. | Compound . | 1 μM . | 10 μM . | 100 μM . |
|---|---|---|---|---|
| ST1926 |  | 72.0 ± 2.3* | > 95 | > 95 |
| CD437 |  | 35.1 ± 3.0* | 80.0 ± 2.5* | > 95 |
| ST1898 |  | 5.0 ± 2.6 | 25.7 ± 6.5* | 40.1 ± 16.5* |
| ST1879 |  | 2.5 ± 0.8 | 33.3 ± 19.7* | > 95 |
| ST2474 |  | 9.0 ± 4.1 | 58.8 ± 1.8* | 88.2 ± 14.3* |
| ST2306 |  | 5.6 ± 0.1 | 43.9 ± 2.3* | 93.3 ± 1.9* |
| ST2307 |  | 6.2 ± 3.0 | 41.6 ± 4.6* | 46.5 ± 3.8* |
| ST2142 |  | < 1 | 9.9 ± 6.7* | 48.4 ± 6.6* |
| ST2064 |  | < 1 | 2.2 ± 1.7 | 70.4 ± 18.1* |
| ST2475 |  | < 1 | 1.6 ± 1.6 | 26.6 ± 5.9* |
| ST1927 |  | 3.0 ± 1.0 | 2.9 ± 0.8 | 4.1 ± 2.6* |
| ST2016 |  | < 1 | < 1 | Necrosis |
| ST2060 |  | < 1 | < 1 | 1.8 ± 1.3 |
| ST2062 |  | < 1 | < 1 | 1.7 ± 0.8 |
| ST2065 |  | < 1 | < 1 | 1.3 ± 0.3 |
| ST2067 |  | < 1 | 1.1 ± 0.4 | < 1 |
The apoptotic index of each compound was measured at 3 different concentrations, as indicated, following incubation of NB4 cells for 4 hours. The notations < 1 and > 95 indicate an apototic index below the limit of detection of the assay and evidence of nuclear fragmentation in practically the totality of the cell population, respectively.
Significantly higher than NB4 cells treated with vehicle (P < .01 according to the Student t test). The apoptotic index in vehicle-treated NB4 cells varies between 3% and 5% (mean, n = 3) according to the experiment, with a standard deviation of less than or equal to 10% of the mean value.
Figure 1A-B demonstrates that ST1926-induced apoptosis and cytotoxicity are dose dependent and the calculated effective concentrations (EC50s) for the two effects are similar. The two values are aproximately 6- and 9-fold lower, respectively, than those observed in the case of CD437. The NB4.437r cell line is selectively resistant to CD437.2 As shown in Figure 1C-D, the NB4.437r cell line is resistant to the apoptotic and cytotoxic actions of ST1926 as well. This indicates that ST1926 and CD437 are not only structurally related molecules but they also belong to the same pharmacologic class of agents. ST1926 is apoptotic and cytotoxic on a large spectrum of cancerous and leukemic cells, including freshly isolated AML blasts in primary culture (data not shown). However, there are examples of cells that are refractory to the compound. Figure 2 illustrates two examples each of sensitive and refractory cell lines. Challenge of KG1 and HL-60 myeloblasts with increasing concentrations of ST1926 for 6 hours results in apoptosis (Figure 2A-B). This is followed by a significant drop in the number of viable cells, which is evident after 24 or 48 hours. In both cell lines, ST1926 is more apoptotic and cytotoxic than CD437. Treatment of U937 and Kasumi blasts for 6 hours with either ST1926 or CD437 is not accompanied by a significant apoptotic response (Figure 2C-D), though protracted incubation with the 2 agents eventually results in cell death. Interestingly, even in U937 and Kasumi cells, the cytotoxic effect is more evident in the case of ST1926 than CD437.
Apoptotic and cytotoxic effect of ST1926 in NB4 and NB4.437r cells: comparison with CD437. NB4 (A,B) or NB4.437r (C,D) cells (300 000/mL) were treated with vehicle and the indicated concentrations of ST1926 and CD437 for 4 hours (A) or 6 hours (C) in the case of the measurement of the apoptotic index or for 24 hours in the case of the determination of the number of viable cells (B, D). Results are expressed as the mean ± SD of 3 replicate cultures. The data are representative of 2 independent experiments. In A and B, experimental points were fitted using the sigmoidal dose-response equation (variable slope) and statistically analyzed with the GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego, CA). Calculation of the EC50 values and statistical analysis of the data were performed with the same program. The EC50 value for the apoptotic effect of ST11926 (0.2 μM; interval of confidence 0.08-0.67, n = 3) is significantly lower (P < .03, following application of the F test) than that of CD437 (1.1 μM; interval of confidence 0.82-1.51, n = 3). Similarly, the EC50 value for the cytotoxic effect of ST11926 (0.1 μM; interval of confidence 0.05-0.19, n = 3) is significantly lower (P < .01, following application of the F test) than that of CD437 (0.9 μM; interval of confidence 0.43-2.07, n = 3). *Significantly higher or lower than the corresponding vehicle-treated group (P < .01 according to the Student t test).
Apoptotic and cytotoxic effect of ST1926 in NB4 and NB4.437r cells: comparison with CD437. NB4 (A,B) or NB4.437r (C,D) cells (300 000/mL) were treated with vehicle and the indicated concentrations of ST1926 and CD437 for 4 hours (A) or 6 hours (C) in the case of the measurement of the apoptotic index or for 24 hours in the case of the determination of the number of viable cells (B, D). Results are expressed as the mean ± SD of 3 replicate cultures. The data are representative of 2 independent experiments. In A and B, experimental points were fitted using the sigmoidal dose-response equation (variable slope) and statistically analyzed with the GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego, CA). Calculation of the EC50 values and statistical analysis of the data were performed with the same program. The EC50 value for the apoptotic effect of ST11926 (0.2 μM; interval of confidence 0.08-0.67, n = 3) is significantly lower (P < .03, following application of the F test) than that of CD437 (1.1 μM; interval of confidence 0.82-1.51, n = 3). Similarly, the EC50 value for the cytotoxic effect of ST11926 (0.1 μM; interval of confidence 0.05-0.19, n = 3) is significantly lower (P < .01, following application of the F test) than that of CD437 (0.9 μM; interval of confidence 0.43-2.07, n = 3). *Significantly higher or lower than the corresponding vehicle-treated group (P < .01 according to the Student t test).
Comparative apoptotic and cytotyoxic effects of ST1926 and CD437 in myeloid leukemia cell lines. KG1 (A), HL-60 (B), U937 (C), and Kasumi (D) cells (300 000/mL) were treated with vehicle or the indicated concentrations of ST1926 and CD437 for the indicated amount of time. The apoptotic index (left panels) or the number of viable cells (right panels) was determined on aliquots of the cell cultures. Results are expressed as the mean ± SD of 3 replicate cultures. The data are representative of 2 independent experiments. *Significantly higher or lower than the corresponding vehicle-treated group (P < .01 according to the Student t test).
Comparative apoptotic and cytotyoxic effects of ST1926 and CD437 in myeloid leukemia cell lines. KG1 (A), HL-60 (B), U937 (C), and Kasumi (D) cells (300 000/mL) were treated with vehicle or the indicated concentrations of ST1926 and CD437 for the indicated amount of time. The apoptotic index (left panels) or the number of viable cells (right panels) was determined on aliquots of the cell cultures. Results are expressed as the mean ± SD of 3 replicate cultures. The data are representative of 2 independent experiments. *Significantly higher or lower than the corresponding vehicle-treated group (P < .01 according to the Student t test).
In the limited series presented, NB4 is the cell line that is most sensitive to the apoptotic action of ST1926. The phenomenon may relate to the presence of the promyelocytic leukemia–retinoic acid receptor alpha (PML-RARα) oncogenic protein in the APL-derived cell line. However, this is unlikely, as ST1926 and CD437 seem to belong to the same pharmacologic class of agents, and PML-RARα does not play any significant role in CD437-induced apoptosis.2
The molecular target of ST1926 apoptotic activity in myeloid leukemia cells is similar to the ligand-binding domain of RARγ
As a first step toward the definition of the intracellular target(s) of RRM activity, we looked for antagonists of ST1926 among the inactive members of the congener series. Figure 3 demonstrates that preincubation of NB4 cells with two concentrations of ST2062, ST2064, or ST2067 (10 μM and 100 μM) has no or minimal (ST2067) effect as to the number and proportion of blasts undergoing apoptosis following treatment with ST1926. However, preincubation with a 100-fold molar excess of ST2065 over ST1926 suppresses the apoptotic response observed in NB4 cells, suggesting an antagonistic action.
Effect of ST2062, ST2064, ST2065, and ST2067 on ST1926-dependent apoptosis in NB4 cells. NB4 cells (300 000/mL) were pretreated for 1 hour with vehicle, ST2062 (A), ST2064 (A), ST2065 (B), and ST2067 (B) at the indicated concentrations before addition of vehicle or ST1926 for an additional 6 hours. The apoptotic index was determined on aliquots of the cell cultures. Results are expressed as the mean ± SD of 3 replicate cultures. The data are representative of at least 2 independent experiments. °Significantly higher than the corresponding vehicle-treated group (P < .01 according to the Student t test). *Significantly lower than the corresponding ST1926-treated group (P < .01 according to the Student t test).
Effect of ST2062, ST2064, ST2065, and ST2067 on ST1926-dependent apoptosis in NB4 cells. NB4 cells (300 000/mL) were pretreated for 1 hour with vehicle, ST2062 (A), ST2064 (A), ST2065 (B), and ST2067 (B) at the indicated concentrations before addition of vehicle or ST1926 for an additional 6 hours. The apoptotic index was determined on aliquots of the cell cultures. Results are expressed as the mean ± SD of 3 replicate cultures. The data are representative of at least 2 independent experiments. °Significantly higher than the corresponding vehicle-treated group (P < .01 according to the Student t test). *Significantly lower than the corresponding ST1926-treated group (P < .01 according to the Student t test).
CD437 was originally developed as a selective RARγ agonist,14-16 although the process of apoptosis triggered by CD437 in NB4 and HL-60 is not dependent on the nuclear retinoic acid receptor.2,17 Indeed, NB4 and HL-60 blasts do not express detectable amounts of RARγ and antagonists of the receptor do not block CD437-induced apoptosis in the two cell lines.2,17,27 Nevertheless, we deemed it interesting to compare the RARγ transactivation properties of ST1926 and selected congeners to that of CD437. As shown in Figure 4A, in COS-7 cells transfected with the different RAR isoforms, CD437 transactivates RARγ in a dose-dependent fashion. Whereas RARα is activated by CD437 only at the highest concentration considered (10–6 M), the RRM is not an efficient ligand of RARβ. ST1926 maintains RARγ agonistic activity, although the selectivity of this compound for the receptor isoform is lower than that of CD437 (Figure 4B). Interestingly, Figure 4C-D indicates that there is an excellent correlation between the RARγ-transactivating potential and the apoptotic activity of ST1926 and congeners. The finding does not imply that RARγ mediates the apoptotic process triggered by RRMs in myelod leukemia blasts. Rather, the correlation suggests that the primary molecular target of ST1926 has structural similarity with the ligand-binding domain of RARγ.
Comparative RARα, RARβ, and RARγ transactivation of ST1926, CD437, and congeners: correlation with the apoptotic potential. COS-7 cells (150 000/well) were cotransfected with the indicated RAR subtype (0.05 μg) (A,B) or RARγ (C), a retinoic acid–dependent reporter construct (DR5-tk-CAT; 1 μg) and the β-galactosidase expression vector pCH110 (0.5 μg). Twenty-four hours after transfection, cells were treated for an additional 24 hours with vehicle or the indicated concentration of ST1926 (A) and CD437 (B) or the indicated compound (1 μM) (C). At the end of the experiment, cells were harvested and homogenized, and CAT as well as β-galactosidase activities determined. The results are expressed in fold induction of CAT activity following normalization for the amount of β-galactosidase expressed. CAT activity measured in cells treated with vehicle is considered as 1.0. Panel D illustrates the linear correlation between the RARγ transactivating activity of each RRM, as determined at a concentration of 1 μM (C), and the apoptotic potential, as determined at a concentration of 10 μM (Table 1). Results are expressed as the mean ± SD of 3 replicate cultures. The data are representative of at least 2 independent experiments. *Significantly higher than the corresponding vehicle-treated group (P < .01 according to the Student t test).
Comparative RARα, RARβ, and RARγ transactivation of ST1926, CD437, and congeners: correlation with the apoptotic potential. COS-7 cells (150 000/well) were cotransfected with the indicated RAR subtype (0.05 μg) (A,B) or RARγ (C), a retinoic acid–dependent reporter construct (DR5-tk-CAT; 1 μg) and the β-galactosidase expression vector pCH110 (0.5 μg). Twenty-four hours after transfection, cells were treated for an additional 24 hours with vehicle or the indicated concentration of ST1926 (A) and CD437 (B) or the indicated compound (1 μM) (C). At the end of the experiment, cells were harvested and homogenized, and CAT as well as β-galactosidase activities determined. The results are expressed in fold induction of CAT activity following normalization for the amount of β-galactosidase expressed. CAT activity measured in cells treated with vehicle is considered as 1.0. Panel D illustrates the linear correlation between the RARγ transactivating activity of each RRM, as determined at a concentration of 1 μM (C), and the apoptotic potential, as determined at a concentration of 10 μM (Table 1). Results are expressed as the mean ± SD of 3 replicate cultures. The data are representative of at least 2 independent experiments. *Significantly higher than the corresponding vehicle-treated group (P < .01 according to the Student t test).
ST1926 and CD437 have similar effects on the gene expression profile of NB4 cells
Using oligonucleotide microarrays, we evaluated the expression profile of approximately 12 000 genes in NB4 cells treated for 4 hours with ST1926 and CD437 at approximately equipotent concentrations (Table 2). Only 8 genes are significantly up-regulated by either ST1926 or CD437. Five of these are common targets of ST1926 and CD437, one (UDP gycosyltransferase B15) is specifically regulated by ST1926, whereas 2 (ribosomal protein L35 and coronin) are selectively modulated by CD437. ST1926 and/or CD437 reduce the expression of at least 184 genes, 62% of which are down-regulated by both adamantyl-retinoids while the remaining 38% is a specific target of either compound. The results support the idea that the processes of apoptosis induced by ST1926 and CD437 are similar. Although the data presented are the result of a single experiment, it is reassuring that the majority (approximately 80%) of the genes down-regulated with 0.2 μM ST1926 are the same as those down-regulated by 1 μM ST1926 in a separate experiment (data not shown). A high proportion of down-regulated genes codes for products that fall within 3 main functional classes: mitochondrial, ribosomal, and translation-related proteins. The down-regulation of genes involved in protein synthesis is consistent with the fact that de novo protein synthesis is not required for the process of apoptosis activated by the 2 RRMs (Mologni et al3 and data not shown). We validated the results of the microarray analysis for a randomly selected sample of 6 transcripts by Northern blot analysis. As shown in Figure 5, the transcripts coding for the translation initiation factor 4A, the POU domain transcription factor 1, the ribosomal proteins L24 and L36a as well as the subunit VIIa of cytochrome c oxidase show the expected down-regulation upon treatment of NB4 cells for 4 hours with ST1926 (0.2 μM) and CD437 (1 μM). Consistent with the results obtained with the microarrays, the mRNAs encoding translation initiation factor 4A, the POU domain transcription factor 1 and the ribosomal protein L36a are down-regulated more efficiently by ST1926 than CD437. As anticipated, the serine/threonine kinase 3 mRNA is up-regulated by CD437 and ST1926, though this last compound is active only at 1 μM.
Gene profiling in NB4 cells treated with ST1926 and CD437
Down regulated proteins/genes . | Accession no. . | ST1926 (Ratio) . | CD437 (Ratio) . |
|---|---|---|---|
| Ribosome | |||
| Ribosomal protein 113a | X56932 | 0.67 | 0.46* |
| Ribosomal protein L32 | NM_000994 | 0.47* | 0.71 |
| Ribosomal protein L39 | NM_001000 | 0.49* | 0.57* |
| Ribosomal protein 136a-like | NM_001001 | 0.40* | 0.29* |
| Ribosomal protein S2 | NM_002952 | 0.50* | 0.60* |
| Ribosomal protein s27a | NM_002954 | 0.27* | 0.76 |
| Ribosomal protein S26 | NM_001029 | 0.33* | 0.64 |
| Ribosomal protein L6 | NM_000970 | 0.34* | 0.62 |
| Ribosomal protein L8 | NM_000973 | 0.40* | 0.51* |
| Ribosomal protein L11 | NM_000975 | 0.62 | 0.31* |
| Ribosomal protein L13 | NM_000977 | 0.56* | 0.50* |
| Ribosomal protein L24 | NM_000986 | 0.42* | 0.46* |
| Ribosomal protein L29 | NM_000992 | 0.18* | 1.12 |
| Ribosomal protein 136a | NM_021029 | 0.30* | 0.53* |
| Ribosomal protein, large P2 | NM_001004 | 1.49 | 0.42* |
| Ribosomal protein S10 | NM_001014 | 0.34* | 0.49* |
| Ribosomal protein S15 | NM_001018 | 0.42* | 0.60* |
| Ribosomal protein S16 | NM_001020 | 0.66 | 0.49* |
| Ribosomal protein S17 | NM_001021 | 0.71 | 0.48* |
| Ribosomal protein L41 | NM_021104 | 0.36* | 0.70 |
| Ribosomal protein L10 | NM_006013 | 0.46* | 0.57* |
| Ribosomal protein L19 | NM_000981 | 0.48* | 0.56* |
| Ribosomal protein 118a | NM_000980 | 0.49* | 0.62 |
| Ribosomal protein, large, P1 | NM_001003 | 0.47* | 0.51* |
| Ribosomal protein S27 | NM_001030 | 0.57* | 0.42* |
| Protein synthesis | |||
| Translation elongation factor 2 | NM_001961 | 0.40* | 0.38* |
| Translation initiation factor 5A | NM_001970 | 0.49* | 0.47* |
| Translation initiation factor 4A, isoform 1 | NM_001416 | 0.28* | 0.40* |
| Translation initiation factor 4 gamma, 2 | NM_001418 | 0.26* | 0.76 |
| Translation initiation factor 4B | NM_001417 | 0.49* | 0.33* |
| Translation initiation factor 4A, isoform 2 | NM_001967 | 0.32* | 0.46* |
| Translation initiation factor 3, subunit 7 | NM_003753 | 0.30* | 0.41* |
| Translation initiation factor 3, subunit 4 | NM_003755 | 0.15* | 0.56* |
| Translation initiation factor 3, subunit 2 | NM_003757 | 0.21* | 0.28* |
| Nascent-polypeptide-associated complex alpha | NM_005594 | 0.50* | 0.50* |
| Translation initiation factor 3, subunit 6 | NM_001568 | 0.39* | 0.64 |
| Mitochondria | |||
| H+ transporting, V0 subunit c | NM_001694 | 0.80 | 0.44* |
| H+ transporting, mitochondrial F1 complex, _ | NM_004046 | 0.37* | 0.59* |
| H+ transporting, mitochondrial F1 complex, _ | NM_001686 | 0.37* | 0.54* |
| H+ transporting, mitochondrial F1 complex, _1 | NM_005174 | 0.64 | 0.26* |
| H+ transporting, mitochondrial F0 complex, c | NM_001689 | 2.00 | 0.57* |
| H+ transporting, mitochondrial F0 complex, F6 | NM_001685 | 0.34* | 0.47* |
| H+ transporting, mitochondrial F0 complex, g | NM_006476 | 0.74 | 0.52* |
| H+ transporting, mitochondrial F1 complex, O | NM_001697 | 1.12 | 0.43* |
| H+ transporting, mitochondrial F0 complex, e | NM_007100 | 0.39* | 0.64 |
| Ubiquinol-cytochrome c reductase core protein II | NM_003366 | 0.59* | 0.26* |
| Ubiquinol-cytochrome c reductase hinge protein | NM_006004 | 0.32* | 0.48* |
| Ubiquinol-cytochrome c reductase binding prot. | NM_006294 | 0.58* | 1.00 |
| Translocase of mitochondrial membr. 8 hom. B | NM_012459 | 0.52* | 0.34* |
| Cytochrome c oxidase subunit Vic | NM_004374 | 0.60* | 0.35* |
| Mitochondrial adenine translocator, member 6 | J03592 | 0.29* | 0.33* |
| Cytochrome c oxidase subunit viia polypeptide 2 | NM_001865 | 0.14* | 0.41* |
| Ubiquinol-cytochrome c reductase binding prot. | NM_006294 | 0.58* | 1.00 |
| Mitochondrial adenine nucleotide translocator, 5 | NM_001152 | 1.42 | 0.21* |
| Cytochrome c oxidase subunit viia, 2 like | NM_004718 | 0.34* | 0.40* |
| Cytochrome c oxidase subunit Vb | NM_001862 | 0.34* | NC |
| NADH-coenzyme Q reductase | NM_004552 | 0.42* | 0.42* |
| Cytochrome c oxidase subunit viib | NM_001866 | 0.42* | 0.46* |
| Mitochondrial adenine nucleotide translocator, 5 | NM_001152 | 1.42 | 0.21* |
| Mitochondrial phosphate carrier, member 3 | NM_002635 | 0.32* | 0.76* |
| Mitochondrial carrier homologue 2 | NM_014342 | 0.44* | 0.38* |
| Mitochondrial solute carrier | NM_016612 | 0.45* | 0.39* |
| Cytochrome c oxidase subunit viic | NM_001867 | 0.60* | 0.42* |
| NADH dehydrogenase 1 beta subcomplex, 9 | NM_005005 | 0.55* | 0.22* |
| Apoptosis | |||
| Death-associated protein kinase 3 | NM_001348 | 0.45* | 0.83 |
| Apoptosis inhibitor 5 | NM_006595 | 0.82 | 0.23* |
| FK506 binding protein 5 | NM_004117 | 0.56* | 0.39* |
| Transcription and nuclear | |||
| Purine-rich element binding protein B | NM_033224 | 0.33* | 0.41* |
| POU domain, class 3, transcription factor 1 | NM_002699 | 0.29* | 0.32* |
| Corepressor/HDAC3 complex subunit | NM_024665 | 0.46* | 0.42* |
| CCAAT/enhancer binding protein, beta | NM_005194 | 0.48* | 0.30* |
| Cardiac-specific homeo box | NM_004387 | 0.57* | 0.36* |
| Paired-like homeodomain transcription factor 1 | NM_001288 | 0.43* | 0.47* |
| RING1 and YY1 binding protein | NM_001762 | 0.44* | 0.39* |
| Nuclease sensitive element binding protein 1 | NM_012234 | 0.50* | 0.02* |
| Sex-determining region Y-box 4 | NM_003107 | 0.35* | 0.43* |
| Sex-determining region Y-box 11 | NM_003108 | 0.37* | 0.32* |
| Interleukin enhancer binding factor 2, 45 kDa | NM_004515 | 0.50* | 0.44* |
| High-mobility group protein 1 | NM_002128 | 0.75 | 0.48* |
| High-mobility group protein 17 | NM_002266 | 0.54* | 0.15* |
| H2A histone family, member Z | NM_002106 | 0.54* | 1.15 |
| Sin3-associated polypeptide, 18 kDa | NM_005870 | 0.42* | 0.45* |
| Single-stranded DNA binding protein | NM_003143 | 0.17* | 0.25* |
| Nucleophosmin | NM_018285 | 0.50* | 0.48* |
| Nucleolar protein family A, member 3 | NM_003860 | 0.26* | 0.56* |
| Thymosin, beta 4 | NM_021109 | 0.32* | 0.91 |
| Thymosin, beta 10 | NM_004559 | 0.41* | 0.60* |
| SP140 nuclear body protein | M92381 | 0.45* | 0.49* |
| Ornithine decarboxylase 1 | NM_002539 | 0.54* | 0.40* |
| Proteasome degradation pathway | |||
| Ubiquitin-conjugating enzyme E2N | NM_003348 | 0.52 | 0.16* |
| Ubiquitin-conjugating enzyme E2M | NM_003969 | 0.45* | 0.41* |
| Ubiquitin-like 5 | NM_024292 | 0.37* | 0.42* |
| Ubiquitin A-52 residue ribosomal protein fusion | NM_003333 | 0.47* | 0.44* |
| Ring-box 1 | NM_014248 | 0.15* | 0.32* |
| Proteasome inhibitor subunit 1 (PI31) | NM_006814 | 0.56* | 0.28* |
| Proteasome subunit, beta type, 1 | NM_002793 | 0.41* | 0.33* |
| Proteasome subunit, alpha type, 4 | D00763 | 0.35* | 0.36* |
| Proteasome 26S subunit, non-atpase, 3 | NM_002809 | 0.38* | 0.55* |
| RNA and splicing | |||
| Heterogeneous nuclear ribonucleoprotein A1 | NM_002136 | 0.76 | 0.37* |
| Heterogeneous nuclear ribonucleoprotein C | NM_004500 | 0.70 | 0.40* |
| Splicing factor, arginine/serine-rich 3 | NM_003017 | 0.43* | 0.53* |
| Small nuclear ribonucleoprotein polypeptide G | NM_003096 | 0.31* | 0.34* |
| Small nuclear ribonucleoprotein polypeptide F | NM_003095 | 0.43* | 0.43* |
| Small nuclear ribonucleoprotein D2 | NM_004597 | 0.35* | 0.40* |
| Small nuclear ribonucleoprotein, B and B1 | NM_003091 | 0.43* | 0.49* |
| RNA polymerase II polypeptide G | NM_002696 | 0.49* | 0.43* |
| RNA binding motif protein, X chromosome | NM_002139 | 0.73 | 0.38* |
| Poly(A) binding protein, cytoplasmic 1 | NM_002568 | 0.49* | 0.39* |
| Poly(rc) binding protein 1 | NM_006196 | 0.77 | 0.30* |
| Poly(rc) binding protein 2 | NM_005016 | 0.47* | 0.42* |
| Lysyl-trna synthetase | NM_005548 | 0.56* | 0.41* |
| Asparaginyl-trna synthetase | NM_004539 | 0.59* | 0.49* |
| 5′-3′ exoribonuclease 2 | NM_012255 | 0.72 | 0.10* |
| Signal transduction | |||
| Adaptor-related protein complex 2, sigma 1 | AJ010148 | 0.35* | 0.43* |
| Adaptor-related protein complex 2, mu 1 | NM_004068 | 0.47* | 0.38* |
| ADP-ribosylation factor 1 | M36340 | 0.34* | 0.55* |
| ADP-ribosylation factor-like 2 | NM_001667 | 0.89 | 0.47* |
| fms-related tyrosine kinase 3 ligand | U04806 | 0.73 | 0.46* |
| Rho GDP dissociation inhibitor (GDI) beta | L20688 | 0.63 | 0.43* |
| ras-like protein VTS58635 | NM_033315 | 0.80 | 0.38* |
| G protein-coupled receptor 6 | NM_005284 | 0.67 | 0.48* |
| Ion channels | |||
| Voltage-dependent anion channel 2 | NM_003375 | 0.49* | 0.57* |
| Chloride intracellular channel 1 | NM_001288 | 0.43* | 0.47* |
| Structural proteins | |||
| Tubulin, alpha, ubiquitous | K00558 | 0.69 | 0.60* |
| Tubulin, alpha 3 | NM_006009 | 0.68 | 0.42* |
| Tubulin, alpha | K00558 | 0.72 | 0.48* |
| Actin, beta | X00351 | 0.47* | 1.44 |
| Actin-related protein 2/3 complex, subunit 2 | NM_005731 | 0.23* | 0.51* |
| Actin-related protein 2/3 complex, subunit 3 | NM_005719 | 0.45* | 0.40* |
| Profilin 1 | NM_005022 | 0.44* | 0.53* |
| Cofilin 1 (nonmuscle) | NM_005507 | 0.43* | 0.72 |
| Sentrin | NM_003352 | 0.78 | 0.23* |
| Integral membrane protein 2B | NM_021999 | 0.52* | 0.22* |
| Dynein, cytoplasmic, light polypeptide | NM_003746 | 0.25* | 0.80 |
| Kinesin family member 3A | NM_007054 | 0.47* | 0.07* |
| Integrin, alpha 11 | NM_012211 | 0.36* | 0.10* |
| Vesicle-associated membrane protein 8 | NM_003761 | 0.31* | 0.65 |
| Lectin, galactoside-binding, soluble, 1 | NM_002305 | 0.43* | 0.46* |
| Metabolism | |||
| Phytanoyl-coa hydroxylase | NM_006214 | 0.86 | 0.47* |
| Aldolase A, fructose-bisphosphate | NM_000034 | 0.71 | 0.41* |
| UDP-glucose ceramide glucosyltransferase | NM_003358 | 0.50* | 0.26* |
| Enolase 1, (alpha) | NM_001428 | 0.42* | 0.38* |
| Aldo-keto reductase family 1, member B1 | NM_001628 | 0.30* | 0.30* |
| Hydroxyacyl-coenzyme A dehydrogenase, alpha | NM_000182 | 0.73 | 0.47* |
| Enolase 1, (alpha) | NM_005945 | 0.49* | 0.48* |
| Arylsulfatase A | NM_000487 | 0.58* | 0.27* |
| Tyrosine 3-monooxygenase, epsilon polypeptide | NM_006761 | 0.24* | 0.63 |
| Phosphoglycerate mutase 1 | NM_002629 | 0.67 | 0.35* |
| Mannosidase, alpha, class 1B, member 1 | NM_007230 | 0.53* | 0.31* |
| Tyrosine 3-monooxygenase, zeta polypeptide | M86400 | 0.53* | 0.46* |
| Cell cycle and mitosis | |||
| Cyclin-dependent kinase 4 | M14505 | 0.42* | 0.45* |
| CDC10 cell division cycle 10 homologue | S72008 | 0.56* | 0.20* |
| S-phase kinase-associated protein 1A (p19A) | NM_006930 | 0.12* | 0.35* |
| Mitotic centromere-associated kinesin | NM_006845 | 0.60* | 0.02* |
| Karyopherin alpha 2 | NM_002266 | 0.54* | 0.15* |
| Oxidation | |||
| Thioredoxin | NM_003329 | 0.36* | 0.48* |
| Peroxiredoxin 4 | NM_006406 | 0.56 | 2.45† |
| Superoxide dismutase 1, soluble | K00065 | 0.27* | 0.38* |
| 15 kDa selenoprotein | NM_004261 | 0.28* | 0.35* |
| Microsomal glutathione S-transferase 1 | J03746 | 0.55* | 0.37* |
| Miscellaneous | |||
| MHC, class I, C | M11886 | 0.77 | 0.38* |
| Adenylate kinase 2 | NM_001625 | 0.36* | 0.21* |
| Neuropathy target esterase | NM_006702 | 0.70 | 0.03* |
| Roundabout homologue 4 | NM_019055 | 0.38* | 0.29* |
| Hypermethylated in cancer 1 | NM_006497 | 0.40* | 0.41* |
| 6.2 kDa protein | NM_019059 | 0.47* | 0.22* |
| Ferritin, heavy polypeptide 1 | NM_002032 | 0.73 | 0.49* |
| Nonmetastatic cells 1, protein (NM23A) | X17620 | 0.50* | 0.35* |
| Anti-Mullerian hormone | NM_000479 | 0.40* | 0.24* |
| 16.7 kDa protein | NM_016139 | 0.48* | 0.65 |
| Proteoglycan 1, secretory granule | NM_002727 | 0.64 | 0.38* |
| Peptidyl-prolyl cis/trans isomerase NIMA-inter. 1 | U49070 | 0.57* | 0.50* |
| Cystatin F (leukocystatin) | NM_003650 | 0.61 | 0.36* |
| Glucose regulated protein, 58 kda | NM_005313 | 0.16* | 0.34* |
| Signal sequence receptor, beta | NM_003145 | 0.49* | 0.44* |
| Tumor protein, translationaly controlled 1 | NM_003295 | 0.24* | 0.70 |
| Deleted in split-hand/split-foot 1 region | NM_006304 | 0.59* | 0.30* |
| Ubiquitously expressed transcript | NM_004182 | 0.42* | 0.08* |
| BCR protein, uterine leiomyoma, 1 | NM_018648 | 0.71 | 0.48* |
| Multiple endocrine neoplasia I | NM_000244 | 0.59* | 0.37* |
| Small inducible cytokine A5 (RANTES) | M21121 | NC | 0.37* |
| Enhancer of rudimentary homolog | NM_004450 | 0.75 | 0.50* |
| Pyrophosphatase | NM_031210 | 0.19* | 0.39* |
| S100 calcium binding protein P | NM_005980 | 0.37* | 0.46* |
| Overexpressed breast tumor protein | NM_013397 | 0.21* | 0.47* |
| Ferritin, light polypeptide | NM_000146 | 0.60 | 0.44* |
| Adenomatous polyposis coli-like | NM_005883 | 1.24 | 0.15* |
| Proteolipid protein 2 (colonic epithelium-enriched) | NM_002668 | 0.80 | 0.46* |
| Topoisomerase (DNA) II binding protein | NM_007027 | 0.62 | 0.09* |
| Lsm3 protein | NM_014463 | 0.40* | 0.42* |
| ATPase, Ca++ transporting, plasma membrane 2 | L20977 | 0.43* | 0.65 |
| Stomatin (EPB72)-like 2 | NM_013442 | 0.66 | 0.43* |
| Signal recognition particle 72 kda | NM_001222 | 0.42* | 0.64 |
| Glutamic acid-rich protein-like 3 | NM_031286 | 0.86 | 0.12* |
| Singed-like (fascin homologue, sea urchin) | NM_003088 | 0.73 | 0.43* |
| PAP-1 binding protein | NM_031288 | 1.10 | 0.13* |
| SET translocation (myeloid leukemia-associated) | NM_003011 | 0.94 | 0.54* |
| Putative proteins | |||
| Hypothetical protein MGC2803 | NM_024038 | 0.27* | 0.62 |
| Hypothetical protein HSPC194 | NM_016462 | 0.47* | 0.39* |
| KIAA0101 gene product | NM_014736 | 0.24* | 0.40* |
| Hypothetical protein MGC5499 | NM_024055 | 0.28* | 0.23* |
| Chromosome 2 open reading frame 6 | NM_018221 | 0.28* | 0.22* |
| Hypothetical protein FLJ10815 | NM_018231 | 0.43* | 0.21* |
| JM5 protein | NM_007075 | 0.69 | 0.10* |
| Chromosome 15 open reading frame 12 | NM_018285 | 0.50* | 0.48* |
| GNAS complex locus | M14631 | 0.56* | 0.49* |
| Hypothetical protein DC50 | NM_031210 | 0.19* | 0.39* |
| Chromosome 11 open reading frame 10 | AF086763 | 0.54* | 0.53* |
| Vesicle-associated membrane protein 8 | NM_003761 | 0.31* | 0.65 |
| Peptidyl-prolyl cis/trans isomerase NIMA-inter. 1 | U49070 | 0.57* | 0.50* |
| Chaperons and heat shock | |||
| Hsp40 homologue, subfamily A, member 1 | NM_001539 | 0.62 | 0.25* |
| Heat shock 70 kDa protein 8 | Y00371 | 0.68 | 0.47* |
| Heat shock 90 kDa protein 1, alpha | X07270 | 0.23* | 0.47* |
| Chaperonin containing TCP1, subunit 2 (beta) | NM_006431 | 0.43* | 0.50* |
| Hsp70 interacting protein | NM_003932 | 0.34* | 0.49* |
| Chaperonin containing TCP1, subunit 3 | NM_005998 | 0.52* | 0.36* |
| Chaperonin containing TCP1, subunit 6A (zeta 1) | NM_001762 | 0.44* | 0.39* |
| Membrane receptors | |||
| Growth hormone secretagogue receptor | NM_004122 | 0.65 | 0.42* |
| Somatostatin receptor 3 | NM_001051 | 0.48* | 0.61 |
| Up-regulated proteins/genes | |||
| Ribosomal protein L35 | NM_007209 | NC | 7.13† |
| Activated RNA polymerase II cofactor 4 | NM_006713 | 5.29† | 9.51† |
| Coronin, actin binding protein, 1A | NM_007074 | NC | 4.27† |
| Myeloid/lymphoid or mixed-lineage leukemia 4 | NM_014727 | 2.76† | 1.82 |
| Hypothetical protein FLJ10707 | NM_018187 | 5.59† | 3.99† |
| Interleukin 8 | Y00787 | 2.02† | 11.07† |
| Serine/threonine kinase 3 | NM_006281 | 2.41† | 7.11† |
| UDP glycosyltransferase 2 family, B15 | NM_001076 | 2.94† | 1.39 |
Down regulated proteins/genes . | Accession no. . | ST1926 (Ratio) . | CD437 (Ratio) . |
|---|---|---|---|
| Ribosome | |||
| Ribosomal protein 113a | X56932 | 0.67 | 0.46* |
| Ribosomal protein L32 | NM_000994 | 0.47* | 0.71 |
| Ribosomal protein L39 | NM_001000 | 0.49* | 0.57* |
| Ribosomal protein 136a-like | NM_001001 | 0.40* | 0.29* |
| Ribosomal protein S2 | NM_002952 | 0.50* | 0.60* |
| Ribosomal protein s27a | NM_002954 | 0.27* | 0.76 |
| Ribosomal protein S26 | NM_001029 | 0.33* | 0.64 |
| Ribosomal protein L6 | NM_000970 | 0.34* | 0.62 |
| Ribosomal protein L8 | NM_000973 | 0.40* | 0.51* |
| Ribosomal protein L11 | NM_000975 | 0.62 | 0.31* |
| Ribosomal protein L13 | NM_000977 | 0.56* | 0.50* |
| Ribosomal protein L24 | NM_000986 | 0.42* | 0.46* |
| Ribosomal protein L29 | NM_000992 | 0.18* | 1.12 |
| Ribosomal protein 136a | NM_021029 | 0.30* | 0.53* |
| Ribosomal protein, large P2 | NM_001004 | 1.49 | 0.42* |
| Ribosomal protein S10 | NM_001014 | 0.34* | 0.49* |
| Ribosomal protein S15 | NM_001018 | 0.42* | 0.60* |
| Ribosomal protein S16 | NM_001020 | 0.66 | 0.49* |
| Ribosomal protein S17 | NM_001021 | 0.71 | 0.48* |
| Ribosomal protein L41 | NM_021104 | 0.36* | 0.70 |
| Ribosomal protein L10 | NM_006013 | 0.46* | 0.57* |
| Ribosomal protein L19 | NM_000981 | 0.48* | 0.56* |
| Ribosomal protein 118a | NM_000980 | 0.49* | 0.62 |
| Ribosomal protein, large, P1 | NM_001003 | 0.47* | 0.51* |
| Ribosomal protein S27 | NM_001030 | 0.57* | 0.42* |
| Protein synthesis | |||
| Translation elongation factor 2 | NM_001961 | 0.40* | 0.38* |
| Translation initiation factor 5A | NM_001970 | 0.49* | 0.47* |
| Translation initiation factor 4A, isoform 1 | NM_001416 | 0.28* | 0.40* |
| Translation initiation factor 4 gamma, 2 | NM_001418 | 0.26* | 0.76 |
| Translation initiation factor 4B | NM_001417 | 0.49* | 0.33* |
| Translation initiation factor 4A, isoform 2 | NM_001967 | 0.32* | 0.46* |
| Translation initiation factor 3, subunit 7 | NM_003753 | 0.30* | 0.41* |
| Translation initiation factor 3, subunit 4 | NM_003755 | 0.15* | 0.56* |
| Translation initiation factor 3, subunit 2 | NM_003757 | 0.21* | 0.28* |
| Nascent-polypeptide-associated complex alpha | NM_005594 | 0.50* | 0.50* |
| Translation initiation factor 3, subunit 6 | NM_001568 | 0.39* | 0.64 |
| Mitochondria | |||
| H+ transporting, V0 subunit c | NM_001694 | 0.80 | 0.44* |
| H+ transporting, mitochondrial F1 complex, _ | NM_004046 | 0.37* | 0.59* |
| H+ transporting, mitochondrial F1 complex, _ | NM_001686 | 0.37* | 0.54* |
| H+ transporting, mitochondrial F1 complex, _1 | NM_005174 | 0.64 | 0.26* |
| H+ transporting, mitochondrial F0 complex, c | NM_001689 | 2.00 | 0.57* |
| H+ transporting, mitochondrial F0 complex, F6 | NM_001685 | 0.34* | 0.47* |
| H+ transporting, mitochondrial F0 complex, g | NM_006476 | 0.74 | 0.52* |
| H+ transporting, mitochondrial F1 complex, O | NM_001697 | 1.12 | 0.43* |
| H+ transporting, mitochondrial F0 complex, e | NM_007100 | 0.39* | 0.64 |
| Ubiquinol-cytochrome c reductase core protein II | NM_003366 | 0.59* | 0.26* |
| Ubiquinol-cytochrome c reductase hinge protein | NM_006004 | 0.32* | 0.48* |
| Ubiquinol-cytochrome c reductase binding prot. | NM_006294 | 0.58* | 1.00 |
| Translocase of mitochondrial membr. 8 hom. B | NM_012459 | 0.52* | 0.34* |
| Cytochrome c oxidase subunit Vic | NM_004374 | 0.60* | 0.35* |
| Mitochondrial adenine translocator, member 6 | J03592 | 0.29* | 0.33* |
| Cytochrome c oxidase subunit viia polypeptide 2 | NM_001865 | 0.14* | 0.41* |
| Ubiquinol-cytochrome c reductase binding prot. | NM_006294 | 0.58* | 1.00 |
| Mitochondrial adenine nucleotide translocator, 5 | NM_001152 | 1.42 | 0.21* |
| Cytochrome c oxidase subunit viia, 2 like | NM_004718 | 0.34* | 0.40* |
| Cytochrome c oxidase subunit Vb | NM_001862 | 0.34* | NC |
| NADH-coenzyme Q reductase | NM_004552 | 0.42* | 0.42* |
| Cytochrome c oxidase subunit viib | NM_001866 | 0.42* | 0.46* |
| Mitochondrial adenine nucleotide translocator, 5 | NM_001152 | 1.42 | 0.21* |
| Mitochondrial phosphate carrier, member 3 | NM_002635 | 0.32* | 0.76* |
| Mitochondrial carrier homologue 2 | NM_014342 | 0.44* | 0.38* |
| Mitochondrial solute carrier | NM_016612 | 0.45* | 0.39* |
| Cytochrome c oxidase subunit viic | NM_001867 | 0.60* | 0.42* |
| NADH dehydrogenase 1 beta subcomplex, 9 | NM_005005 | 0.55* | 0.22* |
| Apoptosis | |||
| Death-associated protein kinase 3 | NM_001348 | 0.45* | 0.83 |
| Apoptosis inhibitor 5 | NM_006595 | 0.82 | 0.23* |
| FK506 binding protein 5 | NM_004117 | 0.56* | 0.39* |
| Transcription and nuclear | |||
| Purine-rich element binding protein B | NM_033224 | 0.33* | 0.41* |
| POU domain, class 3, transcription factor 1 | NM_002699 | 0.29* | 0.32* |
| Corepressor/HDAC3 complex subunit | NM_024665 | 0.46* | 0.42* |
| CCAAT/enhancer binding protein, beta | NM_005194 | 0.48* | 0.30* |
| Cardiac-specific homeo box | NM_004387 | 0.57* | 0.36* |
| Paired-like homeodomain transcription factor 1 | NM_001288 | 0.43* | 0.47* |
| RING1 and YY1 binding protein | NM_001762 | 0.44* | 0.39* |
| Nuclease sensitive element binding protein 1 | NM_012234 | 0.50* | 0.02* |
| Sex-determining region Y-box 4 | NM_003107 | 0.35* | 0.43* |
| Sex-determining region Y-box 11 | NM_003108 | 0.37* | 0.32* |
| Interleukin enhancer binding factor 2, 45 kDa | NM_004515 | 0.50* | 0.44* |
| High-mobility group protein 1 | NM_002128 | 0.75 | 0.48* |
| High-mobility group protein 17 | NM_002266 | 0.54* | 0.15* |
| H2A histone family, member Z | NM_002106 | 0.54* | 1.15 |
| Sin3-associated polypeptide, 18 kDa | NM_005870 | 0.42* | 0.45* |
| Single-stranded DNA binding protein | NM_003143 | 0.17* | 0.25* |
| Nucleophosmin | NM_018285 | 0.50* | 0.48* |
| Nucleolar protein family A, member 3 | NM_003860 | 0.26* | 0.56* |
| Thymosin, beta 4 | NM_021109 | 0.32* | 0.91 |
| Thymosin, beta 10 | NM_004559 | 0.41* | 0.60* |
| SP140 nuclear body protein | M92381 | 0.45* | 0.49* |
| Ornithine decarboxylase 1 | NM_002539 | 0.54* | 0.40* |
| Proteasome degradation pathway | |||
| Ubiquitin-conjugating enzyme E2N | NM_003348 | 0.52 | 0.16* |
| Ubiquitin-conjugating enzyme E2M | NM_003969 | 0.45* | 0.41* |
| Ubiquitin-like 5 | NM_024292 | 0.37* | 0.42* |
| Ubiquitin A-52 residue ribosomal protein fusion | NM_003333 | 0.47* | 0.44* |
| Ring-box 1 | NM_014248 | 0.15* | 0.32* |
| Proteasome inhibitor subunit 1 (PI31) | NM_006814 | 0.56* | 0.28* |
| Proteasome subunit, beta type, 1 | NM_002793 | 0.41* | 0.33* |
| Proteasome subunit, alpha type, 4 | D00763 | 0.35* | 0.36* |
| Proteasome 26S subunit, non-atpase, 3 | NM_002809 | 0.38* | 0.55* |
| RNA and splicing | |||
| Heterogeneous nuclear ribonucleoprotein A1 | NM_002136 | 0.76 | 0.37* |
| Heterogeneous nuclear ribonucleoprotein C | NM_004500 | 0.70 | 0.40* |
| Splicing factor, arginine/serine-rich 3 | NM_003017 | 0.43* | 0.53* |
| Small nuclear ribonucleoprotein polypeptide G | NM_003096 | 0.31* | 0.34* |
| Small nuclear ribonucleoprotein polypeptide F | NM_003095 | 0.43* | 0.43* |
| Small nuclear ribonucleoprotein D2 | NM_004597 | 0.35* | 0.40* |
| Small nuclear ribonucleoprotein, B and B1 | NM_003091 | 0.43* | 0.49* |
| RNA polymerase II polypeptide G | NM_002696 | 0.49* | 0.43* |
| RNA binding motif protein, X chromosome | NM_002139 | 0.73 | 0.38* |
| Poly(A) binding protein, cytoplasmic 1 | NM_002568 | 0.49* | 0.39* |
| Poly(rc) binding protein 1 | NM_006196 | 0.77 | 0.30* |
| Poly(rc) binding protein 2 | NM_005016 | 0.47* | 0.42* |
| Lysyl-trna synthetase | NM_005548 | 0.56* | 0.41* |
| Asparaginyl-trna synthetase | NM_004539 | 0.59* | 0.49* |
| 5′-3′ exoribonuclease 2 | NM_012255 | 0.72 | 0.10* |
| Signal transduction | |||
| Adaptor-related protein complex 2, sigma 1 | AJ010148 | 0.35* | 0.43* |
| Adaptor-related protein complex 2, mu 1 | NM_004068 | 0.47* | 0.38* |
| ADP-ribosylation factor 1 | M36340 | 0.34* | 0.55* |
| ADP-ribosylation factor-like 2 | NM_001667 | 0.89 | 0.47* |
| fms-related tyrosine kinase 3 ligand | U04806 | 0.73 | 0.46* |
| Rho GDP dissociation inhibitor (GDI) beta | L20688 | 0.63 | 0.43* |
| ras-like protein VTS58635 | NM_033315 | 0.80 | 0.38* |
| G protein-coupled receptor 6 | NM_005284 | 0.67 | 0.48* |
| Ion channels | |||
| Voltage-dependent anion channel 2 | NM_003375 | 0.49* | 0.57* |
| Chloride intracellular channel 1 | NM_001288 | 0.43* | 0.47* |
| Structural proteins | |||
| Tubulin, alpha, ubiquitous | K00558 | 0.69 | 0.60* |
| Tubulin, alpha 3 | NM_006009 | 0.68 | 0.42* |
| Tubulin, alpha | K00558 | 0.72 | 0.48* |
| Actin, beta | X00351 | 0.47* | 1.44 |
| Actin-related protein 2/3 complex, subunit 2 | NM_005731 | 0.23* | 0.51* |
| Actin-related protein 2/3 complex, subunit 3 | NM_005719 | 0.45* | 0.40* |
| Profilin 1 | NM_005022 | 0.44* | 0.53* |
| Cofilin 1 (nonmuscle) | NM_005507 | 0.43* | 0.72 |
| Sentrin | NM_003352 | 0.78 | 0.23* |
| Integral membrane protein 2B | NM_021999 | 0.52* | 0.22* |
| Dynein, cytoplasmic, light polypeptide | NM_003746 | 0.25* | 0.80 |
| Kinesin family member 3A | NM_007054 | 0.47* | 0.07* |
| Integrin, alpha 11 | NM_012211 | 0.36* | 0.10* |
| Vesicle-associated membrane protein 8 | NM_003761 | 0.31* | 0.65 |
| Lectin, galactoside-binding, soluble, 1 | NM_002305 | 0.43* | 0.46* |
| Metabolism | |||
| Phytanoyl-coa hydroxylase | NM_006214 | 0.86 | 0.47* |
| Aldolase A, fructose-bisphosphate | NM_000034 | 0.71 | 0.41* |
| UDP-glucose ceramide glucosyltransferase | NM_003358 | 0.50* | 0.26* |
| Enolase 1, (alpha) | NM_001428 | 0.42* | 0.38* |
| Aldo-keto reductase family 1, member B1 | NM_001628 | 0.30* | 0.30* |
| Hydroxyacyl-coenzyme A dehydrogenase, alpha | NM_000182 | 0.73 | 0.47* |
| Enolase 1, (alpha) | NM_005945 | 0.49* | 0.48* |
| Arylsulfatase A | NM_000487 | 0.58* | 0.27* |
| Tyrosine 3-monooxygenase, epsilon polypeptide | NM_006761 | 0.24* | 0.63 |
| Phosphoglycerate mutase 1 | NM_002629 | 0.67 | 0.35* |
| Mannosidase, alpha, class 1B, member 1 | NM_007230 | 0.53* | 0.31* |
| Tyrosine 3-monooxygenase, zeta polypeptide | M86400 | 0.53* | 0.46* |
| Cell cycle and mitosis | |||
| Cyclin-dependent kinase 4 | M14505 | 0.42* | 0.45* |
| CDC10 cell division cycle 10 homologue | S72008 | 0.56* | 0.20* |
| S-phase kinase-associated protein 1A (p19A) | NM_006930 | 0.12* | 0.35* |
| Mitotic centromere-associated kinesin | NM_006845 | 0.60* | 0.02* |
| Karyopherin alpha 2 | NM_002266 | 0.54* | 0.15* |
| Oxidation | |||
| Thioredoxin | NM_003329 | 0.36* | 0.48* |
| Peroxiredoxin 4 | NM_006406 | 0.56 | 2.45† |
| Superoxide dismutase 1, soluble | K00065 | 0.27* | 0.38* |
| 15 kDa selenoprotein | NM_004261 | 0.28* | 0.35* |
| Microsomal glutathione S-transferase 1 | J03746 | 0.55* | 0.37* |
| Miscellaneous | |||
| MHC, class I, C | M11886 | 0.77 | 0.38* |
| Adenylate kinase 2 | NM_001625 | 0.36* | 0.21* |
| Neuropathy target esterase | NM_006702 | 0.70 | 0.03* |
| Roundabout homologue 4 | NM_019055 | 0.38* | 0.29* |
| Hypermethylated in cancer 1 | NM_006497 | 0.40* | 0.41* |
| 6.2 kDa protein | NM_019059 | 0.47* | 0.22* |
| Ferritin, heavy polypeptide 1 | NM_002032 | 0.73 | 0.49* |
| Nonmetastatic cells 1, protein (NM23A) | X17620 | 0.50* | 0.35* |
| Anti-Mullerian hormone | NM_000479 | 0.40* | 0.24* |
| 16.7 kDa protein | NM_016139 | 0.48* | 0.65 |
| Proteoglycan 1, secretory granule | NM_002727 | 0.64 | 0.38* |
| Peptidyl-prolyl cis/trans isomerase NIMA-inter. 1 | U49070 | 0.57* | 0.50* |
| Cystatin F (leukocystatin) | NM_003650 | 0.61 | 0.36* |
| Glucose regulated protein, 58 kda | NM_005313 | 0.16* | 0.34* |
| Signal sequence receptor, beta | NM_003145 | 0.49* | 0.44* |
| Tumor protein, translationaly controlled 1 | NM_003295 | 0.24* | 0.70 |
| Deleted in split-hand/split-foot 1 region | NM_006304 | 0.59* | 0.30* |
| Ubiquitously expressed transcript | NM_004182 | 0.42* | 0.08* |
| BCR protein, uterine leiomyoma, 1 | NM_018648 | 0.71 | 0.48* |
| Multiple endocrine neoplasia I | NM_000244 | 0.59* | 0.37* |
| Small inducible cytokine A5 (RANTES) | M21121 | NC | 0.37* |
| Enhancer of rudimentary homolog | NM_004450 | 0.75 | 0.50* |
| Pyrophosphatase | NM_031210 | 0.19* | 0.39* |
| S100 calcium binding protein P | NM_005980 | 0.37* | 0.46* |
| Overexpressed breast tumor protein | NM_013397 | 0.21* | 0.47* |
| Ferritin, light polypeptide | NM_000146 | 0.60 | 0.44* |
| Adenomatous polyposis coli-like | NM_005883 | 1.24 | 0.15* |
| Proteolipid protein 2 (colonic epithelium-enriched) | NM_002668 | 0.80 | 0.46* |
| Topoisomerase (DNA) II binding protein | NM_007027 | 0.62 | 0.09* |
| Lsm3 protein | NM_014463 | 0.40* | 0.42* |
| ATPase, Ca++ transporting, plasma membrane 2 | L20977 | 0.43* | 0.65 |
| Stomatin (EPB72)-like 2 | NM_013442 | 0.66 | 0.43* |
| Signal recognition particle 72 kda | NM_001222 | 0.42* | 0.64 |
| Glutamic acid-rich protein-like 3 | NM_031286 | 0.86 | 0.12* |
| Singed-like (fascin homologue, sea urchin) | NM_003088 | 0.73 | 0.43* |
| PAP-1 binding protein | NM_031288 | 1.10 | 0.13* |
| SET translocation (myeloid leukemia-associated) | NM_003011 | 0.94 | 0.54* |
| Putative proteins | |||
| Hypothetical protein MGC2803 | NM_024038 | 0.27* | 0.62 |
| Hypothetical protein HSPC194 | NM_016462 | 0.47* | 0.39* |
| KIAA0101 gene product | NM_014736 | 0.24* | 0.40* |
| Hypothetical protein MGC5499 | NM_024055 | 0.28* | 0.23* |
| Chromosome 2 open reading frame 6 | NM_018221 | 0.28* | 0.22* |
| Hypothetical protein FLJ10815 | NM_018231 | 0.43* | 0.21* |
| JM5 protein | NM_007075 | 0.69 | 0.10* |
| Chromosome 15 open reading frame 12 | NM_018285 | 0.50* | 0.48* |
| GNAS complex locus | M14631 | 0.56* | 0.49* |
| Hypothetical protein DC50 | NM_031210 | 0.19* | 0.39* |
| Chromosome 11 open reading frame 10 | AF086763 | 0.54* | 0.53* |
| Vesicle-associated membrane protein 8 | NM_003761 | 0.31* | 0.65 |
| Peptidyl-prolyl cis/trans isomerase NIMA-inter. 1 | U49070 | 0.57* | 0.50* |
| Chaperons and heat shock | |||
| Hsp40 homologue, subfamily A, member 1 | NM_001539 | 0.62 | 0.25* |
| Heat shock 70 kDa protein 8 | Y00371 | 0.68 | 0.47* |
| Heat shock 90 kDa protein 1, alpha | X07270 | 0.23* | 0.47* |
| Chaperonin containing TCP1, subunit 2 (beta) | NM_006431 | 0.43* | 0.50* |
| Hsp70 interacting protein | NM_003932 | 0.34* | 0.49* |
| Chaperonin containing TCP1, subunit 3 | NM_005998 | 0.52* | 0.36* |
| Chaperonin containing TCP1, subunit 6A (zeta 1) | NM_001762 | 0.44* | 0.39* |
| Membrane receptors | |||
| Growth hormone secretagogue receptor | NM_004122 | 0.65 | 0.42* |
| Somatostatin receptor 3 | NM_001051 | 0.48* | 0.61 |
| Up-regulated proteins/genes | |||
| Ribosomal protein L35 | NM_007209 | NC | 7.13† |
| Activated RNA polymerase II cofactor 4 | NM_006713 | 5.29† | 9.51† |
| Coronin, actin binding protein, 1A | NM_007074 | NC | 4.27† |
| Myeloid/lymphoid or mixed-lineage leukemia 4 | NM_014727 | 2.76† | 1.82 |
| Hypothetical protein FLJ10707 | NM_018187 | 5.59† | 3.99† |
| Interleukin 8 | Y00787 | 2.02† | 11.07† |
| Serine/threonine kinase 3 | NM_006281 | 2.41† | 7.11† |
| UDP glycosyltransferase 2 family, B15 | NM_001076 | 2.94† | 1.39 |
NB4 cells (0.5 × 106/mL) were treated for 4 hours with medium alone and medium containing 0.2 μM ST1926 or 1 μM CD437. At the end of the incubation, total RNA was extracted and the corresponding poly(A+) fraction isolated. Each RNA preparation was processed in parallel and corresponded to a pool of 3 separate culture flasks per each experimental situation. 33P-labeled single-strand cDNA probes from medium-, ST1926-, and CD437-treated RNAs were synthesized and hybridized to 12K human ATLAS plastic filters in parallel. Filters were washed and exposed for 1 week prior to determination of the intensity of each spot on the array with the use of the ATLAS 2.7 imaging software. With this tool, the average intensity of each couple of spots corresponding to a single gene was automatically subtracted from the local background and normalized on the basis of the average intensity of all the arrayed spots. The results are expressed as the ratio between the intensity of each spot in the ST1926 and CD437 arrays and the corresponding spot in the control array (hybridized with RNA extracted from medium treated cells). The Genbank accession number for each gene is indicated in parentheses and the genes are classified according to their function or subcellular localization. NC indicates not calculated.
Equal or below the threshold value of 0.4 set for a significant down-regulation of gene expression. † Equal or above the threshold value of 2.0 set for a significant up-regulation of gene expression.
ST1926 and CD437 activate p38 and JNK but the 2 MAP kinases are not involved in the RRM-dependent process of apoptosis
Phosphorylation and activation of p38 and JNK are purported to play a role in the CD437-dependent cytotoxicity.2,3,11,22 To define the role of MAP kinases in NB4 cells, we evaluated the effects of CD437and ST1926 at concentrations of 0.2 μM and 1 μM. The former concentration is apoptotic in the case of ST1926 and inactive in the case of CD437, whereas the latter one is fully apoptic in both cases. Figure 6A demonstrates that ERK is constitutively expressed in its phosphorylated form. Treatment of the leukemic blasts with CD437 or ST1926, at the selected concentrations, does not affect the total amounts or the phosphorylation levels of ERK. Inhibition of extracellular signal–regulated kinase (ERK) phosphorylation by U0126 does not change the level of NB4 apoptosis or cytotoxicity associated with ST1926 or CD437 treatment (Figure 6D). This indicates that ERK is not involved in RRM-induced apoptosis of NB4 cells. As shown in Figure 6B, phosphorylation of p38 is increased only when ST1926 and CD437 are present in the growth medium at concentrations of 1 μM. Hence, treatment of NB4 cells with a fully apoptotic concentration of ST1926 (0.2 μM) is not associated with elevated phosphorylation of p38. The Jun-phosphorylating activity of JNK is increased by the highest concentration of CD437 considered and is significantly affected by ST1926 only at 1 μM and following treatment for 2 hours (Figure 6C). ST1926 and CD437 do not have any detectable effect on the amounts of p38 or JNK (data not shown) proteins expressed in NB4 cells. Taken together, these data indicate that ST1926 is less potent than CD437 in phosphorylating p38 and JNK. In addition, the results do not support a direct relationship between phosphorylation of the 2 MAP kinases and RRM-dependent apoptosis. This is also consistent with the fact that coincubation of NB4 cells with p38 or JNK inhibitors, like PD169366 and SP600125, at concentrations specifically inhibiting the phosphorylation of the two proteins (Pisano et al24 and data not shown), has a modest impact on the apoptosis and the cytotoxicity of ST1926 and CD437 (Figure 6E-F).
Influence of ST1926 and CD437 on the phosphorylation of MAP kinases and effect of MAP kinase inhibitors on RRM-dependent apoptosis. NB4 cells were treated for the indicated amount of time with vehicle and the indicated concentrations of ST1926 and CD437. Total cellular extracts were subjected to Western blot analysis using polyclonal antibodies recognizing ERK (A) and p38 (B). In the case of panel C, we measured the level of JNK kinase activity on JNK immunoprecipitates using c-Jun as a substrate. Equivalent amounts of immunoprecipitates were electrophoresed on SDS-PAGE, blotted on nitrocellulose, and challenged with antibodies recognizing the phosphorylated form of c-JUN (serine/63) or JNK. The blots shown are representative of at least 2 independent experiments. NB4 cells were treated for 6 hours with vehicle (DMSO), ST1926 (1 μM), CD437 (1 μM), the indicated concentrations of the ERK inhibitor, U0126, and the indicated combinations (D), the p38 inhibitor, PD169316, and the indicated combinations (E), or the JNK inhibitor, SP600125, and the indicated combinations (F). Aliquots of the extracts were used for the determination of the apoptotic index (left-most panels) and the number of viable cells (rightmost panels). The numbers above the columns indicate percentages of viable cells. Less than 100 = below the limit of detection of the assay. Results are the mean ± SD of 3 separate culture dishes and are representative of at least 2 independent experiments. *Significantly higher or lower than the corresponding vehicle-treated group (P < .01 according to the Student t test).
Influence of ST1926 and CD437 on the phosphorylation of MAP kinases and effect of MAP kinase inhibitors on RRM-dependent apoptosis. NB4 cells were treated for the indicated amount of time with vehicle and the indicated concentrations of ST1926 and CD437. Total cellular extracts were subjected to Western blot analysis using polyclonal antibodies recognizing ERK (A) and p38 (B). In the case of panel C, we measured the level of JNK kinase activity on JNK immunoprecipitates using c-Jun as a substrate. Equivalent amounts of immunoprecipitates were electrophoresed on SDS-PAGE, blotted on nitrocellulose, and challenged with antibodies recognizing the phosphorylated form of c-JUN (serine/63) or JNK. The blots shown are representative of at least 2 independent experiments. NB4 cells were treated for 6 hours with vehicle (DMSO), ST1926 (1 μM), CD437 (1 μM), the indicated concentrations of the ERK inhibitor, U0126, and the indicated combinations (D), the p38 inhibitor, PD169316, and the indicated combinations (E), or the JNK inhibitor, SP600125, and the indicated combinations (F). Aliquots of the extracts were used for the determination of the apoptotic index (left-most panels) and the number of viable cells (rightmost panels). The numbers above the columns indicate percentages of viable cells. Less than 100 = below the limit of detection of the assay. Results are the mean ± SD of 3 separate culture dishes and are representative of at least 2 independent experiments. *Significantly higher or lower than the corresponding vehicle-treated group (P < .01 according to the Student t test).
ST1926 treatment of cells results in rapid accumulation of intracellular calcium
Accumulation of calcium in the cytosol induces PCD in certain cell types.29,30 Figure 7A demonstrates that apoptotic concentrations of ST1926 induce a rapid accumulation of calcium in NB4 cells preloaded with the cytosolic calcium indicator FURA-2. The increase of intracellular calcium triggered by ST1926 is immediate, long lasting, and dose dependent. Typically, the cytosolic concentration of calcium in ST1926-callenged NB4 cells rises from 20 nM to 600 to 700 nM, as determined by calibrating the FURA-2 fluorescence signals.28 ST1926 is more powerful than CD437 in causing calcium mobilization, which indicates that the parameter correlates with the apoptotic potency of the 2 compounds. The correlation is evident across the whole series of ST1926 congeners. As illustrated in Figure 7B, ST1879, a weaker apoptotic molecule than ST1926 or CD437, is also less active in terms of calcium mobilization. The two nonapoptotic RRMs, ST2188 and ST2062, do not cause detectable increases in cytosolic calcium. Figure 7C demonstrates that the putative antagonist ST2065 suppresses the calcium-mobilizing effect of ST1926. Interestingly, treatment of NB4 cells with ST2065 is associated with a drop in the baseline levels of cytosolic calcium. As shown in Figure 7D, doxorubicin, etoposide, and the synthetic retinoid, fenretinide, compounds that induce apoptosis in NB4 cells at the concentrations considered,2 are devoid of calcium-mobilizing activity. The protein kinase C inhibitor staurosporine is a weak calcium-mobilizing agent.
Effect of RRMs alone or in combination and of known apoptotic agents on the levels of intracellular calcium. Following preloading with the calcium fluorescent indicator FURA-2, NB4 cells (1 × 106/mL) were resuspended in calcium-containing phosphate-buffered saline (PBS) with calcium chloride at a concentration of 1.26 mM. Cells were placed in a cuvette under stirring at 37°C and changes in fluorescence were measured continuously with the use of a spectrophotofluorometer. The arrows indicate the addition of the various compounds at the concentrations shown (A, B). In panel C, ST1926 was used at a concentration of 1 μM, whereas ST2065 was added at a concentration of 100 μM. The left arrow indicates the addition of ST2065, whereas the right arrow indicates the addition of ST1926. In panel D, the concentrations considered are: ST1926, 1 μM; staurosporine, 1 μM; doxorubicin, 1 μM; etoposide, 1 μM; and fenretinide, 10 μM. Each tracing is representative of at least 3 independent experiments run in triplicate.
Effect of RRMs alone or in combination and of known apoptotic agents on the levels of intracellular calcium. Following preloading with the calcium fluorescent indicator FURA-2, NB4 cells (1 × 106/mL) were resuspended in calcium-containing phosphate-buffered saline (PBS) with calcium chloride at a concentration of 1.26 mM. Cells were placed in a cuvette under stirring at 37°C and changes in fluorescence were measured continuously with the use of a spectrophotofluorometer. The arrows indicate the addition of the various compounds at the concentrations shown (A, B). In panel C, ST1926 was used at a concentration of 1 μM, whereas ST2065 was added at a concentration of 100 μM. The left arrow indicates the addition of ST2065, whereas the right arrow indicates the addition of ST1926. In panel D, the concentrations considered are: ST1926, 1 μM; staurosporine, 1 μM; doxorubicin, 1 μM; etoposide, 1 μM; and fenretinide, 10 μM. Each tracing is representative of at least 3 independent experiments run in triplicate.
Given the correlation between the calcium-mobilizing and the apoptotic activities of the RRM series, we investigated whether calcium mobilization is different in cells with different levels of sensitivity to the apoptotic action of ST1926. As shown in Figure 8A, HL-60 and KG1 cells, which are sensitive to the apoptogenic action of ST1926 (Figure 2), respond to the compound with an increase in cytosolic calcium. In these cell lines, the elevation of intracellular calcium has the same kinetic profile and is of the same order of magnitude as that observed in NB4 cells. Figure 8B demonstrates that the U937 cell line, which is refractory to ST1926-induced apoptosis, is also resistant to the calcium-mobilizing activity of the RRM. On the other hand, the two ST1926-resistant cell lines, NB4.437r and Kasumi, show a prompt elevation of cytosolic calcium levels upon challenge with the compound. Figure 8C demonstrates that there is no significant difference between the dose-dependent accumulation of cytosolic calcium in the parental NB4 relative to the RRM-resistant NB4.437r cell line. Thus, elevation of cytosolic calcium by ST1926 and congeners is necessary but not sufficient to trigger apoptosis.
Effect of ST1926 on the levels of intracellular calcium in CD437-resistant NB4.437r blasts and other myeloid cell lines. Following prelodaing with the calcium fluorescent indicator FURA-2, NB4, NB4.437r, HL-60 KG1 (A) as well as Kasumi and U937 (B) cells (1 × 106/mL) were resuspended in PBS with calcium chloride at a concentr ation of 1.26 mM. Cells were placed in a cuvette under stirring at 37°C and changes in fluorescence were measured continuously with the use of a spectrophotofluorometer. The concentration of ST1926 used in the experiments illustrated in (A) and (B) is 1 μM. The dose-response curve for the increase of intracellular calcium in NB4 and NB4.437r cells is shown in panel C. Each tracing is representative of at least 2 independent experiments run in triplicate. *Significantly higher than the vehicle-treated group (P < .01 according to the Student t test).
Effect of ST1926 on the levels of intracellular calcium in CD437-resistant NB4.437r blasts and other myeloid cell lines. Following prelodaing with the calcium fluorescent indicator FURA-2, NB4, NB4.437r, HL-60 KG1 (A) as well as Kasumi and U937 (B) cells (1 × 106/mL) were resuspended in PBS with calcium chloride at a concentr ation of 1.26 mM. Cells were placed in a cuvette under stirring at 37°C and changes in fluorescence were measured continuously with the use of a spectrophotofluorometer. The concentration of ST1926 used in the experiments illustrated in (A) and (B) is 1 μM. The dose-response curve for the increase of intracellular calcium in NB4 and NB4.437r cells is shown in panel C. Each tracing is representative of at least 2 independent experiments run in triplicate. *Significantly higher than the vehicle-treated group (P < .01 according to the Student t test).
Cytosolic accumulation of calcium is predominantly the consequence of a block in the uptake of the cation by the mitochondria
To define the origin of calcium accumulation in the cytosol of ST1926-treated NB4 cells, first we entertained the possibility that the cation come from the extracellular compartment. We added nickel salts to the growth medium, as this blocks the influx of extracellular calcium through plasma membrane calcium channels.31 However, nickel salts do not affect the cytosolic calcium rise afforded by ST1926 (Figure 9A). Similarly, preincubation of NB4 cells with verapamil, a calcium channel blocker,32 does not have any significant effect on ST1926-induced calcium mobilization (Figure 9B). Pertussis toxin, a powerful inhibitor of G-protein–dependent, membrane-receptor–coupled calcium channels,33 is equally ineffective (data not shown). More important, resuspension of NB4 cells in calcium-free medium and subsequent challenge with ST1926 results in a level of cytosolic calcium accumulation that is virtually identical to that observed in cells kept in calcium-containing medium (Figure 9C-D). Surprisingly, preincubation of NB4 cells with nicardipine, a calcium channel blocker of the dihydropyridine type, reduces the ST1926-dependent effect on calcium dose dependently. Reduction is evident regardless of the presence of calcium in the growth medium. Complete inhibition of intracellular calcium accumulation is observed at relatively high concentrations of nicardipine (100 μM; Figure 9E). This along with the fact that nicardipine is active in the absence of extracellular calcium indicates that the dihydropyridine acts on an as-yet-unidentified intracellular target. Similar effects are observed upon preincubation of NB4 cells with two other dihydropyridines, like nitrendipine and nimodipine (Figure 9F).
Effect of ST1926, nicardipine, and the combination of the 2 agrents on the levels of intracellular calcium in NB4 cells incubated in the presence or absence of calcium in the medium. Following prelodaing with the calcium fluorescent indicator FURA-2, NB4 cells (1 × 106/mL) were resuspended in calcium-containing (+Ca++) or calcium-free (-Ca++) PBS. Cells were placed in a cuvette under stirring at 37°C and changes in fluorescence were measured continuously with the use of a spectrophotofluorometer. NB4 cells were preincubated with 5 mM nickel chloride (Ni++; A); 100 μM verapamil (B); the indicated concentrations of nicardipine (C-E); or nitrendipine, nicardipine, and nimodipine (F) prior to addition of ST1926 at the concentration of 1 μM. The left arrow indicates the addition of Ni++, verapamil, or nicardipine, whereas the right arrow indicates the addition of ST1926. Each tracing is representative of at least 2 independent experiments run in triplicate. In panel F, the results are expressed as the mean ± SD of 3 replicate cultures. °Significantly higher than the vehicle-treated group (P < .01 according to the Student t test). *Significantly lower than the ST1926-treated group (P < .01 according to the Student t test).
Effect of ST1926, nicardipine, and the combination of the 2 agrents on the levels of intracellular calcium in NB4 cells incubated in the presence or absence of calcium in the medium. Following prelodaing with the calcium fluorescent indicator FURA-2, NB4 cells (1 × 106/mL) were resuspended in calcium-containing (+Ca++) or calcium-free (-Ca++) PBS. Cells were placed in a cuvette under stirring at 37°C and changes in fluorescence were measured continuously with the use of a spectrophotofluorometer. NB4 cells were preincubated with 5 mM nickel chloride (Ni++; A); 100 μM verapamil (B); the indicated concentrations of nicardipine (C-E); or nitrendipine, nicardipine, and nimodipine (F) prior to addition of ST1926 at the concentration of 1 μM. The left arrow indicates the addition of Ni++, verapamil, or nicardipine, whereas the right arrow indicates the addition of ST1926. Each tracing is representative of at least 2 independent experiments run in triplicate. In panel F, the results are expressed as the mean ± SD of 3 replicate cultures. °Significantly higher than the vehicle-treated group (P < .01 according to the Student t test). *Significantly lower than the ST1926-treated group (P < .01 according to the Student t test).
The endoplasmic reticulum (ER) is an important intracellular calcium store.29 To evaluate whether the rise of cytosolic calcium by ST1926 results from an inhibition of the cation uptake by the ER, cells were pretreated with thapsygargin or TBHQ, two effective sarco-endoplasmic reticulum ATPase (SERCA) inhibitors.34,35 Incubation of NB4 cells with different concentrations of the two inhibitors has no significant effect on calcium mobilization by ST1926 (data not shown). Notice that the concentrations of thapsygargin and TBHQ considered effectively block the ER calcium pump in NB4 cells.36 The results obtained suggest that the ER is not involved in calcium mobilization by ST1926.
Uptake by the mitochondria is another mechanism controlling the intracellular levels of calcium.37 The process requires oxidative phosphorylation, an intact mitochondrial membrane potential, and is energy-dependent.38,39 Antimycin A is a selective inhibitor of the mitochondrial complex III and an uncoupler of oxidative phosphorylation, particularly when the compound is used in combination with oligomycin, an ATP synthase inhibitor.40 As expected, pretreatment of NB4 cells with antimycin A/oligomycin (Anti/Oligo) results in an immediate calcium transient, which leads to an increased steady-state level of cytosolic calcium (Figure 10A). The Anti/Oligo combination reduces the ST1926-induced cytosolic accumulation of calcium by approximately 30% ± 2% (n = 3). As shown in Figure 10B, a similar effect is observed when the Anti/Oligo couple is substituted by FCCP, a different oxidative phosphorylation uncoupler, acting by dissipating the electrogenic potential of the mitochondrial membrane.41,42 In this case, a 29% ± 1% and a 51% ± 2% (n = 3) inhibition of intracellular calcium accumulation are observed when FCCP is used at concentrations of 5 μM and 10 μM, respectively. Our results are consistent with the fact that at least part of the cytosolic calcium rise afforded by ST1926 results from an inhibition of the mitochondrial reuptake of the cation. As illustrated in Figure 10C, cyclosporin A, a blocker of the mitochondrial transition pore,40 has only a modest effect on the cytosolic calcium rise caused by ST1926 in NB4 cells. Interestingly, following 3 hours of coincubation, cyclosporin A (5 μM) reduces the apoptotic index of NB4 cells treated with 1 μM ST1926 by 60% ± 1% (P < .01 according to the Student t test, n = 3) or with 1 μM CD437 by 54% ± 12% (P < .01 according to the Student t test, n = 3). This suggests that opening of the transition pore lays downstream of CD437 and possibly ST1926 calcium mobilization.
Effect of antimycin/oligomycin, FCCP, and cyclosporine A on the calcium-mobilizing activity of ST1926 in NB4 cells. Following preloading with the calcium fluorescent indicator FURA-2, NB4 cells (1 × 106/mL) were resuspended in PBS with calcium chloride at a concentr ation of 1.26 mM. Cells were placed in a cuvette under stirring at 37°C and changes in fluorescence were measured continuously with the use of a spectrophotofluorometer. Prior to addition of ST1926 at the concentration of 1 μM, NB4 cells were preincubated with vehicle (black tracing) or the following stimuli (gray tracings): a mixture of antimycin (1 μg/mL) and oligomycin (1 μg/mL) (Anti/Oligo; A), FCCP at the indicated concentrations (B), or cyclosporin A at the indicated concentrations (C). Each tracing is representative of at least 2 independent experiments run in triplicate.
Effect of antimycin/oligomycin, FCCP, and cyclosporine A on the calcium-mobilizing activity of ST1926 in NB4 cells. Following preloading with the calcium fluorescent indicator FURA-2, NB4 cells (1 × 106/mL) were resuspended in PBS with calcium chloride at a concentr ation of 1.26 mM. Cells were placed in a cuvette under stirring at 37°C and changes in fluorescence were measured continuously with the use of a spectrophotofluorometer. Prior to addition of ST1926 at the concentration of 1 μM, NB4 cells were preincubated with vehicle (black tracing) or the following stimuli (gray tracings): a mixture of antimycin (1 μg/mL) and oligomycin (1 μg/mL) (Anti/Oligo; A), FCCP at the indicated concentrations (B), or cyclosporin A at the indicated concentrations (C). Each tracing is representative of at least 2 independent experiments run in triplicate.
Dihydropyridines and the intracellular calcium chelator BAPTA inhibit the process of apoptosis set in motion by ST1926 and CD437
We evaluated whether high concentrations of dihydropyridines had any effect on the process of apoptosis triggered by ST1926. Figure 11A shows typical flow cytometry profiles of NB4 cells treated with ST1926 (1 μM), nicardipine (100 μM) or the combination of the two compounds for 4 hours and subsequently stained with PI and fluorescently labeled annexin V (AX) to determine the proportion of viable (PI–/AX–), apoptotic (PI–/AX+), necrotic (PI+/AX–), and necrotic/apoptotic (PI+/AX+) cells. As illustrated in Figure 11B, treatment of NB4 blasts with ST1926 leads to a dramatic increase in the proportion of apoptotic cells and a corresponding decrease in the percentage of viable cells. By contrast, the RRM has minimal effects on the number of necrotic or necrotic/apoptotic cells. Nicardipine, at a concentration capable of suppressing the intracellular rise of calcium afforded by ST1926 (100 μM), inhibits the apoptotic effect of the RRM almost completely. The calcium blocker is not apoptotic on its own (at least up to 6 hours) and does not alter the proportion of necrotic cells regardless of the presence of ST1926 in the medium. As illustrated in Figure 9C, the protective effect of nicardipine (100 μM) is confirmed by measurement of the apoptotic index. A similar inhibition of ST1926-induced apoptosis is observed when nicardipine is substituted by the other dihydropyridine, nitrendipine, but not by verapamil (data not shown). In addition, at 100 μM both nicardipine and nitrendipine diminish the apoptotic effect afforded by CD437. The inhibitory effect of nicardipine and nitrendipine is dose dependent and correlates with the ability of the 2 dihydropyridines to decrease the calcium-mobilizing effect of the RRMs (Figure 9C-E). Significantly, the concentration of nicardipine (100 μM) causing inhibition of ST1926-induced apoptosis is also capable of blocking the activation of the effector caspase-3 afforded by the RRM, as documented by Figure 11D.
Effect of dyhydropyridines, nicardipine, and nitrendipine on ST1926- and CD437-dependent apoptosis and cytochromec release from mitochondria in NB4 cells. NB4 cells (150 000/well) were treated with vehicle, the indicated dihydropyridines, ST1926 or CD437, or the indicated mixtures of calcium blockers and RRMs for 6 hours. (A) A typical cytometric profile of cells treated with vehicle (control), ST1926 (1 μM), nicardipine (100 μM), or the combination of the 2 compounds. The profile shows the level of propidium iodide cell positivity (PI+, ordinate axis) and annexin-V positivity (Annexin+, abscissa). The lower left, lower right, upper left, and upper right quadrants indicate the percentage of viable, apoptotic, necrotic, and necrotic+apoptotic cells, respectively. (B) A quantitative summary of an experiment performed as in panel A. The results are expressed as the mean ± SD of 3 replicate cultures. The data presented are representative of 2 independent experiments. *Significantly lower than the corresponding vehicle-treated group (P < .01 according to the Student t test). °Significantly higher than the corresponding vehicle-treated group (P < .01 according to the Student t test). §Significantly lower than the corresponding ST1926-treated group (P < .01 according to the Student t test). (C) Following treatment with ST1926 and CD437 in the presence and absence of the indicated concentrations of dihydropyridines, aliquots of the cultures were subjected to the determination of the apoptotic index by scoring the percentage of cells showing morphologic signs of nuclear fragmentation upon DAPI staining. The results are expressed as the mean ± SD of 3 replicate cultures. The data presented are representative of 2 independent experiments. *Significantly lower than the corresponding ST1926- or CD437-treated group (P < .01 according to the Student t test). °Significantly higher than the corresponding vehicle-treated group (P < .01 according to the Student t test). (D) Cells were treated for 6 hours with 10 μMor100 μM nicardipine in the presence of medium or medium containing 1 μM ST1926. The level of caspase-3 was measured following incubation of cell extracts with the fluorogenic peptide substrate DEVD-amc. The results are expressed as the mean ± SD of 3 replicate cultures. °Significantly lower than the corresponding ST1926- or CD437-treated group (P < .01 according to the Student t test). In panel E, cells were treated with vehicle (control), ST1926 (1 μM), nitrendipine (100 μM), or the combination of the 2 compounds for 4 hours. Cells were harvested, homogenized, and fractionated in a mitochondrial and a cytosolic fraction or left unfractionated (total). Equivalent aliquots of the various fractions were subjected to Western blot analysis with antibodies directed against the indicated proteins.
Effect of dyhydropyridines, nicardipine, and nitrendipine on ST1926- and CD437-dependent apoptosis and cytochromec release from mitochondria in NB4 cells. NB4 cells (150 000/well) were treated with vehicle, the indicated dihydropyridines, ST1926 or CD437, or the indicated mixtures of calcium blockers and RRMs for 6 hours. (A) A typical cytometric profile of cells treated with vehicle (control), ST1926 (1 μM), nicardipine (100 μM), or the combination of the 2 compounds. The profile shows the level of propidium iodide cell positivity (PI+, ordinate axis) and annexin-V positivity (Annexin+, abscissa). The lower left, lower right, upper left, and upper right quadrants indicate the percentage of viable, apoptotic, necrotic, and necrotic+apoptotic cells, respectively. (B) A quantitative summary of an experiment performed as in panel A. The results are expressed as the mean ± SD of 3 replicate cultures. The data presented are representative of 2 independent experiments. *Significantly lower than the corresponding vehicle-treated group (P < .01 according to the Student t test). °Significantly higher than the corresponding vehicle-treated group (P < .01 according to the Student t test). §Significantly lower than the corresponding ST1926-treated group (P < .01 according to the Student t test). (C) Following treatment with ST1926 and CD437 in the presence and absence of the indicated concentrations of dihydropyridines, aliquots of the cultures were subjected to the determination of the apoptotic index by scoring the percentage of cells showing morphologic signs of nuclear fragmentation upon DAPI staining. The results are expressed as the mean ± SD of 3 replicate cultures. The data presented are representative of 2 independent experiments. *Significantly lower than the corresponding ST1926- or CD437-treated group (P < .01 according to the Student t test). °Significantly higher than the corresponding vehicle-treated group (P < .01 according to the Student t test). (D) Cells were treated for 6 hours with 10 μMor100 μM nicardipine in the presence of medium or medium containing 1 μM ST1926. The level of caspase-3 was measured following incubation of cell extracts with the fluorogenic peptide substrate DEVD-amc. The results are expressed as the mean ± SD of 3 replicate cultures. °Significantly lower than the corresponding ST1926- or CD437-treated group (P < .01 according to the Student t test). In panel E, cells were treated with vehicle (control), ST1926 (1 μM), nitrendipine (100 μM), or the combination of the 2 compounds for 4 hours. Cells were harvested, homogenized, and fractionated in a mitochondrial and a cytosolic fraction or left unfractionated (total). Equivalent aliquots of the various fractions were subjected to Western blot analysis with antibodies directed against the indicated proteins.
The relocation of cytochrome c from the mitochondria into the cytosol is a central event in the apoptotic pathway activated by cytotoxic drugs and CD437 in particular.2,3 Treatment of NB4 cells with ST1926 results in the release of cytochrome c into the cytosol and in a concomitant depletion of the protein pool associated with mitochondria (Figure 11E). These effects are not accompanied by variations in the total amount of cytochrome c, demonstrating that the protein relocates from the mitochondrial to the cytosolic compartment. The intracellular relocation is suppressed by cotreatment of cells with nitrendipine at concentrations of the dihydropyridine that inhibit both ST1926-dependent calcium mobilization and apoptosis.
To further support the significance of cytosolic calcium mobilization in the process of apoptosis triggered by ST1926 and CD437, we used BAPTA, a powerful intracellular calcium chelator. Preloading of NB4 cells with different concentrations of BAPTA (10 μM-100 μM) results in a dose-dependent inhibition of the FURA-2 fluorescence signals activated by ST1926 (Figure 12A) and CD437 (data not shown). Figure 12B demonstrates that, in our experimental conditions, BAPTA has a slight apoptotic effect on NB4 cells at all the concentrations tested. Significantly, however, the same concentrations of BAPTA (50 μM-100 μM) suppressing the RRM-dependent FURA-2 signal inhibit the process of apoptosis set in motion by both ST1926 and CD437. As illustrated in Figure 12C, this effect is associated with a substantial decrease in the levels of caspase-3 activation afforded by treatment of NB4 cells with the 2 RRMs.
Effect of ST1926, BAPTA, and the combination of the 2 agents on the levels of intracellular calcium, apoptosis, and caspase-3 activation in NB4 cells. (A) Following preloading with the calcium fluorescent indicator FURA-2, NB4 cells (1 × 106/mL) were resuspended in calcium-containing PBS. Cells were placed in a cuvette under stirring at 37°C and changes in fluorescence were measured continuously with the use of a spectrophotofluorometer. NB4 cells were incubated with the indicated concentrations of BAPTA prior to addition of ST1926 at the concentration of 1 μM. The left arrow indicates the addition of BAPTA, whereas the right arrow indicates the addition of ST1926. Each tracing is representative of at least 2 independent experiments run in triplicate. (B) NB4 cells (150 000/well) were treated with vehicle, the indicated concentrations of BAPTA, ST1926 (1 μM), or CD437 (1 μM), and mixtures of the intracellular calcium chelator and RRMs for 6 hours. Following treatment, aliquots of the cultures were subjected to the determination of the apoptotic index by scoring the percentage of cells showing morphologic signs of nuclear fragmentation upon DAPI staining. The results are expressed as the mean ± SD of 3 replicate cultures. The data are representative of 2 independent experiments. *Significantly lower than the corresponding ST1926- or CD437-treated group (P < .01 according to the Student t test). (C) Cells were treated for 6 hours with BAPTA (50 μM or 100 μM) in the presence of medium or medium containing 1 μM ST1926. The level of caspase-3 was measured following incubation of cell extracts with the fluorogenic peptide substrate DEVD-amc. The results are expressed as the mean ± SD of 3 replicate cultures. *Significantly lower than the corresponding ST1926- or CD437-treated group (P < .01 according to the Student t test).
Effect of ST1926, BAPTA, and the combination of the 2 agents on the levels of intracellular calcium, apoptosis, and caspase-3 activation in NB4 cells. (A) Following preloading with the calcium fluorescent indicator FURA-2, NB4 cells (1 × 106/mL) were resuspended in calcium-containing PBS. Cells were placed in a cuvette under stirring at 37°C and changes in fluorescence were measured continuously with the use of a spectrophotofluorometer. NB4 cells were incubated with the indicated concentrations of BAPTA prior to addition of ST1926 at the concentration of 1 μM. The left arrow indicates the addition of BAPTA, whereas the right arrow indicates the addition of ST1926. Each tracing is representative of at least 2 independent experiments run in triplicate. (B) NB4 cells (150 000/well) were treated with vehicle, the indicated concentrations of BAPTA, ST1926 (1 μM), or CD437 (1 μM), and mixtures of the intracellular calcium chelator and RRMs for 6 hours. Following treatment, aliquots of the cultures were subjected to the determination of the apoptotic index by scoring the percentage of cells showing morphologic signs of nuclear fragmentation upon DAPI staining. The results are expressed as the mean ± SD of 3 replicate cultures. The data are representative of 2 independent experiments. *Significantly lower than the corresponding ST1926- or CD437-treated group (P < .01 according to the Student t test). (C) Cells were treated for 6 hours with BAPTA (50 μM or 100 μM) in the presence of medium or medium containing 1 μM ST1926. The level of caspase-3 was measured following incubation of cell extracts with the fluorogenic peptide substrate DEVD-amc. The results are expressed as the mean ± SD of 3 replicate cultures. *Significantly lower than the corresponding ST1926- or CD437-treated group (P < .01 according to the Student t test).
ST1926 is active in vivo
To determine the in vivo antileukemic activity of ST1926, SCID mice were inoculated intraperitoneally with NB4 cells. Table 3 demonstrates that intraperitoneal administration of ST1926 is accompanied by a significant increase in the overall survival of leukemia-bearing animals. The antileukemic effect of the adamantyl-retinoid at a dosage of 40 mg/kg is of the same order of magnitude as that of ATRA at 50 mg/kg. When the 2 compounds are coadministered to a leukemia-bearing animal an additive effect is observed. This indicates that the 2 compounds act through different mechanisms. When administered orally, ST1926 results in a significant and dose-dependent increase in the life span of NB4-bearing SCID mice. Treated animals do not show any sign of overt toxicity and ST1926 treatment is accompanied by a modest decrease in body weight which is maximal at the 2 highest concentrations considered (40 mg/kg and 50 mg/kg). At the doses and with the routes of administration considered, the effect of ST1926 on transplanted NB4 cells is purely apoptotic and does not involve cytodifferentiation, as demonstrated by short term in vivo experiments (data not shown). These results demonstrate that the RRM has a favorable pharmacokinetic and toxicity profile, and can be used for the treatment of AML either alone or in combination with ATRA.
Antileukemic activity of ST1926 and ATRA in NB4-bearing SCID mice
Treatment . | No. of animals . | Dosage (mg/kg) . | BWL, % max . | MST (range) . | MST . | ILS, % . |
|---|---|---|---|---|---|---|
| Vehicle, orally | 8 | 0 | 0 | (29-35) | 34 | — |
| ST1926, orally | 8 | 30 | 6 | (39-48) | 41* | 21 |
| ST1926, orally | 8 | 40 | 17 | (36-50) | 46* | 3 |
| ST1926, orally | 8 | 50 | 15 | (13-50) | 49† | 44 |
| ATRA, orally | 8 | 40 | 0 | (28-43) | 34.5 | — |
| Vehicle, interperitoneally | 8 | 0 | 0 | (30-39) | 35.5 | — |
| ST1926, interperitoneally | 8 | 50 | 9 | (21-63) | 55.5* | 56 |
| ATRA, interperitoneally | 8 | 40 | 0 | (50-86) | 55* | 55 |
| ATRA + ST1926, interperitoneally | 8 | 40 + 50 | 14 | (56-84) | 67*‡ | 89 |
Treatment . | No. of animals . | Dosage (mg/kg) . | BWL, % max . | MST (range) . | MST . | ILS, % . |
|---|---|---|---|---|---|---|
| Vehicle, orally | 8 | 0 | 0 | (29-35) | 34 | — |
| ST1926, orally | 8 | 30 | 6 | (39-48) | 41* | 21 |
| ST1926, orally | 8 | 40 | 17 | (36-50) | 46* | 3 |
| ST1926, orally | 8 | 50 | 15 | (13-50) | 49† | 44 |
| ATRA, orally | 8 | 40 | 0 | (28-43) | 34.5 | — |
| Vehicle, interperitoneally | 8 | 0 | 0 | (30-39) | 35.5 | — |
| ST1926, interperitoneally | 8 | 50 | 9 | (21-63) | 55.5* | 56 |
| ATRA, interperitoneally | 8 | 40 | 0 | (50-86) | 55* | 55 |
| ATRA + ST1926, interperitoneally | 8 | 40 + 50 | 14 | (56-84) | 67*‡ | 89 |
SCID mice were xenografted with NB4 cells intraperitoneally and treated as stated in “Materials and methods” with the indicated dosages of ATRA and/or ST1346 5 times per week for 3 weeks. The effect on body weight is expressed as BWL, % (maximum body weight loss during treatments), and is calculated as: 100 - (average BW dayx/average BW day1 × 100). In addition, ILS, % was evaluated (increase life span of treated mice vs untreated mice) calculated as: 100 - [(median survival time (MST) of treated mice/median survival time of untreated mice) × 100].
BWL indicates body weight loss; MST, median survival time; ILS, increased life span; —, not applicable.
P < .001 versus vehicle (Mann-Whitney).
P < .01 versus vehicle (Mann-Whitney).
P < .05 versus ATRA (Mann-Whitney).
Discussion
In this article, we report on a novel chemical series of apoptotic compounds designed after the structure of the RRM prototype, CD437.4,7-13 The series includes a molecule, ST2065, with antagonistic properties and a compound, ST1926, which is more powerful and has cytotoxic activity on a larger spectrum of myeloid cell lines than CD437. ST1926 is a bona fide RRM and is likely to act on the same molecular determinants as CD437, at least on the basis of the cross-resistance observed in NB4.437r cells. Like CD437 and unlike ATRA, ST1926 is a poor activator of gene expression. Rather, it represses the transcription of a large set of genes that are similarly down-regulated by CD437. Among these, protein translation and mitochondrial genes stand out. A salient and clinically significant feature of ST1926 is the promising antileukemic activity in vivo, which is evident in the aggressive model of the SCID mouse that received a transplant of NB4 cells. Importantly, the antileukemic effect is observed following oral administration of ST1926.
Although a systematic analysis of the chemical functions responsible for the apoptotic activity of RRMs is beyond the scope of this study, analysis of our series permits us to draw a few conclusions. When the adamantyl ring of ST1926 and CD437 is substituted by a cyclohexane (ST2306), a significant reduction in the apoptotic potential is observed. A similar effect is evident if we block the hydroxyl residue on the phenyl ring adjacent to the adamantyl group or the carboxylic function on the other side of the molecule. Introduction of a heterocyclic ring in the center of the molecule (ST2060, ST2062, ST2065, and ST2067) suppresses the apoptotic activity. Interestingly, when this is done in the context of a molecule that maintains the adamantyl, hydroxyl, and carboxylic functions described above, a putative RRM antagonist is produced (ST2065). Indeed, when NB4 cells are pretreated with a molar excess of ST2065, the apoptotic activity of ST1926 is inhibited. It is likely that the inhibitory effect is the result of a bona fide antagonistic blockade of an unknown intracellular target of ST1926, CD437, and congeners. The primary molecular target of ST1926 in myeloid leukemia cells is unlikely to be a nuclear retinoic acid receptor of the RAR or RXR subtype. Although we have not formally excluded this possibility with the use of specific antagonists, experiments performed with RNA transcription and protein synthesis inhibitors are against it. In fact, similar to what was observed in the case of CD437, actinomycin D and cycloheximide do not block the apoptogenic response to ST1926 in NB4 cells (data not shown), suggesting that gene expression phenomena, such as those activated by RARs or RXRs, are not involved in the apoptogenic process triggered by the RRM. As to the characteristics of the putative RRM binding protein present in NB4 and other AML cell lines, this is likely to have a conformation similar to that of the RARγ ligand-binding domain. In fact, there is a strict correlation between the RARγ transactivating and the apoptotic activities of the various members of the chemical series analyzed.
The molecular mechanisms underlying the activity of ST1926 and RRMs have been explored in some detail. Our data indicate that the activation of p38 and JNK is not necessary for the process of RRM-dependent apoptosis in myeloid cells. A pivotal result stemming from the analysis of the present chemical series relates to the calcium-mobilizing properties of ST1926 and CD437. Treatment of AML cells with apoptotic concentrations of the two RRMs results in an immediate rise in the cytosolic levels of calcium. The increase is dose dependent and correlates with the apoptotic potential of ST1926, CD437, and all the other members of the series considered. Calcium mobilization is suppressed by inhibitors of ST1926- and CD437-dependent apoptosis like the putative RRM antagonist ST2065, by the intracellular calcium chelator, BAPTA, and by calcium blockers of the dihydropyridine type. Although intracellular calcium is known to represent a determinant of the process of PCD induced by different types of stimuli,29,30 such a rapid mobilizing effect in myeloid leukemia cells is a peculiarity of ST1926 and congeners that is not shared by other common chemotherapeutic and apoptotic agents. The event is necessary but not sufficient for the process of apoptosis activated by RRMs. In fact, myeloid cell lines naturally refractory to or induced to be resistant to RRMs, such as Kasumi and NB4.437r,2 are still responsive to the calcium-mobilizing activity of ST1926. Incidentally, our data demonstrate that the molecular determinants underlying the selective resistance to RRMs in NB4.437r cells lay downstream of calcium mobilization and upstream of cytochrome c release from the mitochondria.
A key question relates to the origin of the cytosolic calcium rise produced by ST1926 and active congeners. This is not extracellular and the result of an influx of the cation through voltage-activated or receptor-dependent channels.43 In fact, the amplitude and the kinetics of the cytosolic calcium rise afforded by RRMs in NB4 or NB4.437r cells are not affected by (1) calcium elimination from the cell growth medium, (2) addition of the competing anion, nickel, to the extracellular compartment, (3) pretreatment of cells with the calcium blocker, verapamil, or with pertussis toxin, an inhibitor of membrane receptor–coupled G-proteins. Similarly, the results obtained with SERCA inhibitors suggest that the ER is not involved in the RRM-induced calcium effect. On the other hand, the observations made with uncouplers of the oxidative phosphorylation are consistent with the view that the steady-state levels of cytosolic calcium are increased by RRMs largely because of an inhibition of the energy-dependent calcium uptake into mitochondria. Disruption of the calcium reuptake process is immediate and precedes opening of the mitochondrial transition pore and release of cytochrome c into the cytosol.
Surprisingly, the calcium-mobilizing effect of ST1926 and congeners in NB4 cells is inhibited by calcium channel blockers of the dihydropyridine type. This effect is unrelated to the ability of these compounds to block the influx of calcium from outside the cell, as inhibition is observed also in calcium-free medium. Thus, we propose that RRMs increase the amount of cytosolic calcium by inhibiting the mitochondrial uptake through interference with an unidentified molecular determinant(s) that is sensitive to dihydropyridines. These last agents block the opening of the mitochondrial transition pore, the release of cytochrome c into the cytosol, and the subsequent activation of effector caspases.
In conclusion, the results contained in the present report contribute to the elucidation of the molecular determinants and mechanisms underlying the pharmacologic activity of RRMs. We provide evidence for a central role of intracellular calcium mobilization in the rapid process of apoptosis triggered by RRMs in myeloid cells. More important, we describe ST1926 as a novel and orally active agent, which is currently under clinical development.
Prepublished online as Blood First Edition Paper, September 4, 2003; DOI 10.1182/blood-2003-05-1577.
Supported by grants to E.G. from the Associazione Italiana per la Ricerca contro il Cancro (AIRC), the “Istituto Superiore di Sanità”, the “Progetto Finalizzato Oncologia” (Consiglio Nazionale Delle Ricerche e Ministero dell'Università e della Ricerca Scientifica e Tecnologica [CNR-MURST], the “Fondo D'Investimento per la Ricerca Biotecnologica” (FIRB), and Sigma-Tau S.p.A., and by the Weizmann-Pasteur-Negri Foundation.
Several of the authors (R.Z., F.F.F., I.C., P.C., and C.P.) are employed by Sigma-Tau Industrie Farmaceutiche Riunite S.p.A., whose potential products were studied in the present work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Michel Lanotte (Unitè INSERM 301, “Genetique cellulaire et moleculaire de Leucemies,” Centre G. Hayem, Hopital St Louis, Paris, France) for supplying us with the NB4 cell line. We are grateful to Dr Eugenio Erba (Istituto di Ricerche Farmacologiche “Mario Negri”) for the assistance in performing experiments involving flow cytometry. The expert technical assistance of Silvia Mattavelli (Istituto di Ricerche Farmacologiche “Mario Negri”) is acknowledged. We thank Dr Mario Salmona and Prof Silvio Garattini (Istituto di Ricerche Farmacologiche “Mario Negri”) for critical reading of the manuscript.

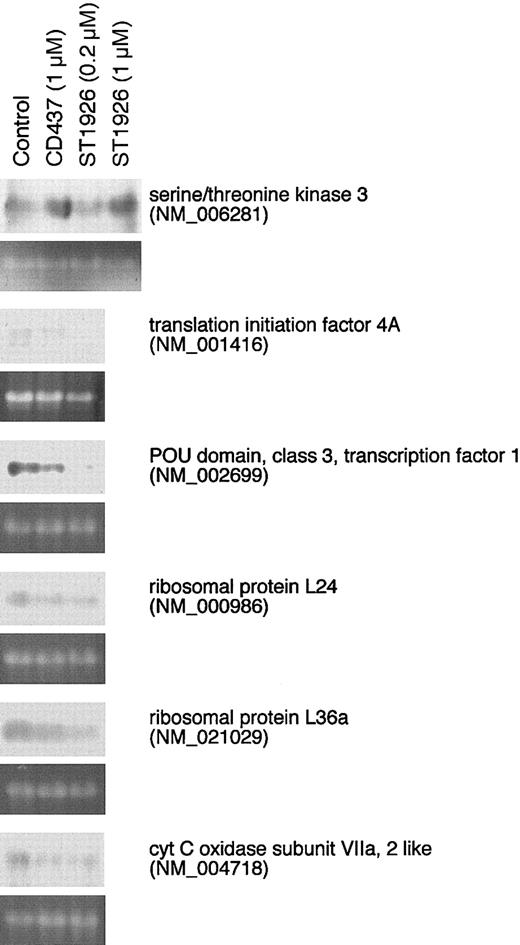
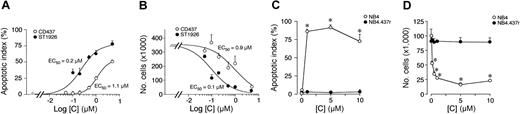
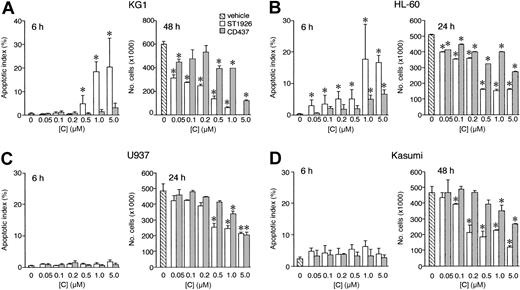
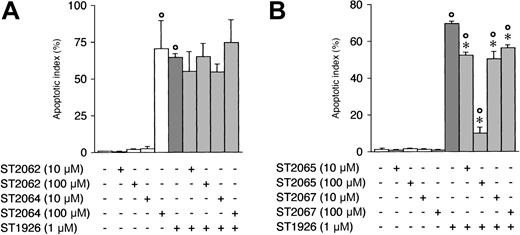
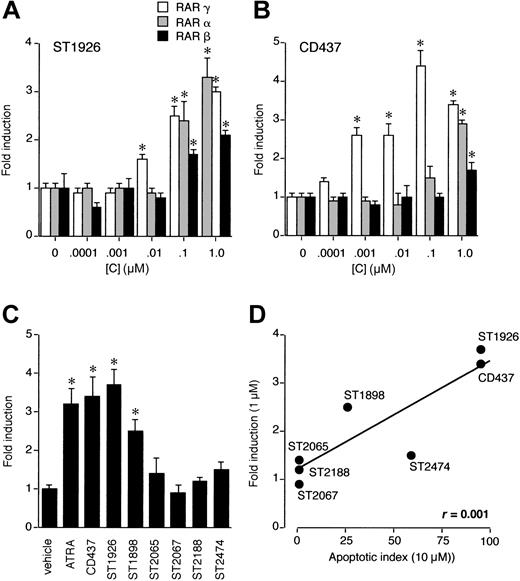
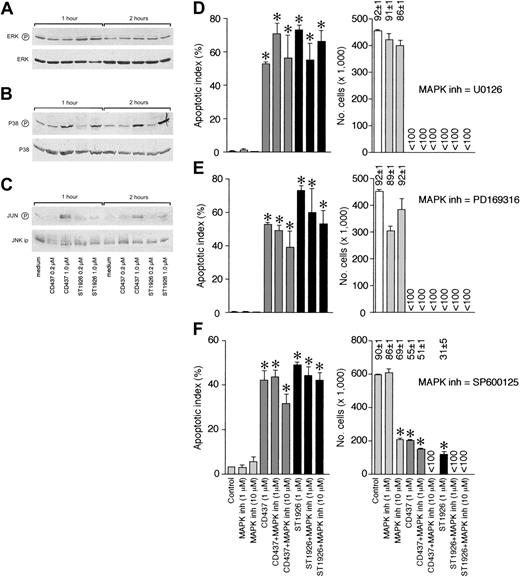
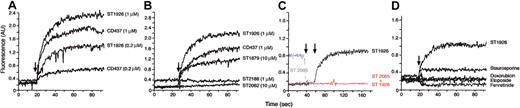
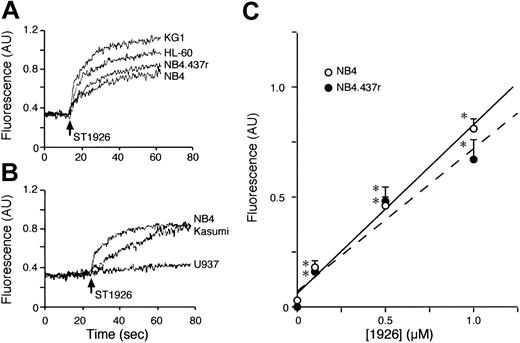
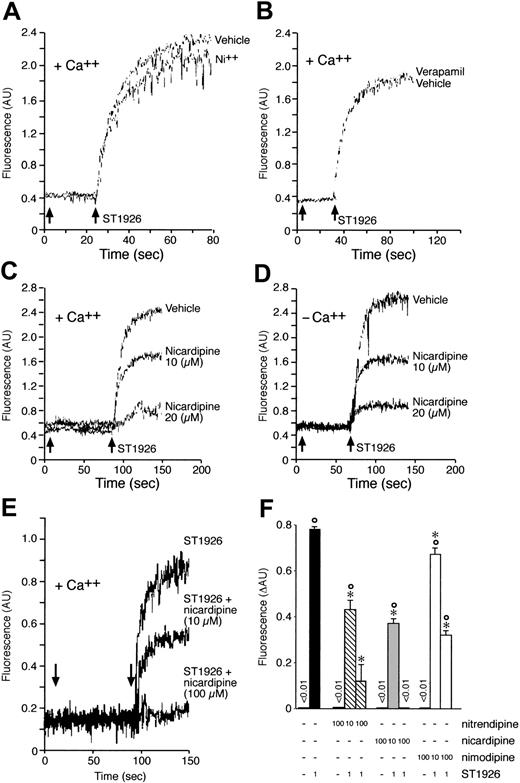
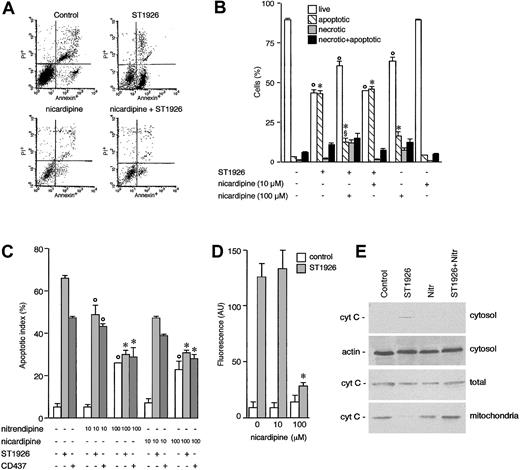
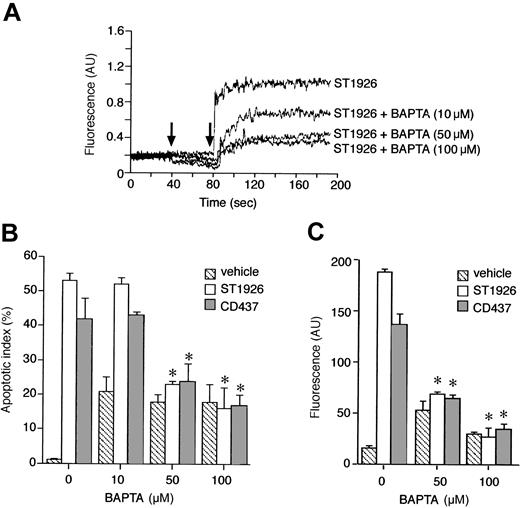
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal