Abstract
Translocations involving the abl locus on chromosome 9 fuses the tyrosine kinase c-ABL to proteins harboring oligomerization interfaces such as BCR or TEL, enabling these ABL-fusion proteins (X-ABL) to transform cells and to induce leukemia. The ABL kinase activity is blocked by the ABL kinase inhibitor STI571 which abrogates transformation by X-ABL. To investigate the role of oligomerization for the transformation potential of X-ABL and for the sensitivity to STI571, we constructed ABL chimeras with oligomerization interfaces of proteins involved in leukemia-associated translocations such as BCR, TEL, PML, and PLZF. We assessed the capacity of these chimeras to form high molecular weight (HMW) complexes as compared with p185(BCR-ABL). There was a direct relationship between the size of HMW complexes formed by these chimeras and their capacity to induce factor independence in Ba/F3 cells, whereas there was an inverse relationship between the size of the HMW complexes and the sensitivity to STI571. The targeting of the oligomerization interface of p185(BCR-ABL) by a peptide representing the coiled coil region of BCR reduced its potential to transform fibroblasts and increased sensitivity to STI571. Our results indicate that targeting of the oligomerization interfaces of the X-ABL enhances the effects of STI571 in the treatment of leukemia caused by X-ABL.
Introduction
One of the hallmarks of BCR-ABL–induced transformation is the constitutively activated ABL kinase activity. The phenotype of hematopoietic cells induced by the deregulated kinase activity of BCR-ABL is characterized by a reduced susceptibility to a variety of proapoptotic stimuli, including growth factor deprivation.1 It has been reported that one key event leading to ABL kinase activation is the oligomerization of ABL mediated by the N-terminal coiled coil region of BCR (BCC) in the t(9;22) fusion proteins.2,3
There is consensus, that BCC is indispensable for the complete transformation capacity of BCR-ABL, because its deletion inhibits the transformation of immortal Rat-1 cells or NIH3T3 cells.2,4 In contrast, the effect of the deletion of BCC on the capacity of BCR-ABL to mediate factor-independent growth of hematopoietic progenitor cells is controversially discussed. It has been reported that BCC is essential in mediating growth factor independence,2,5,6 but more recent studies showed that BCR-ABL lacking BCC (ΔCC-BCR-ABL) confers factor independence, but growth rates of ΔCC-BCR-ABL–transduced hematopoietic cell lines are slightly lower than those of wild-type (wt) BCR-ABL–expressing cells.4 Nevertheless, ΔCC-BCR-ABL–expressing cells were uniformly reported to exhibit a decrease in overall tyrosine phosphorylation of BCR-ABL targets such as CRK, AKT, STAT5, and SHC, as well as decrease of BCR-ABL autophosphorylation levels as compared with wt BCR-ABL–expressing cells.2,4
Data showing that the deletion of BCC diminishes the transformation capacity of BCR-ABL without completely abolishing it are corroborated by the observation that wt p210(BCR-ABL) induces almost always a chronic myelogenous leukemia (CML)–like disease, whereas p210(BCR-ABL) lacking BCC causes a malignant T-cell disease.4,7
First, structural studies demonstrated that the coiled coil of BCC spans from amino acid (aa) 28 to 682 and that residues beyond position 65 contribute to the structural stability and solubility of the α-helical structures.8 Moreover, BCC mediates the formation of homo-tetramers and aa's 1 to 63 are folded to a functional oligomerization interface.2 Crystal structure analyses of the coiled coil region using a peptide spanning aa's 1 to 72 revealed that residues 5 to 15 and residues 28 to 67 form helices separated by a flexible loop.8
Fusion of isolated BCCs spanning aa's 1 to 63 as well as aa's 1 to 77 to the ABL portion of BCR-ABL results in constitutive activation of the ABL kinase, leading to factor independence but not to transformation of fibroblasts.2,4,7 It has been shown that the overexpression of BCR inhibits the transforming activity of p210(BCR-ABL).9,10 Furthermore, expression of the coiled coil domain flanked by an additional BCR sequence of aa's 1 to 160 reverted factor independence of p210(BCR-ABL)-positive 32D cells.11 These data indicate that targeting of the oligomerization interface may be a possibility to reduce the transformation potential of BCR-ABL.
The role of the oligomerization domains in the oncogenic activation of ABL has been confirmed by the t(12;22)–derived translocation product TEL-ABL.12 TEL is a member of the Ets-family of transcription factors.12-15 The TEL oligomerization interface is represented by a helix-loop-helix (HLH) domain, whose deletion abolishes the transformation capacity of TEL-ABL.12
Transformation of cells by activated ABL can be prevented by STI571, a highly selective inhibitor of the ABL kinase activity. STI571 competes with adenosine triphosphate (ATP) for the active site of the ABL kinase. Thus, STI571 inhibits the transformation potential of the CML-associated p210(BCR-ABL), as well as of the acute lymphocytic leukemia (ALL)–associated p185(BCR-ABL). By receiving treatment with STI571 almost all patients with CML in chronic phase achieve a hematologic remission. With prolonged therapy a considerable percentage of these patients experiences cytogenetic and/or molecular remissions.16 Moreover, 60% to 70% of patients with CML in blast crisis as well as patients with Philadelphia chromosome–positive (Ph+) ALL reach complete hematologic remissions on STI571 monotherapy. Nevertheless, remissions in this poor prognosis group are only of short duration (2-3 months), and on relapse the diseases are resistant to further treatment with STI571.17-19 One of the major mechanisms for resistance is the development of mutations in the ATP-binding site, resulting in a decreased affinity of the fusion proteins to STI571.20,21 The observed primary and secondary resistance to STI571 demonstrates the necessity to investigate additional molecular targets that are essential in the transformation process and thus can be exploited for therapeutic intervention.
Hence, we evaluated the possibility to interfere with the oncogenic potential of p185(BCR-ABL), the BCR-ABL fusion protein associated with high-risk Ph+ ALL rapidly acquiring resistance to STI571, by disrupting its oligomerization capacity. Therefore, we analyzed the functional relevance of the chimeric products containing the oligomerization interfaces of known translocation partners of ABL (the coiled coil of BCR and the HLH of TEL), as well as the role of other leukemia-associated fusion proteins (the coiled coil of PML and the POZ [poxvirus and zinc finger] domain of PLZF) when fused to theABL portion of BCR-ABL. Thus, we demonstrate a direct relationship between the size of HMW complexes formed by the different chimeras, as well as by p185(BCR-ABL), and their individual capacity to induce factor-independent growth. In addition, an inverse relationship between the size of the HMW complexes and sensitivity treatment with STI571 was revealed. Finally, the targeting of the oligomerization interface of p185(BCR-ABL) by a fusion peptide containing the aa's 1 to 63 of BCR reduced the potential to transform fibroblasts and increased sensitivity to STI571.
Materials and methods
Retroviral vectors and p185(BCR-ABL) mutant constructs
The mutant constructs are based on the p185(BCR-ABL) cDNA published,22 and all polymerase chain reaction (PCR) products were controlled for the presence of mutations by sequencing.
To generate the p185(BCR-ABL) mutant lacking the coiled coil region (ΔCCp185) the sequence encoding the first 63 amino acids of p185(BCR-ABL) was substituted by a Kozak-ATG. The following primers were used: 5′-atctacctgcagacgacgatggccaag-3′ (start codon underlined) and 5′-atggcccttgcggatccgctcg-3′ (BamHI site underlined). ΔCCp185 was constructed using the unique BamHI site of p185(BCR-ABL) cDNA.
BCC-ABL was cloned using a 2-step PCR-based cloning strategy. Wt p185(BCR-ABL) cDNA was used as a template. The first PCR was performed with primers Pr1, 5′-ccaccatggtggacccggtgggcttc-3′ (start codon underlined), and Pr2, 5′-cgctgaagggcttcttccagcaacgtctgcaggt-3′ (ABL sequences underlined, BCR sequences in italics). The product of this PCR was used as “megaprimer” in a second PCR together with the primer Pr3, 5′-aggcccatggtaccaggagtg-3′, spanning the KpnI site of ABL. The PCR product was ligated with pCR2.1 (Invitrogen, Karlsruhe, Germany) and subcloned into pcDNA3 p185(BCR-ABL) using KpnI.
For cloning the PML-CC and TEL-HLH, a PCR product spanning from the breakpoint to the KpnI site in ABL was ligated into the pCR2.1 vector, which was previously modified by the introduction of an EcoRV site. The following primers were used: Pr4, 5′-catggagacggatatcaagcccttcagcgg-3′ (EcoRV underlined), and primer Pr3. The HLH region of TEL was obtained with PCR on the first strand cDNA from HL-60 cells followed by ligation into pCR2.1 with primers Pr5, 5′-gcgcacctgagcatggagccaatttactgga-3′ (HLH start atg underlined), and Pr6, 5′-ggtgaaaaagaatcccgggtttcctctgc-3′ (SmaI site underlined). pCR2.1-EcoRV-ABL-KpnI was digested with EcoRV and KpnI, and the resulting fragment was cloned into SmaI-digested PCR2.1-TEL-HLH. The resulting construct was cloned into p185(BCR-ABL) cDNA using KpnI to generate HLH-ABL. The PML-CC region was obtained by PCR on previously published PML-RAR cDNA.23 We introduced a Kozak-atg (underlined) with the primer Pr7, 5′-ccgccatgacgcaggcgctgcag-3′ and a SnaBI site (underlined), with the primer Pr8, 5′-agagctgaggttacgtaggcggaccttgaactc-3′. The PCR product was cloned into pCR2.1 (pCR2.1-PML-CC-SnaBI). pCR2.1-EcoRV-ABL-KpnI was digested with EcoRV and KpnI, and the resulting fragment was cloned into PCR2.1-PML-CC-SnaBI digested with SnaBI. The resulting construct was cloned into p185(BCR-ABL) cDNA using KpnI to generate PCC-ABL.
PCR2.1-EcoRV-ABL-KpnI was digested with EcoRV and KpnI, and the resulting fragment was cloned into EcoRV and KpnI-cut pENTR1A (Invitrogen) (pENTR-EcoRV-ABL-KpnI). To generate PLZF-POZ ABL a previously published cDNA of PLZF24 was digested with HindIII and StuI. After Klenow fill-in the fragment was ligated with EcoRV-digested pENTR-EcoRV-ABL-KpnI (pENTR-PLZF-POZ-ABL-KpnI). The p185(BCR-ABL) cDNA was subcloned into pENTR1A with EcoRI. The pENTR-p185(BCR-ABL) was digested with KpnI and ligated with KpnI-cut pENTR-PLZF-POZ-ABL-KpnI to generate PLZF-POZ-ABL.
For further subcloning of the constructs in different expression vectors the “Gateway” recombination system (Invitrogen) was used. Therefore, all constructs were cloned in the vector pENTR1A and shuttled with the “LR-clonase” enzyme kit (GIBCO/Invitrogen, Karlsruhe, Germany) into the retroviral PINCO vector25 and pCDNA3 previously converted into Gateway destination vectors according to the manufacturer's instructions.
To generate the BCR-CC-Flag fusion peptide, a PCR product using primer Pr1 and primer Pr9, 5′-tcacttgtcatcgtcatccttgtagtccatcagcaacgtctgcaggtagatc-3′ (Flag-tag underlined, stop codon in italics), was generated and ligated with pCR2.1. With the use of EcoRI, BCR-CC-FLAG was subcloned into pENTR1A (pE-CC-FLAG) and into pGEX4T3.
For the CC-GFP (green fluorescent protein) fusion, a coiled coil region lacking the STOP codon was generated by PCR with 3′-primer Pr10, 5′-ggattcccaccatggtggacccgg-3′, and Pr11, 5′-ggattcagcaacgtctgcagg-3′ (BamHI sites in italics), and cloned into pCR2.1. GFP was cut out of PINCO-cmyc-GFP (kindly provided by Pier Giuseppe Pelicci, IEO, Milan, Italy) and ligated with pENTR1A using BamHI and NotI. The BCR-CC was introduced in pENTR1A-GFP with BamHI (pE-CC-GFP). GFP was deleted from PINCO using NotI and BstBI, and p185(BCR-ABL) was cloned into the same PINCO with EcoRI. The resulting vector was converted into a Gateway destination vector at the HindIII-site (pIDEp185). pE-CC-Flag and pE-CC-GFP were recombined with pIDEp185 (pIDEp185/BCCFLAG and pIDEp185/BCCGFP).
Cell lines and cell culture
Ba/F3 cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS; GIBCO BRL, Karlsruhe, Germany) containing 10 ng/mL interleukin 3 (IL-3) (Cell Concepts, Umkirch, Germany). Phoenix and Rat-1 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FCS. STI571 (1 μM) was added to Ba/F3 cells plated at 100 000 cells/mL.
Retroviral infection and transformation assays
Retroviral supernatants using the ecotropic Phoenix packaging cell line were obtained as described.25 The infection was performed by culturing the Ba/F3 target cells (100 000 cells/mL) in the viral supernatant for 3 hours at 37°C in 24-well plates in the presence of 4 μg/mL polybrene (Sigma-Aldrich, Steinheim, Germany). Cells were centrifuged at 2200 rpm for 45 minutes. Infection was repeated 4 times. Infection efficiency was measured by fluorescence-activated cell sorting (FACS) analysis of GFP-positive cells. For IL-3 withdrawal the cells were washed twice with phosphate-buffered saline (PBS) and plated at 100 000 cells/mL. Rat-1 cells were plated in 24-well plates (20 000 cells/well) and were infected as described earlier but in the absence of polybrene. Cells (5000) were plated in 1 mL methylcellulose, and colonies were counted at day 14.
Inhibitory concentration 50% (IC50)
To study the IC50, STI571 was added to the medium of Ba/F3 cells expressing the mutant constructs in concentrations ranging from 0.25 to 4 μM in the absence of IL-3. After 72 hours, cell death was measured with 7-AAD (7-aminoactinomycin D) staining as previously described. Briefly, 200 000 cells were collected and stained with 20 μg/mL 7-AAD (Fluka, Neu-Ulm, Germany) in PBS for 20 minutes. Cells were analyzed immediately by FACS scan analysis. The IC50 was estimated by a graphical interpolation of the resulting dose-response curve.
Western blot
Western blot analysis was performed according to widely diffused protocols using the anti-ABL antibody (α-ABL; Santa Cruz Biotechnology, Santa Cruz, CA), antiphosphotyrosine antibody (α-pY) (Upstate-Biotechnology, Lake Placid, NY), and antiphospho–c-ABL (Tyr245) (α-pABL) (Cell Signaling Technology, Frankfurt, Germany). Blocking was performed in 5% low-fat dry milk, washing was carried out in tris (tris(hydroxymethyl)aminomethane)–buffered saline (TBS) containing 0.1% Tween20 (TBS-T). Antibody incubations were performed in either 5% low-fat dry milk (α-pY) or TBS-T (α-ABL and α-pABL).
“Pull-down” assays
The glutathione S-transferase (GST)–BCC fusion proteins were expressed in Escherichia coli BL21 cells by induction with isopropyl thiogalactoside (IPTG) for 3 hours at 37°C. Extracts were prepared after cell lysis by sonication/detergent treatment. Cell lysates were cleared by centrifugation (10 minutes at 10 000g) and incubated for 2 hours at 4°C with glutathione-Sepharose beads (Amersham/Pharmacia Biotech, Freiburg, Germany). The beads were washed twice with E1A buffer (HEPES 50 mM, pH 7.8, NaCl 150 mM, EDTA 5 mM, DTT [dithiothreitol] 1 mM, NP40 0.1%) and quantified on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) by comparison to a standard curve of bovine serum albumin (BSA). For the pull-down experiments cells were resuspended in E1A buffer (NaCl, 150 mM). Cell extracts were prepared after cell lysis by sonication and clarification by centrifugation (10 minutes at 10 000g). Total protein (1 mg) was then incubated with GST or GST-BCC fusion protein (approximately 10 μg) bound to glutathione Sepharose for 1 hour at 4°C. The bound proteins were eluted by boiling in 30 μL 2 × SDS-PAGE loading buffer, resolved by SDS-PAGE, and visualized by Western blotting with the α-ABL antibody described earlier.
Immunofluorescence
Cells were fixed on coverslips and stained with an α-ABL antibody and Cy-3–conjugated goat-α-mouse secondary antibody as described before.26 Pictures were taken by a Axioplan II–imaging microscope (Zeiss, Göttingen, Germany) and digitalized by the Axiovision software (Zeiss). For further elaboration, images were introduced in Adobe Photoshop 6.01 (Adobe Systems, San Jose, CA) as TIF files. All pictures were modified applying identical parameters for contrast and resolution.
Characterization of HMW complexes
The formation of high molecular weight complexes was detected as previously described.27 Briefly, COS-1 cells were transiently transfected by electroporation with cytomegalovirus (CMV)–driven expression vectors encoding c-DNAs for the different ABL chimeric products. Cytosolic extracts were prepared from 1 to 5 × 108 cells and fractionated at 4°C by high-performance liquid chromatography (HPLC) using a superose 6 HR 10/30 size exclusion column (Pharmacia, Uppsala, Sweden). The flow rate was 0.4 mL/min. Fractions of 0.4 mL were pulled, concentrated, and analyzed by Western blot as described in “Western blot.”
Results
HMW complexes formed by BCR-ABL and its mutants
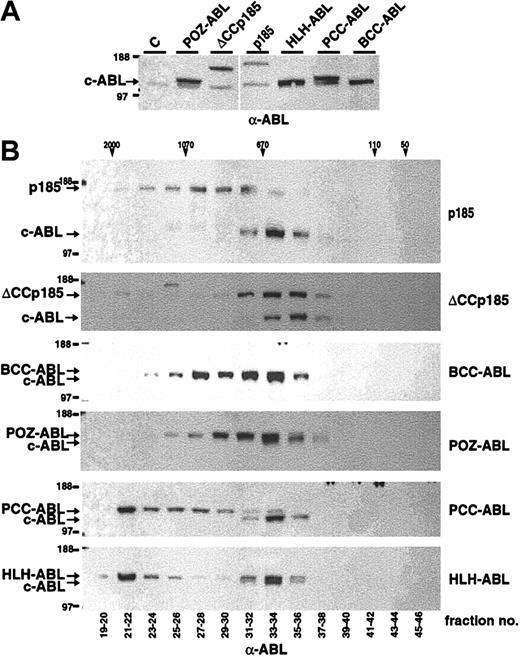
It has been shown that the chemical dimerization of c-ABL leads to its oncogenic activation.28 To further elucidate the role of dimerization/oligomerization for the transforming activity of the ABL portion of BCR-ABL (#ABL) we fused #ABL to the oligomerization domains of TEL, PML, PLZF, and BCR (Figure 1). In detail, the helix-loop-helix domain of TEL aa's 51 to 124 (HLH),12 the coiled coil–containing sequence of PML, aa's 221 to 361 of PML (PCC),29,30 PLZF aa's 1 to 125 (POZ),31 and the coiled coil of BCR aa's 1 to 63 (BCC) were fused to #ABL (Figure 1). The ability of these #ABL chimeras to form oligomers was investigated by size exclusion HPLC analysis, thus revealing their quaternary conformation reflected by the size of HMW complexes. Therefore, Cos-1 cells were transfected with CMV-driven expression vectors of p185(BCR-ABL), BCC-ABL, HLH-ABL, PCC-ABL, and POZ-ABL. We extended the analysis to p185(BCR-ABL) lacking the coiled coil region (ΔCCp185). Cells were lysed 36 to 48 hours after transfection, and lysates were separated by size exclusion HPLC. The single HPLC elution fractions were then subjected to Western blot analysis using an α-ABL antibody. As depicted in Figure 2, p185(BCR-ABL) was found in HMW complexes peaking in fractions corresponding to an apparent molecular weight of 670 to 1070 kDa. In contrast, the endogenous ABL (MW, 145 kDa) was found in HMW complexes corresponding to an apparent molecular weight of 400 to 670 kDa. These results sustain the data that the oligomerization capacity of ABL is increased by the fusion with BCR. Accordingly, deletion of the coiled coil region of BCR shifted the HMW formed by p185(BCR-ABL) to fractions of about 670 kDa, resulting in an elution profile of ΔCCp185 comparable to that of endogenous ABL. Of note, ΔCCp185-related bands, although very weak, were detectable in fractions corresponding to MW of 2000 kDa. In addition, although BCC-ABL as well as POZ-ABL exhibited patterns similar to p185(BCR-ABL), both HLH-ABL and PCC-ABL were mainly contained in HPLC fractions eluting in HMW complexes more than 670 kDa and peaked in the fraction of 2000 kDa.
Modular organization of BCR-ABL constructs. (A) The following cDNAs were used in the study: wild-type p185(BCR-ABL); a deletion-mutant of the N-terminal coiled coil oligomerization domain (ΔCCp185); a fusion of the N-terminal coiled coil domain (CC) of BCR (aa 1-63) to the ABL portion (#ABL) of BCR-ABL (BCC-ABL); a fusion of the helix-loop-helix (HLH) dimerization domain of TEL (aa 52-124) to #ABL (HLH-ABL); a fusion of the coiled coil domain oligomerization domain of PMLII (aa 221-383) to #ABL (PCC-ABL); and a fusion of the PLZF-POZ oligomerization domain (aa 1-125) to #ABL. Schematical representations of wild-type TEL; TEL-ABL; PML and PLZF are also added to this figure. (B) The provirus used for transduction of the cell line. The transgenes are driven by the 5′-LTR (long-term repeat), whereas the GFP-reporter gene is under the control of the CMV promoter (CMV-P). (C) Provirus for the coexpression of p185(BCR-ABL) and the isolated BCR coiled coil (aa 1-63) fused to either GFP (BCC-GFP) or to a Flag-tag (BCC-Flag).
Modular organization of BCR-ABL constructs. (A) The following cDNAs were used in the study: wild-type p185(BCR-ABL); a deletion-mutant of the N-terminal coiled coil oligomerization domain (ΔCCp185); a fusion of the N-terminal coiled coil domain (CC) of BCR (aa 1-63) to the ABL portion (#ABL) of BCR-ABL (BCC-ABL); a fusion of the helix-loop-helix (HLH) dimerization domain of TEL (aa 52-124) to #ABL (HLH-ABL); a fusion of the coiled coil domain oligomerization domain of PMLII (aa 221-383) to #ABL (PCC-ABL); and a fusion of the PLZF-POZ oligomerization domain (aa 1-125) to #ABL. Schematical representations of wild-type TEL; TEL-ABL; PML and PLZF are also added to this figure. (B) The provirus used for transduction of the cell line. The transgenes are driven by the 5′-LTR (long-term repeat), whereas the GFP-reporter gene is under the control of the CMV promoter (CMV-P). (C) Provirus for the coexpression of p185(BCR-ABL) and the isolated BCR coiled coil (aa 1-63) fused to either GFP (BCC-GFP) or to a Flag-tag (BCC-Flag).
Formation of high molecular weight complexes by BCR-ABL mutants. (A) Expression in Cos-1 cells was controlled by Western blot analysis using an α-ABL antibody. (B) Cos-1 cell lysates were fractionated by size-exclusion HPLC. The fractions indicated on the bottom were analyzed by Western blot using an α-ABL antibody. Molecular weights (MWs) corresponding to HPLC fractions are given on the top. Expression of the endogenous ABL and the transgenes is indicated by arrows.
Formation of high molecular weight complexes by BCR-ABL mutants. (A) Expression in Cos-1 cells was controlled by Western blot analysis using an α-ABL antibody. (B) Cos-1 cell lysates were fractionated by size-exclusion HPLC. The fractions indicated on the bottom were analyzed by Western blot using an α-ABL antibody. Molecular weights (MWs) corresponding to HPLC fractions are given on the top. Expression of the endogenous ABL and the transgenes is indicated by arrows.
Taken together, these data indicate that the capacity of ABL to form HMW complexes is dependent on the type of oligomerization interface. Interestingly, the HLH oligomerization interface of TEL and the coiled coil domain of BCR, both partners of ABL in leukemia-associated fusion proteins, result in different HMW patterns.
Oligomerization interfaces derived from different proteins activate the ABL kinase and induce factor independence in Ba/F3 cells but are unable to transform Rat-1 fibroblasts
To investigate whether the type of oligomerization interface and the corresponding size of HMW complexes influences the transformation potential of ABL fusion proteins, we compared the capacity of the #ABL chimeras to mediate factor independence with that of p185(BCR-ABL). Therefore, the fusion constructs HLH-ABL, PCCABL, POZ-ABL, BCC-ABL, and p185(BCR-ABL) were expressed in the IL-3–dependent Ba/F3 cells by retroviral infection. Seventy-two hours after infection IL-3 was withdrawn. Expression of the transgenes and their autophosphorylation was studied by Western blot analysis using α-ABL and α-pY antibodies, respectively (Figure 3A).
Induction of factor-independent growth in Ba/F3 cells by the expression of #ABL chimeras. (A) Expression levels and tyrosine phosphorylation of the transgenes expressed in Ba/F3 cells. (B) Factor-independent growth of Ba/F3 cells expressing the indicated transgenes. (C) Effect of 1 μM STI571 on IL-3–independent growth of Ba/F3 cells expressing p185(BCR-ABL) or the indicated #ABL chimeras (mean of 3 independent experiments ± SD). (Left) Without STI571. (Right) With STI571.
Induction of factor-independent growth in Ba/F3 cells by the expression of #ABL chimeras. (A) Expression levels and tyrosine phosphorylation of the transgenes expressed in Ba/F3 cells. (B) Factor-independent growth of Ba/F3 cells expressing the indicated transgenes. (C) Effect of 1 μM STI571 on IL-3–independent growth of Ba/F3 cells expressing p185(BCR-ABL) or the indicated #ABL chimeras (mean of 3 independent experiments ± SD). (Left) Without STI571. (Right) With STI571.
As detailed in Figure 3B, Ba/F3 cells expressing HLH-ABL proliferated in the absence of IL-3 comparable to p185(BCR-ABL)-expressing Ba/F3 cells. In contrast, BCC-ABL experienced an exponential growth phase after about 2 days. The delay of PCC-ABL– and POZ-ABL–expressing cells to proliferate is associated with a lower expression level (Figure 3A), because the proliferating cell populations expressed the transgene and assessment of GFP expression confirmed a similar infection rate of the different constructs (data not shown). After selection by IL-3 withdrawal, no significant differences regarding protein expression and phosphorylation status of the transgenes were revealed (Figure 3A).
The ABL kinase inhibitor STI571 selectively inhibits the deregulated ABL kinase activity in the leukemia-associated ABL fusion proteins and is able to abolish the transformation induced by ABL-derived oncoproteins.32 Because there were apparently no differences in phosphorylation between the #ABL chimeras after selection by IL-3 withdrawal, we investigated whether there exists a relationship between response to STI571 and the capacity of different fusion products to form HMW complexes. Thus, p185(BCR-ABL), BCC-ABL, HLH-ABL, PCC-ABL, and POZ-ABL were expressed in Ba/F3 cells by retroviral infection. After 7 days of IL-3 withdrawal the cells were seeded in the absence or in the presence of 1 μM STI571. Proliferation was monitored for 4 days by dye exclusion assay. As shown in Figure 3C, in the absence of STI571 no significant difference regarding growth kinetics of the Ba/F3 populations expressing the different constructs was seen. In contrast, growth of Ba/F3 cells expressing either p185(BCR-ABL), HLH-ABL, or PCC-ABL was only slightly delayed by 1 μM STI571, whereas the growth of BCC-ABL– and POZ-ABL–expressing Ba/F3 cells was completely abolished by STI571.
Taken together, these data indicate that different oligomerization interfaces fused to #ABL interfere with the sensitivity to STI571.
#ABL chimeras are unable to transform Rat-1 cells
To further disclose the transformation capacity of the #ABL chimeras we investigated their ability to mediate anchorage-independent growth of immortalized fibroblasts in semisolid medium. Hence, we transduced Rat-1 cells with the different fusion constructs. Infection efficiency was assessed by the percentage of GFP-positive cells as determined by FACS analysis. Cells (5000) were plated 48 hours after infection in methylcellulose, and the colonies were counted at day 14 after plating. All experiments were performed in triplicate, and the number of colonies was normalized to the infection efficiency. Table 1 shows that in contrast to p185(BCR-ABL) none of the other #ABL chimeras were able to form anchorage-independent Rat-1 colonies.
Rat-1 colony-formation assay with #ABL chimeras
Construct . | Normalized colony no. . | SD . |
|---|---|---|
| Control | 0.0 | ± 0.0 |
| p185 | 195.9 | ± 21.9 |
| BCC-ABL | 0.0 | ± 0.0 |
| HLH-ABL | 0.0 | ± 0.0 |
| PCC-ABL | 0.0 | ± 0.0 |
| POZ-ABL | 0.0 | ± 0.0 |
Construct . | Normalized colony no. . | SD . |
|---|---|---|
| Control | 0.0 | ± 0.0 |
| p185 | 195.9 | ± 21.9 |
| BCC-ABL | 0.0 | ± 0.0 |
| HLH-ABL | 0.0 | ± 0.0 |
| PCC-ABL | 0.0 | ± 0.0 |
| POZ-ABL | 0.0 | ± 0.0 |
Rat-1 cells were infected with #ABL chimeras as indicated and plated as triplicates. Colonies were counted on day 14. The number of colonies was normalized with the infection efficiency.
Taken together, these data indicate that the expression of #ABL chimeras leads to factor independence of hematopoietic cells with variable efficiency but is unable to transform immortalized fibroblasts.
Deletion of aa's 1 to 63 abolishes the transformation of fibroblasts but not of hematopoietic progenitors by p185(BCR-ABL)
Recently, it has been shown that a p210(BCR-ABL) mutant lacking the N-terminal coiled coil of BCR mediates factor independence in hematopoietic progenitor cells and induces a T-cell leukemia in a transduction/transplantation mouse model.7 Because we found that ΔCCp185 has a decreased capacity compared with wt p185(BCR-ABL) to form HMW complexes and that sensitivity to STI571 varies with the oligomerization interface fused toABL and the resulting HMW pattern, we investigated the effects of the targeting of the coiled coil region of BCR-ABL on transformation activity of p185(BCR-ABL) and STI571 sensitivity.
To this end, we compared the capacity of ΔCCp185 with that of p185(BCR-ABL) to mediate factor independence in Ba/F3 cells. In contrast to mock-infected control cells, Ba/F3 cells transduced with ΔCCp185 proliferated in the absence of IL-3 with a delay of about 2 days as compared with Ba/F3 cells expressing p185(BCR-ABL) (Figure 4B). Both p185(BCR-ABL) and ΔCCp185 appeared fully phosphorylated before and after IL-3 withdrawal (Figure 4A).
Induction of factor-independent growth in Ba/F3 cells by the expression of the oligomerization interface-deficient ΔCCp185. (A) Ba/F3 cells were infected with a control vector, p185(BCR-ABL) or ΔCCp185. Western blot analysis of expression levels and phosphorylation state was performed using the α-ABL and α-pY antibodies as indicated. (B) Factor-independent growth of Ba/F3 cells expressing the indicated transgenes. (C) Effect of 1 μM STI571 on IL-3–independent growth of Ba/F3 cells expressing p185(BCR-ABL) or ΔCCp185 (mean of 3 independent experiments ± SD). (Left) Without STI571. (Right) With STI571.
Induction of factor-independent growth in Ba/F3 cells by the expression of the oligomerization interface-deficient ΔCCp185. (A) Ba/F3 cells were infected with a control vector, p185(BCR-ABL) or ΔCCp185. Western blot analysis of expression levels and phosphorylation state was performed using the α-ABL and α-pY antibodies as indicated. (B) Factor-independent growth of Ba/F3 cells expressing the indicated transgenes. (C) Effect of 1 μM STI571 on IL-3–independent growth of Ba/F3 cells expressing p185(BCR-ABL) or ΔCCp185 (mean of 3 independent experiments ± SD). (Left) Without STI571. (Right) With STI571.
To determine whether the delayed growth kinetics of the oligomerization-deficient BCR-ABL mutants signify a reduced transforming potential, Ba/F3 cells were infected with p185(BCR-ABL) or ΔCCp185 and were treated with 1 μM STI571 after 7 days of IL-3 withdrawal. Proliferation was assessed by dye exclusion for an additional 4 days. Without STI571 no difference in growth kinetics between Ba/F3 cells expressing p185(BCR-ABL) or ΔCCp185 existed (Figure 4C). On exposure to 1 μM STI571, growth of Ba/F3 cells expressing ΔCCp185 was significantly inhibited as compared with p185(BCR-ABL)-positive Ba/F3 cells.
In summary, these data indicate that targeting the oligomerization interface is able to reduce the transformation capacity of BCR-ABL.
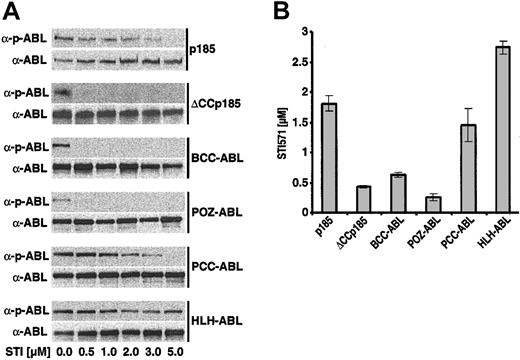
Capacity of STI571 to inhibit the autophosphorylation of #ABL chimeras and the IC50 for STI571 depends on their oligomerization interface
The direct relationship between the type of oligomerization interface fused to #ABL and sensitivity to STI571 prompted us to investigate whether there are differences regarding the influence of STI571 on autophosphorylation of the #ABL chimeras as well as of ΔCCp185 as compared with the wt p185(BCR-ABL). Therefore, IL-3–independent Ba/F3 cells, expressing p185(BCR-ABL), BCCABL, HLH-ABL, PCC-ABL, POZ-ABL, and ΔCCp185 were exposed to increasing concentrations of STI571 (0.5-5 μM) for 6 hours. Autophosphorylation was assessed by Western blot analysis detecting both the ABL signal as well as phospho-ABL signal as described previously.33 The relation between the ABL and phospho-ABL signals revealed a progressive decrease of p185(BCR-ABL) autophosphorylation with increasing concentrations of STI571 up to 3 μM, and only 5 μM was able to completely dephosphorylate p185(BCR-ABL) (Figure 5A). In contrast, autophosphorylation of ΔCCp185, BCC-ABL, as well as of POZ-ABL was already completely inhibited by 0.5 μM STI571 (Figure 5A). The response of PCC-ABL to STI571 was comparable to that of p185(BCR-ABL), whereas autophosphorylation of HLH-ABL was only slightly decreased in the presence of 5 μM STI571.
Influence of STI571 on the autophosphorylation of #ABL chimeras and the IC50 for STI571 of Ba/F3 cells expressing the #ABL chimeras. (A) Autophosphorylation of p185(BCR-ABL) and the different #ABL chimeras in the presence of increasing concentrations of STI571 detected by the relationship between the amount of ABL and phosphorylated ABL detected by α-ABL and α-pABL, respectively. (B) IC50 for STI571 of Ba/F3 cells expressing p185(BCR-ABL) and the different #ABL chimeras. Data presented are the mean of 2 independent experiments ± SD.
Influence of STI571 on the autophosphorylation of #ABL chimeras and the IC50 for STI571 of Ba/F3 cells expressing the #ABL chimeras. (A) Autophosphorylation of p185(BCR-ABL) and the different #ABL chimeras in the presence of increasing concentrations of STI571 detected by the relationship between the amount of ABL and phosphorylated ABL detected by α-ABL and α-pABL, respectively. (B) IC50 for STI571 of Ba/F3 cells expressing p185(BCR-ABL) and the different #ABL chimeras. Data presented are the mean of 2 independent experiments ± SD.
Next, we defined the IC50 for STI571 of Ba/F3 cells expressing p185(BCR-ABL) and the ABL chimeras. Thus, we exposed these cells to increasing concentrations of STI571 (0.25-4 μM). After 72 hours, the percentage of apoptosis was assessed by FACS analysis of cells stained with 7-AAD as described elsewhere.22 The resulting dose-response curves were used to estimate the IC50 for STI571 of Ba/F3 cells expressing different #ABL chimeras. As shown in Figure 5B, p185(BCR-ABL)-expressing Ba/F3 cells showed an IC50 of about 1.8 μM STI571, whereas BCC-ABL–, POZABL–, and ΔCCp185-expressing cells showed an IC50 of about 0.7, 0.3 and 0.5, respectively. Of note, HLH-ABL–expressing Ba/F3 cells showed a significantly higher IC50 of 2.6 μM. The IC50 of PCC-ABL–expressing Ba/F3 cells of 1.5 μM was closer to that of p185(BCR-ABL).
Taken together, these data further confirm the relationship between the capacity of #ABL chimeras to form HMW complexes and the capacity of STI571 to reduce their transformation potential.
Isolated BCC interacts with BCR-ABL in vitro as well as in vivo
It has been shown that overexpression of full-length BCR or of the aa's1 to 160 abolishes the transformation capacity of p210(BCR-ABL).9-11 We hypothesized that these effects are attributable to an inhibition of autophosphorylation by interfering with the homo-oligomerization of BCR-ABL. To test this hypothesis we assessed whether the transforming potential of p185(BCR-ABL) is affected by targeting the coiled coil region of p185(BCR-ABL) through a peptide of the aa's 1 to 63 of BCR known to form a functional oligomerization interface.2
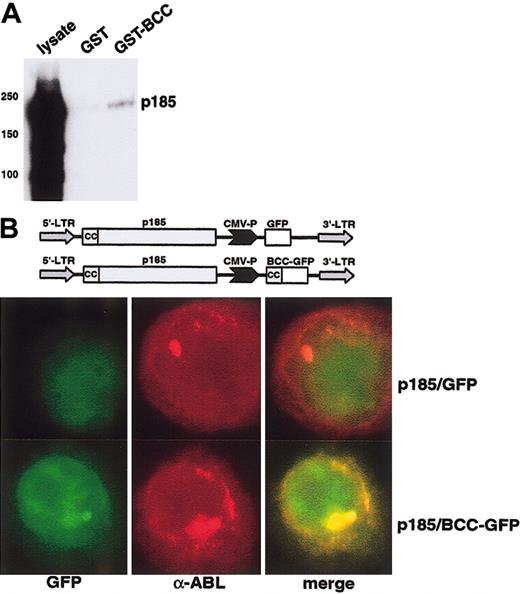
To investigate the capacity of an isolated BCC peptide to bind to p185(BCR-ABL) we performed pull-down experiments by incubating cell lysates of Ba/F3 cells expressing p185(BCR-ABL) with the GST-BCC fusion peptide coupled to Sepharose. After intense washing the complexes were dissolved in an SDS-PAGE and analyzed by an α-ABL Western blot analysis. As depicted in Figure 6A, GST-BCC was able to precipitate BCR-ABL from the cell lysates.
In vitro and in vivo interaction of aa 1 to 63 of BCR (BCC) with p185(BCR-ABL). (A) Pull down of p185(BCR-ABL) by GST-BCC. p185(BCR-ABL)-expressing Ba/F3 cells were incubated with GST and GST-BCC bound to Sepharose beads. The precipitated complexes were resolved on SDS-PAGE and blotted with an α-ABL antibody. (B) Localization of the BCC-GFP fusion peptide in p185(BCR-ABL)-expressing Ba/F3 cells. Cells were stained with the α-ABL antibody α-ABL; red fluorochrome), or GFP, (BCC-GFP expression; green fluorochrome). Colocalization images were obtained by electronic superimposition of the images (merge). Colocalization of fluorochromes yields a yellow color. The retroviral construct is shown at the top. Original magnification, × 100.
In vitro and in vivo interaction of aa 1 to 63 of BCR (BCC) with p185(BCR-ABL). (A) Pull down of p185(BCR-ABL) by GST-BCC. p185(BCR-ABL)-expressing Ba/F3 cells were incubated with GST and GST-BCC bound to Sepharose beads. The precipitated complexes were resolved on SDS-PAGE and blotted with an α-ABL antibody. (B) Localization of the BCC-GFP fusion peptide in p185(BCR-ABL)-expressing Ba/F3 cells. Cells were stained with the α-ABL antibody α-ABL; red fluorochrome), or GFP, (BCC-GFP expression; green fluorochrome). Colocalization images were obtained by electronic superimposition of the images (merge). Colocalization of fluorochromes yields a yellow color. The retroviral construct is shown at the top. Original magnification, × 100.
To confirm the interaction between p185(BCR-ABL) and BCC in vitro, we sought to disclose whether BCC can colocalize to p185(BCR-ABL) in vivo. Hence, we performed immunofluorescence experiments on Ba/F3 cells transduced with pIDEp185/BCC-GFP, a PINCO-p185(BCR-ABL)–derived bi-cistronic retroviral vector in which the GFP was substituted by a GFP-BCC fusion construct (Figure 6B). The α-ABL staining (red fluorochrome) of the Ba/F3 cells expressing p185(BCR-ABL) revealed a cytoplasmic staining pattern characterized by the existence of 1 to 2 big foci/cell and several small aggregates which were not detectable in Ba/F3 wt cells (data not shown). The foci appeared in cells expressing high levels of p185(BCR-ABL). The GFP-derived signal (green fluorochrome) was diffused throughout the cell, and superimposition with GFP (green fluorochrome) and α-ABL staining (red fluorochrome) revealed no colocalization between GFP and BCR-ABL. In contrast, the GFP-BCC–derived signal (green fluorochrome) in the Ba/F3 cells transduced with pIDEp185/BCC-GFP revealed a pattern similar to the α-ABL staining pattern (red fluorochrome). Superimposition of the BCC-GFP signal and the α-ABL staining showed a nearly complete colocalization, proving that BCC-GFP is localized in cytosolic aggregates formed by BCR-ABL.
Taken together, these data show that the aa's 1 to 63 of BCRare able to interact with p185(BCR-ABL).
Expression of BCC reduces the transforming potential of p185(BCR-ABL)
The capacity of BCC-GFP to interact with BCR-ABL in vivo raised the question of whether this interaction is able to interfere with the transformation activity of p185(BCR-ABL).
Therefore, we expressed p185(BCR-ABL) together with GFP or BCC-GFP in Rat-1 cells by retroviral infection using PINCO, PINCO-p185(BCR-ABL), and pIDE-p185/BCC-GFP. Infection effi-ciency was assessed by the percentage of GFP-positive cells as determined by FACS analysis. Cells (5000) were plated 48 hours after infection in methylcellulose, and colonies were counted at day 14 after plating. All experiments were performed in triplicate, and the number of colonies was normalized to the infection efficiency. The expression of BCC-GFP significantly decreased (by about one third) the number of colonies induced by p185(BCR-ABL) (Table 2).
Influence of the expression of BCC-GFP on colony formation in Rat-1 by p185(BCR-ABL)
Construct . | Normalized colony no. . | SD . |
|---|---|---|
| Control | 0.0 | ± 0.0 |
| p185 | 195.9 | ± 21.9 |
| p185/GFP-BCC | 112.2 | ± 4.9 |
Construct . | Normalized colony no. . | SD . |
|---|---|---|
| Control | 0.0 | ± 0.0 |
| p185 | 195.9 | ± 21.9 |
| p185/GFP-BCC | 112.2 | ± 4.9 |
Rat-1 cells were infected with p185(BCR-ABL) in presence of either GFP or BCC-GFP as indicated and plated as triplicates. Colonies were counted on day 14. The colony numbers were normalized with the infection efficiency.
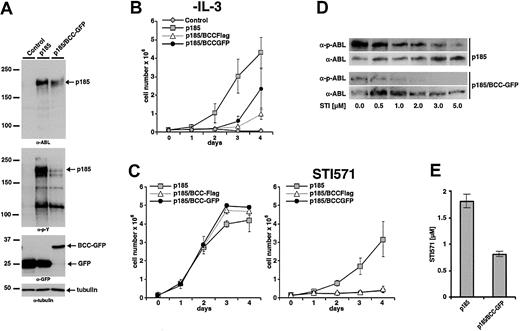
The effect of expression of BCC on p185(BCR-ABL)-mediated factor independence was investigated in Ba/F3 cells as described earlier. To exclude that any biologic effect of BCC-GFP could be due to the GFP portion of the fusion construct, a BCC-FLAG fusion in which a FLAG-tag of 11 aa's was fused to BCC was included in the analysis. Expression of the transgenes and autophosphorylation of p185(BCR-ABL) were studied by Western blot analysis using α-ABL and α-pY, respectively (Figure 7A). Expression of BCC-FLAG as well as of BCC-GFP delayed the factor-independent growth of Ba/F3 cells in the presence of p185(BCR-ABL), although both constructs were unable to block proliferation completely (Figure 7B). Of note, as shown in Figure 7A expression of BCC-GFP strongly reduced the phosphorylation of p185(BCR-ABL).
Inhibitory effects of BCC peptides on the growth of p185(BCR-ABL)-expressing Ba/F3 cells in the presence and absence of STI571. (A) Coexpression of p185(BCR-ABL) and GFP or p185(BCR-ABL) and BCC-GFP in Ba/F3 cells. Expression levels and phosphorylation state were assessed by Western blot analysis using the indicated antibodies. Tubulin was used as a loading control. (For schematic representation of the proviruses used in this experiment see Figure 1.) (B) Factor-independent growth of Ba/F3 cells expressing the indicated transgenes. (C) Effect of 1 μM STI571 on IL-3–independent growth of Ba/F3 cells expressing p185(BCR-ABL) and GFP or BCC-GFP or BCC-FLAG. (Left) Without STI571. (Right) With STI571. (D) Autophosphorylation of p185(BCR-ABL) in the presence and absence of BCC-GFP. Effect of increasing concentrations of STI571 on BCR-ABL autophosphorylation detected by the relationship between the amount of ABL and phosphorylated ABL detected by α-ABL and α-pABL, respectively. (E) IC50 for STI571 of Ba/F3 cell expressing p185(BCR-ABL) in the absence or presence of BCC-GFP. Data presented in panels B, C, and E are the mean of 3 independent experiments ± SD.
Inhibitory effects of BCC peptides on the growth of p185(BCR-ABL)-expressing Ba/F3 cells in the presence and absence of STI571. (A) Coexpression of p185(BCR-ABL) and GFP or p185(BCR-ABL) and BCC-GFP in Ba/F3 cells. Expression levels and phosphorylation state were assessed by Western blot analysis using the indicated antibodies. Tubulin was used as a loading control. (For schematic representation of the proviruses used in this experiment see Figure 1.) (B) Factor-independent growth of Ba/F3 cells expressing the indicated transgenes. (C) Effect of 1 μM STI571 on IL-3–independent growth of Ba/F3 cells expressing p185(BCR-ABL) and GFP or BCC-GFP or BCC-FLAG. (Left) Without STI571. (Right) With STI571. (D) Autophosphorylation of p185(BCR-ABL) in the presence and absence of BCC-GFP. Effect of increasing concentrations of STI571 on BCR-ABL autophosphorylation detected by the relationship between the amount of ABL and phosphorylated ABL detected by α-ABL and α-pABL, respectively. (E) IC50 for STI571 of Ba/F3 cell expressing p185(BCR-ABL) in the absence or presence of BCC-GFP. Data presented in panels B, C, and E are the mean of 3 independent experiments ± SD.
As shown earlier, IL-3–independent Ba/F3 cells expressing ΔCCp185 exhibited an increased sensitivity to STI571 as compared with p185(BCR-ABL)-expressing Ba/F3 cells. To test the hypothesis that the expression of BCC recapitulates the deletion of BCC in BCR-ABL, we expressed p185(BCR-ABL) in Ba/F3 cells in the presence of BCC-FLAG or BCC-GFP. After 7 days of IL-3 withdrawal cells were seeded in the absence or in the presence of 1 μM STI571. Proliferation was assessed by the dye exclusion assay for an additional 4 days. On exposure to 1 μM STI571 only factor-independent Ba/F3 cells expressing p185(BCR-ABL) were able to proliferate, whereas the presence of either BCC-FLAG or BCC-GFP completely abolished proliferation.
To confirm the influence of the expression of GFP-BCC on the autophosphorylation of p185(BCR-ABL), these cells were incubated with 0.5 to 5 μM STI571. After 6 hours the autophosphorylation of p185(BCR-ABL) was assessed as described earlier. As depicted in Figure 7D, the coexpression of BCC-GFP decreased autophosphorylation of p185(BCR-ABL). A further confirmation is provided by the decrease of the IC50 of p185(BCR-ABL)-expressing Ba/F3 cells in response to STI571 in the presence of BCC-GFP with respect to p185(BCR-ABL)-expressing cells (0.8 and 1.8 μM STI571, respectively) (Figure 7E).
Taken together, these data show that the interaction with BCC reduces the transformation activity of p185(BCR-ABL) and increases sensitivity to STI571.
Discussion
Aberrant oligomerization is a common feature of leukemia-associated translocation products. Indeed, oligomerization leads to activation of kinases such as ABL and platelet-derived growth factor receptor (PDGFR)2,5,12,34 or to the transformation of an activator into a repressor of transcription as is the case in the AML-associated translocation products AML-1/ETO, PML/RAR, and PLZF/RAR.27,35
The leukemic phenotype induced by PML/RAR, characterized by the differentiation block at the promyelocytic stage of the myeloid precursors, depends on the presence of the coiled coil oligomerization interface located in the PML portion resulting in the PML/RAR HMW complexes.27,30,35,36 The formation of HMW complexes leads to aberrant recruitment of histone-deacetylase (HDAC) activity to the RAR portion of the fusion protein, considered to be the key event by which PML/RAR represses transcription of all trans-retinoic acid (t-RA) target genes, thus blocking differentiation of hematopoietic cells. The release of the HDAC recruited to the fusion protein by pharmacologic dosages of t-RA then allows the transcription of t-RA target genes and differentiation of the leukemic clone.37
Our data presented here reveal a relationship between the capacity to form HMW complexes and the determination of the malignant phenotype induced by activated ABL. The #ABL chimeras can be subdivided into 2 groups: one forming protein complexes of high MW such as HLH-ABL and PCC-ABL, and one forming protein complexes with a lower MW, such as BCC-ABL, POZ-ABL, and ΔCC-p185. The complexes formed by HLH-ABL and PCC-ABL were even of higher MW than that formed by p185(BCR-ABL). Of note, also p185(BCR-ABL) lacking the oligomerization interface was still able to form HMW complexes even to a lesser extent than p185(BCR-ABL). One can speculate that this complex formation is due to phosphotyrosine-independent binding of the BCR–SH-2 (Src homology domain 2) binding domain to SH-2 of the ABL portion.38,39 This may be the reason why deletion of BCC does not abolish completely the autophosphorylation of p185(BCR-ABL), confirming previously published data.4,7,40 This hypothesis is supported by the finding that TEL-ABL lacking the HLH oligomerization interface is not autophosphorylated and, therefore, unable to mediate factor independence or transformation of fibroblasts.12
In our system it was impossible to determine how many monomers of p185(BCR-ABL) or #ABL chimeras were present in the HMW complexes, because the endogenous c-ABL was not present as a monomer of its theoretical MW of 145 kDa but appeared in the elution fractions corresponding to an MW more than 500 kDa. This finding is most likely due to the intrinsic capacity of c-ABL to oligomerize.41 Nevertheless, p185(BCR-ABL) forms HMW complexes eluting between 670 and 1400 kDa, similar to p210(BCR-ABL).42
The displayed differences in sensitivity to STI571 also allow classification of the #ABL chimeras into 2 groups: one group exhibiting high and one group characterized by a markedly decreased sensitivity, as proven by the IC50 for STI571. The group with the low sensitivity to STI571 corresponds to the #ABL chimeras forming high MW complexes, whose autophosphorylation was hardly affected by exposure even to high doses of STI571. In contrast, high sensitivity is associated with #ABL chimeras forming low MW complexes, whose autophosphorylation is abolished already by low dosages of STI571. The low sensitivity group behaves comparable to p185(BCR-ABL)-expressing cells, and the high sensitivity group exhibits a comparable response to STI571, as does the ΔCCp185 mutant. Of note, the highest HMW complexes were not formed by the BCR-ABL–derived BCC-ABL but by the TEL-ABL–derived HLH-ABL, leading to the highest resistance to STI571. Sensitivity to STI571 was not predictable by differences in the phosphorylation status of the #ABL chimeras in the absence of STI571.
In comparison to previously published data on CML- and Ph+ ALL-derived cells and primary blasts,43,44 high IC50 for STI571 of our Ba/F3 cells expressing p185(BCR-ABL) can most likely be attributed to the fact that we expressed p185(BCR-ABL) in our Ba/F3 cells by retroviral infection with a high-titer viral supernatant resulting in a considerably high expression of the transgenes, which overcomes, at least to some extent, the competition of STI571 for the ATP binding site of BCR-ABL.
From our data one could hypothesize that the capacity of STI571 to interfere with the autophosphorylation of leukemic ABL fusion proteins is diminished with the increase of the number of molecules present in the HMW complexes, indicating that a dimer or a monomer might be more sensitive to STI571 than oligomers.
Furthermore, our data clearly demonstrate that activation of ABL, as assessed by its autophosphorylation, is sufficient to transform hematopoietic progenitor cells but not to satisfy classical transformation parameters such as focus-formation and anchorage-independent growth of fibroblasts. None of the #ABL chimeras was able to transform RAT-1 cells independently of their ability to form HMW complexes and their autophosphorylation status as already known for BCC-ABL.2 Nevertheless, the capacity of BCC-ABL to transform hematopoietic cells is proven by its ability to induce a malignant T-cell disease in mice similar to ΔCC-p210.4,7
Because interference with the capacity of BCR-ABL to oligomerize reduces its transformation potential and increases sensitivity to STI571, we directly targeted the main oligomerization interface of BCR-ABL by a BCC peptide containing exactly the coiled coil structure representing the oligomerization domain of BCR. Thus, we show that the isolated BCC is able to interact directly with p185(BCR-ABL) in vitro as well as in vivo.
Regarding the transformation potential, this interaction is not able to completely recapitulate the phenotype of ΔCCp185-expressing cells. In contrast to ΔCCp185-expressing RAT-1 cells, which are anchorage dependent, expression of BCC does not completely abolish but significantly reduces anchorage-independent growth of p185(BCR-ABL)-expressing fibroblasts. With regard to factor independence, the p185(BCR-ABL)-expressing Ba/F3 cells expressing BCC behave similar to ΔCC-p185–expressing cells on IL-3 withdrawal. The presence of BCC leads to a decrease of autophosphorylation of p185(BCR-ABL) as well as the overall phosphorylation status of the transformed cells.
Furthermore, sensitivity of p185(BCR-ABL)-expressing cells to STI571 is strongly increased by the presence of BCC, as proven by the significant reduction of the IC50 of STI571 by the coexpression of BCC. Of note, coexpression of BCC strongly increases the effectiveness of STI571 to inhibit autophosphorylation of p185(BCR-ABL). Potentially, the binding between BCC and p185(BCR-ABL) interferes with the formation of HMW complexes, thereby diminishing autophosphorylation as efficiently as the deletion of BCC in ΔCCp185.
Taken together, our results presented here suggest that the targeting of the BCR-ABL oligomerization interface is not suffi-cient to overcome its transforming potential. Although targeting of the BCR-ABL oligomerization interface interferes with the kinetics of transformation, it is not able to finally prevent transformation. Nevertheless, it renders cells more sensitive to other “molecular-targeting agents” such as STI571. This is of particular importance considering the high frequency of resistance to STI571 occurring, especially in patients with Ph+ ALL.20,45 Therefore, these cells may not only exhibit increased sensitivity to STI571 but also to other signal transduction modifiers such as Src inhibitors or farnesyl transferase inhibitors.
Prepublished online as Blood First Edition Paper, June 26, 2003; DOI 10.1182/blood-2003-03-0811.
Supported by a fellowship from Deutsche José Carreras Leukämie-Stiftung e.V. (DJCLS; 2001/NAT-1) (T.B.); a contribution from Associazione Italiana per la Ricerca sul Cancro (AIRC) and from the Istituto Pasteur Fondazione Cenci Bolognetti, Università di Roma “La Sapienza” (C.N.); and grants from “Deutsche Forschungsgemeinschaft” (DFG; Ru 728/2-2) (M.R.), Dr. Mildred Scheel-Stiftung der Deutschen Krebshilfe e.V. (10-1916) (M.R.), and Wilhelm Sander-Stiftung e.V. (2001.026.1) (M.R.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal