Abstract
Immunoglobulin variable heavy chain gene (VH) mutation status and VDJ rearrangement structure were analyzed in 141 patients with mantle cell lymphoma (MCL) and correlated with biologic and clinical characteristics; 29% of the MCLs displayed mutated VH using a 98% germline homology cutoff. Striking differences occurred in the VH mutation subgroups with respect to the use of specific V genes. Rearrangements involving V4-34 and V3-21 were almost exclusively unmutated, whereas rearrangements using V4-59 and V3-23 were typically mutated. Significant association occurred between mutated VH with shorter CDR3 lengths and the use of JH4b. V3-21 and V4-59 were involved in highly characteristic rearrangements, implying that antigen specificity might have been involved in MCL development. There was no evidence for isotype switch recombination or Bcl-6 expression in any MCL. ZAP70 expression was not different in VH-mutated or -unmutated MCL. Although the deletions 11q– and 17p– showed a balanced distribution, an overrepresentation was observed for trisomies +3q, +8q, and tetraploidy in the VH-unmutated subgroup and +12q in the VH-mutated subgroup. Clinically, mutated VH was associated with a higher rate of complete remission, but there was no correlation between VH mutation status and other clinical characteristics or overall survival.
Introduction
Mantle cell lymphoma (MCL) follows an aggressive clinical course, with poor response to chemotherapy and a median survival time of 3 to 4 years.1-3 The genetic hallmark of MCL is the translocation (11;14)(q13;q32), leading to the overexpression of cyclin D1.4-6 Clonal genomic aberrations in addition to t(11;14) can be identified in most MCLs.7-9 The pattern of genomic aberrations in MCL shows similarities to chronic lymphocytic leukemia (CLL) with a high frequency of the deletions 13q– and 11q–.9 Prognostic relevance was demonstrated for 17p/p53 abnormalities10-12 and for INK4a/ARF deletions,13-15 and complex karyotypes or tetraploid chromosome clones were associated with aggressive variants of MCL.16,17 ATM gene inactivation was observed in MCL18-20 and was associated with increased chromosomal imbalances.18
The cellular origin of MCL appeared to be related to the pre-germinal–center stage of B-cell development based on the observation of an unmutated VH status in early studies.21,22 However, recently several groups have reported mutated VH in a sizable proportion of MCL.23-27 Similarly, chronic lymphocytic leukemia (CLL) has been divided into 2 subtypes, 1 with unmutated and 1 with mutated VH, which are characterized by a markedly different clinical outcome.28,29 Biologically, the 2 CLL subtypes showed different gene expression signatures30-33 and distinct genomic aberrations of the tumor cells.34-36
The current study aimed at the evaluation of VH mutation status and VDJ structure in MCL in relation to other biologic characteristics, such as genomic aberration, isotype switch recombination, expression of ZAP70, and Bcl-6 and to clinical parameters such as disease manifestation and outcome.
Patients, materials, and methods
Samples
One hundred forty-one MCL patient samples were collected at the Universities of Würzburg (n = 83), Heidelberg (n = 34), and Ulm (n = 24) between October 1979 and December 2002 at the time of diagnosis (n = 129) or during the course of the disease (n = 12). DNA and RNA were extracted from frozen tissue blocks (n = 95) and from peripheral blood or bone marrow (n = 46) obtained for diagnostic procedures after informed consent. Approval was obtained from the institutional review boards of the Universities of Ulm, Würzburg, and Heidelberg for these studies.
Diagnosis
Histologic diagnosis according to the criteria of the World Health Organization (WHO) classification was made at German reference pathology institutions (G.O., T.F.E.B., P.M., H.K.M.-H.) in 123 patients (87.2%). The remaining, mostly leukemia patients, were included based on t(11;14) positivity and typical immunophenotype. Overall, the presence of t(11;14) by interphase fluorescence in situ hybridization (FISH) or classical cytogenetics or the expression of cyclin D1 by immunohistochemistry was demonstrated in 127 patients (90.1%). In the remaining 12 patients, t(11;14) or cyclin D1 expression was not evaluated, and for 2 patients findings were negative for t(11;14). These patients were included because of their classical morphology and typical immunophenotype, including CD23 negativity. The blastoid variant of MCL was diagnosed in 7 patients.
Clinical features and treatment
Clinical characteristics at the time of diagnosis were available for a subset of patients, as detailed in Table 1. Median follow-up time for the patients with survival information (n = 105) was 43.8 months. Twelve patients underwent 1 or more therapy regimens before study. Median time to first treatment was 1 month. Therapy extended beyond a single clinical trial and was heterogeneous.
Clinical and treatment characteristics
Clinical feature at diagnosis . | No. patients . | All patients . | Percentage . |
|---|---|---|---|
| Male sex | 101 | 130 | 77.7 |
| Stages III/IV | 98 | 108 | 81.5 |
| B symptoms | 46 | 101 | 45.5 |
| Bulk, 10 cm or more | 22 | 90 | 24.4 |
| Leukemic manifestation | 47 | 99 | 47.5 |
| IPI score 3-5 | 43 | 78 | 55.1 |
| Bone marrow involvement | 76 | 102 | 74.5 |
| Palpable lymphadenopathy | 79 | 103 | 76.7 |
| Splenomegaly | 60 | 101 | 59.4 |
| Gastrointestinal involvement | 24 | 102 | 23.5 |
| LDH level elevated | 32 | 86 | 37.2 |
| Treatment | |||
| Treated | 99 | 104 | 95.2 |
| Chemotherapy | 88 | 99 | 88.9 |
| Polychemotherapy | 77 | 88 | 87.5 |
| Anthracycline | 35 | 86 | 40.7 |
| Single-agent therapy | 11 | 88 | 12.5 |
| Radiotherapy | 15 | 99 | 15.2 |
| Resection/splenectomy | 18 | 99 | 18.2 |
| CR after initial therapy | 22 | 93 | 23.7 |
| ASCT* | 14 | 104 | 13.5 |
Clinical feature at diagnosis . | No. patients . | All patients . | Percentage . |
|---|---|---|---|
| Male sex | 101 | 130 | 77.7 |
| Stages III/IV | 98 | 108 | 81.5 |
| B symptoms | 46 | 101 | 45.5 |
| Bulk, 10 cm or more | 22 | 90 | 24.4 |
| Leukemic manifestation | 47 | 99 | 47.5 |
| IPI score 3-5 | 43 | 78 | 55.1 |
| Bone marrow involvement | 76 | 102 | 74.5 |
| Palpable lymphadenopathy | 79 | 103 | 76.7 |
| Splenomegaly | 60 | 101 | 59.4 |
| Gastrointestinal involvement | 24 | 102 | 23.5 |
| LDH level elevated | 32 | 86 | 37.2 |
| Treatment | |||
| Treated | 99 | 104 | 95.2 |
| Chemotherapy | 88 | 99 | 88.9 |
| Polychemotherapy | 77 | 88 | 87.5 |
| Anthracycline | 35 | 86 | 40.7 |
| Single-agent therapy | 11 | 88 | 12.5 |
| Radiotherapy | 15 | 99 | 15.2 |
| Resection/splenectomy | 18 | 99 | 18.2 |
| CR after initial therapy | 22 | 93 | 23.7 |
| ASCT* | 14 | 104 | 13.5 |
LDH indicates lactate dehydrogenase.
†Median age of 111 patients was 63 years (quartiles: 55, 70).
High-dose chemotherapy and autologous stem cell transplantation (ASCT).
Genomic aberrations
A previously described probe set37 was used for FISH.7-9 In addition to t(11;14)(q13;q32), this allowed screening for the following partial deletions and trisomies: add(3q26), del(6q27), add(8q24), del(11q22-q23), add(12q13), del(13q14), and del(17p13). FISH analysis was performed in a subset of 87 patients based on the availability of material.
Identification and sequencing of clonal VDJ rearrangements and isotypes
Genomic DNA and total RNA were isolated with Trizol (Gibco BRL, Eggenstein, Germany). Multiplex polymerase chain reaction (PCR) amplification and subsequent direct sequencing of the clonal VDJ rearrangement was performed as previously described.34 Sequence analysis with the corresponding leader region primers was successful in 86.5% of the rearrangements. In all patients at least 1 independent PCR product was sequenced for verification. The VDJ nucleotide sequences were aligned to current databases (EMBL/GenBank V-base directory). Homology to the nearest germline gene was calculated in relation to the sequenced fragment. D segments were assigned according to the criteria described by Corbett et al.38 To determine the involved constant region isotype, reverse transcription of RNA to cDNA was carried out from 2 μg RNA with oligo dT primer (GeneAmp Kit, RNA PCR System; Applied Biosystems, Weiterstadt, Germany). Fluorochrome-labeled μ, γ, and α isotype-specific CH primers, described previously,39 were used for GeneScan analysis after multiplex PCR amplification, followed by direct sequencing.
Bcl-6 staining
ZAP70RQ-PCR
Only patients in whom more than 70% of the cells carried clonal genomic aberrations as detected by FISH, indicating a high tumor cell load, were selected for real-time quantitative reverse transcription PCR (RQ-PCR) analysis as described previously.42 Serial dilutions of cDNA from Jurkat cells were used to obtain a calibration graph. Phosphoglycerate kinase I (PGK) gene expression served as endogenous control for normalization. Primers were (for ZAP70) 5′-GTTGACTCATCCTCAGAGACGAATC (sense) and 5′-AGGTTATCGCGCTTCAGGAA (antisense) and (for PGK) 5′-ACCGAATCACCGACCTCTCTC (sense) and 5′-ATTGAAGTCGACTCTCATAACGACC (antisense) using SYBR Green. To quantify the amount of contaminating T cells, with a constitutively high ZAP70 expression, RQ-PCR was performed for CD3G (Assay-on-Demand; Applied Biosystems).
Statistical analysis
Fisher exact test was used for pairwise comparison of distributions of the categorical variables. To account for the higher risk for false-positive results from multiple testing, the method of Holm was used43 for the differential distribution of VH, JH, and D families and genes with respect to VH mutation status and for testing the prognostic relevance of clinical features with respect to overall survival (OS). Survival-time distributions from the date of diagnosis were plotted using Kaplan-Meier estimates and likelihood-ratio tests using Cox partial likelihood to study the prognostic impact of various parameters. Mann-Whitney U tests were used to determine the significance of location differences in the distribution of age and CDR3 lengths in the VH subgroups. Estimation of the location difference was performed using the Hodges-Lehmann estimates. Odds ratios, together with their 95% confi-dence limits, were used to compare the risk for genomic aberrations between the VH subgroups. The association of CDR3 length and number of aberrations was estimated using the Spearman rank correlation coefficient. All data were analyzed using GraphPad Prism version 3.0 (GraphPad Software, San Diego, CA) and R version 1.6.2.44 Two-sided tests were used. An (adjusted) P value (P ≤ .05) was considered significant.
Results
VHmutation status andVDJrearrangement structure in MCL
From the 141 MCL patients, 148 VDJ rearrangements were amplified. All rearrangements except 1 (stop codon) were potentially productive. Five patients had 2 distinct VDJ rearrangements, and 1 had 3 rearrangements. All patients with accessory rearrangements were t(11;14) positive, and there was no evidence of different tumor subclones based on genomic aberrations (data not shown). VH mutation status was concordant between the different rearrangements in 4 of the 6 patients.
Using a homology cutoff value of 98% to the nearest germline gene, we observed mutated VH in 41 (29.1%) and unmutated VH in 100 (70.9%) of the 141 MCL patients. The germline homology rate ranged from 91.2% to 100% (median, 99.3%), with a median homology of 96.9% in the mutated group. Eight patients had less than 95% and 23 patients had less than 97% homology to the nearest germline gene. Among the 14 patients not analyzed (n = 12) or negative (n = 2) for t(11;14) or cyclin D1, 4 patients had mutated VH, and the median VH homology was 99.3%. The 12 patients treated before the study showed a median VH homology of 99.3%; 3 had mutated VH. Twelve patients fulfilled the criteria described by Orchard et al24 as having “nonnodal MCL” because of their leukemic manifestation and absence of lymphadenopathy. Of these, 5 (41.7%) had somatic mutations compared with 36 (27.9%) of 129 in the “nodal” group (P = .33).
VHfamilies and genes
The VH families VH1, VH3, and VH4 accounted for 91.2% of all rearrangements observed in this series of 141 MCLs (16.2%, 46.6%, and 28.4%, respectively). Although the VH3 and VH4 families showed a balanced distribution among the VH mutation subgroups (VH mutated vs unmutated: 52.4% vs 44.3% and 35.7% vs 25.5%), VH1 family members were more frequently found in VH-unmutated rearrangements (7.1% vs 19.8%). The 5 most frequently used individual VH genes (V4-34, V3-21, V1-08, V3-23, and V4-59) were found in 52% of all VDJ rearrangements (54.6% of all patients). V4-34 and V3-21 were overrepresented in the VH-unmutated group, and V3-23 and V4-59 were typically found in mutated MCL (Table 2). The V1-69 gene was used in 4 (2.7%) MCL patients only, all of them VH-unmutated. VH gene use among the 14 patients not analyzed (n = 12) or negative (n = 2) for t(11;14) or cyclin D1 was similar to the entire cohort (V3-21, 3 patients; V4-34, 2 patients; V3-23, 1 patient).
Frequency of VH,D, and JH genes*and VDJ structure in 148 rearrangements of 141 MCL patients: incidence and distribution according to VH mutation status
. | Overall . | VH mutated . | VH unmutated . | Differential distribution, adjusted P† . |
|---|---|---|---|---|
| VH gene, n | 148 | 42 | 106 | |
| V4-34 | 23 (15.5) | 2 (4.8) | 21 (19.8) | .05 |
| V3-21 | 18 (12.1) | 0 (0) | 18 (17) | .008 |
| V1-08 | 15 (10.1) | 3 (7.1) | 12 (11.3) | .56 |
| V3-23 | 11 (7.4) | 7 (16.7) | 4 (3.8) | .04 |
| V4-59 | 10 (6.8) | 9 (21.4) | 1 (0.9) | .005 |
| Other | 71 (48) | 21 (50) | 50 (47.2) | — |
| JH gene, n | 145 | 42 | 103 | |
| JH4b | 62 (42.8) | 26 (61.9) | 36 (35) | .02 |
| JH5b | 29 (20) | 6 (14.3) | 23 (22.3) | .72 |
| JH6b | 22 (15.2) | 6 (14.3) | 16 (15.5) | 1.0 |
| JH6c | 14 (9.7) | 2 (4.8) | 12 (11.7) | .35 |
| Other | 18 (12.4) | 2 (4.8) | 16 (15.5) | — |
| D gene, n | 94 | 18 | 76 | |
| D2-2 | 16 (17) | 3 (16.7) | 13 (17.1) | 1.0 |
| D3-3 | 14 (14.9) | 1 (5.6) | 13 (17.1) | .58 |
| Other | 64 (68.1) | 14 (77.8) | 50 (65.8) | — |
| CDR3, n | 148 | 42 | 106 | |
| Median length in | ||||
| codons | ||||
| [quartiles] | 13 [10, 17] | 11 [9, 14] | 15 [11, 18] | < .001‡ |
. | Overall . | VH mutated . | VH unmutated . | Differential distribution, adjusted P† . |
|---|---|---|---|---|
| VH gene, n | 148 | 42 | 106 | |
| V4-34 | 23 (15.5) | 2 (4.8) | 21 (19.8) | .05 |
| V3-21 | 18 (12.1) | 0 (0) | 18 (17) | .008 |
| V1-08 | 15 (10.1) | 3 (7.1) | 12 (11.3) | .56 |
| V3-23 | 11 (7.4) | 7 (16.7) | 4 (3.8) | .04 |
| V4-59 | 10 (6.8) | 9 (21.4) | 1 (0.9) | .005 |
| Other | 71 (48) | 21 (50) | 50 (47.2) | — |
| JH gene, n | 145 | 42 | 103 | |
| JH4b | 62 (42.8) | 26 (61.9) | 36 (35) | .02 |
| JH5b | 29 (20) | 6 (14.3) | 23 (22.3) | .72 |
| JH6b | 22 (15.2) | 6 (14.3) | 16 (15.5) | 1.0 |
| JH6c | 14 (9.7) | 2 (4.8) | 12 (11.7) | .35 |
| Other | 18 (12.4) | 2 (4.8) | 16 (15.5) | — |
| D gene, n | 94 | 18 | 76 | |
| D2-2 | 16 (17) | 3 (16.7) | 13 (17.1) | 1.0 |
| D3-3 | 14 (14.9) | 1 (5.6) | 13 (17.1) | .58 |
| Other | 64 (68.1) | 14 (77.8) | 50 (65.8) | — |
| CDR3, n | 148 | 42 | 106 | |
| Median length in | ||||
| codons | ||||
| [quartiles] | 13 [10, 17] | 11 [9, 14] | 15 [11, 18] | < .001‡ |
All values in parentheses are percentages.—indicates not done.
Gene frequency in 10 or more patients.
To account for multiple testing, family/genewise P values were adjusted according to Holm.43
Estimated difference, 3 codons (95% confidence limits, 2-4 codons).
JHfamilies and genes
Most MCL patients used the JH families JH4 (43.5%), JH5 (20%), and JH6 (24.8%), accounting for 88.3% of all rearrangements. JH4b, JH5b, JH6b, and JH6c were the most commonly used individual JH genes. JH4b was significantly overrepresented in VH-mutated MCL (Table 2).
Dfamilies and genes
D segments were assignable in 43% of the VH-mutated and in 72% of the VH-unmutated rearrangements (63.5% of all rearrangements). The most frequent D families were D2 and D3 (26.6% and 38.3%), and the most common individual D genes were D2-2 and D3-3 (Table 2). In unmutated rearrangements, where the rate of assignable D segments was high, a higher frequency of the long D families (D2 + D3) was observed compared with mutated ones (data not shown; VH mutated vs unmutated, 44.4% vs 69.7%).
CDR3length
There was a highly significant difference in the distribution of CDR3 lengths between the VH mutation subgroups. VH-mutated rearrangements were characterized by shorter median lengths of the CDR3 (11 codons) compared with VH-unmutated rearrangements (15 codons) (P < .001).
Characteristic VDJ rearrangements
Strong correlations between V3-21, V4-34, V3-23, and V4-59 and VH mutation status led to further examination of the most common individual VDJ rearrangements (Table 3).45,46 V3-21, which was found only in VH-unmutated rearrangements, was associated with longer CDR3, JH6 family members, and D3-3 segment. The mostly VH-mutated rearrangements using V4-59 were characterized by short CDR3 lengths and showed a high incidence of 6q deletions. V4-34 rearrangements were frequently associated with the use of JH5b, and V1-08 rearrangements were associated with a high incidence of trisomy 3q. Characteristics of these rearrangements and their relation to CD5+/IgM+ normal B cells and CLL is shown in Table 3.
Frequency and characteristics of the most commonly used VH genes in MCL compared with normal CD5+ IgM+ B cells and CLL
. | V1-08 . | V3-21 . | V3-23 . | V4-34 . | V4-59 . |
|---|---|---|---|---|---|
| Frequency in CD5+ B cells, %* | — | — | 13.9 | 3.5 | 6.3 |
| No. VH mutated | — | — | 9 of 20 | 0 of 5 | 0 of 9 |
| Frequency in CLL, %† | 1 | 4.7 | 4.7 | 7.8 | 2.1 |
| No. VH mutated | 0 of 3 | 8 of 14 | 8 of 14 | 19 of 23 | 3 of 8 |
| Frequency in MCL, % | 10.1 | 12.1 | 7.4 | 15.5 | 6.8 |
| No. VH mutated | 3 of 5 | 0 of 18 | 7 of 11 | 2 of 23 | 9 of 10 |
| Median CDR3 length, codons | 13 | 18 | 16 | 14 | 10 |
| Frequent D segment | — | D3-3: 5 of 15 | — | — | — |
| Frequent JH | JH4b: 6 of 15;JH5b: 6 of 15 | JH6: 12 of 17 | — | JH5b: 9 of 23;JH4b: 9 of 23 | JH4b: 7 of 10 |
| Frequent genomic aberration | +3q: 10 of 14 | — | — | — | 6q-: 4 of 6 |
| Patients with CR | 2 of 7 | 3 of 10 | 1 of 7 | 4 of 20 | 5 of 8 |
| Median OS, mo (no. patients) | 43.3 (8) | 103.4 (12) | 33.4 (8) | 34.5 (21) | 45.1 (9) |
. | V1-08 . | V3-21 . | V3-23 . | V4-34 . | V4-59 . |
|---|---|---|---|---|---|
| Frequency in CD5+ B cells, %* | — | — | 13.9 | 3.5 | 6.3 |
| No. VH mutated | — | — | 9 of 20 | 0 of 5 | 0 of 9 |
| Frequency in CLL, %† | 1 | 4.7 | 4.7 | 7.8 | 2.1 |
| No. VH mutated | 0 of 3 | 8 of 14 | 8 of 14 | 19 of 23 | 3 of 8 |
| Frequency in MCL, % | 10.1 | 12.1 | 7.4 | 15.5 | 6.8 |
| No. VH mutated | 3 of 5 | 0 of 18 | 7 of 11 | 2 of 23 | 9 of 10 |
| Median CDR3 length, codons | 13 | 18 | 16 | 14 | 10 |
| Frequent D segment | — | D3-3: 5 of 15 | — | — | — |
| Frequent JH | JH4b: 6 of 15;JH5b: 6 of 15 | JH6: 12 of 17 | — | JH5b: 9 of 23;JH4b: 9 of 23 | JH4b: 7 of 10 |
| Frequent genomic aberration | +3q: 10 of 14 | — | — | — | 6q-: 4 of 6 |
| Patients with CR | 2 of 7 | 3 of 10 | 1 of 7 | 4 of 20 | 5 of 8 |
| Median OS, mo (no. patients) | 43.3 (8) | 103.4 (12) | 33.4 (8) | 34.5 (21) | 45.1 (9) |
Features with strong associations in MCL are shown in bold letters. — indicates no frequent D, JH, or genomic aberration was assignable.
VH gene use in CD5+/IgM+ B cells from healthy donors according to Brezinschek et al45 ; the frequency of V1-08 and V3-21 was not reported.
VH gene use in CLL, as reported by Kröber et al.46
Biologic background (Bcl-6, isotype switch, ZAP70)
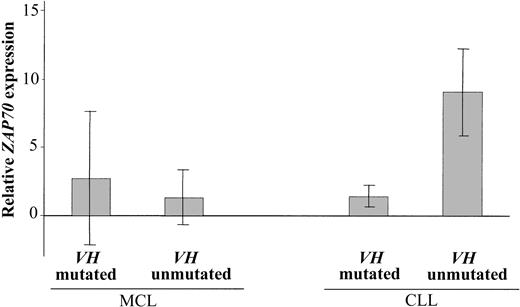
Immunohistochemical staining for Bcl-6 was performed in VH-mutated (n = 9) and VH-unmutated (n = 9) MCL. All rearrangements were immunohistochemically negative for Bcl-6. In 11 MCLs (VH mutated, n = 5; VH unmutated, n = 6) only μ constant region genes were detected, indicating no evidence for class switch recombination. ZAP-70 expression was investigated in patients with VH-mutated (n = 11) and VH-unmutated (n = 11) MCL and in patients with VH-mutated (n = 3) and VH-unmutated (n = 3) CLL. In CLL, the relative ZAP70 expression level was higher in all VH-unmutated patients than in VH-mutated patients (Table 4; Figure 1). In contrast to CLL, no differential expression of ZAP70 was found in the MCL subgroups defined by VH mutation status. Some MCL patients exhibited high ZAP70 expression, but this was mostly because of a high proportion of contaminating T cells indicated by high CD3 expression.
Relative ZAP70 and CD3 expression levels in patients with VH-mutated and VH-unmutated MCL and CLL
. | VH-mutated MCL . | . | VH-unmutated MCL . | . | ||
|---|---|---|---|---|---|---|
. | ZAP70 . | CD3 . | ZAP70 . | CD3 . | ||
| Patients with MCL | ||||||
| 1 | 0.17 | 0.03 | 0.72 | 1.67 | ||
| 2 | 2.89 | 4.17 | 1.52 | 1.09 | ||
| 3 | 16.48 | 0.51 | 7.30 | 5.08 | ||
| 4 | 7.99 | 7.11 | 0.53 | 0.02 | ||
| 5 | 0.25 | 0.22 | 0.60 | 0.01 | ||
| 6 | 0.30 | 1.43 | 1.48 | 0.12 | ||
| 7 | 0.16 | 0.41 | 2.17 | 0.58 | ||
| 8 | 1.12 | 0.02 | 0.02 | 0.15 | ||
| 9 | 0.04 | 0.29 | 0.03 | 0.06 | ||
| 10 | 0.17 | 4.84 | 0.07 | 0.27 | ||
| 11 | 0.01 | 0.25 | 0.00 | 0.04 | ||
| Median | 0.25 | 0.41 | 0.6 | 0.15 | ||
| Quartiles | 0.165-2.00 | 0.235-2.80 | 0.05-1.50 | 0.05-0.835 | ||
| Patients with CLL | ||||||
| 1 | 1.93 | 3.02 | 4.45 | 0.38 | ||
| 2 | 0.31 | 2.54 | 11.42 | 1.07 | ||
| 3 | 2.09 | 1.80 | 11.05 | 0.29 | ||
. | VH-mutated MCL . | . | VH-unmutated MCL . | . | ||
|---|---|---|---|---|---|---|
. | ZAP70 . | CD3 . | ZAP70 . | CD3 . | ||
| Patients with MCL | ||||||
| 1 | 0.17 | 0.03 | 0.72 | 1.67 | ||
| 2 | 2.89 | 4.17 | 1.52 | 1.09 | ||
| 3 | 16.48 | 0.51 | 7.30 | 5.08 | ||
| 4 | 7.99 | 7.11 | 0.53 | 0.02 | ||
| 5 | 0.25 | 0.22 | 0.60 | 0.01 | ||
| 6 | 0.30 | 1.43 | 1.48 | 0.12 | ||
| 7 | 0.16 | 0.41 | 2.17 | 0.58 | ||
| 8 | 1.12 | 0.02 | 0.02 | 0.15 | ||
| 9 | 0.04 | 0.29 | 0.03 | 0.06 | ||
| 10 | 0.17 | 4.84 | 0.07 | 0.27 | ||
| 11 | 0.01 | 0.25 | 0.00 | 0.04 | ||
| Median | 0.25 | 0.41 | 0.6 | 0.15 | ||
| Quartiles | 0.165-2.00 | 0.235-2.80 | 0.05-1.50 | 0.05-0.835 | ||
| Patients with CLL | ||||||
| 1 | 1.93 | 3.02 | 4.45 | 0.38 | ||
| 2 | 0.31 | 2.54 | 11.42 | 1.07 | ||
| 3 | 2.09 | 1.80 | 11.05 | 0.29 | ||
Relative expression of ZAP70 and CD3 in patients with VH-mutated and -unmutated (n = 22) MCL. For comparison, data from patients with VH-mutated and -unmutated (n = 6) CLL are shown. ZAP70 expression levels are presented as relative expression levels after normalization on the endogenous control gene PGK.
Mean ZAP70 expression levels. Mean relative ZAP70 expression levels in patients with VH-mutated MCL (n = 11) and -unmutated MCL (n = 11) and in patients with VH-mutated CLL (n = 3) and -unmutated CLL (n = 3). Mean relative ZAP70 expression levels are indicated by columns; error bars represent the corresponding standard deviation.
Mean ZAP70 expression levels. Mean relative ZAP70 expression levels in patients with VH-mutated MCL (n = 11) and -unmutated MCL (n = 11) and in patients with VH-mutated CLL (n = 3) and -unmutated CLL (n = 3). Mean relative ZAP70 expression levels are indicated by columns; error bars represent the corresponding standard deviation.
Genomic aberrations
Through interphase FISH, 73 (87%) of 87 patients with MCL exhibited genomic abnormalities in addition to t(11;14). In 29 patients (33.7%), a complex karyotype with 3 or more aberrations was detected, and tetraploidy or partial tetraploidy was detected in 11 patients (13.8%) (Table 5). The overall incidence of genomic abnormalities in the VH-mutated and VH-unmutated subgroups was similar, as was the incidence of the deletions 13q–, 11q–, and 17p–. Trisomies +3q and +8q were more frequently observed in VH-unmutated patients, whereas trisomy +12q was overrepresented in VH-mutated patients (Table 5). Tetraploidy occurred predominantly in VH-unmutated patients. The total number of genomic aberrations was inversely correlated with CDR3 length (Spearman rho = –0.26; P = .02)—that is, rearrangements with a long CDR3 were characterized by a low number of genomic aberrations.
Genomic aberrations in MCL overall and according to VH mutation status
Aberration . | Patients with aberrations (%) . | VH-mutated (%) . | VH-unmutated (%) . | Odds ratios VH-mutated/VH-unmutated (95% confidence interval) . |
|---|---|---|---|---|
| 3q trisomy | 36 of 86 (41.9) | 6 of 24 (25) | 30 of 62 (48.4) | 0.36 (0.12-1.02) |
| 8q trisomy | 19 of 87 (21.8) | 2 of 25 (8) | 17 of 62 (27.4) | 0.23 (0.05-1.08) |
| 12q trisomy | 14 of 86 (16.3) | 7 of 24 (29.2) | 7 of 62 (11.3) | 3.24 (0.99-10.5) |
| 6q deletion | 15 of 87 (17.2) | 6 of 25 (24) | 9 of 62 (14.5) | 1.86 (0.58-5.92) |
| 11q deletion | 35 of 87 (40.2) | 11 of 25 (44) | 24 of 62 (38.7) | 1.24 (0.49-3.19) |
| 13q deletion | 43 of 87 (49.4) | 10 of 25 (40) | 33 of 62 (53.2) | 0.59 (0.23-1.50) |
| 17p deletion | 18 of 86 (18) | 7 of 25 (28) | 11 of 61 (18) | 1.77 (0.59-5.26) |
| Tetraploidy | 12 of 87 (13.8) | 1 of 25 (4) | 11 of 62 (17.7) | 0.19 (0.02-1.58) |
| Complex* | 29 of 86 (33.7) | 5 of 24 (20.8) | 24 of 62 (38.7) | 0.42 (0.14-1.26) |
| No aberration | 13 of 86 (13) | 3 of 24 (12.5) | 10 of 62 (16.1) | 0.74 (0.19-2.97) |
Aberration . | Patients with aberrations (%) . | VH-mutated (%) . | VH-unmutated (%) . | Odds ratios VH-mutated/VH-unmutated (95% confidence interval) . |
|---|---|---|---|---|
| 3q trisomy | 36 of 86 (41.9) | 6 of 24 (25) | 30 of 62 (48.4) | 0.36 (0.12-1.02) |
| 8q trisomy | 19 of 87 (21.8) | 2 of 25 (8) | 17 of 62 (27.4) | 0.23 (0.05-1.08) |
| 12q trisomy | 14 of 86 (16.3) | 7 of 24 (29.2) | 7 of 62 (11.3) | 3.24 (0.99-10.5) |
| 6q deletion | 15 of 87 (17.2) | 6 of 25 (24) | 9 of 62 (14.5) | 1.86 (0.58-5.92) |
| 11q deletion | 35 of 87 (40.2) | 11 of 25 (44) | 24 of 62 (38.7) | 1.24 (0.49-3.19) |
| 13q deletion | 43 of 87 (49.4) | 10 of 25 (40) | 33 of 62 (53.2) | 0.59 (0.23-1.50) |
| 17p deletion | 18 of 86 (18) | 7 of 25 (28) | 11 of 61 (18) | 1.77 (0.59-5.26) |
| Tetraploidy | 12 of 87 (13.8) | 1 of 25 (4) | 11 of 62 (17.7) | 0.19 (0.02-1.58) |
| Complex* | 29 of 86 (33.7) | 5 of 24 (20.8) | 24 of 62 (38.7) | 0.42 (0.14-1.26) |
| No aberration | 13 of 86 (13) | 3 of 24 (12.5) | 10 of 62 (16.1) | 0.74 (0.19-2.97) |
Three or more aberrations.
Clinical and treatment characteristics
The clinical parameters of the MCL patients, their significance with respect to OS in the entire cohort, and the distribution in the VH-mutated and -unmutated subgroups are shown in Table 6. Factors associated with inferior clinical outcome were IPI score 3 or higher, B symptoms, and age older than 60. Estimated median OS time of all patients was 39 months. Achieving complete remission (CR) after initial chemotherapy was associated with better survival times (Table 6). A higher rate of complete responders was observed in the VH-mutated group, though treatment modalities were comparable in the 2 VH groups (P = .031; Table 6). Five of 8 patients with V4-59 use achieved CR after initial therapy (Table 3). There was no association of any other clinical feature with VH mutation status.
Distribution of clinical and treatment characteristics in MCL according to VH mutation status
Clinical feature . | Significance for OS, adjusted P* . | VH-mutated (%) . | VH-unmutated (%) . | Differential distribution, P . |
|---|---|---|---|---|
| IPI score 3-5 | .04 | 13 of 22 (59.1) | 29 of 56 (51.8) | .62 |
| B symptoms | .04 | 12 of 29 (41.4) | 34 of 72 (47.2) | .66 |
| Older than 60 y | .05 | 18 of 32 (56.3) | 45 of 79 (57) | 1.0 |
| Bulk, 10 cm or more | .07 | 9 of 27 (33.3) | 13 of 63 (20.6) | .28 |
| LDH level elevated | .13 | 9 of 25 (36) | 23 of 61 (37.7) | 1.0 |
| Leukemic manifestation | .20 | 16 of 30 (53.3) | 31 of 69 (44.9) | .51 |
| Female sex | .27 | 7 of 37 (18.9) | 22 of 93 (23.7) | .65 |
| Splenomegaly | .34 | 14 of 29 (48.3) | 46 of 72 (63.9) | .18 |
| Bone marrow involvement | .64 | 20 of 29 (69) | 55 of 72 (76.4) | .46 |
| Stage III/IV | .67 | 30 of 32 (93.8) | 68 of 76 (89.5) | .72 |
| Palpable lymphadenopathy | .93 | 18 of 24 (75) | 56 of 79 (70.9) | .80 |
| Gastrointestinal involvement | .93 | 4 of 25 (16) | 20 of 77 (26) | .42 |
| CR after initial therapy† | .002 | 10 of 25 (40) | 12 of 68 (17.6) | .031 |
Clinical feature . | Significance for OS, adjusted P* . | VH-mutated (%) . | VH-unmutated (%) . | Differential distribution, P . |
|---|---|---|---|---|
| IPI score 3-5 | .04 | 13 of 22 (59.1) | 29 of 56 (51.8) | .62 |
| B symptoms | .04 | 12 of 29 (41.4) | 34 of 72 (47.2) | .66 |
| Older than 60 y | .05 | 18 of 32 (56.3) | 45 of 79 (57) | 1.0 |
| Bulk, 10 cm or more | .07 | 9 of 27 (33.3) | 13 of 63 (20.6) | .28 |
| LDH level elevated | .13 | 9 of 25 (36) | 23 of 61 (37.7) | 1.0 |
| Leukemic manifestation | .20 | 16 of 30 (53.3) | 31 of 69 (44.9) | .51 |
| Female sex | .27 | 7 of 37 (18.9) | 22 of 93 (23.7) | .65 |
| Splenomegaly | .34 | 14 of 29 (48.3) | 46 of 72 (63.9) | .18 |
| Bone marrow involvement | .64 | 20 of 29 (69) | 55 of 72 (76.4) | .46 |
| Stage III/IV | .67 | 30 of 32 (93.8) | 68 of 76 (89.5) | .72 |
| Palpable lymphadenopathy | .93 | 18 of 24 (75) | 56 of 79 (70.9) | .80 |
| Gastrointestinal involvement | .93 | 4 of 25 (16) | 20 of 77 (26) | .42 |
| CR after initial therapy† | .002 | 10 of 25 (40) | 12 of 68 (17.6) | .031 |
To account for multiple testing of clinical variables, P values were adjusted according to Holm.43
Treatment modalities among patients achieving CR (VH-mutated vs -unmutated): radiotherapy, 2 versus 3; chemotherapy, 8 versus 11; polychemotherapy, 8 versus 10; anthracyclines, 4 versus 7; ASCT, 1 versus 2. P values not adjusted.
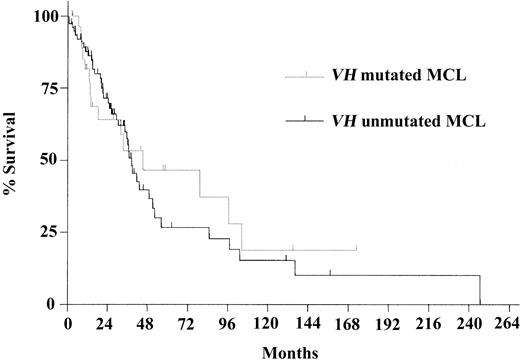
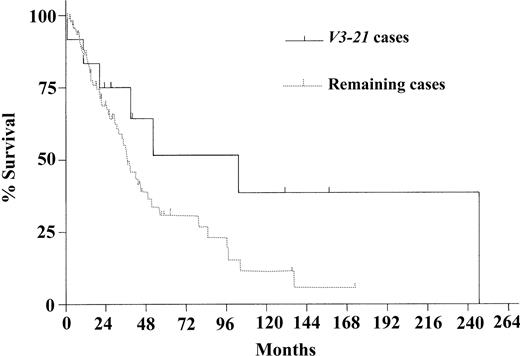
Survival probability was not different when comparing the VH mutation subgroups (median OS in VH-mutated and VH-unmutated groups, 45.1 and 38.4 months, respectively [P = .67]; Figure 2). Restricting the analysis to the patients with t(11;14) or cyclin D1 positivity (n = 98) showed a median survival time of 45.1 months in the VH-mutated and 38.4 months in the VH-unmutated group (P = .57). Similarly, none of the frequently used VH genes (V4-34, V1-08, V3-23, and V4-59) were correlated with differences in survival probability except for the group of patients using V3-21, which showed a trend toward longer survival time (median, 103.4 vs 36.6 months; P = .08; Figure 3). Even after exclusion of the patients using V3-21 from the unmutated VH group, the difference between the VH subgroups was not significant (median OS, 45.1 vs 36.6 months; P = .28). Regarding the subgroups of nodal and nonnodal MCL, as described by Orchard et al,24 there was no significant difference in OS time (37.0 and 97.3 months, respectively; P = .56).
Kaplan-Meier survival curves. Curves compare VH-mutated and -unmutated MCL. Median survival times were 45.1 months in the VH-mutated subgroup (n = 30) and 38.4 months in the VH-unmutated subgroup (n = 75) (P = .67).
Kaplan-Meier survival curves. Curves compare VH-mutated and -unmutated MCL. Median survival times were 45.1 months in the VH-mutated subgroup (n = 30) and 38.4 months in the VH-unmutated subgroup (n = 75) (P = .67).
Kaplan-Meier survival estimates. Estimates compare V3-21 patients with the remaining MCL patients. Median survival times were 103.4 months in the V3-21 group (n = 12) and 36.6 months in the group with the remaining patients (n = 93) (P = .08).
Kaplan-Meier survival estimates. Estimates compare V3-21 patients with the remaining MCL patients. Median survival times were 103.4 months in the V3-21 group (n = 12) and 36.6 months in the group with the remaining patients (n = 93) (P = .08).
Discussion
VH mutation status and VDJ rearrangement structure were analyzed in relation to biologic and clinical characteristics in a large series of MCL patients. Clinical characteristics and genomic aberrations were analyzable only in subsets of the 141 patients, but these subsets were comparable to the entire cohort with respect to VH mutation status and VDJ gene distribution; therefore, selection biases in the subsets seemed unlikely.
Mutated VH genes were detected in 41 (29%) of the 141 MCL patients in this series, thus confirming the presence of a significant fraction of somatically VH-mutated MCL, as recently described.23-27 Therefore, it appears that MCL, similar to CLL, can arise from the malignant transformation of a pre-germinal–center B cell with unmutated VH or a post-germinal–center B cell with mutated VH. However, the mean rate of VH mutations in the entire MCL cohort (1.4%) and in the VH-mutated subgroup (3.7%) was lower than in CLL (2.9% and 6.1%, respectively)34 and compared with typically VH-mutated lymphomas such as diffuse large B-cell lymphoma (overall, 9.6%).47
To further elucidate the cellular nature of the 2 VH mutation status MCL subtypes, we studied a number of markers related to B-cell maturation. There was no evidence for class switch recombination, which is frequently observed in CLL with mutated VH.39 Bcl-6, a marker expressed by germinal center cells, was uniformly negative in both VH subtypes of MCL, arguing against a germinal-center B cell as progenitor for VH-mutated MCL, which is in line with the lack or low frequency of ongoing VH mutation in MCL.24,26,48 In CLL, both VH mutation status subgroups appear similar and more related to post-germinal–center (memory) B cells based on their gene expression signatures, but they can be distinguished by differential expression of B-cell receptor (BCR)–signaling-associated genes such as ZAP70.30-33 In contrast, and in line with a recent immunohistochemistry study,49 ZAP70 was expressed mostly at low levels independently of VH mutation status, arguing against a pathogenic role of differential BCR signaling capacity between the 2 MCL subtypes, as recently documented for CLL.32,33
Despite these similarities, a number of differences were observed between the 2 MCL VH mutation subgroups. The most frequently used VH genes—V4-34, V3-21, V3-23, and V4-59—were associated with characteristic features. Only 5 individual VH genes accounted for more than half of all rearrangements, and 4 of these genes showed significant associations with VH mutation status. Rearrangements using V3-21 were unmutated and were associated with long CDR3 regions and with the use of D3-3 and JH6. V4-59, in contrast to another recent report,25 was typically VH-mutated and related to short CDR3 regions. These observations indicate that antigen specificity may be involved in MCL development, as discussed recently.23-27
The comparison of VDJ structure and VH status between MCL and CLL revealed striking differences. V4-34, one of the most commonly used genes in both diseases, was typically mutated in CLL but was unmutated in MCL, whereas V3-21, mostly mutated in CLL, was unmutated in all MCL patients.28,29,50 V1-69, by far the most frequently used unmutated VH gene in CLL, was rarely used in MCL, whereas V1-08, one of the most common VH genes in MCL, was rarely used in CLL. These findings could reflect a different immunogenetic background triggering the pathogenesis of both diseases.
The oncogenetic hallmark of MCL is t(11;14), leading to cyclin D1 overexpression. However, additional genomic aberrations appear to be required for malignant transformation51,52 and were, indeed, observed in 87% of our patients. When comparing the 2 VH-mutation subtypes of MCL, there was no difference in the overall presence of genomic aberrations, whereas all trisomies (+3q, +8q, and +12q) and tetraploidy showed an imbalanced distribution, indicating distinct oncogenetic pathways. However, the deletions 17p– and 11q– were observed at a similar incidence in VH-mutated and -unmutated MCL, which is in marked contrast to CLL, in which most patients with 17p– and virtually all patients with 11q– had VH unmutated rearrangements.34-36 This is of note because the inactivation of p53 and, particularly, ATM have been viewed as pathogenic events exclusively observed in pre-germinal–center B cells based on the CLL data.34-36 The inactivation of ATM because of the deletion of 1 allele and the mutation of the remaining allele are frequent events in MCL,18-20 but they appear to occur in the pre-germinal– and in the post-germinal–center B-cell–derived variant.
Clinically, VH mutation status gained great interest as a marker separating 2 subgroups of CLL with markedly different outcomes.28,29,34-36 In the current MCL series, there was no association between VH mutation status and the clinical features studied with the exception of a higher CR rate in the VH-mutated subgroup, possibly reflecting a different in vivo chemosensitivity. However, this did not translate into clinical outcome because there was no significant association between VH mutation status and OS, which is in agreement with other recent reports.24-26 No correlation of a specific VH gene with survival probability was observed except for a trend toward a better outcome for MCL patients using V3-21 as described.25,26 This is in contrast to CLL, in which the use of V3-21, independently of its mutation status, appears to be an adverse prognostic factor.46,49 The collection of cases under study was not obtained in a single clinical trial and is therefore prone to selection bias. However, factors such as age, IPI score, B symptoms, and response to treatment showed the expected relation with outcome, indicating that the series may represent a valid collection for the evaluation of prognostic factors. Therefore, and in agreement with other recent studies,24-26 the current findings do not argue for a strong impact of VH mutation status or VDJ structure on the clinical behavior in MCL.
An explanation for this, particularly when comparing MCL with CLL, may be that the effect of VH mutation status and VDJ structure in MCL may be overruled by the cell-cycle deregulation subsequent to cyclin D1 overexpression. Accordingly, Rosenwald et al15 recently described a correlation between expression levels of cyclin D1 and other “proliferation signature” genes and survival probability in MCL. Analogously, Korz et al,42 in a comprehensive RQ-PCR study, demonstrated the predominance of cell-cycle deregulation in MCL compared with impaired apoptosis control in CLL. Therefore, t(11;14) leading to cyclin D1 overexpression and the subsequent cell-cycle deregulation may be the dominating factors with respect to biologic and clinical behaviors of MCL.
Prepublished online as Blood First Edition Paper, July 3, 2003; DOI 10.1182/blood-2003-05-1383.
Supported by grants from the University of Ulm (P766), European Community (EC) (QLG1-CT-2000-00687), Bundesministerium für Bildung und Forschung (BMBF) (01KW9938/4, NGFN01GR0101), and Sander Stiftung (2001.004.1).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Sabrina Kless, Sandrine Sander, and Lars Bullinger for their excellent technical assistance and contributions to the FISH analyses.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal