Abstract
Leukemic B-chronic lymphoproliferative disorders (B-CLPDs) are generally believed to derive from a monoclonal B cell; biclonality has only occasionally been reported. In this study, we have explored the incidence of B-CLPD cases with 2 or more B-cell clones and established both the phenotypic differences between the coexisting clones and the clinicobiologic features of these patients. In total, 53 B-CLPD cases with 2 or more B-cell clones were studied. Presence of 2 or more B-cell clones was suspected by immunophenotype and confirmed by molecular/genetic techniques in leukemic samples (n = 42) and purified B-cell subpopulations (n = 10). Overall, 4.8% of 477 consecutive B-CLPDs had 2 or more B-cell clones, their incidence being especially higher among hairy cell leukemia (3 of 13), large cell lymphoma (2 of 10), and atypical chronic lymphocytic leukemia (CLL) (4 of 29). In most cases the 2 B-cell subsets displayed either different surface immunoglobulin (sIg) light chain (n = 37 of 53) or different levels of the same sIg (n = 9 of 53), usually associated with other phenotypic differences. Compared with monoclonal cases, B-CLL patients with 2 or more clones had lower white blood cell (WBC) and lymphocyte counts, more frequently displayed splenomegaly, and required early treatment. Among these, the cases in which a CLL clone coexisted with a non-CLL clone were older and more often displayed B symptoms, a monoclonal component, and diffuse infiltration of bone marrow and required early treatment more frequently than cases with monoclonal CLL or 2 CLL clones.
Introduction
B-cell chronic lymphoproliferative disorders (B-CLPDs) are a heterogeneous group of diseases that result from the proliferation and accumulation of mature-appearing aberrant B lymphocytes arrested at a given stage of differentiation.1-5 B-CLPDs are generally believed to result from the monoclonal expansion of a single transformed B lymphocyte.6 Therefore, the tumoral B lymphocytes constitute a clone in which all cells are related by possessing the original transforming mutation, possessing identical VDJ rearrangements of the immunoglobulin heavy chain (IgH) gene, and showing restricted Ig light chain expression. Additionally, distinctive genetic and phenotypic features may be gained during the evolution of the disease. In this sense, the presence of 2 or more morphologically different populations of neoplastic lymphocytes in the same patient, detected either simultaneously or at different time points, is usually interpreted as reflecting either different maturation stages or subclone formation within the original malignant tumor stem cell line.7 Although most lymphoproliferative disorders are considered to be monoclonal, biclonality has been occasionally reported in the literature,8-19 frequently corresponding to rare cases of B-cell non-Hodgkin lymphomas (NHLs), which more commonly develop in immunocompromised individuals.8-11 In most instances these reports describe the coexistence of 2 distinct diseases such as mantle cell lymphoma (MCL) and follicular lymphoma (FL),17 FL and small lymphocytic lymphoma (SLL),17 MCL and chronic lymphocytic leukemia/SLL (CLL/SLL),17 SLL and large cell lymphoma (LCL),18 and hairy cell leukemia (HCL) and CLL/SLL.19 Therefore, these cases would not correspond to oligoclonal expansions of B cells, which have long been claimed to precede the monoclonal disease state.12
Despite this, to the best of our knowledge, no study has been reported so far in which either the presence of 2 or more different B-cell clones has been systematically analyzed in a large series of patients with leukemic B-CLPD or the clinical and biologic characteristics of patients with 2 or more B-cell clones have been compared with those of monoclonal cases.
The aim of the present study was to establish the incidence of cases with 2 or more B-cell clones based on a large series of 477 consecutive leukemic B-CLPD patients, to establish the different phenotypic patterns of the coexisting neoplastic B-cell clones, and to compare the clinical and biologic disease features of cases with 2 or more B-cell clones versus monoclonal B-CLPD patients.
Patients, materials, and methods
Patients
A total of 507 newly diagnosed untreated patients suffering from leukemic B-CLPD were analyzed in this study. From them, 477 corresponded to consecutive B-CLPDs studied in our laboratory and the other 30 cases to patients suspected of having 2 or more clones by phenotypic criteria, who were referred to our center for this specific reason. Diagnosis of B-CLPD was established in all cases on the basis of clinical, morphologic, immunophenotypic, molecular and, whenever available, also histologic criteria, according to the World Health Organization (WHO) classification.20 In 36% of the cases diagnosis was established in a routine blood cell analysis. Distribution according to diagnosis was as follows: 399 cases were CLL/SLL (for simplicity they were referred as CLL) (365 were typical [typCLL] and 34 atypical [atypCLL]); prolymphocytic leukemia (PLL), 5; hairy cell leukemia (HCL), 14; lymphoplasmacytic lymphoma (LPL), 16; splenic marginal zone lymphoma (SMZL), 16; FL, 25; MCL, 20; and LCL, 10 cases; 2 B-CLPD patients could not be further classified. Mean age of the patients was 70 ± 11 years (median, 71 years; range, 19-95 years); 314 were males and 193 were females. In all cases, samples were obtained for this study after informed consent according to the Ethics Committee of the University Hospital of Salamanca. At the moment of closing of this study, the median follow-up of the whole series was 28 months (range, 3-75 months): 29 months for consecutively studied patients and 21 months for referred cases.
Cytomorphologic studies
Either peripheral blood (PB) and/or bone marrow (BM) smears from EDTA (ethylenediaminetetraacetic acid)–-anticoagulated samples, prepared immediately after sampling, were stained with May-Grünwald-Giemsa (MGG) and cell morphology was analyzed by optical microscopy. The samples were examined by 2 independent observers and lymphocytes classified into the following categories: (1) small lymphocytes, (2) cleaved lymphocytes, (3) large nongranular lymphocytes, (4) large granular lymphocytes, (5) prolymphocytes, and (6) lymphoplasmacytoid cells. The presence or absence of “smudged cells” was also evaluated in each sample.
Immunophenotypic analyses
PB (n = 417) and BM (n = 80) samples were collected in tubes containing K3 EDTA as anticoagulant, while ascitic fluid (n = 1) was placed in a tube containing heparin. Lymph node samples (n = 9) were collected in an isotonic saline buffer, cut into small pieces that were then placed in RPMI medium (BioWhittaker, Walkersville, MD), and gently dispersed into single cell suspensions; after 2 washing steps, the cell pellet was resuspended in phosphate-buffered saline (PBS) at a concentration of around 2 × 106 cells per 100 μL. All samples were stained using a direct immunofluorescence stain-and-then-lyse technique previously described in detail21 with the following 3-color combinations of monoclonal antibodies (moAb) conjugated with fluorescein isothiocyanate (FITC)/phycoerythrin (PE)/and the PE–cyanine 5 (PE/Cy5) fluorochrome tandem: FMC7/CD5/CD19, CD22/CD23/CD19, CD103/CD25/CD19, CD10/CD11c/CD19, and sIgκ/sIgλ/CD19. Directly conjugated moAb reagents were purchased from Becton Dickinson Biosciences (BDB, San Jose, CA): CD5-PE, CD10-PE, CD11c-FITC, CD22-FITC, CD23-PE, CD25-PE, and anti-κ–FITC/anti-λ–PE; from Caltag Laboratories (San Francisco, CA): CD19-PE/Cy5; from Immunotech (Marseille, France): FMC7-FITC; and from Immunoquality Products (Gröningen, The Netherlands): CD103-FITC. Data acquisition was performed using CellQuest software on a FACSort flow cytometer (BDB). In those samples with a low percentage of B lymphocytes, a selective acquisition of CD19+ gated cells was also made as previously described in detail.21 The median percentage of CD19+ clonal B cells within the lymphocyte gate was 62% (range, 3.4%-95%). For data analysis, the Paint-A-Gate PRO software program (BDB) was used. B lymphocytes were identified according to their SSClo/int/CD19+ distribution, and their percentage was calculated after excluding cell debris and platelets according to conventional procedures.21
Assessment of clonality by molecular biology
Genomic DNA preparation and Southern blot analysis. High molecular weight DNA was isolated by standard proteinase K digestion, phenolchloroform extraction, and ethanol precipitation.22 For Southern blot analysis, 10 μg DNA was digested to completion with the BglII and BamHI/HindIII restriction enzymes, size fractionated in 0.7% agarose gels, and blotted onto nylon membranes as previously described.23 The blots were hybridized with the IgHJ6 digoxigenin-labeled probe.
PCR amplification and heteroduplex analysis. For amplification of complete VDJH gene rearrangements 3 different sets of family-specific primers and 1 JH consensus primer were used in 3 different multiplexed polymerase chain reactions (PCRs) covering the 3 framework regions (FRs). Amplification of incomplete DJH gene rearrangements was performed in 2 different reactions using family-specific primers for DH1 to DH6 and DH7 families, respectively, together with the consensus JH primer. All primers had been newly designed during the BIOMED-2 concerted action “PCR-based clonality studies for early diagnosis of lymphoproliferative disorders” (PL96-3936).24 All reactions were carried out in 50 μL containing 0.1 μg DNA samples and 10 pmol of each primer.
For heteroduplex analysis, PCR products were denatured at 94°C for 10 minutes and subsequently cooled at 4°C for 60 minutes to induce duplex formation.23-25 The heteroduplexes and/or homoduplexes generated were immediately loaded on a 10% nondenaturing polyacrylamide gel in 1 × Tris (tris(hydroxymethyl)aminomethane)–acetate–EDTA (TAE) buffer, run at room temperature (RT), and visualized by ethidium bromide staining. The presence of one or more clear bands within the expected size indicated clonal rearrangements, while polyclonal rearrangements appeared as a rough smear in the gel.23-25
Sequencing and analysis of Ig genes. PCR products were eluted from the polyacrylamide gels and directly sequenced in an automated ABI 377 DNA sequencer using Big-Dye terminators (Applied Biosystems, Foster City, CA). To avoid nucleotide misinterpretations due to Taq errors, all products were sequenced at least twice from different PCR reactions using VH/DH and/or JH primers. Germ line VH, DH, and JH segments from complete VDJH gene rearrangements were identified by comparison with the V base26 and IGMT database27 using online DNAPLOT (MRC Center for Protein Engineering, Cambridge, United Kingdom). DH and JH germ line segments from incomplete DJH gene rearrangements were identified using BLAST search in the DH-JH germ line locus sequence (accession no. EMB/X97051).
Molecular biclonalilty/triclonalilty was defined when 3 or 4 unrelated VDJH, DJH gene rearrangements and/or translocations involving the 14q32 IGH gene locus were found. Secondary rearrangements to a pre-existent VDJH or DJH gene rearrangement were not taken into account for definition of biclonalilty/triclonalilty.
Cytogenetic analysis
Cytogenetic studies were carried out in BM samples from 20 of the 53 multiclonal cases, after culture with TPA (12-O-tetradecanoylphorbol-13-acetate) for 72 hours, according to previously reported methods.28 In addition, conventional interphase fluorescence in situ hybridization (iFISH) for the translocations t(11;14), t(14;18), and t(11;18) was performed in FACS-purified subsets of clonal B cells in a subgroup of 6 of these cases using Vysis (Downers Grove, IL) LSI IgH/CCND1 dual-color, dual-fusion translocation probe; LSI IgH/Bcl2 dual-color, dual-fusion translocation probe; and LSI MALT1 dual-color Breakapart probe.
Flow cytometry sorting of B-cell subsets
In 10 cases in which different B-cell subsets displaying aberrant phenotypes were identified, specific fluorescence-activated cell sorting (FACS) of each of the 2 B-cell subpopulations was performed in a FACS Vantage flow cytometer (BDB). Discrimination between the 2 cell subsets was based on their distinct expression for 1 or more B-cell markers, according to well-established methods. Molecular and cytogenetic clonality based on IgH gene rearrangements as well as on the presence of different chromosome translocations was further assessed in the sorted B cells using the techniques described elsewhere in this paper. The purity of the sorted fractions was 98% ± 1.6% (range, 95%-99.5%).
Statistical methods
Either relative frequencies or mean values and their standard deviation, as well as range, were calculated for each categorical or continuous variable under study using the SPSS software program (SPSS 10.0, Chicago, IL). To explore for the potential statistical significance of differences found between groups of B-CLPD patients, the Student t and χ2 tests (SPSS 10.0.) were used for continuous and categorical variables, respectively. Curves representing the cases requiring treatment within each subgroup of patients were plotted according to the method of Kaplan and Meier, and the comparison between curves was performed using the log-rank test. P < .05 was considered to be associated with statistical significance.
Results
Diagnosis of B-CLPD with 2 or more B-cell clones
The presence of 2 or more B-cell clones was primarily suspected based on immunophenotypic studies in a total of 53 B-CLPD cases. From these patients, 23 were from the group of 477 B-CLPD cases consecutively analyzed in our laboratory and 30 corresponded to selected patients whose samples were specifically referred to our laboratory for confirmation of suspected presence of 2 or more B-cell clones. In all these 53 cases, 2 different neoplastic B-cell populations were observed, which showed a different pattern of expression for 1 or more phenotypic markers. Clonality molecular studies devoted to confirm the existence of 2 different tumoral B-cell clones—based on the coexistence of 3 or more distinct rearrangements of the IgH gene—were carried out in a total of 44 of these 53 cases. In the remaining 9 cases, molecular studies could not be performed due to shortage of the sample. In all 44 cases studied, molecular techniques confirmed the presence of at least 2 different B-cell clones, based on the criteria defined above (Table 1). In addition, in 10 of these cases, molecular analysis and/or iFISH studies were performed individually on each of the 2 phenotypically distinct B-cell subpopulations purified by fluorescence-activated cell sorting, confirming that each of the 2 B-cell fractions had different IgH gene rearrangements and/or chromosome translocations (Figure 1). To exclude the presence of 2 or more B-cell clones in the remaining 454 cases that showed a uniform antigenic profile (continuous expression of markers), we randomly selected and studied a cohort of 54 cases. None of them showed 3 or more IgH rearrangements. The distribution of the 53 cases with 2 or more B-cell clones according to their diagnosis was as follows: B-CLL, 33 cases (24 typical and 9 atypical); HCL, 4 patients; LPL, 4 cases; SMZL, 3 cases; FL, 3 patients; MCL, 2 cases; and LCL, 2 cases; the remaining 2 B-CLPD patients could not be further subclassified.
Molecular characteristics of B-CLPD with 2 or more B-cell clones
. | . | Molecular studies . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|
| . | . | Southern blot . | . | PCR . | . | |||
| Case no. . | Diagnosis . | Pattern of IgH gene rearrangement . | No. of clones . | Pattern of IgH gene rearrangement . | No. of clones . | |||
| 1 | typCLL | RR RG | 2 | RR RG | 2 | |||
| 2 | typCLL | RR RR | 2 | RR RG | 2 | |||
| 3 | typCLL | RR RG | 2 | RR RR | 2 | |||
| 4 | typCLL | RR RG | 2 | RR RG | 2 | |||
| 5 | typCLL | RR RR RG | 3 | RR RG | 2 | |||
| 6 | typCLL | NA | NA | NA | NA | |||
| 7 | typCLL | RR RR | 2 | RR RR | 2 | |||
| 8 | typCLL | RR | 1 | RR RG | 2 | |||
| 9 | typCLL | RR | 1 | RR RR | 2 | |||
| 10 | typCLL | RR | 1 | RR RR | 2 | |||
| 11 | typCLL | RR RR | 2 | RR RG | 2 | |||
| 12 | typCLL | RR RG | 2 | RR RR | 2 | |||
| 13 | typCLL | RR RG | 2 | RR | 1 | |||
| 14 | typCLL | NA | NA | RR RG | 2 | |||
| 15 | typCLL | RR | 1 | RR RR | 2 | |||
| 16 | typCLL | NA | NA | RR RR | 2 | |||
| 17 | typCLL | NA | NA | RR RG | 2 | |||
| 18 | typCLL | NA | NA | RR RG | 2 | |||
| 19 | typCLL | NA | NA | RR RG | 2 | |||
| 20 | typCLL | NA | NA | RG RG | 2 | |||
| 21 | typCLL | NA | NA | RR RG | 2 | |||
| 22 | typCLL | NA | NA | RR RR | 2 | |||
| 23 | typCLL | NA | NA | NA | NA | |||
| 24 | typCLL | NA | NA | NA | NA | |||
| 25 | atypCLL | RR | 1 | RR RG | 2 | |||
| 26 | atypCLL | RR RG | 2 | RR | 1 | |||
| 27 | atypCLL | NA | NA | NA | NA | |||
| 28 | atypCLL | RR RR | 2 | RR RR | 2 | |||
| 29 | atypCLL | RR RR | 2 | RR RR RR | 3 | |||
| 30 | atypCLL | RR | 1 | RR RG | 2 | |||
| 31 | atypCLL | NA | NA | RR RR | 2 | |||
| 32 | atypCLL | RR | 1 | RR RG | 2 | |||
| 33 | atypCLL | NA | NA | NA | NA | |||
| 34 | HCL | RR RG | 2 | NA | NA | |||
| 35 | HCL | NA | NA | NA | NA | |||
| 36 | HCL | NA | NA | RR RR | 2 | |||
| 37 | HCL | NA | NA | RR RG | 2 | |||
| 38 | LPL | NA | NA | RR RG | 2 | |||
| 39 | LPL | RR | 1 | RR RR | 2 | |||
| 40 | LPL | NA | NA | NA | NA | |||
| 41 | LPL | NA | NA | RR RG | 2 | |||
| 42 | SMZL | RR | 1 | RR RR | 2 | |||
| 43 | SMZL | NA | NA | RR RG | 2 | |||
| 44 | SMZL | NA | NA | NA | NA | |||
| 45 | FL | NA | NA | NA | NA | |||
| 46 | FL | RR RR | 2 | RR RG | 2 | |||
| 47 | FL | GG | 0 | RR RR | 2 | |||
| 48 | MCL | RR RR | 2 | RR | 1 | |||
| 49 | MCL | RR RR RR | 3 | RR RR | 2 | |||
| 50 | LCL | RR RR | 2 | RR | 1 | |||
| 51 | LCL | RG | 1 | RR RR | 2 | |||
| 52 | Unclassifiable | RR RR | 2 | RR RG | 2 | |||
| 53 | Unclassifiable | RR | 1 | RR RG | 2 | |||
. | . | Molecular studies . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|
| . | . | Southern blot . | . | PCR . | . | |||
| Case no. . | Diagnosis . | Pattern of IgH gene rearrangement . | No. of clones . | Pattern of IgH gene rearrangement . | No. of clones . | |||
| 1 | typCLL | RR RG | 2 | RR RG | 2 | |||
| 2 | typCLL | RR RR | 2 | RR RG | 2 | |||
| 3 | typCLL | RR RG | 2 | RR RR | 2 | |||
| 4 | typCLL | RR RG | 2 | RR RG | 2 | |||
| 5 | typCLL | RR RR RG | 3 | RR RG | 2 | |||
| 6 | typCLL | NA | NA | NA | NA | |||
| 7 | typCLL | RR RR | 2 | RR RR | 2 | |||
| 8 | typCLL | RR | 1 | RR RG | 2 | |||
| 9 | typCLL | RR | 1 | RR RR | 2 | |||
| 10 | typCLL | RR | 1 | RR RR | 2 | |||
| 11 | typCLL | RR RR | 2 | RR RG | 2 | |||
| 12 | typCLL | RR RG | 2 | RR RR | 2 | |||
| 13 | typCLL | RR RG | 2 | RR | 1 | |||
| 14 | typCLL | NA | NA | RR RG | 2 | |||
| 15 | typCLL | RR | 1 | RR RR | 2 | |||
| 16 | typCLL | NA | NA | RR RR | 2 | |||
| 17 | typCLL | NA | NA | RR RG | 2 | |||
| 18 | typCLL | NA | NA | RR RG | 2 | |||
| 19 | typCLL | NA | NA | RR RG | 2 | |||
| 20 | typCLL | NA | NA | RG RG | 2 | |||
| 21 | typCLL | NA | NA | RR RG | 2 | |||
| 22 | typCLL | NA | NA | RR RR | 2 | |||
| 23 | typCLL | NA | NA | NA | NA | |||
| 24 | typCLL | NA | NA | NA | NA | |||
| 25 | atypCLL | RR | 1 | RR RG | 2 | |||
| 26 | atypCLL | RR RG | 2 | RR | 1 | |||
| 27 | atypCLL | NA | NA | NA | NA | |||
| 28 | atypCLL | RR RR | 2 | RR RR | 2 | |||
| 29 | atypCLL | RR RR | 2 | RR RR RR | 3 | |||
| 30 | atypCLL | RR | 1 | RR RG | 2 | |||
| 31 | atypCLL | NA | NA | RR RR | 2 | |||
| 32 | atypCLL | RR | 1 | RR RG | 2 | |||
| 33 | atypCLL | NA | NA | NA | NA | |||
| 34 | HCL | RR RG | 2 | NA | NA | |||
| 35 | HCL | NA | NA | NA | NA | |||
| 36 | HCL | NA | NA | RR RR | 2 | |||
| 37 | HCL | NA | NA | RR RG | 2 | |||
| 38 | LPL | NA | NA | RR RG | 2 | |||
| 39 | LPL | RR | 1 | RR RR | 2 | |||
| 40 | LPL | NA | NA | NA | NA | |||
| 41 | LPL | NA | NA | RR RG | 2 | |||
| 42 | SMZL | RR | 1 | RR RR | 2 | |||
| 43 | SMZL | NA | NA | RR RG | 2 | |||
| 44 | SMZL | NA | NA | NA | NA | |||
| 45 | FL | NA | NA | NA | NA | |||
| 46 | FL | RR RR | 2 | RR RG | 2 | |||
| 47 | FL | GG | 0 | RR RR | 2 | |||
| 48 | MCL | RR RR | 2 | RR | 1 | |||
| 49 | MCL | RR RR RR | 3 | RR RR | 2 | |||
| 50 | LCL | RR RR | 2 | RR | 1 | |||
| 51 | LCL | RG | 1 | RR RR | 2 | |||
| 52 | Unclassifiable | RR RR | 2 | RR RG | 2 | |||
| 53 | Unclassifiable | RR | 1 | RR RG | 2 | |||
typCLL indicates typical chronic lymphocytic leukemia/small lymphocytic lymphoma; R, rearranged allele; G, germinal configuration; NA, not analyzed; atyp CLL, atypical CLL/small lymphocytic lymphoma; HCL, hairy cell leukemia; LPL, lymphoplasmacytic lymphoma; SMZL, splenic marginal zone lymphoma; FL, follicular lymphoma; MCL, mantle cell lymphoma; and LCL, large cell lymphoma.
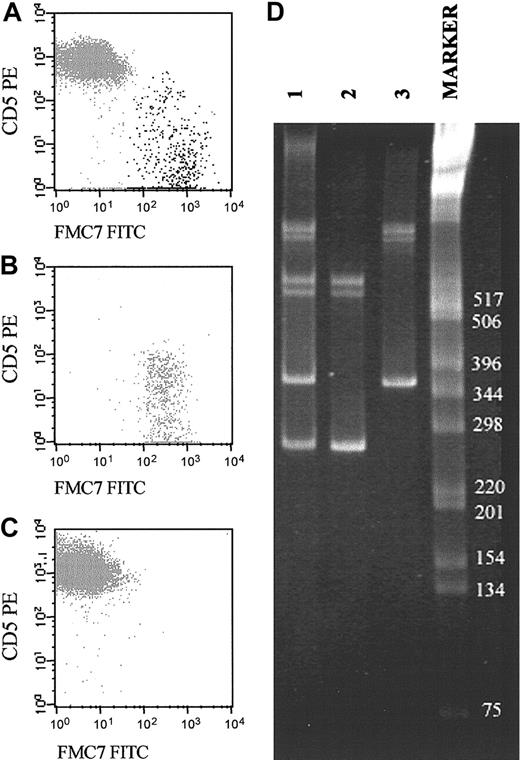
Representative example of a biclonal B-CLPD case initially suspected because of the presence of 2 phenotypically different B-cell populations (CD19+/CD5+/FMC7– and CD19+/CD5–/FMC7+). (A-C) The reactivity of both B-cell subsets for FMC7 and CD5 on CD19+ gated cells before (A) and after (B-C) sorting of each B-cell subset. CD5–FMC7+ cells are shown in panel B, and CD5+FMC7– cells are shown in panel C. (D) Molecular amplification of VDJH rearrangements of the IgH genes in the whole PB samples (lane 1) and the purified fractions of CD5–/FMC7+/CD19+ (lane 2) and CD5+/FMC7–/CD19+ (lane 3) FACS-sorted B cells are displayed. As shown, the 2 sorted B-cell populations had different VDJH gene rearrangements. The numbers to the right of the blot indicate kilodaltons.
Representative example of a biclonal B-CLPD case initially suspected because of the presence of 2 phenotypically different B-cell populations (CD19+/CD5+/FMC7– and CD19+/CD5–/FMC7+). (A-C) The reactivity of both B-cell subsets for FMC7 and CD5 on CD19+ gated cells before (A) and after (B-C) sorting of each B-cell subset. CD5–FMC7+ cells are shown in panel B, and CD5+FMC7– cells are shown in panel C. (D) Molecular amplification of VDJH rearrangements of the IgH genes in the whole PB samples (lane 1) and the purified fractions of CD5–/FMC7+/CD19+ (lane 2) and CD5+/FMC7–/CD19+ (lane 3) FACS-sorted B cells are displayed. As shown, the 2 sorted B-cell populations had different VDJH gene rearrangements. The numbers to the right of the blot indicate kilodaltons.
Incidence of B-CLPD with 2 or more B-cell clones
The overall incidence of cases with 2 or more B-cell clones found was 4.8% (23 of the 477 consecutive B-CLPD patients analyzed). As shown in Table 2, the incidence of patients with 2 or more B-cell clones within each diagnostic group was rather variable. Accordingly, a significantly (P ≤ .05) higher percentage of HCL (23.1%), LCL (20%), and atypCLL patients (13.8%) showed 2 different neoplastic B-cell clones, whereas this was a much less frequent finding among patients with SMZL (7.1%), MCL (5.3%), and typCLL (3.4%). None of the PLL, FL, and LPL patients studied consecutively showed the presence of 2 or more B-cell clones.
Incidence of B-CLPD with 2 or more B-cell clones within each diagnostic group for the 477 consecutively studied patients
Diagnostic group . | No. of biclonal cases/total cases . | % of cases . |
|---|---|---|
| typCLL | 12/353 | 3.4 |
| atypCLL | 4/29 | 13.8 |
| PLL | 0/5 | 0 |
| HCL | 3/13 | 23.1 |
| LPL | 0/12 | 0 |
| SMZL | 1/14 | 7.1 |
| FL | 0/22 | 0 |
| MCL | 1/19 | 5.3 |
| LCL | 2/10 | 20 |
| Total | 23/477 | 4.8 |
Diagnostic group . | No. of biclonal cases/total cases . | % of cases . |
|---|---|---|
| typCLL | 12/353 | 3.4 |
| atypCLL | 4/29 | 13.8 |
| PLL | 0/5 | 0 |
| HCL | 3/13 | 23.1 |
| LPL | 0/12 | 0 |
| SMZL | 1/14 | 7.1 |
| FL | 0/22 | 0 |
| MCL | 1/19 | 5.3 |
| LCL | 2/10 | 20 |
| Total | 23/477 | 4.8 |
typCLL versus atypCLL, P = .03; typCLL versus HCL, P = .01; typCLL versus LCL, P = .05; and HCL versus FL, P = .04. PLL indicates prolymphocytic leukemia. Other abbreviations are explained in Table 1.
Differential antigen expression in B-CLPD with 2 or more B-cell clones
The specific phenotypic differences observed for each B-cell clone in B-CLPD cases with 2 or more B-cell clones are detailed in Table 3 and summarized in Table 4. As shown in Table 3, in 24 of 53 cases (45%), B cells displayed both a different phenotype and a distinct surface immunoglobulin (sIg) light chain in each of the 2 populations, 1 being sIgκ+ and the other sIgλ+. In 13 cases (25%) although the sIg light chain isotype was also different, the expression of all antigens studied was identical in the 2 B-cell populations. In another 9 cases (17%), the sIg light chain isotype was the same in the 2 B-cell populations but could be distinguished based on either a different intensity of sIg expression alone (n = 1) or the intensity of sIg expression and coexisting differential expression of other phenotypic markers (n = 8). In the remaining 7 patients (13%), the pattern of expression of sIg light chain was identical in the 2 B-cell populations that could be specifically identified based on the pattern of expression of the other markers and/or their light scatter properties. Representative examples of these phenotypic differences are illustrated in Figure 2.
Phenotypic characteristics differentially expressed by each of the neoplastic B-populations found in B-CLPD with 2 or more B-cell clones
Case no. . | Diagnosis . | Morphology: no. of major lymphocyte populations . | Phenotype of population 1 (% from the total leukocytes; compatible diagnosis) . | Phenotype of population 2 (% from the total leukocytes; compatible diagnosis) . |
|---|---|---|---|---|
| 1 | typCLL | 3 | CD19+ FMC7- CD5+ CD22- CD23+d CD11c-/+het slg- (28%; B-CLL) | CD19++ FMC7++ CD5- CD22+ CD23- CD11c- k+ (6.7%; LPL) |
| 2 | typCLL | 2 | CD19+ FMC7+d CD5+ CD22+d CD23+ CD25+d (39%; B-CLL) | CD19++ FMC7++ CD5- CD22+ CD23- CD25- (6%; SMZL) |
| 3 | typCLL | 2 | FSC/SSClo CD25+d slg- (25%; B-CLL) | FSC/SSCint CD25+ λ+d (29%; B-CLL) |
| 4 | typCLL | 3 | CD23+ κ+d (50%; B-CLL) | CD23- λ+ (9%; MALT) |
| 5 | typCLL | 1 | κ+d (34%; B-CLL) | λ+ (13%; B-CLL) |
| 6 | typCLL | 2 | FSC/SSClo FMC7+d CD5+ CD22+d CD23+ CD25+ CD11c+het slg- (15%; B-CLL) | FSC/SSCint FMC7+ CD5++ CD22+ CD23- CD25- CD11c- λ+d (12%; MCL) |
| 7 | typCLL | 2 | FSC/SSClo CD19+d FMC7+d CD5+ CD22+d CD23+ CD25+d CD11c- λ+d (24%; B-CLL) | FSC/SSCint CD19+ FMC7++ CD5- CD22++ CD23- CD25- CD11c++ κ+d (22%; SMZL) |
| 8 | typCLL | 1 | FSC/SSClo CD5+ CD11c- λ+d (9%; B-CLL) | FSC/SSCint CD5++ CD11c+ κ+d (10%; B-CLL) |
| 9 | typCLL | 1 | κ+ (25%; B-CLL) | λ+ (8%; B-CLL) |
| 10 | typCLL | 2 | CD5++ κ+d (63%; B-CLL) | CD5+ λ+ (2%; B-CLL) |
| 11 | typCLL | 2 | κ+d (72%; B-CLL) | λ+ (7%; B-CLL) |
| 12 | typCLL | NA | κ+ (2.4%; B-CLL) | λ+ (38%; B-CLL) |
| 13 | typCLL | 2 | FSC/SSClo CD5++ CD25- CD11c+d (36%; B-CLL) | FSC/SSCint CD5+++ CD25+d CC11c+ (37%; B-CLL) |
| 14 | typCLL | 2 | CD5- CD11c- (31%; B-CLL) | CD5+ CD11c+ (7%; B-CLL) |
| 15 | typCLL | 2 | λ+ (48%; B-CLL) | κ+ (6%; B-CLL) |
| 16 | typCLL | 3 | CD19+ FMC7- (18%; B-CLL) | CD19++ FMC7+ (44%; B-CLL) |
| 17 | typCLL | NA | λ+d (38%; B-CLL) | κ+d (8%; B-CLL) |
| 18 | typCLL | NA | FSC/SSClo CD5++ CD25+d λ+ (39%; B-CLL) | FSC/SSCint CD5+++ CD25+ κ+d (16%; B-CLL) |
| 19 | typCLL | NA | κ+ (8%; B-CLL) | λ+ (20%; B-CLL) |
| 20 | typCLL | NA | FMC7++ CD5- CD22+ CD23- CD25- κ++ (9.7%; LPL) | FMC7- CD5++ CD22+d CD23++ CD25+ λ++ (55%; B-CLL) |
| 21 | typCLL | 2 | FSC/SSClo CD19+ FMC7- CD5++ CD22+d CD23+ CD25+ CD11c-/+het λ+ (36%; B-CLL) | FSC/SSCint CD19++ FMC7+ CD5+ CD22+ CD23- CD25- CD11c+ κ++ (8%; MCL) |
| 22 | typCLL | 2 | FMC7- CD5+++ CD22+d CD23+ κ+ (28%; B-CLL) | FMC7++ CD5++ CD22+ CD23- κ++ (8.5%; MCL) |
| 23 | typCLL | NA | FSC/SSClo CD19+ CD5+ CD22- CD23+d CD25- λ+d (10%; B-CLL) | FSC/SSCint CD19++ CD5++ CD22+d CD23+ CD25+ κ+d (45%; B-CLL) |
| 24 | typCLL | NA | κ+ (31%; B-CLL) | λ+ (42%; B-CLL) |
| 25 | atypCLL | 2 | FSC/SSClo CD19+d FMC7- CD5+ CD23+ slg- (3%; B-CLL) | FSC/SSCint CD19+ FMC7++ CD5- CD23+d κ+ (9%; LPL) |
| 26 | atypCLL | 2 | FSC/SSClo CD19+ FMC7+d CD5+ CD23+ CD25+ κ+ (23.2%; B-CLL) | FSC/SSCint CD19++ FMC7++ CD5+d CD23+d CD25- κ++ (62.8%; B-CLL) |
| 27 | atypCLL | ≥3 | CD5- CD23- CD11c- λ+ (7.4%; LPL) | CD5+ CD23+ CD11c++ κ+d (2.3%; B-CLL) |
| 28 | atypCLL | NA | FSC/SSClo CD19+d FMC7+d CD5++ CD22+d CD23++ CD25++ κ+ (10%; B-CLL) | FSC/SSCint CD19+ FMC7++ CD5+ CD22+ CD23+ CD25+ λ++ (15%; MALT) |
| 29 | atypCLL | 2 | CD19+ FMC7- CD5+d CD22+d CD23+ κ+d (60%; B-CLL) | CD19++ FMC7+ CD5- CD22+ CD23- λ+ (7%; FL) |
| 30 | atypCLL | 2 | CD5++ κ+d (16%; B-CLL) | CD5+ κ+ (42%; MCL) |
| 31 | atypCLL | 2 | λ+ (56%; B-CLL) | κ+d (8%; B-CLL) |
| 32 | atypCLL | 3 | FSC/SSClo CD19+d FMC7- CD5+ CD22+d CD103- CD11c- slg- (3.8%; B-CLL) | FSC/SSCint CD19+ FMC7++ CD5- CD22+ CD103+ CD11c+ λ+ (25.4%; HCL) |
| 33 | atypCLL | NA | κ+ (4%; MALT) | λ+ (20%; MALT) |
| 34 | HCL | 2 | FSC/SSClo CD103- CD25- CD11c++ λ+ (3.7%; SMZL) | FSC/SSCint CD103+ CD25+ CD11c+++ κ+ (4.5%; HCL) |
| 35 | HCL | NA | FSC/SSClo CD19+d FMC7- CD5+ CD22+d CD23+ CD103- CD25+d CD11c-/+het κ+d (0.8%; B-CLL) | FSC/SSCint CD19+ FMC7++ CD5- CD22+ CD23- CD103+ CD25- CD11c++ κ+ (1.1%; HCL) |
| 36 | HCL | 1 | CD19+ FMC7-/+het CD11c+d κ+ (30.5%; MALT) | CD19++ FMC7+++ CD11c++ κ+d (6.3%; HCLv) |
| 37 | HCL | 2 | CD103+ (22%; SMZL) | CD103++ (5%; HCL) |
| 38 | LPL | NA | CD19+ FMC7+ CD5+ CD22+d CD23+ CD25++ CD11c- (6.5%; B-CLL) | CD19++ FMC7++ CD5- CD22+ CD23- CD25- CD11c-/+het (38%; LPL) |
| 39 | LPL | NA | λ+d (21%; B-CLL) | λ++ (0.6%; LPL) |
| 40 | LPL | 3 | FSC/SSClo FMC7- CD5+ κ++ (1.5%; MCL) | FSC/SSCint FMC7+ CD5- λ++ (4.7%; LCL) |
| 41 | LPL | 2 | λ+ (46%; LPL) | κ+ (6%; LPL) |
| 42 | SMZL | 2 | FSC/SSClo CD19+d FMC7+d CD5+ CD22+d CD23+ CD25+ slg- (0.6%; B-CLL) | FSC/SSCint CD19+ FMC7++ CD5- CD22+ CD23- CD25- λ+d (3.7%; SMZL) |
| 43 | SMZL | 2 | CD5+ (60%; MALT) | CD5- (6%; SMZL) |
| 44 | SMZL | 2 | κ++ (29%; SMZL) | λ++ (25%; SMZL) |
| 45 | FL | NA | CD5+ CD22+d CD23- κ++ (57%; MCL) | CD5- CD22+ CD23-/+het κ+ (9%; FL) |
| 46 | FL | 2 | FSC/SSClo CD5- CD23- CD103- CD25- CD11c- λ+ (6%; FL) | FSC/SSCint CD5+ CD23+ CD103+ CD25+ CD11c+++ κ+ (47%; LCL) |
| 47 | FL | NA | κ+ (4.5%; FL) | λ+ (2.2%; FL) |
| 48 | MCL | 2 | FSC/SSClo CD19+ FMC7+ CD22+ CD23+ κ+ (12%; MALT) | FSC/SSCint CD19+d FMC7++ CD22+d CD23- λ+ (66%; MCL) |
| 49 | MCL | NA | FSC/SSClo FMC7+hom CD5+d κ+d (11%; MCL) | FSC/SSCint FMC7+het CD5+ κ+ (22%; MCL) |
| 50 | LCL | NA | FSC/SSClo CD22+d CD10- CD11c- κ+d (23%; B-CLL) | FSC/SSCint CD22+ CD10+d CD11c+ slg- (12%; LCL) |
| 51 | LCL | 2 | FSC/SSClo CD19+d FMC7- CD22- CD25- CD11c- κ+d (60%; LPL) | FSC/SSCint CD19+ FMC7+het CD22+d CD25+d CD11c+d κ+ (14%; LCL) |
| 52 | Unclassifiable | 2 | FSC/SSClo FMC7+d CD5+ CD22+d CD23+ CD25+ λ+ (42%; B-CLL) | FSC/SSCint FMC7++ CD5- CD22+ CD23- CD25- κ+ (36%; SMZL) |
| 53 | Unclassifiable | 2 | CD5- CD23- λ+ (18%; LPL) | CD5++ CD23+ κ+d (3.5%; B-CLL) |
Case no. . | Diagnosis . | Morphology: no. of major lymphocyte populations . | Phenotype of population 1 (% from the total leukocytes; compatible diagnosis) . | Phenotype of population 2 (% from the total leukocytes; compatible diagnosis) . |
|---|---|---|---|---|
| 1 | typCLL | 3 | CD19+ FMC7- CD5+ CD22- CD23+d CD11c-/+het slg- (28%; B-CLL) | CD19++ FMC7++ CD5- CD22+ CD23- CD11c- k+ (6.7%; LPL) |
| 2 | typCLL | 2 | CD19+ FMC7+d CD5+ CD22+d CD23+ CD25+d (39%; B-CLL) | CD19++ FMC7++ CD5- CD22+ CD23- CD25- (6%; SMZL) |
| 3 | typCLL | 2 | FSC/SSClo CD25+d slg- (25%; B-CLL) | FSC/SSCint CD25+ λ+d (29%; B-CLL) |
| 4 | typCLL | 3 | CD23+ κ+d (50%; B-CLL) | CD23- λ+ (9%; MALT) |
| 5 | typCLL | 1 | κ+d (34%; B-CLL) | λ+ (13%; B-CLL) |
| 6 | typCLL | 2 | FSC/SSClo FMC7+d CD5+ CD22+d CD23+ CD25+ CD11c+het slg- (15%; B-CLL) | FSC/SSCint FMC7+ CD5++ CD22+ CD23- CD25- CD11c- λ+d (12%; MCL) |
| 7 | typCLL | 2 | FSC/SSClo CD19+d FMC7+d CD5+ CD22+d CD23+ CD25+d CD11c- λ+d (24%; B-CLL) | FSC/SSCint CD19+ FMC7++ CD5- CD22++ CD23- CD25- CD11c++ κ+d (22%; SMZL) |
| 8 | typCLL | 1 | FSC/SSClo CD5+ CD11c- λ+d (9%; B-CLL) | FSC/SSCint CD5++ CD11c+ κ+d (10%; B-CLL) |
| 9 | typCLL | 1 | κ+ (25%; B-CLL) | λ+ (8%; B-CLL) |
| 10 | typCLL | 2 | CD5++ κ+d (63%; B-CLL) | CD5+ λ+ (2%; B-CLL) |
| 11 | typCLL | 2 | κ+d (72%; B-CLL) | λ+ (7%; B-CLL) |
| 12 | typCLL | NA | κ+ (2.4%; B-CLL) | λ+ (38%; B-CLL) |
| 13 | typCLL | 2 | FSC/SSClo CD5++ CD25- CD11c+d (36%; B-CLL) | FSC/SSCint CD5+++ CD25+d CC11c+ (37%; B-CLL) |
| 14 | typCLL | 2 | CD5- CD11c- (31%; B-CLL) | CD5+ CD11c+ (7%; B-CLL) |
| 15 | typCLL | 2 | λ+ (48%; B-CLL) | κ+ (6%; B-CLL) |
| 16 | typCLL | 3 | CD19+ FMC7- (18%; B-CLL) | CD19++ FMC7+ (44%; B-CLL) |
| 17 | typCLL | NA | λ+d (38%; B-CLL) | κ+d (8%; B-CLL) |
| 18 | typCLL | NA | FSC/SSClo CD5++ CD25+d λ+ (39%; B-CLL) | FSC/SSCint CD5+++ CD25+ κ+d (16%; B-CLL) |
| 19 | typCLL | NA | κ+ (8%; B-CLL) | λ+ (20%; B-CLL) |
| 20 | typCLL | NA | FMC7++ CD5- CD22+ CD23- CD25- κ++ (9.7%; LPL) | FMC7- CD5++ CD22+d CD23++ CD25+ λ++ (55%; B-CLL) |
| 21 | typCLL | 2 | FSC/SSClo CD19+ FMC7- CD5++ CD22+d CD23+ CD25+ CD11c-/+het λ+ (36%; B-CLL) | FSC/SSCint CD19++ FMC7+ CD5+ CD22+ CD23- CD25- CD11c+ κ++ (8%; MCL) |
| 22 | typCLL | 2 | FMC7- CD5+++ CD22+d CD23+ κ+ (28%; B-CLL) | FMC7++ CD5++ CD22+ CD23- κ++ (8.5%; MCL) |
| 23 | typCLL | NA | FSC/SSClo CD19+ CD5+ CD22- CD23+d CD25- λ+d (10%; B-CLL) | FSC/SSCint CD19++ CD5++ CD22+d CD23+ CD25+ κ+d (45%; B-CLL) |
| 24 | typCLL | NA | κ+ (31%; B-CLL) | λ+ (42%; B-CLL) |
| 25 | atypCLL | 2 | FSC/SSClo CD19+d FMC7- CD5+ CD23+ slg- (3%; B-CLL) | FSC/SSCint CD19+ FMC7++ CD5- CD23+d κ+ (9%; LPL) |
| 26 | atypCLL | 2 | FSC/SSClo CD19+ FMC7+d CD5+ CD23+ CD25+ κ+ (23.2%; B-CLL) | FSC/SSCint CD19++ FMC7++ CD5+d CD23+d CD25- κ++ (62.8%; B-CLL) |
| 27 | atypCLL | ≥3 | CD5- CD23- CD11c- λ+ (7.4%; LPL) | CD5+ CD23+ CD11c++ κ+d (2.3%; B-CLL) |
| 28 | atypCLL | NA | FSC/SSClo CD19+d FMC7+d CD5++ CD22+d CD23++ CD25++ κ+ (10%; B-CLL) | FSC/SSCint CD19+ FMC7++ CD5+ CD22+ CD23+ CD25+ λ++ (15%; MALT) |
| 29 | atypCLL | 2 | CD19+ FMC7- CD5+d CD22+d CD23+ κ+d (60%; B-CLL) | CD19++ FMC7+ CD5- CD22+ CD23- λ+ (7%; FL) |
| 30 | atypCLL | 2 | CD5++ κ+d (16%; B-CLL) | CD5+ κ+ (42%; MCL) |
| 31 | atypCLL | 2 | λ+ (56%; B-CLL) | κ+d (8%; B-CLL) |
| 32 | atypCLL | 3 | FSC/SSClo CD19+d FMC7- CD5+ CD22+d CD103- CD11c- slg- (3.8%; B-CLL) | FSC/SSCint CD19+ FMC7++ CD5- CD22+ CD103+ CD11c+ λ+ (25.4%; HCL) |
| 33 | atypCLL | NA | κ+ (4%; MALT) | λ+ (20%; MALT) |
| 34 | HCL | 2 | FSC/SSClo CD103- CD25- CD11c++ λ+ (3.7%; SMZL) | FSC/SSCint CD103+ CD25+ CD11c+++ κ+ (4.5%; HCL) |
| 35 | HCL | NA | FSC/SSClo CD19+d FMC7- CD5+ CD22+d CD23+ CD103- CD25+d CD11c-/+het κ+d (0.8%; B-CLL) | FSC/SSCint CD19+ FMC7++ CD5- CD22+ CD23- CD103+ CD25- CD11c++ κ+ (1.1%; HCL) |
| 36 | HCL | 1 | CD19+ FMC7-/+het CD11c+d κ+ (30.5%; MALT) | CD19++ FMC7+++ CD11c++ κ+d (6.3%; HCLv) |
| 37 | HCL | 2 | CD103+ (22%; SMZL) | CD103++ (5%; HCL) |
| 38 | LPL | NA | CD19+ FMC7+ CD5+ CD22+d CD23+ CD25++ CD11c- (6.5%; B-CLL) | CD19++ FMC7++ CD5- CD22+ CD23- CD25- CD11c-/+het (38%; LPL) |
| 39 | LPL | NA | λ+d (21%; B-CLL) | λ++ (0.6%; LPL) |
| 40 | LPL | 3 | FSC/SSClo FMC7- CD5+ κ++ (1.5%; MCL) | FSC/SSCint FMC7+ CD5- λ++ (4.7%; LCL) |
| 41 | LPL | 2 | λ+ (46%; LPL) | κ+ (6%; LPL) |
| 42 | SMZL | 2 | FSC/SSClo CD19+d FMC7+d CD5+ CD22+d CD23+ CD25+ slg- (0.6%; B-CLL) | FSC/SSCint CD19+ FMC7++ CD5- CD22+ CD23- CD25- λ+d (3.7%; SMZL) |
| 43 | SMZL | 2 | CD5+ (60%; MALT) | CD5- (6%; SMZL) |
| 44 | SMZL | 2 | κ++ (29%; SMZL) | λ++ (25%; SMZL) |
| 45 | FL | NA | CD5+ CD22+d CD23- κ++ (57%; MCL) | CD5- CD22+ CD23-/+het κ+ (9%; FL) |
| 46 | FL | 2 | FSC/SSClo CD5- CD23- CD103- CD25- CD11c- λ+ (6%; FL) | FSC/SSCint CD5+ CD23+ CD103+ CD25+ CD11c+++ κ+ (47%; LCL) |
| 47 | FL | NA | κ+ (4.5%; FL) | λ+ (2.2%; FL) |
| 48 | MCL | 2 | FSC/SSClo CD19+ FMC7+ CD22+ CD23+ κ+ (12%; MALT) | FSC/SSCint CD19+d FMC7++ CD22+d CD23- λ+ (66%; MCL) |
| 49 | MCL | NA | FSC/SSClo FMC7+hom CD5+d κ+d (11%; MCL) | FSC/SSCint FMC7+het CD5+ κ+ (22%; MCL) |
| 50 | LCL | NA | FSC/SSClo CD22+d CD10- CD11c- κ+d (23%; B-CLL) | FSC/SSCint CD22+ CD10+d CD11c+ slg- (12%; LCL) |
| 51 | LCL | 2 | FSC/SSClo CD19+d FMC7- CD22- CD25- CD11c- κ+d (60%; LPL) | FSC/SSCint CD19+ FMC7+het CD22+d CD25+d CD11c+d κ+ (14%; LCL) |
| 52 | Unclassifiable | 2 | FSC/SSClo FMC7+d CD5+ CD22+d CD23+ CD25+ λ+ (42%; B-CLL) | FSC/SSCint FMC7++ CD5- CD22+ CD23- CD25- κ+ (36%; SMZL) |
| 53 | Unclassifiable | 2 | CD5- CD23- λ+ (18%; LPL) | CD5++ CD23+ κ+d (3.5%; B-CLL) |
+ indicates antigen expression at normal levels; -, absence of expression; het, heterogeneous antigen expression; ++, strong antigen expression; d, dim antigen expression; SSClo, low scatter; SSCint, intermediate scatter; +++, very strong antigen expression; HCLv, HCL variant; and hom, homogeneous antigen expression. Other abbreviations are explained in Table 1.
B-CLPD with 2 or more B-cell clones: phenotypic differences in individual markers between each neoplastic B-cell subpopulation within each diagnostic disease group
Differing phenotypic marker . | Diagnostic group . | . | . | . | . | . | . | . | . | . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | typCLL, n = 24 . | atypCLL, n = 9 . | HCL, n = 4 . | LPL, n = 4 . | SMZL, n = 3 . | FL, n = 3 . | MCL, n = 2 . | LCL, n = 2 . | Unclassifiable, n = 2 . | Total, %; n = 53 . | |||||||||
| slg | 20∥ | 9∥ | 3∥ | 3∥ | 2§ | 3∥ | 2∥ | 2∥ | 2∥ | 87∥ | |||||||||
| CD5* | 13§ | 8∥ | 1¶ | 2§ | 2§ | 2§ | 1§ | 0 | 2∥ | 58§ | |||||||||
| FMC7 | 8¶ | 6§ | 2§ | 2§ | 1¶ | 0 | 2∥ | 1§ | 1§ | 44¶ | |||||||||
| CD23† | 9¶ | 7§ | 2§ | 2§ | 1¶ | 2§ | 1§ | 0 | 2∥ | 49¶ | |||||||||
| CD22 | 8¶ | 5§ | 1¶ | 1¶ | 1¶ | 1¶ | 1§ | 2∥ | 1§ | 39¶ | |||||||||
| CD19 | 6¶ | 5§ | 2§ | 1¶ | 1¶ | 0 | 1§ | 1§ | 0 | 32¶ | |||||||||
| CD25 | 9¶ | 2 | 2§ | 1¶ | 1¶ | 1¶ | 0 | 1§ | 1§ | 34¶ | |||||||||
| CD11c | 7¶ | 2 | 3∥ | 1¶ | 0 | 1¶ | 0 | 2∥ | 0 | 30¶ | |||||||||
| CD103‡ | 0 | 1 | 3∥ | 0 | 0 | 1¶ | 0 | 0 | 0 | 9 | |||||||||
| CD10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1§ | 0 | 2 | |||||||||
| FSC/SSC | 8¶ | 4¶ | 2§ | 1¶ | 0 | 1¶ | 2∥ | 2∥ | 1§ | 40¶ | |||||||||
Differing phenotypic marker . | Diagnostic group . | . | . | . | . | . | . | . | . | . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | typCLL, n = 24 . | atypCLL, n = 9 . | HCL, n = 4 . | LPL, n = 4 . | SMZL, n = 3 . | FL, n = 3 . | MCL, n = 2 . | LCL, n = 2 . | Unclassifiable, n = 2 . | Total, %; n = 53 . | |||||||||
| slg | 20∥ | 9∥ | 3∥ | 3∥ | 2§ | 3∥ | 2∥ | 2∥ | 2∥ | 87∥ | |||||||||
| CD5* | 13§ | 8∥ | 1¶ | 2§ | 2§ | 2§ | 1§ | 0 | 2∥ | 58§ | |||||||||
| FMC7 | 8¶ | 6§ | 2§ | 2§ | 1¶ | 0 | 2∥ | 1§ | 1§ | 44¶ | |||||||||
| CD23† | 9¶ | 7§ | 2§ | 2§ | 1¶ | 2§ | 1§ | 0 | 2∥ | 49¶ | |||||||||
| CD22 | 8¶ | 5§ | 1¶ | 1¶ | 1¶ | 1¶ | 1§ | 2∥ | 1§ | 39¶ | |||||||||
| CD19 | 6¶ | 5§ | 2§ | 1¶ | 1¶ | 0 | 1§ | 1§ | 0 | 32¶ | |||||||||
| CD25 | 9¶ | 2 | 2§ | 1¶ | 1¶ | 1¶ | 0 | 1§ | 1§ | 34¶ | |||||||||
| CD11c | 7¶ | 2 | 3∥ | 1¶ | 0 | 1¶ | 0 | 2∥ | 0 | 30¶ | |||||||||
| CD103‡ | 0 | 1 | 3∥ | 0 | 0 | 1¶ | 0 | 0 | 0 | 9 | |||||||||
| CD10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1§ | 0 | 2 | |||||||||
| FSC/SSC | 8¶ | 4¶ | 2§ | 1¶ | 0 | 1¶ | 2∥ | 2∥ | 1§ | 40¶ | |||||||||
The 2 cases of unclassified B-CLPD were excluded from this table.
Abbreviations are explained in Table 1.
atypCLL versus HCL (P = .05) and LCL (P = .05).
typCLL versus atypCLL (P = .04).
HCL versus typCLL (P = .01) and atypCLL (P = .05).
Phenotypic difference between each B-cell clone present in 50% to 74% of cases.
Phenotypic difference between each B-cell clone present in 25% to 49% of cases.
Phenotypic difference between each B-cell clone present in 75% to 100% of cases.
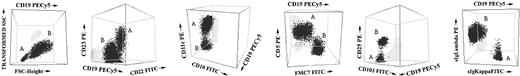
Illustrative 3-dimensional dot plots of samples from patients with several different leukemic B-CLPDs in which 2 distinct neoplastic B-cell clones were detected using immunophenotypic techniques. The neoplastic B-cell clones are named as A (one clone) and B (the second clone); other leukocytes present in the sample are shown as gray events.
Illustrative 3-dimensional dot plots of samples from patients with several different leukemic B-CLPDs in which 2 distinct neoplastic B-cell clones were detected using immunophenotypic techniques. The neoplastic B-cell clones are named as A (one clone) and B (the second clone); other leukocytes present in the sample are shown as gray events.
Overall, within these 53 B-CLPD patients, sIg was the individual antigen most frequently (86%) found to be differentially expressed in the 2 B-cell populations. CD5 and FMC7 expression as well as forward light scatter/sideward light scatter (FSC/SSC) properties were different between the 2 B-cell clones in around half of the patients (53%, 41%, and 41%, respectively). In approximately one third of these patients the 2 B-cell clones showed different reactivity for CD23, CD22, CD19, CD25, and CD11c (39%, 35%, 33%, 33%, and 31% of the patients, respectively). In a lower proportion of patients both clones could be distinguished on the basis of their expression for CD103 (10%) and CD10 (2%) (Table 4). In Table 4 differences in reactivity for individual antigens in the 2 B-cell clones are displayed specifically for each B-CLPD diagnostic group.
In 33 of the 37 cases (89%) in which MGG-stained smears were independently evaluated by 2 different observers, the presence of 2 or more morphologically different populations of lymphocytes was found. In the other 4 cases (11%), a single population of lymphocytes was observed by morphology (Table 3). In 3 of these latter 4 patients, both tumoral clones were phenotypically compatible with typCLL distinguishable by a different pattern of reactivity for CD11c (negative versus positive), CD5 (dim versus strong), and sIg light chain (κ+ versus λ+) in 1 case (case 8) and a distinct sIg light chain isotype in the other 2 patients (cases 5 and 9). In the fourth individual (case 36), corresponding to an HCL, the 2 B-cell clones showed different patterns of expression for several markers, including CD19, FMC7, CD11c, and sIgκ (Table 1).
Clinical and biologic characteristics of patients with 2 or more B-cell clones versus monoclonal B-CLPD cases
To explore whether B-CLPD patients with 2 or more B-cell clones display a different clinical behavior as compared with monoclonal cases, both groups of individuals were compared within each specific diagnostic subgroup. However, due to the relatively low number of cases with 2 or more B-cell clones found in most subgroups, comparison was restricted to B-CLL patients. B-CLPD cases specifically referred to our center for confirmation of the coexistence of 2 or more B-cell clones (n = 30) showed similar clinical and biologic characteristics to those identified within the group of cases consecutively analyzed. Therefore, the 2 cohorts (n = 53) were pooled for comparison with monoclonal cases (n = 454). As seen in Tables 5 and 6, at diagnosis, CLL patients with 2 or more B-cell clones showed a significantly greater incidence of splenomegaly (P = .02) together with significantly lower white blood cell (WBC) (P = .05) and PB absolute lymphocyte counts (P = .01) as compared with monoclonal CLL cases. In addition, CLL cases with 2 or more B-cell clones showed a slightly higher incidence of deaths as compared with monoclonal patients, although no statistically significant differences were found between both groups of patients. Once cases with 2 B-cell populations immunophenotypically compatible with CLL (CLL+CLL) and those who showed one B-cell clone with CLL phenotype and the second one compatible with another B-CLPD (CLL+non-CLL) were separately considered, it was observed that CLL+non-CLL cases had a significantly higher mean age (P = .05), more frequently presented constitutional symptoms (P = .03), had a serum monoclonal component (P = .002), and showed a lower incidence of diffuse pattern of BM involvement (P = .05) as compared with both monoclonal CLL and CLL+CLL cases. No statistically significant differences were found between these groups of patients regarding the other clinical and biologic variables studied at diagnosis.
Clinical characteristics of monoclonal CLL cases versus CLL with 2 or more B-cell clones
. | 2 or more B-cell clones . | . | . | . | |
|---|---|---|---|---|---|
. | CLL + CLL, n = 18 . | CLL + non-CLL, n = 15 . | Monoclonal CLL, n = 366 . | P . | |
| Age, y | 66 ± 11 | 77 ± 8* | 71 ± 10 | .05 | |
| Sex, % | NS | ||||
| Female | 22 | 40 | 38 | ||
| Male | 78 | 60 | 62 | ||
| ECOG greater than 2, % | 7 | 7 | 2 | NS | |
| Binet clinical stage, % | NS | ||||
| A | 69 | 67 | 81 | ||
| B or C | 31 | 33 | 19 | ||
| B symptoms, % | 9 | 50* | 14 | .03 | |
| Adenopathies, % | 56 | 33 | 36 | NS | |
| Hepatomegaly, % | 12 | 7 | 13 | NS | |
| Splenomegaly, % | 37 | 27 | 16* | .02 | |
| Monoclonal component, % | 0 | 15* | 2 | .002 | |
| Diffuse pattern of BM infiltration, % | 44* | 0 | 19 | .05 | |
| % of cases requiring treatment | 47 | 50 | 30 | NS | |
| % of cases requiring treatment 18 mo after diagnosis | 40 | 50 | 26 | .03 | |
| % of cases with associated neoplasias | 0 | 0 | 4 | NS | |
| % deaths due to PD | 6 | 7 | 4 | NS | |
. | 2 or more B-cell clones . | . | . | . | |
|---|---|---|---|---|---|
. | CLL + CLL, n = 18 . | CLL + non-CLL, n = 15 . | Monoclonal CLL, n = 366 . | P . | |
| Age, y | 66 ± 11 | 77 ± 8* | 71 ± 10 | .05 | |
| Sex, % | NS | ||||
| Female | 22 | 40 | 38 | ||
| Male | 78 | 60 | 62 | ||
| ECOG greater than 2, % | 7 | 7 | 2 | NS | |
| Binet clinical stage, % | NS | ||||
| A | 69 | 67 | 81 | ||
| B or C | 31 | 33 | 19 | ||
| B symptoms, % | 9 | 50* | 14 | .03 | |
| Adenopathies, % | 56 | 33 | 36 | NS | |
| Hepatomegaly, % | 12 | 7 | 13 | NS | |
| Splenomegaly, % | 37 | 27 | 16* | .02 | |
| Monoclonal component, % | 0 | 15* | 2 | .002 | |
| Diffuse pattern of BM infiltration, % | 44* | 0 | 19 | .05 | |
| % of cases requiring treatment | 47 | 50 | 30 | NS | |
| % of cases requiring treatment 18 mo after diagnosis | 40 | 50 | 26 | .03 | |
| % of cases with associated neoplasias | 0 | 0 | 4 | NS | |
| % deaths due to PD | 6 | 7 | 4 | NS | |
Results expressed as percentage of cases. Values for age presented as mean ± SD. PD indicates progressive disease. NS indicates that no statistically significant differences were observed between groups (P > .05). ECOG indicates the performance status scaled from 0 to 5 by the Eastern Cooperative Oncology Group.
Statistically significantly different from the other 2 groups of patients.
Biologic characteristics of monoclonal CLL cases versus CLL with 2 or more B-cell clones
. | 2 or more B-cell clones . | . | . | . | |
|---|---|---|---|---|---|
. | CLL + CLL, n = 18 . | CLL + non-CLL, n = 15 . | Monoclonal CLL, n = 366 . | P . | |
| Hemoglobin level, g/dL (range) | 13.2 ± 3 (5.6-17) | 12.2 ± 3 (6.3-15) | 13.8 ± 2.4 (2.1-17.4) | NS | |
| Platelet count, × 109/L (range) | 153.8 ± 81.5 (24-371) | 178.4 ± 94.2 (30-363) | 184.7 ± 78.7 (6-688) | NS | |
| WBC count, × 109/L (range) | 24.4 ± 13 (5.7-62) | 26.1 ± 23.4 (3.2-84.9) | 41.6 ± 57.4* (2.3-450) | ≤ .05 | |
| Lymphocyte count, × 109/L (range) | 17.7 ± 12 (2.9-53) | 17.9 ± 17.6 (0.9-63) | 32.8 ± 50* (0.9-418.5) | .01 | |
| β2-microglobulin level, mg/dL (range) | 2.6 ± 1.5 (1.4-5.9) | 3.1 ± 2.6 (1.3-9.8) | 2.6 ± 1.9 (0.5-16) | NS | |
| LDH level, U/L (range) | 343 ± 121 (149-665) | 1357.3 ± 2163.5 (175-5806) | 418.6 ± 418 (127-7060) | NS | |
| CRP level, mg/L (range) | 1.1 ± 1 (0.2-3.2) | 1 ± 1.3 (0.2-2.5) | 1.7 ± 2.5 (0.1-15.5) | NS | |
| IgG level, mg/dL (range) | 999 ± 201.6 (649-1320) | 1017.6 ± 575 (415-1919) | 1091.1 ± 353.2 (109-2900) | NS | |
| IgM level, mg/dL (range) | 149.4 ± 151 (28.5-488) | 299.1 ± 265.7 (43.6-807) | 100.4 ± 79.1 (4.6-582) | NS | |
| IgA level, mg/dL (range) | 146.5 ± 101.7 (36-414) | 230.1 ± 74.3 (109-315) | 204.2 ± 114.5 (6.6-846) | NS | |
. | 2 or more B-cell clones . | . | . | . | |
|---|---|---|---|---|---|
. | CLL + CLL, n = 18 . | CLL + non-CLL, n = 15 . | Monoclonal CLL, n = 366 . | P . | |
| Hemoglobin level, g/dL (range) | 13.2 ± 3 (5.6-17) | 12.2 ± 3 (6.3-15) | 13.8 ± 2.4 (2.1-17.4) | NS | |
| Platelet count, × 109/L (range) | 153.8 ± 81.5 (24-371) | 178.4 ± 94.2 (30-363) | 184.7 ± 78.7 (6-688) | NS | |
| WBC count, × 109/L (range) | 24.4 ± 13 (5.7-62) | 26.1 ± 23.4 (3.2-84.9) | 41.6 ± 57.4* (2.3-450) | ≤ .05 | |
| Lymphocyte count, × 109/L (range) | 17.7 ± 12 (2.9-53) | 17.9 ± 17.6 (0.9-63) | 32.8 ± 50* (0.9-418.5) | .01 | |
| β2-microglobulin level, mg/dL (range) | 2.6 ± 1.5 (1.4-5.9) | 3.1 ± 2.6 (1.3-9.8) | 2.6 ± 1.9 (0.5-16) | NS | |
| LDH level, U/L (range) | 343 ± 121 (149-665) | 1357.3 ± 2163.5 (175-5806) | 418.6 ± 418 (127-7060) | NS | |
| CRP level, mg/L (range) | 1.1 ± 1 (0.2-3.2) | 1 ± 1.3 (0.2-2.5) | 1.7 ± 2.5 (0.1-15.5) | NS | |
| IgG level, mg/dL (range) | 999 ± 201.6 (649-1320) | 1017.6 ± 575 (415-1919) | 1091.1 ± 353.2 (109-2900) | NS | |
| IgM level, mg/dL (range) | 149.4 ± 151 (28.5-488) | 299.1 ± 265.7 (43.6-807) | 100.4 ± 79.1 (4.6-582) | NS | |
| IgA level, mg/dL (range) | 146.5 ± 101.7 (36-414) | 230.1 ± 74.3 (109-315) | 204.2 ± 114.5 (6.6-846) | NS | |
Results expressed as mean ± standard deviation. NS indicates no statistically significant differences were observed between groups (P > .05); CRP, c-reactive protein; and LDH, lactic dehydrogenase.
Statistically significantly different from the other 2 groups of patients.
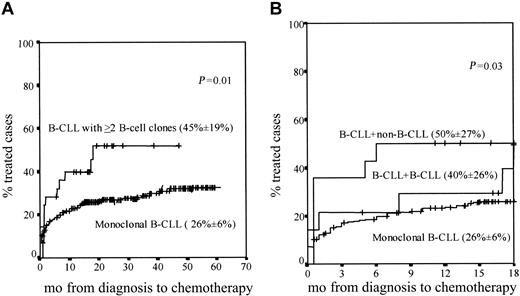
Similar overall survival rates were found in patients with 2 or more B-cell clones as compared with monoclonal CLL cases (data not shown); despite this, statistically significant differences (P = .01) were found between both groups of patients once the percentage of cases requiring treatment during follow-up was considered, this being higher among CLL patients with 2 or more B-cell clones (Figure 3A).
Cases requiring treatment. Curves represent ing the proportion of cases requiring treatment within monoclonal CLL versus CLL patients with 2 or more B-cell clones during both the overall follow-up (A) and the first 18 months after diagnosis (B). In panel B, CLL + CLL and CLL + non-CLL cases are separately represented. The proportion of cases requiring treatment during the first 18 months after diagnosis (± the 95% confidence interval) is in parentheses for each patient group.
Cases requiring treatment. Curves represent ing the proportion of cases requiring treatment within monoclonal CLL versus CLL patients with 2 or more B-cell clones during both the overall follow-up (A) and the first 18 months after diagnosis (B). In panel B, CLL + CLL and CLL + non-CLL cases are separately represented. The proportion of cases requiring treatment during the first 18 months after diagnosis (± the 95% confidence interval) is in parentheses for each patient group.
Interestingly, when patients with 2 or more B-cell clones were divided into the CLL+CLL and CLL+non-CLL subgroups, the proportion of cases that required treatment within the first 18 months after diagnosis was significantly higher (P = .03) among CLL+non-CLL individuals as compared with both CLL+CLL and monoclonal CLL patients (Figure 3B).
Discussion
B-CLPDs are a group of diseases characterized by the proliferation of mature-appearing B cells, which are generally believed to result from the monoclonal expansion of a single transformed precursor cell.1 In fact, in B-CLPD oligoclonality has been usually related to the development of new B-cell subclones resulting from the occurrence of additional genetic abnormalities on the neoplastic cells along the evolution of the disease29,30 ; reports on B-CLPD cases with 2 or more distinct unrelated B-cell clones are scanty in the literature and restricted to between 1 and 3 patients at maximum.12,15,17-19,31,32 In most of these cases, biclonality was suspected based on flow cytometry immunophenotypic studies, which revealed the presence of 2 phenotypically distinct B-cell populations,15,17 frequently displaying a different sIg light chain isotype12,17,18,31,32 ; molecular analysis confirmed the presence of 2 unrelated malignant clones, indicating that these cases corresponded to true biclonal B-CLPD.12,15-17,31,32 Nevertheless, until now the incidence of biclonality among B-CLPD has not been established. The first goal of the present study was to determine this incidence; for that purpose, we have analyzed at diagnosis a large series of 477 consecutive patients suffering from leukemic BCLPD. Our results show that the presence of cases with 2 or more B-cell clones is a relatively frequent finding (4.8%) in B-CLPD patients. The presence of 2 or more B-cell clones was suspected by immunophenotyping and confirmed by molecular analysis of both unfractioned samples and purified B-cell populations. Accordingly, all tested samples with 2 phenotypically different B-cell subsets had 3 or more Ig gene rearrangements, and these were different in each of the B-cell populations identified; in contrast, none of the randomly selected cases with uniform phenotype had more than 2 IgH gene rearrangements. Altogether, these results suggest the utility of the immunophenotypic criteria to detect the presence of 2 or more B-cell clones. In line with previous observations,18,31,32 in most of these patients (66%) the presence of 2 or more B-cell clones was suspected based on the expression of a different sIg light chain isotype by each of the different neoplastic B-cell clones. Interestingly, in some of these cases an sIgκ+/sIgλ+ B-cell ratio within the normal range was observed. Despite this, in these latter cases, a detailed analysis of the immunophenotype of both the sIgκ+ and sIgλ+ B cells for the remaining surface markers tested revealed that both B-cell subsets were immunophenotypically aberrant.21 These results support the notion that the analysis of the sIgκ+/sIgλ+ ratio by itself may not be a useful marker to detect clonality in these patients; in contrast, its combination with the identification of aberrant marker expression would be a useful sensitive and specific tool for the diagnosis of leukemic B-CLPD with 2 or more B-cell clones. The combined use of both sIg light chain together with the detection of phenotypic aberrations is also crucial for the identification of B-CLPD with 2 or more B-cell clones in which the neoplastic B-cell subsets display the same sIg light chain isotype. Accordingly, in 16 cases that showed 2 different neoplastic B-cell subpopulations with an identical sIg light chain isotype by phenotype, the existence of 2 populations that express the same sIg light chain isotype but at different intensities was frequently observed in the context of distinct aberrant phenotypes for each clone.
Previous case reports on biclonal CLPD have suggested the existence of large phenotypic differences between the 2 expanded cell clones.15,17,19 In the present study, a detailed analysis of the phenotypic differences between the coexisting neoplastic cell clones showed that they are relatively variable from one diagnostic subgroup to another, and cases existed in which the phenotypic differences between clones were restricted to a single marker, together with patients displaying more complex phenotypic patterns. Virtually all antigens analyzed were found to be differentially expressed in the 2 coexisting B-cell clones. However, the incidence at which each marker displays a different reactivity in the coexisting B-cell populations varies among the different diagnostic groups. sIg was the individual marker most frequently found to be differentially expressed in the 2 B-cell clones in all diagnostic categories, this including all atypCLL, FL, MCL, and LCL cases with 2 or more B-cell clones. Markers other than sIg showed differential expression in the 2 B-cell clones coexisting in CLPD cases with 2 or more B-cell clones in a variable proportion of patients, depending on the diagnostic subgroup and the individual marker considered. Notably, in all diagnostic groups the 2 B-cell clones detected by phenotype also displayed different FSC/SSC values in a variable proportion of cases, ranging from one fourth of LPL to all MCL and LCL patients analyzed. Interestingly, the presence of both subclones appears to be rather stable because it could be confirmed in sequential studies performed in a subgroup of 8 cases between 3 and 60 months after initial diagnosis (data not shown).
The final goal of our study was to explore whether or not the presence of 2 or more B-cell clones confers a distinct and unique clinical and biologic behavior as compared with monoclonal cases. Due to the relatively low number of cases with 2 or more B-cell clones included in most diagnostic groups, only CLL patients were analyzed in this part of the study. Overall, similar survival rates were found once CLL patients with 2 or more B-cell clones were compared with monoclonal cases (data not shown), probably due to the still relatively short follow-up. Despite this, slightly different clinical and biologic features were detected between monoclonal and CLL patients with 2 or more B-cell clones, especially once cases in which a CLL clone coexisted with a non-CLL clone were considered. Accordingly, a significantly higher incidence of splenomegaly and a higher percentage of cases that required early treatment were found among B-CLPD cases with 2 or more B-cell CLL clones as compared with monoclonal CLL patients. In a similar way, CLL+non-CLL cases more frequently showed B symptoms and a serum monoclonal component than both CLL+CLL and monoclonal CLL patients. Of particular interest was the finding that CLL cases with 2 or more B-cell clones, particularly those in which 1 of the 2 coexisting clones corresponded to a B-CLPD other than CLL, required treatment within the first 18 months significantly more frequently than monoclonal CLL or CLL+CLL cases. This finding would support the hypothesis that, within CLL patients, the presence of 2 or more B-cell clones (one CLL and the second non-CLL) could confer a worse prognosis. It might well be that, in these cases, clinical progression is related mainly to the second B-CLPD (different from CLL) rather than to B-CLL itself.
In summary, in the present study we show that B-CLPD cases with 2 or more B-cell clones are a relatively frequent finding, the phenotypic differences between both B-cell clones involving either the patterns of sIg light chain expression alone or complex combinations of 2 or more markers. From the clinical and biologic point of view, slight differences are observed between monoclonal and B-CLL patients with 2 or more B-cell clones, especially if the latter correspond to cases in which a non-CLL B-cell clone coexists with the CLL clone.
Prepublished online as Blood First Edition Paper, June 26, 2003; DOI 10.1182/blood-2003-01-0045.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal